Abstract
Chikungunya virus (CHIKV) is an arthropod-borne alphavirus. Alphaviruses are positive strand RNA viruses that require a 5´ cap structure to direct translation of the viral polyprotein and prevent degradation of the viral RNA genome by host cell nucleases. Formation of the 5´ RNA cap is orchestrated by the viral protein nsP1, which binds GTP and provides the N-7 methyltransferase and guanylyltransferase activities that are necessary for cap formation. Viruses with aberrant nsP1 activity are unable to replicate effectively suggesting that nsP1 is a promising target for antiviral drug discovery. Given the absence of commercially available antiviral therapies for CHIKV, it is imperative to identify compounds that could be developed as potential therapeutics. This study details a high-throughput screen of 3051 compounds from libraries containing FDA-approved drugs, natural products, and known bioactives against CHIKV nsP1 using a fluorescence polarization-based GTP competition assay. Several small molecule hits from this screen were able to compete with GTP for the CHIKV nsP1 GTP binding site at low molar concentrations. Compounds were also evaluated with an orthogonal assay that measured the ability of nsP1 to perform the guanylation step of the capping reaction in the presence of inhibitor. In addition, live virus assays with CHIKV and closely related alphavirus, Sindbis virus, were used in conjunction with cell toxicity assays to determine the antiviral activity of compounds in cell culture. The naturally derived compound lobaric acid was found to inhibit CHIKV nsP1 GTP binding and guanylation as well as attenuate viral growth in vitro at both 24 hpi and 48 hpi in hamster BHK21 and human Huh 7 cell lines. These data indicate that development of lobaric acid and further exploration of CHIKV nsP1 as a drug target may aid in the progress of anti-alphaviral drug development strategies.
Keywords: nsP1, high throughput screening, lobaric acid, inhibitor, chikungunya virus
1. Introduction
Chikungunya virus (CHIKV) is mosquito-borne arbovirus from the Togaviridae family that is primarily transmitted by Aedes aegypti and Aedes albopictus mosquitos. Once restricted to Africa, Asia, Europe, and parts of the Pacific and Indian Oceans, CHIKV outbreaks were first reported in the Americas in 2013 (Fischer and Staples, 2014). Since that time there have been an estimated 1.7 million suspected cases of CHIKV infection and local transmission of the disease has been reported in 45 countries across the globe (www.cdc.gov). CHIKV infection most often results in mild febrile illness characterized by fever, rash, and arthralgia that generally resolves within two weeks of disease onset (Couderc and Lecuit, 2015; Gasque et al., 2015; Long et al., 2013; Ozden et al., 2007). However, cases of more debilitating joint pain lasting months or even years post infection have been reported (Borgherini et al., 2008; Calabrese, 2008; Dupuis-Maguiraga et al., 2012; Mizuno et al., 2011; Sissoko et al., 2009; Waymouth et al., 2013). Currently there is no commercially available antiviral treatment for people infected with CHIKV and affected persons must rely solely on supportive care. Given the continued spread of CHIKV, concomitant threats from other cocirculating viruses such as Dengue and Zika, increased circulation of Aedes mosquito vectors, and the lack of available antiviral drugs for CHIKV infection, the identification of promising CHIKV drug targets and the pursuit of novel compounds with antiviral activity is imperative (Johansson et al., 2014; Weaver, 2014).
CHIKV is a positive strand RNA virus with an approximately 12 kb genome containing 5´ and 3´ untranslated regions, a 5´ type 0 cap structure, and a 3´ poly A tail (Hefti et al., 1976; Khan et al., 2002). The genome of CHIKV contains two open reading frames. The virus’s nonstructural polyprotein is generated from the first two thirds of the genome, while the last third of the genome is used to code for subgenomic RNA from which the virus’s structural polyprotein is translated (Strauss et al., 1984; Strauss and Strauss, 1994). After a series of protease cleavage steps, the two polyproteins are eventually cleaved into 4 nonstructural proteins (nsP1–4) and 5 structural proteins (C, E3, E2, 6K, and E1) respectively (Kaariainen and Ahola, 2002; Lemm et al., 1994; Shirako and Strauss, 1994). Structural proteins contribute to formation and encapsidation of nascent virions (Jose et al., 2009), while nonstructural proteins orchestrate RNA replication, (Hahn et al., 1989; Lemm et al., 1994; Sawicki and Gomatos, 1976; Shirako and Strauss, 1990). Nonstructural protein nsP4 is the RNA-dependent RNA polymerase that synthesizes new viral RNA from a preexisting viral RNA template (Rubach et al., 2009; Thal et al., 2007). After RNA replication, two nonstructural proteins, nsP1 and nsP2 act as the viral RNA capping enzymes. nsP2 possesses RNA triphosphatase function that removes the terminal phosphate from newly synthesized RNA to generate a diphosphorylated RNA end (Vasiljeva et al., 2000). nsP1 possesses both the guanine-N7-methyltransferase and guanylyltransferase activities that add a methylated guanosine monophosphate to the diphosphorylated RNA to form the type 0 cap on nascent genomic and subgenomic RNAs (Ahola and Kaariainen, 1995; Cross, 1983; Mi and Stollar, 1990; Scheidel and Stollar, 1991). In addition, nsP1 has been shown to be membrane-associated and likely acts as an anchor for linking the viral replication complex (nsP1, nsP2, nsP3, and nsP4) to host cell membranes (Ahola et al., 1999; Laakkonen et al., 1996; Lampio et al., 2000; Spuul et al., 2007).
Capping of viral genomes is critical, as fully formed RNA caps are required for translation of proteins from genomic and subgenomic RNAs, protect viral RNA from degradation by cellular 5´ exonucleases, and protect structural elements in the virus’s 5´UTR that enable evasion of the host’s antiviral response (Hyde, 2014; Strauss and Strauss, 1994). Due to its central role in RNA cap formation, nsP1 has been suggested as a promising antiviral drug target. nsP1’s RNA capping mechanism appears to be distinct from host capping enzymes (Ahola and Kaariainen, 1995; Rupp et al., 2015), providing encouragement that inhibitors of the nsP1 capping reaction may be specific and cause fewer undesirable side effects in host cells compared to canonical capping enzyme inhibitors. Further, it has been shown that mutants lacking either N7-MTase or GTase activity are not viable in cell culture providing evidence that inhibition of nsP1 enzymatic activity is detrimental to viral replication (Ahola et al., 1997). Therefore, nsP1 appears to be an ideal target for anti-CHIKV drug development.
To date, research into the identification of promising small molecule inhibitors of CHIKV nsP1 has been relatively scarce with the exception of recent studies describing the inhibitory activity of a series of [1,2,3]triazolo[4,5-d]pyrimidin-7(6H)-one compounds (Gigante et al., 2017, 2014). Given the immediate need to develop novel therapeutics for CHIKV treatment and lack of effective anti-CHIKV therapeutic options, it is necessary to continue the quest for novel antiviral compounds to feed the drug discovery pipeline (Faqi, 2013). We recently developed a novel and robust high-throughput screening platform for identification of small molecules that can displace or compete with CHIKV nsP1 GTP binding (Bullard-Feibelman et al., 2016). Blocking the CHIKV nsP1 GTP interaction disrupts the RNA capping process, blocks protein translation, and reduces genome stability.
In this report we describe the results of a pilot screen utilizing our CHIKV nsP1 GTP displacement platform against a collection of 3051 bioactive, natural products, and FDA approved molecules. Several small molecule hits were identified from this screen and were demonstrated to inhibit both GTP binding as well as the guanylation step of the nsP1 capping reaction. One of the hits, lobaric acid, was demonstrated to inhibit both Sindbis and CHIKV replication in vitro. These results suggest that identification of lobaric acid and inhibition of nsP1 activity are potential paths forward for anti-alphavirus drug development.
2. Materials and Methods
2.1 Cells and viruses
Baby hamster kidney (BHK17) cells, monkey Vero E6, and human Huh7 cells were obtained from ATCC. Cells were grown in complete Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum (Atlas Biologicals), 25 mM HEPES, and penicillin-streptomycin (cDMEM) in a 37°C incubator with 5% CO2. Sindbis virus (TE/3’2J) stocks were grown on BHK 17 cells and CHIKV (LR2006 OPY1) stocks were grown on Vero E6 cells (Steel et al., 2011). Sindbis and CHIKV viruses were quantified by plaque assay as previously described (Saha et al., 2016; Steel et al., 2011).
2.2 Small molecules
NIH clinical collections 1 and 2 were purchased from the National Institute of Health Common Fund and the Spectrum Collection was purchased from Microsource. Benzbromarone (CID 2333) and pyrantel pamoate (CID 5281033) were purchased from Sigma Aldrich. Garcinolic (CID 6857794) and lobaric (CID 73157) acids were purchased from ChromaDex.
2.3 High throughput screening
High-throughput screening was performed at the High-throughput and High Content Screening Core Facility in the Skaggs School of Pharmacy and Pharmaceutical Science at the University of Colorado’s Anschutz Medical Campus. A fluorescent polarization-based GTP competition assay was developed and described previously (Bullard-Feibelman et al., 2016). The assay was transferred to and adapted for the facilities at the screening core. Assays were plated with a Janus Liquid Handling Robot (Perkin Elmer) and read on an EnVision Multi-Mode Plate Reader (Perkin Elmer). Screening assays were conducted in 384 well black plates (Corning Inc.) and 49.5 µL of master mix (50 mM HEPES pH 6.8, 0.01% NP-40, mM DTT, 500 nM nsP1) was plated before the addition of 0.5 µl small molecules in DMSO, bringing the final concentration of DMSO to 1%. Small molecules were tested in duplicate at a final concentration of 10 µM. To validate the assay with the liquid handling system, positive (GTP) and negative (DMSO) controls were plated on two separate days and the NIH clinical collections 1 and 2 (450 and 281 small molecules respectively) were used to conduct a small validation screen. Z´-factor scores were determined for each plate (Zhang et al., 1999). Following validation, the Spectrum Collection (2320 small molecules) was screened and Z´-factors scores were calculated for each plate.
2.4 Cherrypick and retesting
Strong, medium, and weak hits from the pilot screen were defined as inhibiting GTP binding by >85%, 75–85%, and 50–75%, respectively. Any pan-assay interference compounds were removed from the hit list and hits were prioritized based on Lipinski’s rule of five or number of rotable bonds and polar surface area (Lipinski et al., 2001; Veber et al., 2002). Twenty of the top hits were cherrypicked from the small molecule libraries and retested. The top 4 compounds were reordered from commercial vendors.
2.5 Fluorescence polarization GTP-competition assay
GTP displacement dose response curves were generated for each small molecule as previously described (Bullard-Feibelman et al., 2016). Briefly, small molecules were rehydrated and serially diluted in DMSO in a 24-step 1:2 dilution series starting at 500 µM final concentration. 47.5 µL master mix containing 50 mM HEPES pH 6.8, 0.01% NP-40, 2 mM DTT, 10 nM Bodipy FL GTP-γ-S thioester (ThermoFisher Scientific G22183), and 500 nM CHIKV nsP1 was added to each well in a 384 well black plate. Compounds were added and plates incubated at 22°C for 1 hour in the dark and were then read for fluorescence polarization on a Victor X5 multilabel plate reader (Perkin Elmer) using 488nm excitation/535nm emission filters.
2.6 Guanylation inhibition
Guanylation inhibition reactions were performed in a similar manner as was previously described for dengue NS5 (Geiss et al., 2011). Small molecules were diluted in DMSO in a 1:2 dilution series to give final concentrations ranging from 200-3.1 µM. CHIKV nsP1 was incubated with 1 µM 8-[(6-Amino)hexyl]-amino-GTP – ATTO - 680 (Jena Biosciences) and varying concentrations of small molecule or DMSO in guanylation buffer (5 mM HEPES pH 6.8, 0.1% NP-40, and 500 nM MgCl2) for 1 hour at 37°C in the dark. Reactions were boiled in Laemmli buffer, resolved on 12% SDS-PAGE gels, and nsP1 associated fluorescence was quantified using an Odyssey CLx Imaging System (LI-COR) and Image Studio version 2.1.10 software. Gels were subsequently stained with Coomassie brilliant blue and Coomassie signal was quantified using ImageJ software. Coomassie signal was used to normalize fluorescence signal to control for any variations in protein quantity. Normalized fluorescence from DMSO controls was used to calculate percent of control values for each small molecule concentration.
2.7 Live virus testing
Sindbis virus testing was performed at BSL-2 containment and CHIKV testing was performed at BSL-3 containment. BHK and Huh7 cells were seeded in 12 well plates in cDMEM and allowed to attach overnight. When cells were 50% confluent, small molecules were added at final concentrations ranging from 200 to 2 µM in a final concentration of 1% DMSO. Either Sindbis or CHIKV was added to cells at the same time as compound addition at an MOI of 0.1. Small molecules and virus were incubated on cells for 48 hours. Media samples were taken after 48 hours or after both 24 and 48 hour time periods.
2.8 Cytotoxicity assays
Cell death assays were conducted in white 96 well plates. BHK or Huh7 cells were plated and allowed to attach overnight. Small molecules were dissolved in DMSO and added to cells at final concentrations ranging from 200 µM to 0.4 µM with a final DMSO concentration of 1%. Each concentration was tested in triplicate. Cells were incubated with compound either 24 or 48 hours. Following incubation, cell media was removed and 25 µL each of cDMEM and CellTiter-Glo (Promega) reagents were added. Plates incubated at 37°C for 10 minutes and were then read for luminescence on a Victor X5 multilabel plate reader.
2.9 Statistical analysis
Fluorescence polarization and guanylation inhibition does response curves and Z´-factors were calculated with GraphPad Prism version 7 software. Kis were calculated as previously described for real-time solution-based fluorescence polarization assays (Geiss et al., 2011; Nikolovska-Coleska et al., 2004).
3. Results
3.1 High throughput screen
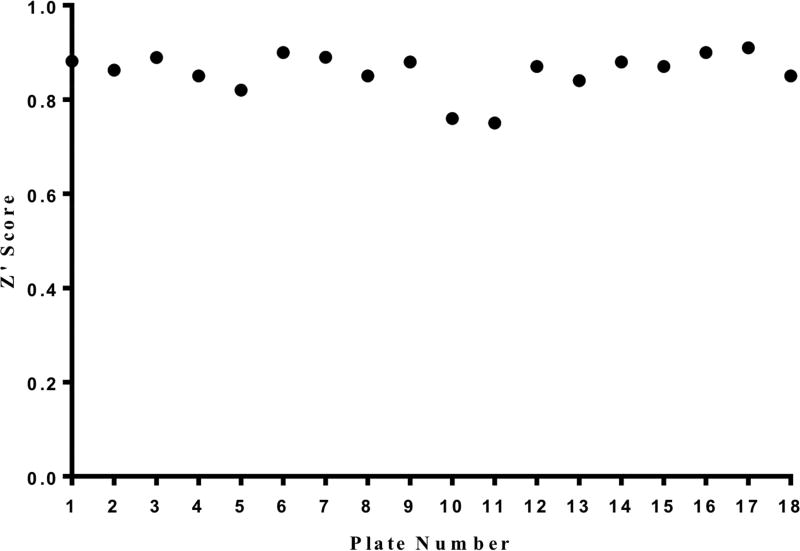
In order to identify novel small molecule inhibitors of the nsP1 RNA capping enzyme from CHIKV, a previously described fluorescence polarization-based GTP competition assay was adapted to a Janus liquid handling system and used to screen 3051 small molecules from three chemical libraries that included known bioactive compounds, approved drugs, and a variety of natural products. Z´-factor scores for each assay plate were >0.75 (average Z’factor = 0.85) indicating a very robust HTS assay (Figure 1). Plates were scanned for fluorescence polarization and total fluorescence signals. Increased total fluorescence was used as an indicator of compounds with intrinsic fluorescence, and fluorescent compounds were removed from the hit list as their fluorescence masks the fluorescence polarization readout during the assay making any FP data unreliable for those compounds. After removal of fluorescent compounds, 101 small molecules retained hit status and could be divided into three subcategories; 1) 35 small molecules were “strong hits” that were able to inhibit >85% of protein activity, 2) 18 small molecules were “medium hits” that were able to knock down 76–85% of protein activity, and 3) 48 small molecules were “weak hits” that were able to inhibit 50–75% activity.
Figure 1. Z´ scores for each plate in CHIKV nsP1 high-throughput pilot screen.
Positive (GTP) and negative (DMSO) control values from each plate in the pilot screen were used to calculate Z´ score. All Z´ scores were > 0.75, and the average Z´ score throughout the screen was 0.85.
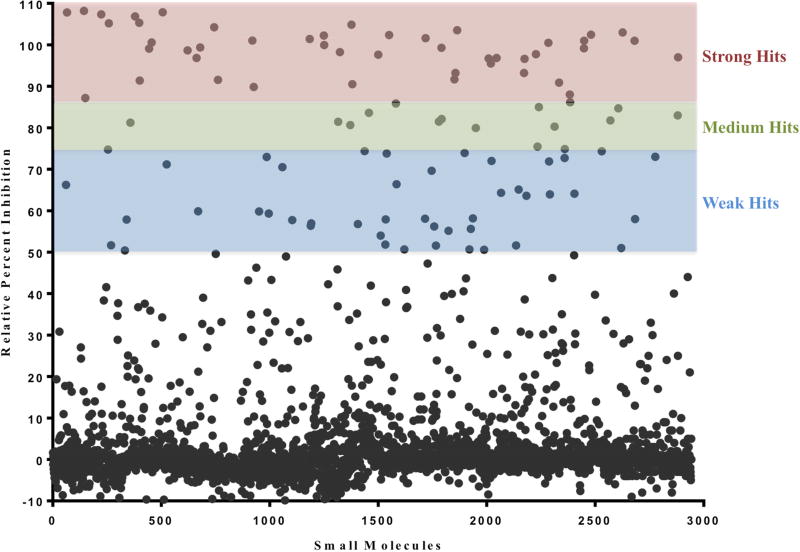
The overall hit rate (inhibition of >50% GTP binding) out of 3051 small molecules was 3.4% (Figure 2). Pan-assay inhibitory compounds (promiscuous hitters) were removed from the hit list and small molecule hits were prioritized based on activity and druglike/leadlike properties. 20 small molecules were cherrypicked and retested at multiple concentrations to determine preliminary dose response relationships and validate HTS assay results (data not shown). All 20 cherrypick compounds displayed similar inhibitory activity upon retest compared to the activity observed in the original screen. Compounds available from commercial vendors and with the greatest apparent affinity for nsP1 were purchased for further testing.
Figure 2. Percent inhibition for each small molecule in the CHIKV nsP1 high-throughput pilot screen.
Recombinant CHIKV nsP1 and GTP-bodipy were complexed and added to assay wells, 10 µM of compound was added to each well, plates were incubated, and total polarization and fluorescence polarization were assessed. Relative percent inhibition (the degree to which each small molecule was able to prohibit nsP1 GTP-binding compared to DMSO control) was calculated for each compound. n=2
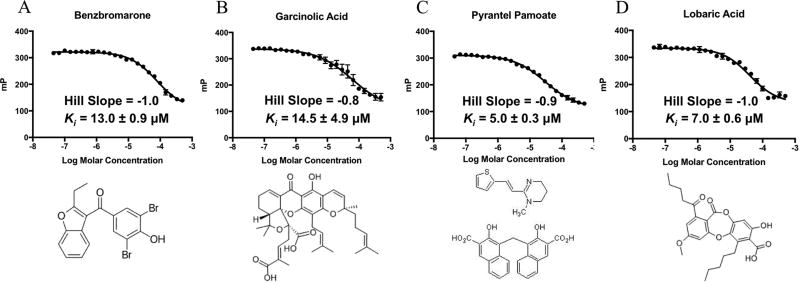
3.2 Small molecule CHIKV nsP1 GTP binding site affinity
We first wanted to quantify the ability of each compound hit to block nsP1 GTP binding and thus determine the inhibitory constant (Ki). In order to determine the Ki of each inhibitor for the CHIKV nsP1 capping enzyme, full dose response curves were generated using a wide range of small molecule concentrations. Kis for inhibitors ranged from 5 µM to 14.5 µM (Figure 3) and lobaric acid and pyrantel pamoate had the greatest GTP-binding inhibition with Kis of and 7.0 ± 0.6 µM and 5.0 ± 0.3 µM respectively. We also tested compounds for specificity for the nsP1 GTP binding site by determining Kis for the flavivirus NS5 protein (Geiss et al., 2011, 2009; Issur et al., 2009; Stahla-Beek et al., 2012), the capping enzyme from another + strand RNA virus family that also binds GTP as a preliminary step in RNA capping (Table 1). Interestingly, while garcinolic acid and benzbromarone display similar and maybe slightly better GTP-binding inhibition with the Dengue NS5 protein, lobaric acid and pyrantel pamoate were both nearly 5X more active against the CHIKV nsP1 enzyme indicating these two small molecules are more selective for the CHIKV nsP1 protein.
Figure 3. Dose response curves and structures for selected hits.
A) GTP displacement dose response curve and structure for benzbromarone. B) GTP displacement dose response curve and structure for garcinolic acid. C) GTP displacement dose response curve and structure for pyrantel pamoate. D) GTP displacement dose response curve and structure for lobaric acid. n=3
Table 1.
Inhibitory activity of small molecules against DENV NS5 and CHIKV nsP1
| DENV NS5 Average Ki | CHIKV nsP1 Average Ki | |
|---|---|---|
| Lobaric Acid | 43.8 ± 7.0 | 7.0 ± 0.6 |
| Garcinolic Acid | 6.9 ± 0.4 | 14.5 ± 4.9 |
| Benzbromarone | 9 ± 0.7 | 13.0 ± 0.9 |
| Pyrantel Pamoate | 38 ± 1.5 | 5.0 ± 0.3 |
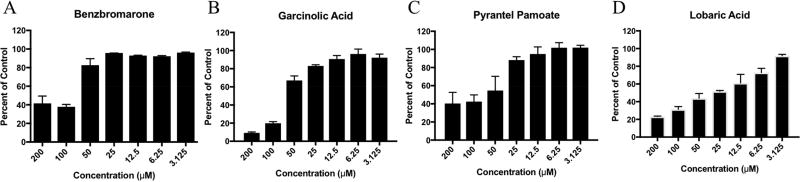
3.3 Small molecule CHIKV nsP1 guanylation inhibition
We determined the ability of CHIKV nsP1 to block formation of the GMP-protein intermediate of the guanylyltransferase reaction using a fluorescently labeled GTP molecule in the presence of varying small molecule concentrations. Disruption of nsP1 guanylation reduces the amount of fluorescent GMP that covalently binds to nsP1, which can be determined by SDS-PAGE gel. Pyrantel pamoate, garcinolic acid, and lobaric acid inhibited CHIKV nsP1 guanlylation activity in a dose dependent manner, while benzbromarone only showed inhibitory activity at high µM concentrations (Figure 4) indicating that this compound’s inhibitory activity was not as likely to translate to a cell-based assay. As such this compound was not evaluated further.
Figure 4. Small molecules inhibit guanylation activity of CHIKV nsP1.
CHIKV nsP1 was incubated with various concentrations of small molecule inhibitors A) benzbromarone, B) garcinolic acid, C) pyrantel pamoate, and D) lobaric acid and fluorescently labeled GTP for 1 hour at 37°C. Samples were boiled then resolved on 12% SDS-page gels. ATTO-680-labeled nsP1 was quantified and indicative of formation of the nsP1-GTP intermediate. Percent of control was calculated for each concentration based on fluorescent signal generated by CHIKV nsP1incubated in the presence of DMSO vehicle alone. n=3
3.4 In vitro antiviral efficacy
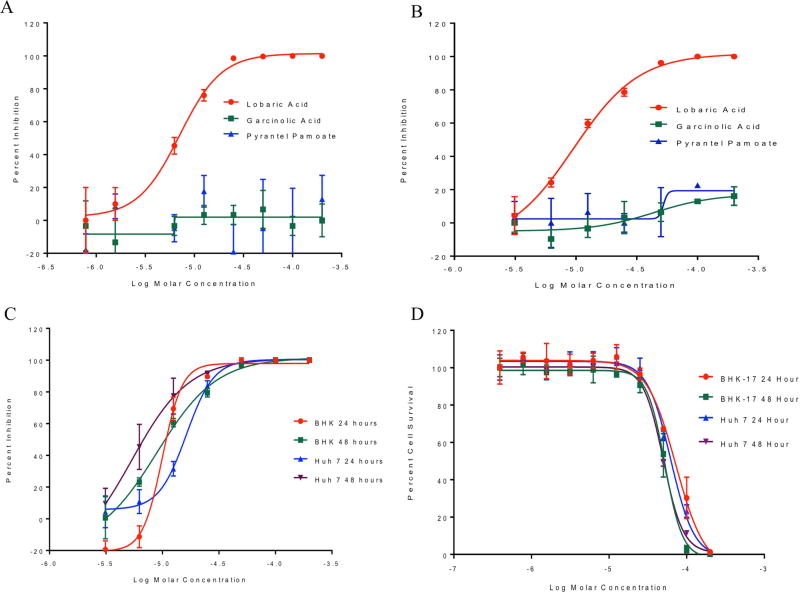
We next determined if pyrantel pamoate, garcinolic acid, and lobaric acid were able to reduce alphavirus replication in cell culture. We first determined if compounds had the ability to interfere with Sindbis virus replication. Sindbis virus is an alphavirus related to CHIKV and can be manipulated in a BSL-2 environment. BHK cells were incubated with varying concentrations of each compound and concurrently infected with Sindbis virus at a multiplicity of infection (MOI) of 0.1. Viral titers were assessed after 48 hours (Figure 5A). Pyrantel pamoate and garcinolic acid did not markedly decrease Sindbis virus replication in vitro, while lobaric acid was able to inhibit virus growth with an EC50 of 5.9 µM ± 1.4 µM. We next determined if the compounds would show antiviral activity against a pathogenic strain of CHIKV. Varying concentrations of pyrantel pamoate, garcinolic acid, and lobaric acid were applied to BHK cells concurrently with CHIKV (strain OPY1) at a MOI of 0.1. Virus and compounds incubated on cells for 48 hours after which viral titers were assessed. Neither pyrantel pamoate nor garcinolic acid were able to reduce CHIKV replication in BHK cells, while lobaric acid inhibited viral replication efficiently after 48 hours (Fig 5B).. To explore the antiviral activity of lobaric acid at multiple time points and in different cell lines, both BHK and Huh 7 cells were treated with varying concentrations of lobaric acid and infected with CHIKV at a MOI of 0.1. Samples were take at 24 and 48 hours post infection and viral titers were determined by plaque assay. Lobaric acid inhibited CHIKV replication in BHK cells at both 24 and 48 hours at similar concentrations (9.9 µM and 8.9 µM respectively). In Huh7 cells lobaric acid displayed greater inhibition at the 48 hour time point with an EC50 of 5.3 µM compared to 16.3 µM at the 24 hour time point (Fig 5C). Cell death assays using BHK and Huh 7 cells revealed that the compound has CC50 values ranging from 50–76 µM (Fig 5D) depending upon time point and cell type making the average selectivity index of lobaric acid nearly 7 (Table 2).
Figure 5. In vitro activity of lobaric acid against Sindbis and CHIKV in BHK and Huh 7 cells.
A) BHK cells were incubated with varying concentrations of compound (lobaric acid, garcinolic acid, or pyrantel pamoate) and Sindbis virus at a MOI of 0.1 for 48 hours. Titers of released virus were assessed by plaque assay and percent inhibition was calculated for each concentration. n=3 B) BHK cells were incubated with varying concentrations of compound (lobaric acid, garcinolic acid, or pyrantel pamoate) and CHIKV at a MOI of 0.1 for 48 hours. Titers of released virus were assessed by plaque assay and percent inhibition was calculated for each concentration. n=3 C) BHK and Huh 7 cells were treated with varying concentrations of lobaric acid and concurrently infected with CHIKV at a MOI of 0.1. Titers of released virus were assessed by plaque assay at 24 and 48 hours and percent inhibition was calculated for each concentration. n=3 D) BHK and Huh 7 cells were grown with varying concentrations of lobaric acid and percent cell survival was determined at 24 and 48 hours. n=3
Table 2.
In vitro efficacy and cytotoxicity of lobaric acid at 24 and 48 hours
| BHK EC50 (µM) |
BHK CC50 (µM) |
BHK SI | Huh 7 EC50 (µM) |
Huh 7 CC50 (µM) |
Huh 7 SI | |
|---|---|---|---|---|---|---|
| 24 hours | 9.9±2.6 | 76.3±2.1 | 7.7 | 16.3±1.2 | 60.5±3.8 | 3.7 |
| 48 hours | 8.9±1.3 | 53.8±8.1 | 6 | 5.3±0.4 | 50±1.3 | 9.4 |
4. Discussion
This study details a high-throughput screening effort that identified pyrantel pamoate, garcinolic acid, and lobaric acid as inhibitors of CHIKV nsP1 GTP-binding and guanylation activities. Pyrantel pamoate is a medication commonly used to treat parasitic worm infections (Moser et al., 2017). It is available as a generic medication and as such is quite inexpensive. Pyrantel pamoate acts by blocking neuromuscular depolarization within the worm, which leads to eventual paralysis. This compound is known to be poorly absorbed in the human intestine, thus unfortunately any potential as an orally bioavailable antiviral would be quite limited. The other compounds that displayed activity against CHIKV nsP1 were garcinolic acid and lobaric acid, which are naturally occurring bioactive phenolic compounds isolated from lichens. Garcinolic acid has previously been shown to increase Ca(2+) uptake of the human adenosine-triphosphategated P2X7 receptor (Fischer et al., 2014). In addition, the cytotoxic properties of this compound in several human cell lines have been explored (Deng et al., 2012). Lobaric acid is isolated from the Stereocaulon genus lichens and has many previously identified biological activities including anti-inflammatory, antiproliferative, antimicrobial, antioxidant and antidiabetic activities (Gissurarson et al., 1997; Hidalgo et al., 2005; Morita et al., 2009; Thadhani and Karunaratne, 2017).
Results of a high-throughput screen identified small molecule inhibitors of the nsP1 capping enzyme from CHIKV. Small molecules from libraries containing known bioactive molecules and FDA-approved drugs were screened against nsP1 with a fluorescence polarization-based GTP competition assay and small molecules benzbromarone, pyrantel pamoate, lobaric acid, and garcinolic acid were able to block CHIKV nsP1 GTP-binding activity. Of the four small molecules tested in the guanylation assay, pyrantel pamoate, garcinolic acid, and lobaric acid were found to inhibit nsP1 guanylation activity in a dose-dependent manner. Further testing of these three compounds in cell-based assays found that only the small molecule lobaric acid was able to attenuate the growth of infectious Sindbis and CHIKV alphaviruses. These data highlight the ability of the nsP1 HTS assay to identify promising inhibitors of nsP1, describe the ability of small molecules garcinolic acid, lobaric acid, and pyrantel pamoate to inhibit necessary steps in the nsP1 capping reaction, and detail the ability of lobaric acid to inhibit alphavirus growth in cell culture. Identification of these small molecules provides a valuable demonstration of nsP1 as an antiviral target and potential compounds that can be further developed for the CHIKV drug discovery pipeline. Given that our screen consisted of 3051 compounds, performing a larger screen of the MLPCN or other large drug libraries may yield a number of additional compounds with antiviral activity that can be further explored. Ultimately, further development of nsP1 and molecules that inhibit its RNA capping function are critical to helping alleviate the serious global public health problem caused by CHIKV infection.
Supplementary Material
Highlights.
-
*
High-throughput fluorescence polarization-based assay identifies inhibitors of the Chikungunya virus nsP1 capping enzyme
-
*
Natural bioactive compounds, lobaric acid and garcinolic acid, inhibit nsP1 activity in the low µM range.
-
*
Lobaric acid reduces Chikungunya virus replication in hamster and human cell lines.
Acknowledgments
The authors would like to acknowledge High-throughput and High Content Screening Core Facility in the Skaggs School of Pharmacy and Pharmaceutical Science at the University of Colorado’s Anschutz Medical Campus and Colorado State University’s Regional Biocontainment facility for providing the BSL-3 laboratory for CHIKV experiments. We would also like to thank Rushika Perera for providing assistance with BSL-3 experiments. This work was supported by a grant from the National Institutes of Health (NIAID R01AI114675) to B.J.G.
Abbreviations
- CHIKV
chikungunya virus
- cDMEM
complete Dulbecco’s Modified Eagle Medium
- MTase
methyltransferase
- GTase
guanylyltransferase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests Statement
The authors declare they have no competing interests.
References
- Ahola T, Kaariainen L. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. U. S. A. 1995;92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahola T, Laakkonen P, Vihinen H, Kaariainen L. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J. Virol. 1997;71:392–397. doi: 10.1128/jvi.71.1.392-397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahola T, Lampio A, Auvinen P, Kaariainen L. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 1999;18:3164–3172. doi: 10.1093/emboj/18.11.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgherini G, Poubeau P, Jossaume A, Gouix A, Cotte L, Michault A, Arvin-Berod C, Paganin F. Persistent Arthralgia Associated with Chikungunya Virus: A Study of 88 Adult Patients on Reunion Island. Clin. Infect. Dis. 2008;47:469–475. doi: 10.1086/590003. [DOI] [PubMed] [Google Scholar]
- Bullard-Feibelman KM, Fuller BP, Geiss BJ. A Sensitive and Robust High-Throughput Screening Assay for Inhibitors of the Chikungunya Virus nsP1 Capping Enzyme. PLoS One. 2016 doi: 10.1371/journal.pone.0158923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese LH. Emerging viral infections and arthritis: the role of the rheumatologist. Nat Clin Pr. Rheum. 2008;4:2–3. doi: 10.1038/ncprheum0679. [DOI] [PubMed] [Google Scholar]
- Couderc T, Lecuit M. Chikungunya virus pathogenesis: From bedside to bench. Antiviral Res. 2015;121:120–131. doi: 10.1016/j.antiviral.2015.07.002. doi: http://dx.doi.org/10.1016/j.antiviral.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Cross RK. Identification of a unique guanine-7-methyltransferase in Semliki Forest virus (SFV) infected cell extracts. Virology. 1983;130:452–463. doi: 10.1016/0042-6822(83)90099-5. [DOI] [PubMed] [Google Scholar]
- Deng Y-X, Pan S-L, Zhao S-Y, Wu M-Q, Sun Z-Q, Chen X-H, Shao Z-Y. Cytotoxic alkoxylated xanthones from the resin of Garcinia hanburyi. Fitoterapia. 2012;83:1548–1552. doi: 10.1016/j.fitote.2012.08.023. [DOI] [PubMed] [Google Scholar]
- Dupuis-Maguiraga L, Noret M, Brun S, Le Grand R, Gras G, Roques P. Chikungunya Disease: Infection-Associated Markers from the Acute to the Chronic Phase of Arbovirus-Induced Arthralgia. PLoS Negl. Trop. Dis. 2012 doi: 10.1371/journal.pntd.0001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faqi AS. Chapter 1 - Introduction. In: Faqi AS, editor. A Comprehensive Guide to Toxicology in Preclinical Drug Development. Academic Press; 2013. pp. 1–2. doi: https://doi.org/10.1016/B978-0-12-387815-1.00001-0. [Google Scholar]
- Fischer M, Staples JE. Notes from the field: chikungunya virus spreads in the Americas - Caribbean and South America, 2013–2014. MMWR. Morb. Mortal. Wkly. Rep. 2014;63:500–501. [PMC free article] [PubMed] [Google Scholar]
- Fischer W, Urban N, Immig K, Franke H, Schaefer M. Natural compounds with P2X7 receptor-modulating properties. Purinergic Signal. 2014;10:313–326. doi: 10.1007/s11302-013-9392-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasque P, Couderc T, Lecuit M, Roques P, Ng LFP. Chikungunya virus pathogenesis and immunity. Vector Borne Zoonotic Dis. 2015;15:241–249. doi: 10.1089/vbz.2014.1710. [DOI] [PubMed] [Google Scholar]
- Geiss BJ, Stahla-Beek HJ, Hannah AM, Gari HH, Henderson BR, Saeedi BJ, Keenan SM. A high-throughput screening assay for the identification of flavivirus NS5 capping enzyme GTP-binding inhibitors: implications for antiviral drug development. J. Biomol. Screen. 2011;16:852–861. doi: 10.1177/1087057111412183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss BJ, Thompson AA, Andrews AJ, Sons RL, Gari HH, Keenan SM, Peersen OB. Analysis of flavivirus NS5 methyltransferase cap binding. J. Mol. Biol. 2009;385:1643–1654. doi: 10.1016/j.jmb.2008.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigante A, Canela M-D, Delang L, Priego E-M, Camarasa M-J, Querat G, Neyts J, Leyssen P, Perez-Perez M-J. Identification of [1,2,3]triazolo[4,5-d]pyrimidin-7(6H)-ones as novel inhibitors of Chikungunya virus replication. J. Med. Chem. 2014;57:4000–4008. doi: 10.1021/jm401844c. [DOI] [PubMed] [Google Scholar]
- Gigante A, Gomez-SanJuan A, Delang L, Li C, Bueno O, Gamo A-M, Priego E-M, Camarasa M-J, Jochmans D, Leyssen P, Decroly E, Coutard B, Querat G, Neyts J, Perez-Perez M-J. Antiviral activity of [1,2,3]triazolo[4,5-d]pyrimidin-7(6H)-ones against chikungunya virus targeting the viral capping nsP1. Antiviral Res. 2017;144:216–222. doi: 10.1016/j.antiviral.2017.06.003. [DOI] [PubMed] [Google Scholar]
- Gissurarson SR, Sigurdsson SB, Wagner H, Ingolfsdottir K. Effect of lobaric acid on cysteinylleukotriene formation and contractile activity of guinea pig taenia coli. J. Pharmacol. Exp. Ther. 1997;280:770–773. [PubMed] [Google Scholar]
- Hahn YS, Strauss EG, Strauss JH. Mapping of RNA- temperature-sensitive mutants of Sindbis virus: assignment of complementation groups A, B, and G to nonstructural proteins. J. Virol. 1989 doi: 10.1128/jvi.63.7.3142-3150.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefti E, Bishop DHL, Dubin DT, Stollar V. 5′ Nucleotide Sequence of Sindbis Viral RNA. J. Virol. 1976 doi: 10.1128/jvi.17.1.149-159.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo ME, Bascunan L, Quilhot W, Fernandez E, Rubio C. Spectroscopic and photochemical properties of the lichen compound lobaric acid. Photochem. Photobiol. 2005;81:1447–1449. doi: 10.1562/2005-05-17-RA-530. [DOI] [PubMed] [Google Scholar]
- Hyde JL. A viral RNA structural element alters host recognition of non-self RNA. 2014 doi: 10.1126/science.1248465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issur M, Geiss BJ, Bougie I, Picard-Jean F, Despins S, Mayette J, Hobdey SE, Bisaillon M. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA. 2009;15:2340–2350. doi: 10.1261/rna.1609709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MA, Powers AM, Pesik N, Cohen NJ, Staples JE. Nowcasting the spread of chikungunya virus in the Americas. PLoS One. 2014;9:e104915. doi: 10.1371/journal.pone.0104915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose J, Snyder JE, Kuhn RJ. A structural and functional perspective of alphavirus replication and assembly. Future Microbiol. 2009 doi: 10.2217/fmb.09.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaariainen L, Ahola T. Functions of alphavirus nonstructural proteins in RNA replication. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:187–222. doi: 10.1016/S0079-6603(02)71044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur P, Lee RCH, Chu JJH. Infectious Viral Quantification of Chikungunya Virus-Virus Plaque Assay. Methods Mol. Biol. 2016;1426:93–103. doi: 10.1007/978-1-4939-3618-2_9. [DOI] [PubMed] [Google Scholar]
- Khan AH, Morita K, Parquet Md, M del C, Hasebe F, Mathenge EGM, Igarashi A. Complete nucleotide sequence of chikungunya virus and evidence for an internal polyadenylation site. J. Gen. Virol. 2002;83:3075–3084. doi: 10.1099/0022-1317-83-12-3075. [DOI] [PubMed] [Google Scholar]
- Laakkonen P, Ahola T, Kaariainen L. The effects of palmitoylation on membrane association of Semliki forest virus RNA capping enzyme. J. Biol. Chem. 1996;271:28567–28571. doi: 10.1074/jbc.271.45.28567. [DOI] [PubMed] [Google Scholar]
- Lampio A, Kilpelainen I, Pesonen S, Karhi K, Auvinen P, Somerharju P, Kaariainen L. Membrane binding mechanism of an RNA virus-capping enzyme. J. Biol. Chem. 2000;275:37853–37859. doi: 10.1074/jbc.M004865200. [DOI] [PubMed] [Google Scholar]
- Lemm JA, Rümenapf T, Strauss EG, Strauss JH, Rice CM. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994 doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Long KM, Whitmore AC, Ferris MT, Sempowski GD, McGee C, Trollinger B, Gunn B, Heise MT. Dendritic cell immunoreceptor regulates Chikungunya virus pathogenesis in mice. J. Virol. 2013;87:5697–5706. doi: 10.1128/JVI.01611-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Stollar V. Both amino acid changes in nsP1 of Sindbis virusLM21 contribute to and are required for efficient expression of the mutant phenotype. Virology. 1990;178:429–434. doi: 10.1016/0042-6822(90)90340-w. [DOI] [PubMed] [Google Scholar]
- Mizuno Y, Kato Y, Takeshita N, Ujiie M, Kobayashi T, Kanagawa S, Kudo K, Lim C-K, Takasaki T. Clinical and radiological features of imported chikungunya fever in Japan: a study of six cases at the National Center for Global Health and Medicine. J. Infect. Chemother. Off. J. Japan Soc. Chemother. 2011;17:419–423. doi: 10.1007/s10156-010-0124-y. [DOI] [PubMed] [Google Scholar]
- Morita H, Tsuchiya T, Kishibe K, Noya S, Shiro M, Hirasawa Y. Antimitotic activity of lobaric acid and a new benzofuran, sakisacaulon A from Stereocaulon sasakii. Bioorg. Med. Chem. Lett. 2009;19:3679–3681. doi: 10.1016/j.bmcl.2009.03.170. [DOI] [PubMed] [Google Scholar]
- Moser W, Schindler C, Keiser J. Efficacy of recommended drugs against soil transmitted helminths: systematic review and network meta-analysis. BMJ. 2017;358:j4307. doi: 10.1136/bmj.j4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolovska-Coleska Z, Wang R, Fang X, Pan H, Tomita Y, Li P, Roller PP, Krajewski K, Saito NG, Stuckey JA, Wang S. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal. Biochem. 2004;332:261–273. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- Ozden S, Huerre M, Riviere J-P, Coffey LL, Afonso PV, Mouly V, de Monredon J, Roger J-C, El Amrani M, Yvin J-L, Jaffar M-C, Frenkiel M-P, Sourisseau M, Schwartz O, Butler-Browne G, Despres P, Gessain A, Ceccaldi P-E. Human muscle satellite cells as targets of Chikungunya virus infection. PLoS One. 2007;2:e527. doi: 10.1371/journal.pone.0000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubach JK, Wasik BR, Rupp JC, Kuhn RJ, Hardy RW, Smith JL. Characterization of purified Sindbis virus nsP4 RNA-dependent RNA polymerase activity in vitro. Virology. 2009;384:201–208. doi: 10.1016/j.virol.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp JC, Sokoloski KJ, Gebhart NN, Hardy RW. Alphavirus RNA synthesis and non-structural protein functions. J. Gen. Virol. 2015 doi: 10.1099/jgv.0.000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Bhagyawant SS, Parida M, Dash PK. Vector-delivered artificial miRNA effectively inhibited replication of Chikungunya virus. Antiviral Res. 2016;134:42–49. doi: 10.1016/j.antiviral.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki DL, Gomatos PJ. Replication of semliki forest virus: polyadenylate in plus-strand RNA and polyuridylate in minus-strand RNA. J. Virol. 1976 doi: 10.1128/jvi.20.2.446-464.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidel LM, Stollar V. Mutations that confer resistance to mycophenolic acid and ribavirin on Sindbis virus map to the nonstructural protein nsP1. Virology. 1991;181:490–499. doi: 10.1016/0042-6822(91)90881-b. [DOI] [PubMed] [Google Scholar]
- Shirako Y, Strauss JH. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 1994 doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirako Y, Strauss JH. Cleavage between nsP1 and nsP2 initiates the processing pathway of Sindbis virus nonstructural polyprotein P123. Virology. 1990;177:54–64. doi: 10.1016/0042-6822(90)90459-5. [DOI] [PubMed] [Google Scholar]
- Sissoko D, Malvy D, Ezzedine K, Renault P, Moscetti F, Ledrans M, Pierre V. Post-epidemic Chikungunya disease on Reunion Island: course of rheumatic manifestations and associated factors over a 15-month period. PLoS Negl. Trop. Dis. 2009;3:e389. doi: 10.1371/journal.pntd.0000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spuul P, Salonen A, Merits A, Jokitalo E, Kaariainen L, Ahola T. Role of the amphipathic peptide of Semliki forest virus replicase protein nsP1 in membrane association and virus replication. J. Virol. 2007;81:872–883. doi: 10.1128/JVI.01785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahla-Beek HJ, April DG, Saeedi BJ, Hannah AM, Keenan SM, Geiss BJ. Identification of a novel antiviral inhibitor of the flavivirus guanylyltransferase enzyme. J. Virol. 2012;86:8730–8739. doi: 10.1128/JVI.00384-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel JJ, Henderson BR, Lama SBC, Olson KE, Geiss BJ. Infectious alphavirus production from a simple plasmid transfection+ Virol. J. 2011;8:356. doi: 10.1186/1743-422X-8-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss EG, Rice CM, Strauss JH. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984;133:92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 1994 doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thadhani VM, Karunaratne V. Potential of Lichen Compounds as Antidiabetic Agents with Antioxidative Properties: A Review. Oxid. Med. Cell. Longev. 2017;2017:2079697. doi: 10.1155/2017/2079697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thal MA, Wasik BR, Posto J, Hardy RW. Template requirements for recognition and copying by Sindbis virus RNA-dependent RNA polymerase. Virology. 2007;358:221–232. doi: 10.1016/j.virol.2006.08.022. [DOI] [PubMed] [Google Scholar]
- Vasiljeva L, Merits A, Auvinen P, Kaariainen L. Identification of a novel function of the alphavirus capping apparatus. RNA 5’-triphosphatase activity of Nsp2. J. Biol. Chem. 2000;275:17281–17287. doi: 10.1074/jbc.M910340199. [DOI] [PubMed] [Google Scholar]
- Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Waymouth HE, Zoutman DE, Towheed TE. Chikungunya-related arthritis: case report and review of the literature. Semin. Arthritis Rheum. 2013;43:273–278. doi: 10.1016/j.semarthrit.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Weaver SC. Arrival of Chikungunya Virus in the New World: Prospects for Spread and Impact on Public Health. PLoS Negl. Trop. Dis. 2014 doi: 10.1371/journal.pntd.0002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Chung, Oldenburg A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.