Abstract
Background
Differences in cardiovascular diseases are evident in men and women throughout life and are mainly attributed to the presence of sex hormones and chromosomes. Epigenetic mechanisms drive the regulation of the biological processes that may lead to CVD and are possibly influenced by sex. In order to gain an overview of the status quo on sex differences in cardiovascular epigenetics, we performed a systematic review.
Materials and methods
A systematic search was performed on PubMed and Embase for studies mentioning cardiovascular disease, epigenetics, and anything related to sex differences. The search returned 3071 publications to be screened. Primary included publications focused on cardiovascular and epigenetics research. Subsequently, papers were assessed for including both sexes in their studies and checked for appropriate sex stratification of results.
Results
Two independent screeners identified 75 papers in the proper domains that had included both sexes. Only 17% (13 papers out of 75) of these publications stratified some of their data according to sex. All remaining papers focused on DNA methylation solely as an epigenetic mechanism. Of the excluded papers that included only one sex, 86% (24 out 28) studied males, while 14% (4 out of 28) studied females.
Conclusion
Our overview indicates that the majority of studies into cardiovascular epigenetics do not show their data stratified by sex, despite the well-known sex differences in CVD. All included and sex-stratified papers focus on DNA methylation, indicating that a lot of ground is still to gain regarding other epigenetic mechanisms, like chromatin architecture, and histone modifications. More attention to sex in epigenetic studies is warranted as such integration will advance our understanding of cardiovascular disease mechanisms in men and women.
Electronic supplementary material
The online version of this article (10.1186/s13293-018-0180-z) contains supplementary material, which is available to authorized users.
Keywords: Cardiovascular, Epigenetic, Sex, Gender, Stratification, DNA methylation, Systematic review
Background
Cardiovascular disease (CVD) is an annual leading cause of mortality across the world. The World Health Organization reported about 17.7 million deaths resulting from CVD in 2015, amounting to a staggering 31% of all the deaths that year [1]. CVD differs in men and women. Macroscopically, men have larger hearts and blood vessels as compared to women [2–4]. Clinically, men tend to develop CVD at a younger age, develop more severe coronary artery disease, and present with heart failure with reduced ejection fraction. Women present with CVD later in life when more comorbidities such as diabetes are present and more often develop non-obstructive coronary artery disease, and they preserve their ejection fraction when heart failure is diagnosed.
The field of epigenetics is rapidly growing, with increasingly more focus and highlight on the link between our epigenetic makeup and CVD etiology and predisposition. As the definition of epigenetics is rather vague and under continuing debate [5], we define epigenetic here as mitotically stable, non-sequence-dependent mechanisms such as DNA methylation and histone modifications that make up the epigenetic landscape. These mechanisms may subsequently influence gene expression, independently of the genetic code. While the amount of research linking epigenetics and CVD is climbing, it is of critical importance that these studies should be stratified according to sex. First, epigenetic mechanisms ensure the inactivation of the second X-chromosome in women, securing dosage compensation of the X-chromosome between men and women [6]. In addition, epigenetic mechanisms control sex-specific gene expression during development in tissue [7]. Furthermore, epigenetic mechanisms set the sex-specific stage for diseases later in life [7]. On top of that, the sex chromosomes contain multiple epigenetic modifiers which are differentially expressed between the sexes, which might influence the autosome in a sex-specific manner [8]. Moreover, steroid sex hormones such as estrogen and testosterone have been shown to affect epigenetic modifications [9, 10]. Another often overlooked reason is the sex-specific impact of the environment on gene expression regulation [7]. To summarize the current knowledge on sex differences in cardiovascular epigenetics, we performed a systematic review of available literature.
Methods
Search criteria and screening process
A systematic search was performed on 14 May, 2017, on PubMed (https://www.ncbi.nlm.nih.gov/pubmed) and Embase (https://embase.com/#search). The exact search strings used can be found in Additional file 1: Table S1. The screening process is depicted in Fig. 1. Two independent researchers (RH and SH) screened all papers found. Whenever there was disagreement about inclusion of papers, a third individual was involved, and the paper was discussed until consensus was reached. Duplicates were removed, and a title/abstract screen was performed to assess primary inclusion based on CVD and epigenetics. Subsequently, a full-text screen was performed to assess whether studies included both men and women and if interaction for sex was tested and whether or not sex-stratified results were presented. Full-texts were also double checked for the proper cardiovascular domain, including papers reporting on CVD (such as heart failure and atherosclerosis), but excluding papers reporting on diseases such as kidney disease. We included papers reporting on cardiovascular function, as well as papers reporting on CVD risk factors that belong to the Framingham Risk Score: age, gender, lipid levels, smoking, blood pressure, and adding BMI, metabolic syndrome, and diabetes. Papers focusing on non-modifiable risk factors such as ethnicity and family history were excluded. We excluded papers reporting on pregnancy-related epigenetics. Telomere length was not considered an epigenetic mechanism in this review. Data extracted for Table 1 included the CVD (risk factor) studied, the epigenetic mechanism, technique for measuring the epigenetic state, size study population and sex, tissue, the gene studied, and the association.
Fig. 1.

Workflow of the systematic search and review. A flow-chart regarding the systematic search and review process is shown, starting with the number of papers found in the top, leading to the number of papers included in the bottom
Table 1.
Summary of included papers performing proper sex stratification in the same order as mentioned in the main text
| Year | First author | CVD/risk factor | Epigenetics | Sample size (% male) | Technique | Tissue | Gene (if applicable) | Association | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| 2017 | Mendelson | BMI | DNA methylation | 3743 (48%) | Illumina Infinium 450K | Whole blood | LGALS3BP (unannotated CpG) | Stronger for ♂ | [12] |
| 2011 | Cash | Factors associated with obesity and CVD | DNA methylation | 355 (25%) | Bisulphite into pyrosequencing | Lymphocytes | Global | More methylation in ♂ | [13] |
| 2013 | Guay | Blood lipid levels | DNA methylation | 98 (62%) | Bisulphite into pyrosequencing | Leukocytes | CETP | Stronger for ♂ | [14] |
| 2014 | Guay | Plasma lipid levels | DNA methylation | 98 (62%) | Bisulphite into pyrosequencing | Leukocytes | Multiple | [15] | |
| 2013 | Zhang | Metabolic syndrome | DNA methylation | 517 (41%) | EpiTYPER | Leukocytes | FABP3 | [16] | |
| 2013 | Johansson | Aging | DNA methylation | 421 (not specified) | Illumina Infinium 450K | Leukocytes | Epigenome-wide | [17] | |
| 2016 | Horvath | Aging | DNA methylation | 4535 (35%) | Illumina Infinium 450K | Blood/saliva/brain | Epigenome-wide | [18] | |
| 2014 | Soriano-Tárrago | Ischemic stroke | DNA methylation | 485 (62%) | LUMA | Whole blood | Global | Hypomethylation for ♂ | [19] |
| 2017 | Lin | Ischemic stroke | DNA methylation | 556 (48%) | Bisulphite into pyrosequencing | Whole blood | MMP2 | Only for ♂ | [20] |
| 2012 | Talens | Myocardial infarction | DNA methylation | 248 (52%) | Mass spectrometry | Leukocytes | INS, GNASAS | Only in MI samples of ♀ | [21] |
| 2013 | Jiang | Coronary heart disease | DNA methylation | 72 (50%) | Bisulphite into pyrosequencing | Whole blood | PLA2G7 | Only for ♀ | [22] |
| 2016 | Guo | Coronary artery disease | DNA methylation | 64 (56%) | Methylation-specific PCR | Whole blood | PTX3 | Only for ♂ | [23] |
| 2014 | Zhang | CVD mortality | DNA methylation | 3588 (44%) | MALDI-TOF | Whole blood | F2RL3 | Stronger for ♂ | [24] |
BMI body mass index, Ref reference
Results
Systematic search
The combined search returned 3071 publications (Fig. 1). After duplicate removal, 2783 publications were left, which were then screened for their title and abstract, yielding 299 papers for the full-text screening. The 2484 excluded papers contain only one domain (cardiovascular, epigenetics, or sex differences) or no domain at all, are reviews/commentaries/book chapters, or are not published in English. Of the 299 primary included papers in cardiovascular epigenetics, 74 publications had included both men and women (Fig. 1). Of the 225 secondary excluded papers, 28 included only one sex and 10 papers did not mention the sex. Of the papers including one sex, 86% (24 out of 28) included only males and 14% (4 out of 28) included only females. We added one extra publication found via other sources to these 74 papers, leading to 75 papers. Startlingly, only 13 papers stratified their data for sex of the 75 publications that included males and females. In the end, these 13 papers were included for the review, all summarized in Table 1. We also excluded three papers from the 75 publications based on diverse criteria for not properly stratifying, although still showing male and female data (Fig. 1).
Review of included papers
All of the included papers look at DNA methylation as an epigenetic mechanism in either blood or leukocytes, with a variety of techniques, such as arrays or bisulphite treatment followed by pyrosequencing (Table 1). Therefore, a small intermezzo about DNA methylation is in place. In mammals, DNA methylation almost exclusively indicates DNA cytosine methylation. Cytosines targetable for methylation are practically always followed by a guanine, giving rise to the CpG dinucleotide [11]. Promoter areas of genes are enriched for CpG dinucleotides, and methylation of promoter regions enriched for CpGs is associated with repression of transcription [11]. DNA methylation plays a key role in maintenance of cell identity as well as in differentiation of cells by repressing transcription of genes which are obsolete in specific lineages [7].
The following section will review the included papers.
Sex differences in the epigenetics of cardiac risk factors
Seven papers looked at one or more risk factors for CVD. All of the seven papers report at least one difference found in epigenetic markers and/or their associations between men and women. Mendelson et al. looked at whole-blood DNA methylation and its relation to BMI [12]. Among 135 discovered CpGs, they report a significant sex interaction for an unannotated CpG positioned closest to the LGALS3BP gene. This CpG was also found in replication cohorts, with larger regression coefficients and smaller p values reported in men as compared to women.
Cash et al. show that LINE-1 methylation, used as a proxy for global DNA methylation, is higher in men as compared to women [13]. Furthermore, stratified analyses show LDL and HDL relationships with LINE-1 methylation only in men and a relationship between BMI and LINE-1 methylation only in women.
Guay et al. studied CETP and LPL promoter methylation and its relationship to blood lipid levels [14]. They report for both sexes negative association between CETP promoter methylation and LDL cholesterol levels (r < − 0.32; p < 0.05) and for men as well associations between CETP promoter methylation and HDL-C (r = − 0.36; p = 0.006), HDL-triglyceride levels (r = 0.59; p < 0.001), and HDL particle size (r = − 0.44, p = 0.019). Methylation of CETP and LPL promoters tend to be higher in women as compared to men.
Another paper by Guay et al. investigated epipolymorphisms in lipoprotein genes and their relationship to plasma lipid levels [15]. ABCG1 showed higher methylation in men as compared to women (p = 0.007), while a CpG in PLTP showed higher methylation in women as compared to that in men (p = 0.025). Higher methylation of CpGC3 in ABCG1 in women was associated with lower TG levels, while in men this methylation was associated with age, a larger waist circumference, and lower total cholesterol levels. Higher methylation of LIPC-CpGA2 in men was associated with lower HDL-C and TG levels, while a positive trend was noticeable for TG levels in women. They also show that epipolymorphisms in ABCG1, LIPC, PLTP, and CETP contributed to variations found in plasma lipid levels, with stronger associations in men, independently of traditional predictors.
Zhang et al. examined DNA methylation in different CpGs of FABP3 and its relationship to insulin, lipids, and cardiovascular phenotypes [16]. They found that multiple different CpGs within FABP3 were influenced by sex, indicating that specific sites within genes are prone to sex differences, which might relate differently to disease outcome.
Johansson et al. looked into the relationship of aging and DNA methylation in white blood cells [17]. They report significant interactions between aging and sex for 163 CpGs, of which 152 CpGs are located on the X-chromosome. It is not reported which CpG shows a significant sex interaction.
Horvath et al. looked at epigenetic aging rates in different tissues by using DNA methylation [18]. They show that men have higher epigenetic aging rates than women in blood, saliva, and brain tissue. Furthermore, epigenetic aging rates were also associated with CVD risk factors, but not with CVD outcomes.
Sex differences in the epigenetics of cardiovascular disease and mortality
Five papers looked directly at CVD outcomes, of which ischemic stroke, myocardial infarction, coronary heart disease and coronary artery disease, and one included paper investigated cardiovascular mortality.
Soriano-Tárrago et al. studied global DNA methylation of ischemic strokes subtypes [19]. No distinct differences were found between the different ischemic stroke subtypes regarding DNA methylation, but global DNA hypomethylation was described in men as compared to women over all samples.
Lin et al. looked at methylation of MMP2 in ischemic stroke [20]. They show that patients suffering from ischemic stroke have lower levels of MMP2 methylation at multiple CpGs as compared to healthy controls. This association was stronger in men (p values ranging from 0.001–0.056) as compared to women (p values ranging from 0.051–0.354).
Talens et al. looked into DNA methylation of loci sensitive to prenatal environment and its relationship to myocardial infarction [21]. DNA methylation of INS and GNASAS in leukocytes were higher in myocardial infarction in females (INS: p = 0.002; GNASAS: p = 0.001), with no associations found in men. This indicates that the sex-specific stage for disease predisposition might be set very early in life.
Jiang et al. focused on PLA2G7 promoter methylation and its relation to coronary heart disease [22]. They describe a signification association between PLA2G7 promoter methylation and coronary heart disease in women (p = 0.003), which was not found in men (p = 0.096). Moreover, this association was independent of age, smoking, hypertension, and diabetes. The PLA2G7 promoter methylation was also associated with total cholesterol levels, triglyceride levels, and apolipoprotein B in women, but not in men.
Guo et al. investigated promoter DNA methylation of PTX3 in coronary artery disease [23]. They show that lower methylation of PTX3 is associated with higher PTX3 plasma levels. In men only, lower PTX3 methylation was associated with a higher neutrophil to lymphocyte ratio (r = − 0.58, p = 0.002).
Zhang et al. studied methylation of F2RL3, an epigenetic biomarker of smoking exposure, and its relationship to mortality outcomes [24]. They found that lower methylation intensity of F2RL3 correlated strongly with mortality outcomes, as well as cardiovascular mortality. The associations found were all much stronger in men as compared to women.
Discussion
In this systematic review of over 3000 publications found by the search, we highlight the insufficient amount of sex stratification performed in cardiovascular epigenetic research and reviewed the available stratified data according to CVD risk factors and their etiology.
Sex stratification in cardiovascular epigenetics
Our overview indicates that the majority of studies into cardiovascular epigenetics do not show their data stratified by sex, despite the well-known sex differences in CVD. Moreover, from the papers describing only one sex, 86% studied men and 14% studied women suggesting that the underrepresentation of women is an ongoing phenomenon, while the knowledge on the female pathology of CVD is limited. Women have not been equally represented in clinical trials for CVD as compared to men [25], as women have been excluded to protect them from any adverse drug reactions after the thalidomide tragedy in the 1950s. On top of that, women are often excluded because of fluctuating hormone levels starting from puberty, which complicate the implementation of acquired data. Another reason why women have been underrepresented is that women develop CVD later in life, while suffering from multiple comorbidities, leading to exclusion of studies. In larger genome-wide association studies, sex chromosomes are often dismissed, as analysis of the sex chromosomes is laborious and requires different algorithms as compared to autosomal chromosomes [26]. Furthermore, most animal studies do not have an equal representation of the sexes [27], while it has been demonstrated that a large proportion of mammalian traits are influenced by sex in wild-type and mutant animals [28].
Evidently, our results (Fig. 1) point to the scarcity of sex stratification in cardiovascular epigenetics. Of the 75 studies that described both sexes to be present in their data, only 13 publications had performed sex stratification in their studies. Often, associations between specific epigenetic markers and cardiovascular phenotypes were found, while only holding true in one of the sexes. Papers that were included primarily, but excluded during the second round, often immediately adjust for the effect of sex, as sex is known to have a major impact on epigenetics. Consequently, the lack of proper stratification can lead to the overlooking of sex-specific epigenetic effects.
Global DNA methylation
Differences in global DNA methylation profiles between the sexes are an area of open debate. Some of the included studies had conflicting conclusions about DNA methylation. For example, Cash et al. demonstrated that there is a higher global DNA methylation in males [13], while Soriano-Tárraga et al. showed that hypomethylation is associated with the male sex [19]. The latter paper looked at methylation in whole blood, while the first studied DNA methylation in lymphocytes, which might potentially explain some of the differences seen here, as the epigenetic makeup is tissue-dependent. Johansson et al. also pointed out a lower median autosomal DNA methylation in men as compared to women, while mean levels of DNA methylation did not differ [17]. This indicates that the distribution of methylation on the autosome might be subject to sex differences, as seen for the sex chromosomes. Interestingly, men showed a higher degree of epigenetic aging based on DNA methylation in three out of the three tissues as compared to women [18]. This points towards a general epigenetic mechanism in different tissues that is influenced by sex. However, zooming in on specific genes affected by the differential methylation might provide more information on epigenetic mechanisms that differ between men and women. The included papers point towards general genome-wide and specific regulatory mechanisms influenced by sex.
Epigenetic mechanisms
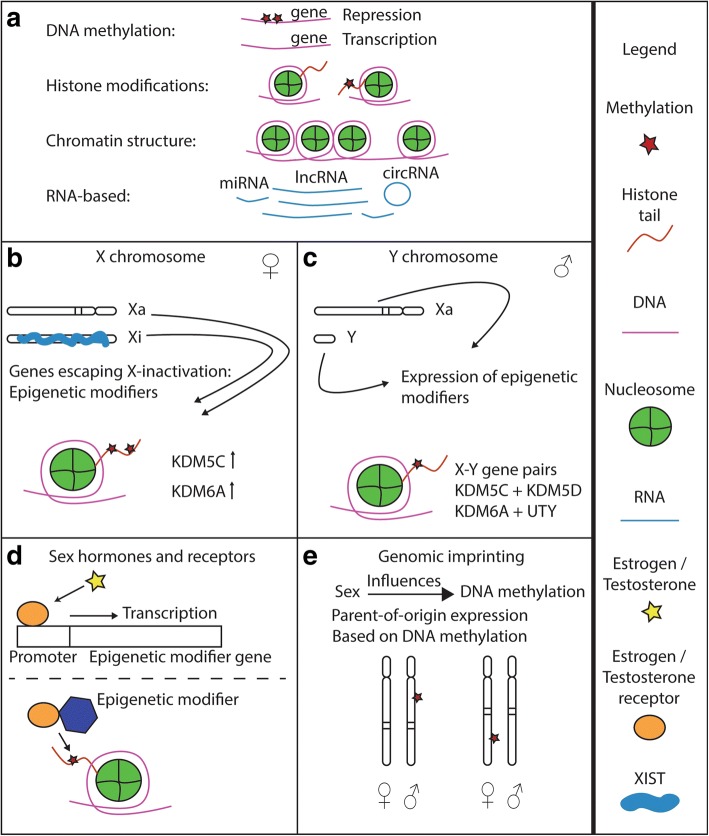
We observed that DNA methylation was the only highlighted epigenetic mechanism in association with cardiovascular phenotypes (see Fig. 2a for a schematic on common epigenetic mechanisms). One reason could be that it is less costly and labor-intensive to investigate genome-wide DNA methylation on widely available arrays, as compared to investigating other epigenetic mechanisms that are not yet available on standard platforms, such as chromatin confirmation capture (3C, 4C, Hi-C) sequencing, assay for transposase accessible (ATAC) sequencing, and chromatin immunoprecipitation (ChIP) sequencing. Nevertheless, the sample size of the included studies ranges from 64 to well over 3000, although the larger epigenome-wide association studies are scarce. Future advances in ChIP sequencing should allow for larger cohort histone modification studies as well. Another epigenetic feature that may be studied on a broader scale in the near future is chromatin organization, with assays such as Hi-C sequencing or ATAC sequencing.
Fig. 2.
Epigenetic mechanisms and sex. a The epigenetic mechanisms as set forth in the main text are schematically depicted here, with a legend on the right. b Possible ways in which sex influences epigenetic mechanisms. Higher expression of X-chromosomal genes that escape inactivation in women might influence epigenetic markers differently as compared to men. c The Y chromosome contains different epigenetic modifiers which might influence the autosome in men, but not in women. d Sex hormones and their receptors can influence the gene expression of epigenetic modifiers, as well as interact with epigenetic modifiers. e Genomic imprinting is an example in which sex influences DNA methylation, as particular maternal or paternal alleles are differentially methylated
Influence of sex on epigenetic mechanisms
There are multiple mechanisms by which sex might directly influence cardiovascular epigenetics (see Fig. 2b-e): (1) The increased expression of X-chromosomal escape genes in women, of which some target epigenetic modifications; (2) the expression of non-pseudo-autosomal Y-chromosomal epigenetic modifiers in men; (3) the (non-)genomic effect of steroid hormones and their receptors on epigenetic regulators, such as DNA methylation enzymes, histone modifiers, and miRNAs; and (4) genomic imprinting, leading for example to DNA methylation of either maternal or paternal alleles. The effect of sex on epigenetic marks might subsequently lead to changes in gene expression, culminating in sex differences in CVD.
Influence of the non-coding genome
The action of microRNAs is considered to be an epigenetic mechanism. Differential miRNA expression is very likely to exist between the sexes, as the X-chromosome contains about 10% of miRNAs harbored in the human genome. This higher density of miRNAs on the X-chromosome as compared to autosomes is noted in other mammalian species as well [29]. This suggests that the X-chromosome has an indispensable role in miRNA-mediated regulation of gene expression, and it might be interesting to find out if any of these miRNAs associate with CVD. A recent review has focused on the potential role of sex-biased miRNAs in the etiology of heart failure with preserved ejection fraction, explaining how sex also affects this epigenetic feature in a syndrome presenting with differential prevalence between the sexes [30]. In addition to miRNAs, the regulatory role of long non-coding RNAs (lncRNA) and circular RNAs should be looked at in a sex-specific way. One of the most studied lncRNAs is XIST, which is crucial for establishing X-chromosomal gene expression dosage compensation in women [6].
Sex stratification and statistical power
Our systematic search revealed that sex stratification is rare in cardiovascular epigenetics, even when sex terms were incorporated in the search string to look for sex-specific epigenetic mechanisms. When sex terms were excluded from the search string, we were returned with twice the number of papers, indicating that the actual definite amount of sex stratification done in cardiovascular epigenetic studies could be even lower. Studies with small sample sizes are limited by their statistical power to detect sex differences, resulting in possible exclusion of sex stratification during their experimental design. CVD with a low prevalence, such as specific gene mutation cardiomyopathies, might be difficult to study in a sex-specific manner, as statistical power will be problematic to acquire. An increase in sample size is not always easy to obtain for power, which warrants different teams to collaborate. Large data sets that permit answers to multiple questions, which can be built as a joint force, will grant more power for testing interactions. Meta-analyses of already existing datasets might give insights to the power issue as well. For future studies on CVD, uniformity in arrays and techniques will aid detection of sex interaction in epigenome-wide designs. Another option might be to let journals require authors to present sex-stratified data in the supplementary material (regardless of significant interactions). Some studies might have investigated the interaction of sex and epigenetic processes, but did not report them in their results. It is valuable to mention a tested but not deviating sex interaction as it points towards similar biology between men and women. A number of excluded studies which did not stratify accordingly for sex mentioned the importance of sex as a variable in epigenetic research and subsequently adjusted for the effect of sex in their model, potentially missing out on mechanisms and drug targets that are sex-specific. Although the knowledge on the importance of sex in biomedical sciences is emerging, a declaration of sex-interaction testing in journals would be incredibly helpful for a wide-scale and fair investigation into the bona fide differences between epigenetic mechanisms in the sexes. The National Institutes of Health in the USA already requires investigators to account for sex and gender, and more measures are being taken to hold scientists more accountable for implementing sex as a biological variable [31].
Limitations
A broad overview of microRNA differences between the sexes was not performed, as the search string limited the amount of papers found on microRNAs, e.g., RNA interference is a subheading under the MeSH term “epigenesis,” but not every paper on microRNAs might be indexed in this fashion. A meta-analysis on transcriptomic differences and thus as well expression of microRNAs between the sexes in different types of cardiovascular tissues would be of value to address this. Although we did not include a publication date criteria, all of the included papers had been published in the last decade, demonstrating that more attention has been given to proper sex stratification recently. However, the status quo is still far from ideal. A limitation of our study is that we limited our search to two databases and did not include pre-print archives or other databases. Therefore, we cannot make statements on the total body of literature available in all systems.
Conclusion
Our systematic review highlighted the lack of stratification in cardiovascular epigenetics, with only 13 out of 75 included publications stratified according to sex. Most of the papers that were excluded regressed out the effect(s) of sex in their studies. Together, our review underscores the acute need to investigate the effects of sex on epigenetic mechanisms as current research reflects the strong influence of sex on epigenetic regulatory processes.
Additional file
Table S1. Search strings used for PubMed and Embase. (DOCX 14 kb)
Acknowledgments
Funding
This study was funded by the Dutch Heart Foundation (2013T084, Queen of Hearts).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- CpG
Cytosine guanine dinucleotide
- CVD
Cardiovascular disease
Authors’ contributions
RH wrote the manuscript, RH and SH performed the screen, SH performed the search, HdR and RH conceptualized the project, and HdR reviewed the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13293-018-0180-z) contains supplementary material, which is available to authorized users.
Contributor Information
Robin J. G. Hartman, Email: r.j.g.hartman-2@umcutrecht.nl
Sarah E. Huisman, Email: sarahhuisman@live.nl
Hester M. den Ruijter, Email: h.m.denruijter-2@umcutrecht.nl
References
- 1.World Health Organization. Noncommunicable diseases. 2017. http://www.who.int/en/news-room/fact-sheets/detail/noncommunicable-diseases. Accessed 27 Feb 2018.
- 2.Gaitskell K, Perera R, Soilleux EJ. Derivation of new reference tables for human heart weights in light of increasing body mass index. J Clin Pathol. 2011;64:358–362. doi: 10.1136/jcp.2010.084574. [DOI] [PubMed] [Google Scholar]
- 3.Hiteshi AK, Li D, Gao Y, Chen A, Flores F, Mao SS, et al. Gender differences in coronary artery diameter are not related to body habitus or left ventricular mass. Clin Cardiol. 2014;37:605–609. doi: 10.1002/clc.22310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi Y, Fearon WF, Honda Y, Tanaka S, Pargaonkar V, Fitzgerald PJ, et al. Effect of sex differences on invasive measures of coronary microvascular dysfunction in patients with angina in the absence of obstructive coronary artery disease. JACC Cardiovasc Interv. 2015;8:1433–1441. doi: 10.1016/j.jcin.2015.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deans C, Maggert KA. What do you mean, “epigenetic”? Genetics. 2015;199:887–896. doi: 10.1534/genetics.114.173492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet. 2011;12:429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- 7.Hochberg Z, Feil R, Constancia M, Fraga M, Junien C, Carel JC, et al. Child health, developmental plasticity, and epigenetic programming. Endocr Rev. 2011;32:159–224. doi: 10.1210/er.2009-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wijchers PJ, Festenstein RJ. Epigenetic regulation of autosomal gene expression by sex chromosomes. Trends Genet. 2011;27:132–140. doi: 10.1016/j.tig.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Mourad R, Hsu P-Y, Juan L, Shen C, Koneru P, Lin H, et al. Estrogen induces global reorganization of chromatin structure in human breast cancer cells. PLoS One. 2014;9:e113354. doi: 10.1371/journal.pone.0113354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dkhil MA, Al-Quraishy S, Abdel-Baki AA, Ghanjati F, Arauzo-Bravo MJ, Delic D, et al. Epigenetic modifications of gene promoter DNA in the liver of adult female mice masculinized by testosterone. J Steroid Biochem Mol Biol. 2015;145:121–130. doi: 10.1016/j.jsbmb.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Illingworth RS, Bird AP. CpG islands—“a rough guide”. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Mendelson MM, Marioni RE, Joehanes R, Liu C, Hedman ÅK, Aslibekyan S, et al. Association of body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: a Mendelian randomization approach. PLoS Med. 2017;14:1–30. doi: 10.1371/journal.pmed.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cash HL, Mcgarvey ST, Houseman EA, Marsit CJ, Hawley NL, Lambert-Messerlian GM, et al. Cardiovascular disease risk factors and DNA methylation at the LINE-1 repeat region in peripheral blood from Samoan Islanders. Epigenetics. 2011;6:1257–1264. doi: 10.4161/epi.6.10.17728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guay SP, Brisson D, Lamarche B, Marceau P, Vohl MC, Gaudet D, et al. DNA methylation variations at CETP and LPL gene promoter loci: new molecular biomarkers associated with blood lipid profile variability. Atherosclerosis. 2013;228:413–420. doi: 10.1016/j.atherosclerosis.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 15.Guay SP, Brisson D, Lamarche B, Gaudet D, Bouchar L. Epipolymorphisms within lipoprotein genes contribute independently to plasma lipid levels in familial hypercholesterolemia. Epigenetics. 2014;9:718–729. doi: 10.4161/epi.27981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Kent JW, Lee A, Cerjak D, Ali O, Diasio R, et al. Fatty acid binding protein 3 (fabp3) is associated with insulin, lipids and cardiovascular phenotypes of the metabolic syndrome through epigenetic modifications in a Northern European family population. BMC Med Genet. 2013;6(6:9):1–14. [DOI] [PMC free article] [PubMed]
- 17.Johansson Å, Enroth S, Gyllensten U. Continuous aging of the human DNA methylome throughout the human lifespan. PLoS One. 2013;8:e67378. doi: 10.1371/journal.pone.0067378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol 2016;17:0–22. doi:10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed]
- 19.Soriano-Tárraga C, Jiménez-Conde J, Giralt-Steinhauer E, Mola M, Ois Á, Rodríguez-Campello A, et al. Global DNA methylation of ischemic stroke subtypes. PLoS One. 2014;9(4):e96543. [DOI] [PMC free article] [PubMed]
- 20.Lin H-F, Hsi E, Huang L-C, Liao Y-C, Juo S-HH, Lin R-T. Methylation in the matrix metalloproteinase-2 gene is associated with cerebral ischemic stroke. J Investig Med. 2017;65:794–799. doi: 10.1136/jim-2016-000277. [DOI] [PubMed] [Google Scholar]
- 21.Talens RP, Jukema JW, Trompet S, Kremer D, Westendorp RGJ, Lumey LH, et al. Hypermethylation at loci sensitive to the prenatal environment is associated with increased incidence of myocardial infarction. Int J Epidemiol. 2012;41:106–115. doi: 10.1093/ije/dyr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang D, Zheng D, Wang L, Huang Y, Liu H, Xu L, et al. Elevated PLA2G7 gene promoter methylation as a gender-specific marker of aging increases the risk of coronary heart disease in females. PLoS One. 2013;8:1–7. doi: 10.1371/journal.pone.0059752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo TM, Huang LL, Liu K, Ke L, Luo ZJ, Li YQ, et al. Pentraxin 3 (PTX3) promoter methylation associated with PTX3 plasma levels and neutrophil to lymphocyte ratio in coronary artery disease. J Geriatr Cardiol. 2016;13:712–717. doi: 10.11909/j.issn.1671-5411.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Yang R, Burwinkel B, Breitling LP, Holleczek B, Schöttker B, et al. F2RL3 methylation in blood DNA is a strong predictor of mortality. Int J Epidemiol. 2014;43:1215–1225. doi: 10.1093/ije/dyu006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melloni C, Berger JS, Wang TY, Gunes F, Stebbins A, Pieper KS, et al. Representation of women in randomized clinical trials of cardiovascular disease prevention. Circ Cardiovasc Qual Outcomes. 2010;3:135–142. doi: 10.1161/CIRCOUTCOMES.110.868307. [DOI] [PubMed] [Google Scholar]
- 26.König IR, Loley C, Erdmann J, Ziegler A. How to include chromosome X in your genome-wide association study. Genet Epidemiol. 2014;38:97–103. doi: 10.1002/gepi.21782. [DOI] [PubMed] [Google Scholar]
- 27.Beery AK, Zucker I. Sex bias in neuroscience and biomedical research. Neurosci Biobehav Rev. 2011;35:565–572. doi: 10.1016/j.neubiorev.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karp NA, Mason J, Beaudet AL, Benjamini Y, Bower L, Braun RE, et al. Prevalence of sexual dimorphism in mammalian phenotypic traits. Nat Commun. 2017;8:15475. doi: 10.1038/ncomms15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo X, Su B, Zhou Z, Sha J. Rapid evolution of mammalian X-linked testis microRNAs. BMC Genomics. 2009;10:1–8. doi: 10.1186/1471-2164-10-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Florijn BW, Bijkerk R, van der Veer E, van Zonneveld AJ. Gender and cardiovascular disease: are sex-biased miRNA networks a driving force behind heart failure with preserved ejection fraction in women? Cardiovasc Res. 2017; 10.1093/cvr/cvx223. [DOI] [PubMed]
- 31.Ventura-Clapier R, Dworatzek E, Seeland U, Kararigas G, Arnal JF, Brunelleschi S, et al. Sex in basic research: concepts in the cardiovascular field. Cardiovasc Res. 2017;113:711–724. doi: 10.1093/cvr/cvx066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Search strings used for PubMed and Embase. (DOCX 14 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article.