Abstract
IgLON family is a subgroup of cell adhesion molecules which is known to have diverse roles in neuronal development. IgLONs are characterized by possessing 3 Ig-like C2 domains, which play a part in mediating various cellular interactions. Recently, IgLONs have been shown to be expressed at the blood-brain barrier (BBB). However, our understanding of the genetic divergence patterns and evolutionary rates of these proteins in relation to their functions, in general, and at the BBB, in particular, remains inadequate. In this study, 12 species were explored to shed more light on the phylogenetic origins, structure, functional specificity, and divergence of this family. A total of 40 IgLON genes were identified from vertebrates and invertebrates. The absence of IgLON family genes in Hydra vulgaris and Nematostella vectensis but not in Drosophila melanogaster suggests that this family appeared during the time of divergence of Arthropoda 455 Mya. In general, IgLON genes have been subject to strong positive selection in vertebrates. Our study, based on IgLONs’ structural similarity, suggests that they may play a role in the evolutionary changes in the brain anatomy towards complexity including regulating neural growth and BBB permeability. IgLONs’ functions seem to be performed through complex interactions on the level of motifs as well as single residues. We identified several IgLON motifs that could be influencing cellular migration and proliferation as well as BBB integrity through interactions with SH3 or integrin. Our motif analysis also revealed that NEGR1 might be involved in MAPK pathway as a form of a signal transmitting receptor through its motif (KKVRVVVNF). We found several residues that were both positively selected and with highly functional specificity. We also located functional divergent residues that could act as drug targets to regulate BBB permeability. Furthermore, we identified several putative metalloproteinase cleavage sites that support the ectodomain shedding hypothesis of the IgLONs. In conclusion, our results present a bridge between IgLONs’ molecular evolution and their functions.
Keywords: IgLON, evolution, drug targets, brain-blood barrier
Introduction
Understanding the role of cell adhesion in the blood-brain barrier (BBB) is crucial in treating various central nervous system (CNS) pathologies. The BBB is a specialized system of brain microvascular endothelial cells (BMVECs) that shield the brain, by controlling the access of different blood-carried components to different brain areas. This is done through permitting the needed nutrients while preventing the access of toxic substances from the blood to the brain, as well as filtering harmful compounds from the brain back to the bloodstream.1,2 This is achieved by various interactions between BMVEC and other components of the neurovascular unit (astrocytes, pericytes, neurons, and basement membrane). Cell adhesion molecules play an important part in controlling these interactions, including adherent junctions and leukocyte migration.2 Studying the evolution of cell adhesion molecules can shed more light on their functional specificity and help build a clearer picture of structure-function relationship within the neurovascular unit. This can lead to controlling BBB permeability and hence enhancing drug delivery into the brain in several CNS diseases (eg, HIV-1 encephalitis, Alzheimer disease, ischaemia, tumours, multiple sclerosis, and Parkinson disease).
The relevance of IgLONs in the BBB is not yet known. IgLONs are a family of 5 glycosylphosphatidylinositol (GPI)-anchored cell adhesion molecules.3 They are formed of 3 Ig-like C2 domains.4 The family includes LSAMP (limbic system–associated membrane protein), NTM (neurotrimin), OPCML (opioid-binding cell adhesion molecule), NEGR1 (neuronal growth regulator 1), and IgLON5.5 IgLONs’ functions include neurite outgrowth regulation,6,7 dendritic arborization,8 and synapse formation.9 IgLONs are known to execute their function through forming homophilic and heterophilic complexes along the cell surface.4,10 Lachesin which is an IgLON homologue11 is required for the function of the BBB in Drosophila.12 In mice, Lsamp, Opcml, Negr1 were shown to be expressed in oligodendrocytes.13 Oligodendrocytes are needed for trophic coupling between cerebral endothelium and other components of the BBB (eg, pericytes and astrocytes).14,15 Opcml was shown to be expressed by cerebral type 1 and 2 astrocytes, which compromise one of the main components of the BBB in rats,16,17 Interestingly, Opcml was also indicated to be stronger in regulating astrocytes’ proliferation and cell size than other IgLON proteins.18 In the cerebellum, Immuno-EM revealed expression of Ntm on the surface of unmyelinated axons but not on astrocytes or oligodendroglia.19 An extensive RNA-Seq study of the mouse cerebral cortex20 concentrated on investigating the expression landscape of endothelial cells, neurons, astrocytes, oligodendrocytes, and microglia constituting the BBB. The study revealed that all IgLON members were expressed in neurons, astrocytes, myelinating oligodendrocytes, and precursor oligodendrocytes, whereas only Iglon5 and Ntm were expressed in microglia. Notably, only Ntm was expressed in endothelial cells. However, IgLONs’ functions and their nature of interactions (eg, homophilic, heterophilic, or both) within the respective components of the BBB are still to be uncovered.
The role of molecular evolution in dictating the functional specificity and divergence of the IgLON in general and with respect to the BBB specifically remains elusive. The evolutionary history of the IgLON is still unknown. NCAM family, which is homologous to IgLON family, was detected in embryonic and adult vertebrates as well as adult mollusc, but not in adult insects, crustacean, or nematodes.21 Conversely, Lachesin was detected in Drosophila.12 Furthermore, it was revealed that near full-length IgLON family members are processed in a metalloproteinase-dependent manner, similar to L1-CAM and NCAM, to facilitate neuronal extension in cortical neurons.10 However, the site of metalloproteinase cleavage has not been characterized.10 Moreover, the selection pressure of several genes belonging to the IgSF (the superfamily encompassing IgLON) has been investigated, suggesting that several IgSF genes were under positive selection in primates.22 Evolutionary selection was not investigated in other vertebrates or invertebrates and the functional specificity of the IgLON family members is still not known. Interestingly, it was shown that both increasing and knocking down of Negr1 expression lead to decreases in locomotor activity and body temperature.23 Functional studies have shown that Lsamp can both promote and inhibit neurite outgrowth7 but no specific amino acids were shown to control its function.
This article studied the relationship between the molecular evolution of the IgLON family and its function. We aligned IgLONs’ sequences from 10 species. This was followed by building a phylogenetic tree using both Bayesian approach and maximum likelihood analysis. Then, we studied positive selection of the IgLON family investigating global, branch, sites and branch-sites models. Next, we explored IgLONs’ similar structures, putative GPI-anchored positions, shared motifs, putative metalloproteinase cleavage sites, and residues with highly functional specificity. Finally, we performed functional divergence analysis. The results of this research allowed us to predict putative relationships between different IgLON groups’ amino acid residues and their functions.
Methods
Database search
The focus of this research was investigating the relationship between IgLONs’ molecular evolution and their functions. IgLON family belongs to the immunoglobulin superfamily. Due to the diverse nature and long evolutionary history of the IgSF,24 we inferred that using DNA data to generate the alignments is likely to misrepresent the mutational history of the IgLONs. We reasoned that studying the protein sequences rather than the DNA sequences could be more informative. Moreover, to make sure that our analysis is a reasonable representation of IgLONs’ evolutionary history, we chose 12 species that span more than 500 million years. Human IgLON protein family was used for BLASTP searches against proteomes of Western gorilla (Gorilla gorilla), Common rat (Rattus norvegicus), House mouse (Mus musculus), Carolina anole (Anolis carolinensis), red junglefowl (Gallus gallus), African clawed frog (Xenopus laevis), Zebrafish (Danio rerio), sea squirt (Ciona intestinalis), common fruit fly (Drosophila melanogaster), sea anemone (Nematostella vectensis), and Hydra (Hydra vulgaris). Whenever one protein possessed more than one transcript, only the longest transcript was used in the analysis. Hence, we did not take into consideration the role of the recently identified 2 promoters’ genomic structure of Lsamp, Ntm, and Opcml.5 Sequences were selected as candidate proteins if their E values were ≤1e-10. Sequences were further filtered for having 3 Ig-like domains using CDD.25
Alignment and phylogenetic analysis
Phylogenetic investigation was done in 3 steps. First, IgLON family amino acid sequences were aligned using MAFFT26 via the iterative refinement method (FFT-NS-i). After that, we employed ProtTest27 to determine the best amino acid replacement model. ProtTest results based on the Akaike information criterion (AIC) suggested that the best substitution model is LG+I+G+F, where LG is the substitution model supplemented by a fraction of invariable amino acids (‘+I’) with each site assigned a probability of belonging to given rate categories (‘+G’) and observed amino acid frequencies (‘+F’).27 The third step included using the protein alignment and the resulting substitution model, in applying 2 different phylogenetic methods to construct the tree, namely, (1) maximum likelihood and (2) Bayesian inference. We performed the maximum likelihood analysis using PHYML28 implemented in Seaview29 with 5 random starting trees. We applied Bayesian inference analysis using MrBayes30 where we implemented a Markov chain Monte Carlo analysis with 1000,000 generations to approximate the posterior probability and a standard deviation of split frequencies <0.01 to indicate convergence as previously described.30
Positive selection analysis
To determine whether members of the IgLON family underwent positive selection during evolution, a maximum likelihood approach was employed. In the first instance, respective complementary DNAs (cDNAs) were downloaded for each of the IgLON protein sequences and aligned according to their codon arrangement. Next, we investigated positive selection using CODEML which is part of PAML v4.4 program suite.31 We used substitution rate ratio (ω) of nonsynonymous (dN) to synonymous (dS) mutations as an indicator of the selection type.31 The models used were basic, branch, branch-site, and sites models. The basic model calculates a global ω ratio averaged over all sites and all lineages in the tree. The branch model permits ω ratio to vary among branches in the phylogeny. Using model 2 option, we calculated 2 ω values, the first is specific for the investigated branch in the tree and the other value is for the rest of the branches in the tree. Branch-site models allow ω to vary both among sites in the protein and across branches on the tree. Branch-site models detect positive selection affecting only a few sites along the tested branches. For site models, 2 tests were performed, namely, (M1a versus M2a) and (M7 versus M8). The nearly neutral model (M1a) covers sites under purifying selection (0 < dN/dS < 1) as well as sites under neutral evolution (dN/dS = 1), whereas the positive selection model (M2a) includes sites that evolved under positive selection (dN/dS > 1). Model M7 uses a beta distribution for ω over sites limited to the interval (0, 1), thus it was used as the null hypothesis. Model M8 adds another site class to M7 with the ω ratio estimated from the data. For each model investigated, a likelihood ratio test (LRT) was performed between the respective model and its neutral counterparts. P values were calculated for the χ2 test according to the following equation; P value = χ2 (2*Δ(ln(LRTmodel) − ln(LRTneutral)), number of degrees of freedom).
Linear motifs, GPI prediction, metalloproteinase cleavage site prediction
To investigate IgLONs’ molecular evolution-function relationship, we used 3 approaches. First, we searched the protein sequences of the IgLONs for linear motifs. These motifs are composed of short stretches of adjacent amino acids that act as putative protein interaction sites. We performed the search using the ELM server http://elm.eu.org/ with a motif cut-off value of 100.32 In the second analysis, putative GPI anchoring positions were predicted using GPI prediction tool http://mendel.imp.ac.at/gpi/cgi-bin/gpi_pred.cgi where a search was performed to locate cleavage site residues that could anchor sites for the GPI signal as described in the work by Pierleoni et al.33 Third, we used the server https://prosper.erc.monash.edu.au to detect putative metalloproteinase cleavage sites.34
Functional specificity
We employed SDPpred35 to identify specificity-determining positions (SDPs) in IgLONs’ multiple alignment sequences. The SDPs are amino acid residues that could be responsible for the functional specificity of IgLON proteins. SDPpred was used to compute a Z score for each alignment column. Z score is an indicator of the probability that the position is a real specificity determinant. The Bernoulli estimator was used within SDPpred to create a recognition cut-off (B-cut-off) to assess the significance of the Z scores. Positions with a score value larger than B-cut-off were accepted to predict higher specificity probability.
Functional divergence estimation
Type I functional divergence between gene clusters of the IgLON family was estimated through posterior analysis using the DIVERGE v2.0 program.36 Functional type I divergence determines amino acid that are highly different in their conservation between 2 groups, indicating that these residues have undergone altered functional constraints.36 IgLONs were clustered into the following clusters: invertebrates, IgLON5 (vertebrates), LSAMP (vertebrates), OPCML (vertebrates), NEGR1 (vertebrates), and NTM (vertebrates). The clusters were pairwise compared. The coefficient of type I functional divergence (θI) was calculated between members of all pairs of clusters. θI values significantly larger than 0 indicated site-specific altered selective constraints or shifts in amino acid physiochemical properties that might have resulted by gene duplication and/or speciation.
Results
Alignment and phylogenetic tree
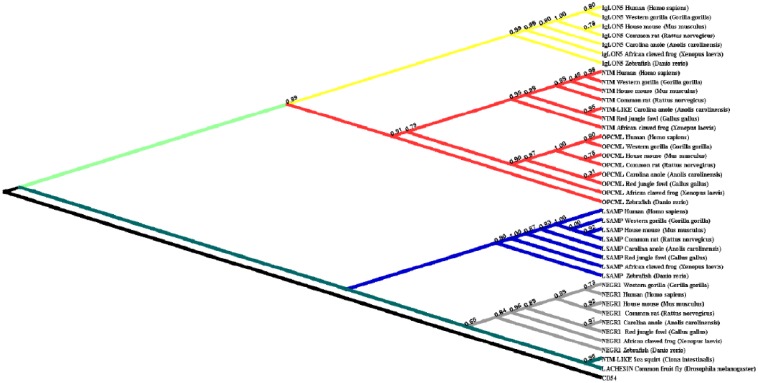
Phylogenetic investigations identified IgLON members in both vertebrates and invertebrates. In invertebrates, Drosophila melanogaster IgLON members seemed to have 1 gene (Figure 1). We also found only 1 paralog in Ciona intestinalis. IgLON evolution in vertebrates followed the typical 2 rounds of duplication (2R) model that started during the Ordovician period, around 450 Mya. The first round resulted in 2 genes. During the second round, one of these genes acted as the potential common ancestor of NEGR1 and LSAMP, whereas the other was potentially the parent of IgLNO5 and the ancestral sequence for NTM and OPCML (Figure 2).
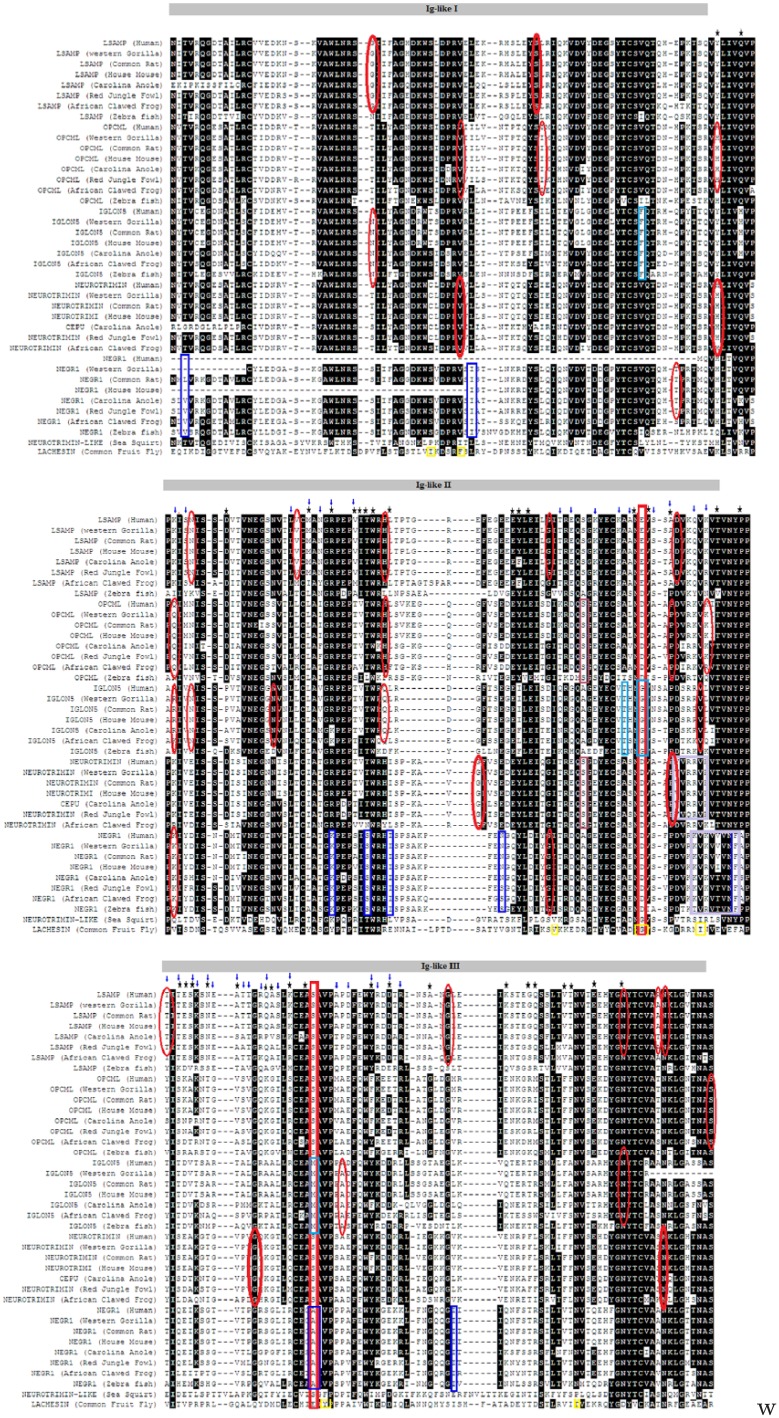
Figure 1.
Alignment of the IgLON family. The 3 Ig-like domains shared by all family members are shown above the alignment. Positive selection on the branch-site levels is indicated by rectangles of different colours. IgLON5 is shown in purple, NEGR1 in blue, OPCML (dark red), NTM (pink), and invertebrates are represented in yellow. Positively selected sites are indicated by blue arrows. Red ellipses were used to identify metalloproteinase putative sites. Black stars indicate functional divergence sites. Motifs are highlighted in magenta squares. Functional specificity sites are shown in red squares at positions 182 and 226.
Figure 2.
Phylogenetic tree for the IgLON family. Only Bayesian results are shown. The tree is rooted on CD54.
Positive selection investigation
Our results suggested that IgLON has a large variation with respect to evolutionary selection (Table 1). We employed global, branch, branch-site, and site models in the CODEML program of PAML v4.431 to examine whether members of the IgLON family underwent positive selection. For the global model, our results indicate that IgLON was under positive selection with ω value of 1.21 and P < .00001. There was variation among branches with only OPCML-NTM appearing to be under negative selection (Table 1). For the branch-site analysis, we found that all the branches expect LSAMP had significantly positively selected sites. Different positively selected sites are highlighted in the alignment (Figure 1).
Table 1.
Likelihood values and parameter estimates for IgLON genes under positive selection.
| Test type | Target | ω | P value |
|---|---|---|---|
| M0-global | All IgLON | 1.21 | <.00001 |
| Branch | Drosophila melanogaster | 1.11 | Not significant |
| IgLON5 | 2.70 | Not significant | |
| OPCML-NTM | 0.54 | <.0001 | |
| LSAMP | 0.97 | Not significant | |
| NEGR1 | 1.21 | <.001 | |
| Branch-site | NEGR1 | >1 | <.0001 |
| IgLON5 | >1 | Not significant | |
| Drosophila melanogaster | >1 | <.0001 | |
| LSAMP | >1 | NA | |
| OPCML-NTM | >1 | Not significant | |
| Sites M1A-M2A | NA | >1 | <.005 |
| Sites M7-M8 | NA | >1 | <.005 |
Structural analysis
The IgLON sequences had a high degree of similarity on different levels, including structural, motif, GPI function, as well as specificity of their residues. On the structural level, all IgLON family members shared 3 Ig-like domains of the C2 type, as highlighted in the alignment (Figure 1). We also identified several motifs that were shared by more than one IgLON group (Table 2). Motif KVTVNYP that acts as SH3 recognition domain was detected in NTM, OPCML, and LSAMP. Motif SEVSNGT/SNGTSRR, which serves as putative CK1 phosphorylation site, was located in LSAMP and invertebrates. In addition, motif TNASIM/SNGTSR that acts as N-glycosylation was detected in IgLON5 and NTM. With respect to GPI anchor positions, we were able to locate putative GPI anchoring sites in all IgLON family members (supplementary Table 1). OPCML and NTM shared several putative metalloproteinase cleavage sites such as V-50, H-94, as well as P-187 (Figure 1, supplementary Figure 1). This is also supported by the observation of putative GPI-linked anchors in all IgLON members (supplementary Table 1).
Table 2.
Motifs positions in each group.
| Family | Motif | Type |
|---|---|---|
| OPCML | KVTVNYP | Recognize SH3 domains |
| IgLON5 | TNASIM/SNGTSR | N-glycosylation motif |
| NEGR1 | PFENGQYL | Motif binds to Dab-like PTB domains. Binding is dependent on the large number of hydrophobic and hydrogen bond contacts between motif and domain with integrin |
| KKVRVVVNF | MAPK interacting molecules (eg, MAPKKs, substrates, phosphatases) carry docking motif that help to regulate specific interaction in the MAPK cascade | |
| LSAMP | SEVSNGT/SNGTSRR | CK1 phosphorylation site |
| KVTVNYP | Recognize SH3 domains | |
| NGR | NGR motif is present in proteins of extracellular matrix which on deamidation forms a biologically active isoDGR motif that binds to various members of integrin family | |
| Invertebrates | RVKVTVN/ | LIG_14-3-3_3 |
| SEVSNGT/SNGTSRR | CK1 phosphorylation site g | |
| VEISSDIS/EVKTTALT/SEVSNGTS | GSK3 phosphorylation recognition site | |
| NTM | TNASIM/SNGTSR | N-glycosylation |
| SEAKGT/SSTLLQEVKTT/STLLQEVKTT | SUMO-1 modification | |
| VRR/RRV | di-Arg ER retrieval and retention (the hydrophobic C-terminal sequence could be then cleaved off and replaced by a GPI anchor) | |
| KVTVNYP | Recognize SH3 domains | |
| KVENRPFL | Motif binds to Dab-like PTB domains. Binding is dependent on the large number of hydrophobic and hydrogen bond contacts between motif and domain with integrin |
Functional specificity
We employed SDPpred to determine positions that were well conserved within IgLON groups but differ between them. The defined amino acid residues might be causing differences in protein functional specificity as well as correct recognition of interaction partners. We observed that these residues could be grouped into 2 locations: location 182 (second Ig-like C2) and 226 (third Ig-like C2). For location 182, LSAMP had (E), OPCML, NTM, and NEGR1 had (D), whereas IgLON5 and invertebrates had (G). For residue number 226, LSAMP, OPCML, NTM, and Ciona intestinalis had (s), whereas IgLON 5 had (M) and NEGR1 (A) (Figure 1).
Functional divergence
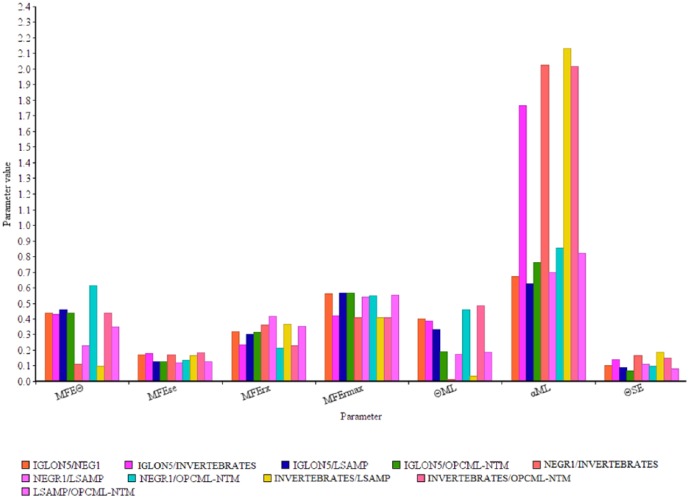
A study of site-specific shifts in evolutionary rates following gene duplication was performed using DIVERGE 2.0.36 We employed this method to investigate whether amino acid substitutions in the IgLON family caused an adaptive functional diversion. Among IgLON family groups, the highest estimate of θI by the model-free method (θMFE) was between NEGR1 and (OPCML-NTM) with the value of 0.61 ± 0.13, the lowest estimate for θMFE was between invertebrates and LSAMP with the value of 0.1 ± 0.16 (Figure 3 and supplementary Table 2). Single amino acid residues responsible for the functional divergence were also predicted based on site-specific profiles (Table 3). The predicted functional sites are not equally distributed throughout the respective IgLON, but instead are clustered between residues 94 and 300. We could not detect any functional sites between NEGR1 and invertebrates, suggesting similar functional divergence patterns.
Figure 3.
Comparison of the values of parameters for functional divergence. Two models are shown for measuring the coefficient of functional divergence θ. MFEƟ is an estimate of θ by the model-free method; MFEse is the standard error of θ estimated by MFE. MFErx is the observed coefficient of correlation between any 2 IgLON groups, whereas MFErmax is the expected maximum coefficient of correlation between any 2 IgLON groups. ƟML is the maximum likelihood model estimate of θ. αML is the maximum likelihood estimate of α (the γ parameter for the among-site rate variation) and SEƟ represents standard error of the maximum likelihood estimate of θ.
Table 3.
Functional divergence sites with MFEƟ > 0.5 and bootstrapped 100 times.
| Residue number | Compared groups | |
|---|---|---|
| 1 | 94 | IgLON5/NEGR1 NEGR1/OPCML-NTM |
| 2 | 98 | NEGR1/OPCML-NTM |
| 3 | 111 | IgLON5/NEGR1 IgLON5/invertebrates IgLON5/LSAMP |
| 4 | 125 | IgLON5/LSAMP |
| 5 | 129 | Invertebrates/OPCML-NTM |
| 6 | 133 | IgLON5/NEGR1 IgLON5/LSAMP Invertebrates/OPCML-NTM |
| 7 | 134 | Invertebrates/OPCML-NTM |
| 8 | 135 | Invertebrates/OPCML-NTM |
| 9 | 136 | IgLON5/invertebrates Invertebrates/OPCML-NTM |
| 10 | 139 | NEGR1/OPCML-NTM |
| 11 | 160 | Invertebrates/OPCML-NTM |
| 12 | 161 | Invertebrates/OPCML-NTM |
| 13 | 163 | Invertebrates/OPCML-NTM |
| 14 | 181 | IgLON5/NEGR1 IgLON5/invertebrates |
| 15 | 183 | Invertebrates/OPCML-NTM |
| 16 | 187 | Invertebrates/OPCML-NTM |
| 17 | 195 | Invertebrates/OPCML-NTM |
| 18 | 199 | IgLON5/NEGR1 NEGR1/OPCML-NTM |
| 19 | 203 | IgLON5/NEGR1 |
| 20 | 204 | NEGR1/OPCML-NTM |
| 21 | 205 | IgLON5/NEGR1 |
| 22 | 208 | NEGR1/OPCML-NTM |
| 23 | 213 | IgLON5/NEGR1 NEGR1/OPCML-NTM |
| 24 | 218 | NEGR1/OPCML-NTM |
| 25 | 219 | IgLON5/LSAMP Invertebrates/OPCML-NTM |
| 26 | 222 | IgLON5/LSAMP IgLON5/OPCML-NTM NEGR1/OPCML-NTM Invertebrates/OPCML-NTM |
| 27 | 225 | IgLON5/NEGR1 NEGR1/OPCML-NTM |
| 28 | 236 | NEGR1/OPCML-NTM |
| 29 | 239 | NEGR1/OPCML-NTM |
| 30 | 246 | IgLON5/NEGR1 IgLON5/invertebrates |
| 31 | 259 | IgLON5/LSAMP NEGR1/OPCML-NTM Invertebrates/OPCML-NTM LSAMP/OPCML-NTM |
| 32 | 263 | Invertebrates/OPCML-NTM |
| 33 | 264 | NEGR1/OPCML-NTM |
| 34 | 269 | Invertebrates/OPCML-NTM |
| 35 | 270 | NEGR1/OPCML-NTM |
| 36 | 274 | IgLON5/LSAMP IgLON5/OPCML-NTM NEGR1/OPCML-NTM |
| 37 | 279 | Invertebrates/OPCML-NTM |
Discussion
Analysis of the IgLON family from a phylogenetic perspective provided the basis for understanding its functional diversity. Phylogenetic analysis was conducted to trace the evolutionary history of the IgLON family in 12 species. Positive selection, functional specificity, and functional divergence were analysed at the amino acid level to investigate the evolutionary drivers of the IgLON family function. We also investigated GPI putative sites, metalloproteinase sites, and motifs. We found that the IgLON family first diverged around the time of emergence of Arthropod around 555 Mya. We identified residues that have a combination of more than one factor (eg, positive selection, functional specificity, and functional divergence). Position 111 seems to be an important residue for determining functional divergence between more than one IgLON group and it could be linked to pathological conditions. We also found putative amino acid residues in OPCML, NTM, LSAMP that support the hypothesis of IgLON controlling axonal growth through of metalloproteinase ectodomain shedding. Our motifs’ analysis hints at a possible IgLONs’ role at the BBB on the level of cellular adhesion between astrocytes, endothelial cells, and leukocytes.
Origin of the IgLON family
Based on the existence of IgLON family members in Drosophila melanogaster and Ciona intestinalis but not in Nematostella vectensis and Hydra vulgaris, our phylogenetic analysis suggests that the IgLON originated by duplication and divergence from a common ancestor around the emergence of Arthropod around 555 Mya. The phylogenetic tree developed in our study included 40 distinct protein sequences of 10 species (Figure 2). The search results identified IgLON homologous genes in all vertebrates as well as invertebrates. Our results are consistent with the findings presented in the work by Garver et al24 that suggest that IgSF family could appear in invertebrates. Our investigation points to the specificity of the IgLON family based on its conservation in vertebrates and several invertebrates.
Putative residues control specific group functions
The variation in the physiological functions of different groups of the IgLON family could be influenced by possessing functional specific residues, functional divergent residues, being subjected to positive selection or a combination of these factors. Although all IgLONs share the same structure (3 Ig-like C2 domains), they seem to be able of doing different functions. For example, quantitative analysis using bromodeoxyuridine immunocytochemistry revealed that the expression of Opcml had a greater inhibitory effect on astrocytic proliferation as compared with Lsamp, Ntm, and Negr1.18 Even more, each IgLON family member could be capable of regulating the development of neuronal projections via cell type–specific, attractive, and repulsive mechanisms.37,38 We identified amino acid residues that might be responsible for correct recognition of interaction partners at 2 locations. At position 182, aspartic acid residue was conserved in OPCML, NTM, and NEGR1; conversely, both IgLON5 and invertebrates had (G), whereas LSAMP had (E). For residue number 226, LSAMP, OPCML, NTM, and Ciona intestinalis (NTM-like) had a conserved serine residue, whereas IgLON5 had (M) and NEGR1 (A). Only Lachesin had (E) (Figure 1). It is interesting to note that NEGR1 branch was subjected to statistically significant positive selection at this specific site (Table 1). Type I functional divergence is the result of the change in evolutionary rate where a site is conserved for one group and is variable in another.39 We identified 3 critical functional divergence type I residues. The first (position 111) could be acting as a functional divergence site between IgLON5 and 3 groups as IgLON5 had residues (P/A/S), whereas the other groups (NEGR1, LSAMP, and invertebrates) had either (D) or (S) (Figure 1 and Table 3). OPCML had an aspartic acid residue as well. We checked the putative effect of the substitution of the human IgLON5 position from P to D using PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/)40 and it seems to have a benign effect. However, substitution of this amino acid to tryptophan or cysteine seems to be damaging with a score of 0.88. We postulate that a mutation of this residue or other functional diverging residues might be linked to autoimmune pathogenic conditions such as IgLON5 encephalopathy.41 The second site of critical functional divergence is at residue 222 and it seems to be critical for 4 groups: IgLON5/LSAMP, IgLON5/OPCML-NTM, NEGR1/OPCML-NTM, and invertebrates/OPCML-NTM. It is worth noting that site 222 was under positive selection. The third site is at position 259 and it was critical between IgLON5/LSAMP, NEGR1/OPCML-NTM, Invertebrates /OPCML-NTM, and LSAMP/OPCML-NTM and was under positive site selection. It was shown that metalloproteinase-dependent shedding of IgLONs from the cell surface of cortical neurons promoted neuronal growth.19,42 IgLON-shed fragments could both provide a growth permissive substrate or could relieve an outgrowth inhibitory signal from the cell surface.42 However, putative metalloproteinase sites have not been identified before.10 We were able to identify several possible metalloproteinase cleavage sites, which were also subjected to functional divergence at 3 different locations (94Ig-1) (OPCML and NTM), 187(Ig-2) (OPCML and NTM), and 279 (I g-3) (which was also positive selected site) (LSAMP and IgLON5) (Figure 1). Strong positive selection drives the rapid gaining of new functions through acquiring beneficial mutations.43 Our investigation revealed several metalloproteinase cleavage sites such as Q-102 (OPCML), K-102 (NEGR1) (Figure 1) which were also under site-positive selection. Even more, branch-sites positive selection for NEGR1 (Table 1) intersects with MAPK motif in NEGR1 suggesting that NEGR1 plays a signal transduction role. This MAPK motif could be functioning as a docking motif that helps to regulate specific interaction in the MAPK cascade which was given by KKVRVVVNF (Figure 1).
IgLONs could be playing a role in the BBB
High similarity in evolutionary patterns, expression profiles, and structure between IgLON groups hints at a possible role in brain morphological evolution. It is worth noting that all organisms that evolved to have a complex nervous system also acquired a BBB.44 However, in higher vertebrates as well as in Danio rerio,45 the BBB is constructed of endothelial cells which form tight junctions, whereas in invertebrates (eg, Drosophila melanogaster), the BBB is exclusively established by glial cells.44 The period of acquiring complex BBB seems to coincide with IgLONs’ duplication events. The only one IgLON family member expressed in Drosophila melanogaster (ie, Lachesin) was shown to be interacting the glial BBB through homophilic binding.12 Conversely, all IgLON members were shown to be expressed in mouse BBB.20
However, their expression patterns varied according to cell type. Only Lsamp was highly expressed (35 FPKM) in astrocytes. Iglon5 and lsamp were highly expressed in precursor oligodendrocytes. Only Iglon5 and Ntm were expressed in microglia, whereas Ntm was solely expressed in endothelial cells20,46 (supplementary Figure 2). We identified an SH3 ligand motif in NTM, OPCML, and LSAMP. SH3 domains are small protein modules of about 50 to 60 residues47 that seem to be playing a part in the BBB integrity. They are expressed on scaffolding proteins, localizing to the tight junctions,48,49 and on endothelial cells.50
It is worth noting that the identified motif given by KVTVNYP was subjected to positive selection site at the ‘K’ position and functional divergence at position ‘P’. IgLONs are known to be capable of forming homophilic and heterophilic interactions.4 Our analysis was not able to predict homophilic or heterophilic interactions based on positive selection, functional specificity, or functional divergence. Thus, a putative interaction could compromise either a heterophilic or a homophilic relationship between NTM, OPCML, or LSAMP and SH3 connecting 2 different types of cells (eg, astrocytes, neurons, oligodendrocytes, and endothelial cells) or 2 cells of the same type. Interestingly, we could also locate putative integrin-binding motifs in NEGR1, NTM, and LSAMP; PFENGQYL (QYL under functional divergence), KVENRPFL, and NGR, respectively (Table 2). BBB astrocytes express β1-integrin,51 whereas lymphocytes migrating the BBB express α4β1-integrin.52 Consequently, NEGR1, NTM, and LSAMP could be linking astrocytes or lymphocytes to other components of the BBB. Hence, NEGR1, NTM, and LSAMP could be used as potential drug targets to control lymphocytes’ migration into the brain, thus replacing drugs with less specificity such as natalizumab.53
Study Limitations
Future studies would take into account the effect of promoter sequence variation on functional divergence and specificity. Our current research investigated the IgLONs’ structure-function relationship based solely on the longest transcript isoform. It was revealed that Ntm and Opcml possess a twin promoter structure5 similar to the genomic organization reported for Lsamp.54 These isoforms differ considerably with respect to their functions. It was demonstrated that Ntm 1b isoform but not isoform 1a increases 1.47-fold compared with healthy controls in the demarcation of the dorsolateral prefrontal cortex of patients with schizophrenia.55 Conversely, Negr1 and Iglon5 transcripts have a single promoter.5 It would be crucial to extend our analysis to study the functional impact of variation between IgLON isoforms in future research.
Conclusions
The IgLON family proposed time of divergence is during the time of radiation of Arthropoda, around 455 Mya. Our study identified several IgLON residues as possible drug targets that could enhance controlling the BBB permeability. We were also able to reveal that NEGR1 might be interacting with the MAPK pathway as a form of signal transmitting receptor through its motif (KKVRVVVNF). In addition, we identified several amino acid residues that could be contributing to IgLONs’ axonal growth activity through controlling contact inhibition and metalloproteinase shedding.
Supplemental Material
Supplemental material, Supplementary_Material for Molecular Evolution and Functional Divergence of the IgLON Family by Norwin Kubick, Desiree Brösamle and Michel-Edwar Mickael in Evolutionary Bioinformatics
Acknowledgments
The authors would like to thank Professor Thomas Burglin for his kind suggestions and directions.
Footnotes
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was partially supported by a Keramina centre grant (StMKR617).
Author Contributions: All authors contributed equally to this work.
References
- 1. Correale J, Villa A. Cellular elements of the blood-brain barrier. Neurochem Res. 2009;34:2067–2077. doi: 10.1007/s11064-009-0081-y. [DOI] [PubMed] [Google Scholar]
- 2. Persidsky Y, Ramirez SH, Haorah J, Kanmogne GD. Blood-brain barrier: structural components and function under physiologic and pathologic conditions. J Neuroimmune Pharmacol. 2006;1:223–236. doi: 10.1007/s11481-006-9025-3. [DOI] [PubMed] [Google Scholar]
- 3. McNamee CJ, Youssef S, Moss D. IgLONs form heterodimeric complexes on forebrain neurons. Cell Biochem Funct. 2011;29:114–119. doi: 10.1002/cbf.1730. [DOI] [PubMed] [Google Scholar]
- 4. Reed J, McNamee C, Rackstraw S, Jenkins J, Moss D. Diglons are heterodimeric proteins composed of IgLON subunits, and Diglon-CO inhibits neurite outgrowth from cerebellar granule cells. J Cell Sci. 2004;117:3961–3973. doi: 10.1242/jcs.01261. [DOI] [PubMed] [Google Scholar]
- 5. Vanaveski T, Singh K, Narvik J, et al. Promoter-specific expression and genomic structure of IgLON family genes in mouse. Front Neurosci. 2017;11:38. doi: 10.3389/fnins.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Akeel M, McNamee CJ, Youssef S, Moss D. DIgLONs inhibit initiation of neurite outgrowth from forebrain neurons via an IgLON-containing receptor complex. Brain Res. 2011;1374:27–35. doi: 10.1016/J.BRAINRES.2010.12.028. [DOI] [PubMed] [Google Scholar]
- 7. Philips M-A, Lilleväli K, Heinla I, et al. Lsamp is implicated in the regulation of emotional and social behavior by use of alternative promoters in the brain. Brain Struct Funct. 2015;220:1381–1393. doi: 10.1007/s00429-014-0732-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pischedda F, Piccoli G. The IgLON family member Negr1 promotes neuronal arborization acting as soluble factor via FGFR2. Front Mol Neurosci. 2015;8:89. doi: 10.3389/fnmol.2015.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hashimoto T, Maekawa S, Miyata S. IgLON cell adhesion molecules regulate synaptogenesis in hippocampal neurons. Cell Biochem Funct. 2009;27:496–498. doi: 10.1002/cbf.1600. [DOI] [PubMed] [Google Scholar]
- 10. Sanz R, Ferraro GB, Fournier AE. IgLON cell adhesion molecules are shed from the cell surface of cortical neurons to promote neuronal growth. J Biol Chem. 2015;290:4330–4342. doi: 10.1074/jbc.M114.628438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pimenta AF, Zhukareva V, Barbe MF, et al. The limbic system-associated membrane protein is an Ig superfamily member that mediates selective neuronal growth and axon targeting. Neuron. 1995;15:287–297. [DOI] [PubMed] [Google Scholar]
- 12. Strigini M, Cantera R, Morin X, Bastiani MJ, Bate M, Karagogeos D. The IgLON protein Lachesin is required for the blood-brain barrier in Drosophila. Mol Cell Neurosci. 2006;32:91–101. doi: 10.1016/j.mcn.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 13. Sharma K, Schmitt S, Bergner CG, et al. Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci. 2015;18:1819–1831. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seo JH, Maki T, Maeda M, et al. Oligodendrocyte precursor cells support blood-brain barrier integrity via TGF-β signaling. PLoS ONE. 2014;9:e103174. doi: 10.1371/journal.pone.0103174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Seo JH, Miyamoto N, Hayakawa K, et al. Oligodendrocyte precursors induce early blood-brain barrier opening after white matter injury. J Clin Invest. 2013;123:782–786. doi: 10.1172/JCI65863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sugimoto C, Maekawa S, Miyata S. OBCAM, an immunoglobulin superfamily cell adhesion molecule, regulates morphology and proliferation of cerebral astrocytes. J Neurochem. 2010;112:818–828. doi: 10.1111/j.1471-4159.2009.06513.x. [DOI] [PubMed] [Google Scholar]
- 17. Guo S, Kim WJ, Lok J, et al. Neuroprotection via matrix-trophic coupling between cerebral endothelial cells and neurons. Proc Natl Acad Sci U S A. 2008;105:7582–7587. doi: 10.1073/pnas.0801105105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sugimoto C, Morita S, Miyata S. Overexpression of IgLON cell adhesion molecules changes proliferation and cell size of cortical astrocytes. Cell Biochem Funct. 2012;30:400–405. doi: 10.1002/cbf.2813. [DOI] [PubMed] [Google Scholar]
- 19. Chen S, Gil O, Ren YQ, Zanazzi G, Salzer JL, Hillman DE. Neurotrimin expression during cerebellar development suggests roles in axon fasciculation and synaptogenesis. J Neurocytol. 2001;30:927–937. [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y, Chen K, Sloan SA, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hall AK, Rutishauser U. Phylogeny of a neural cell adhesion molecule. Dev Biol. 1985;110:39–46. doi: 10.1016/0012-1606(85)90061-2. [DOI] [PubMed] [Google Scholar]
- 22. Ohtani H, Nakajima T, Akari H, Ishida T, Kimura A. Molecular evolution of immunoglobulin superfamily genes in primates. Immunogenetics. 2011;63:417–428. doi: 10.1007/s00251-011-0519-7. [DOI] [PubMed] [Google Scholar]
- 23. Boender AJ, van Gestel MA, Garner KM, Luijendijk MCM, Adan RAH. The obesity-associated gene Negr1 regulates aspects of energy balance in rat hypothalamic areas. Physiol Rep. 2014;2:e12083. doi: 10.14814/phy2.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garver LS, Xi Z, Dimopoulos G. Immunoglobulin superfamily members play an important role in the mosquito immune system. Dev Comp Immunol. 2008;32:519–531. doi: 10.1016/j.dci.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marchler-Bauer A, Anderson JB, DeWeese-Scott C, et al. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 2003;31:383–387. doi: 10.1093/nar/gkg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 28. Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML online – a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 30. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 31. Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 32. Dinkel H, Van Roey K, Michael S, et al. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 2014;42: D259–D266. doi: 10.1093/nar/gkt1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pierleoni A, Martelli P, Casadio R. PredGPI: a GPI-anchor predictor. BMC Bioinformatics. 2008;9:392. doi: 10.1186/1471-2105-9-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Song J, Tan H, Perry AJ, et al. PROSPER: an integrated feature-based tool for predicting protease substrate cleavage sites. PLoS ONE. 2012;7:e50300. doi: 10.1371/journal.pone.0050300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kalinina OV, Novichkov PS, Mironov AA, Gelfand MS, Rakhmaninova AB. SDPpred: a tool for prediction of amino acid residues that determine differences in functional specificity of homologous proteins. Nucleic Acids Res. 2004;32:W424–W428. doi: 10.1093/nar/gkh391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gu X, Velden KV. DIVERGE: phylogeny-based analysis for functional-structural divergence of a protein family. Bioinformatics. 2002;18:500–501. doi: 10.1093/bioinformatics/18.3.500. [DOI] [PubMed] [Google Scholar]
- 37. Gil OD, Zanazzi G, Struyk AF, Salzer JL. Neurotrimin mediates bifunctional effects on neurite outgrowth via homophilic and heterophilic interactions. J Neurosci. 1998;18:9312–9325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lodge AP, Howard MR, McNamee CJ, Moss DJ. Co-localisation, heterophilic interactions and regulated expression of IgLON family proteins in the chick nervous system. Brain Res Mol Brain Res. 2000;82:84–94. [DOI] [PubMed] [Google Scholar]
- 39. Chakrabarti S, Bryant SH, Panchenko AR. Functional specificity lies within the properties and evolutionary changes of amino acids. J Mol Biol. 2007;373:801–810. doi: 10.1016/J.JMB.2007.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2 (Chapter 7: Unit 7.20). Curr Protoc Hum Genet. 2013;76:7.20.1–7.20.41. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Haitao R, Yingmai Y, Yan H, et al. Chorea and parkinsonism associated with autoantibodies to IgLON5 and responsive to immunotherapy. J Neuroimmunol. 2016;300:9–10. doi: 10.1016/J.JNEUROIM.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 42. Sanz RL, Ferraro GB, Girouard M-P, Fournier AE. Ectodomain shedding of limbic system-associated membrane protein (LSAMP) by ADAM Metallopeptidases promotes neurite outgrowth in DRG neurons. Sci Rep. 2017;7:7961. doi: 10.1038/s41598-017-08315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Assis R. Drosophila duplicate genes evolve new functions on the fly. Fly (Austin). 2014;8:91–94. doi: 10.4161/fly.29131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Limmer S, Weiler A, Volkenhoff A, Babatz F, Klämbt C. The Drosophila blood-brain barrier: development and function of a glial endothelium. Front Neurosci. 2014;8:365. doi: 10.3389/fnins.2014.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eliceiri BP, Gonzalez AM, Baird A. Zebrafish model of the blood-brain barrier: morphological and permeability studies. Methods Mol Biol. 2011;686:371–378. doi: 10.1007/978-1-60761-938-3_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Petryszak R, Burdett T, Fiorelli B, et al. Expression Atlas update – a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 2014;42:D926–D932. doi: 10.1093/nar/gkt1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pawson T, Schlessingert J. SH2 and SH3 domains. Curr Biol. 1993;3:434–442. doi: 10.1016/0960-9822(93)90350-W. [DOI] [PubMed] [Google Scholar]
- 48. Bauer H-C, Krizbai IA, Bauer H, Traweger A. ‘You Shall Not Pass’-tight junctions of the blood brain barrier. Front Neurosci. 2014;8:392. doi: 10.3389/fnins.2014.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stamatovic SM, Johnson AM, Keep RF, Andjelkovic AV. Junctional proteins of the blood-brain barrier: new insights into function and dysfunction. Tissue Barriers. 2016;4:e1154641. doi: 10.1080/21688370.2016.1154641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Couraud P-O, Scherman D. Biology and physiology of the blood-brain barrier: transport, cellular interactions, and brain pathologies. In: Cerebral Vascular Biology Symposium. New York: Plenum Press; 1996. [Google Scholar]
- 51. Venkatesan C, Birch D, Peng CY, Kessler JA. Astrocytic β1-integrin affects cellular composition of murine blood brain barrier in the cerebral cortex. Int J Dev Neurosci. 2015;44:48–54. doi: 10.1016/j.ijdevneu.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 52. Martin-Blondel G, Pignolet B, Tietz S, et al. Migration of encephalitogenic CD8 T cells into the central nervous system is dependent on the α4β1-integrin. Eur J Immunol. 2015;45:3302–3312. doi: 10.1002/eji.201545632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Larochelle C, Uphaus T, Broux B, et al. EGFL7 reduces CNS inflammation in mouse. Nat Commun. 2018;9:819. doi: 10.1038/s41467-018-03186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pimenta AF, Levitt P. Characterization of the genomic structure of the mouse limbic system-associated membrane protein (Lsamp) gene. Genomics. 2004;83:790–801. doi: 10.1016/j.ygeno.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 55. Karis K, Eskla K-L, Kaare M, et al. Altered expression profile of IgLON family of neural cell adhesion molecules in the dorsolateral prefrontal cortex of schizophrenic patients. Front Mol Neurosci. 2018;11:8. doi: 10.3389/fnmol.2018.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary_Material for Molecular Evolution and Functional Divergence of the IgLON Family by Norwin Kubick, Desiree Brösamle and Michel-Edwar Mickael in Evolutionary Bioinformatics