Abstract
Push-pull is a canonical computation of excitatory cortical circuits. Here we identify a pull-push inhibitory circuit in frontal cortex that originates in vasoactive intestinal polypeptide (VIP) expressing interneurons. During arousal, VIP cells rapidly and directly inhibit pyramidal neurons; VIP cells also indirectly excite these pyramidal neurons via parallel disinhibition. Thus, arousal exerts a feed-back pull-push influence on excitatory neurons – an inversion of the canonical push-pull of feed-forward input.
An important question in systems neuroscience is how behavioral state modulates the processing of sensory signals in large networks of neurons. To gain further insights into the impact of behavioral state on interactions between different cell types in local cortical circuitry, we employed a novel approach to functional circuit mapping based on examining the covariance of correlation coefficients between responses of genetically defined neurons and changes in locomotion and pupil diameter.
Responses of neurons expressing the calcium indicator GCaMP6 1 were measured in alert, head-fixed mice in a pre-motor region of mouse frontal cortex, referred to as frontal association area (FrA, 2.8mm from Bregma and 0.75mm from the midline. We chose this region because the estimation of correlation coefficients is facilitated when cells show elevated spontaneous activity, and pyramidal neurons in this region show spontaneous activity that is substantially higher than observed in primary sensory cortices. Behavior was monitored by recording pupil diameter and running speed while imaging.
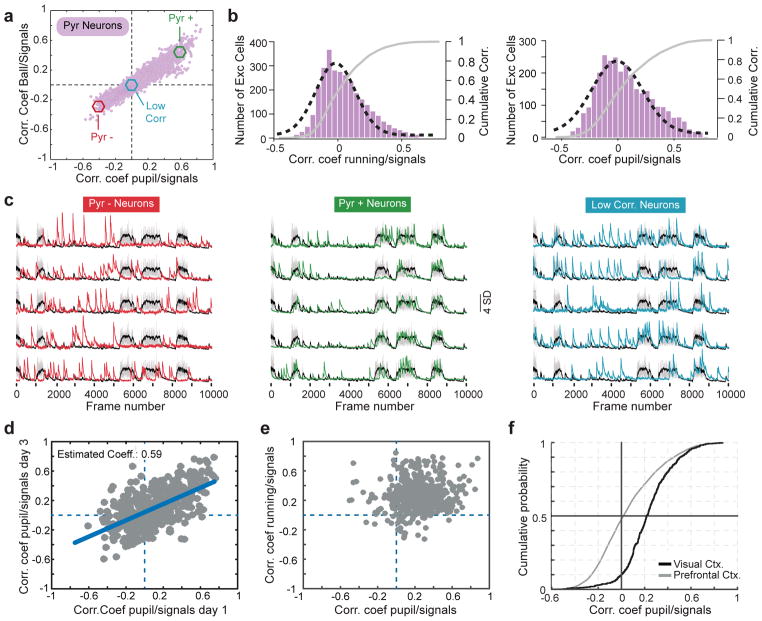
To measure the degree of association between behavior and neural response we computed correlation coefficients between the activity of pyramidal cells in layer 2/3 (L2/3) with pupil size and running speed, which reflect the level of arousal 2–5. We found that 33% of excitatory neurons imaged were significantly (P<0.0001) negatively correlated to both pupil diameter and running, while 43% were significantly positively correlated with both measures (Fig. 1a–c; Supplementary Fig. 1). Similar response profiles were observed using GCaMP6f (Supplementary Fig. 2), and longitudinal imaging showed that neurons stably maintain the sign of their coefficients (Fig. 1d).
Figure 1. A stable subset of excitatory neurons is inhibited during arousal.
a) Ordinate: plot of the correlation coefficients between excitatory response and locomotion. Abscissa: plot of the correlation coefficients between those same neurons and pupil diameter. 3108 neurons, 11 mice, 17 fields of view. Neurons with a P value < 0.0001 for each coefficient: 1175 negatively correlated to pupil diameter; 1193 negatively correlated to running; 1559 positively correlated to pupil diameter; 1643 positively correlated to running; 1350 positively correlated with both pupil and running; 1020 negatively correlated with both pupil and running; Remaining cells were not significantly correlated with either measure.
b) Histograms plotting excitatory correlation coefficients to running (left) and pupil diameter (right). A Gaussian distribution was fitted to each histogram (black dashed line). Gray lines in both show the cumulative distributions.
c) Example traces from 5 excitatory neurons whose activities are negatively (red, left), positively (green, middle), or un-correlated (blue, right) with locomotion/pupil diameter. Cells were taken from the pool of neurons shown in the colored circles in panel c.
d) Plot of correlation coefficients between pyramidal cell responses and pupil diameter for the same 440 pyramidal neurons imaged on two time points that were 3 days apart. 440 neurons, 5 mice, 5 fields of view. Error degrees of freedom = 438; Root mean squared error = 0.198; R-squared = 0.375; t-statistic = 16.2; F-statistic vs. constant model: 263, p = 1.17e-46. Significance of the linear regression was determined using a 2-sided t-statistic with n-2 degrees of freedom.
e) Same plot as in (a), but for pyramidal neurons imaged in primary visual cortex. 518 neurons, 5 mice, 5 fields of view. 53 negatively correlated to pupil diameter; 31 negatively correlated to running; 5 negatively correlated to both pupil diameter and running.
f) Cumulative distributions of coefficients to pupil diameter for neurons in V1 and FrA.
Suppression of L2/3 pyramidal neurons during running has not been reported in visual cortex 2, 5–8. In agreement with these studies, we found that only ~1% of excitatory neurons in V1 were negatively correlated to both locomotion and pupil diameter (Fig. 1e), suggesting that the suppression of pyramidal cells we see in frontal cortex must arise from a unique impact of inhibition in this region (Fig. 1f).
VIP cells are thought to regulate cortical excitability during arousal and locomotion 9 by inhibiting SOM interneurons 10–12; we too found evidence for this interaction in FrA (Supplementary Fig. 3). While this disinhibition may account for the increased excitatory responsiveness of some pyramidal cells during periods of heightened arousal, it cannot drive the suppression of almost 40% of the excitatory network.
To investigate functional interactions between VIP cells, SOM cells, and pyramidal neurons in the same network, we expressed a flexed GCaMP6s in VIP and SOM cells and GCaMP6f in all neurons and again imaged their activities in head-fixed, freely moving mice (Figure 2a; Supplementary Methods). VIP and SOM cells also expressed the red fluorescent protein tdTomato.
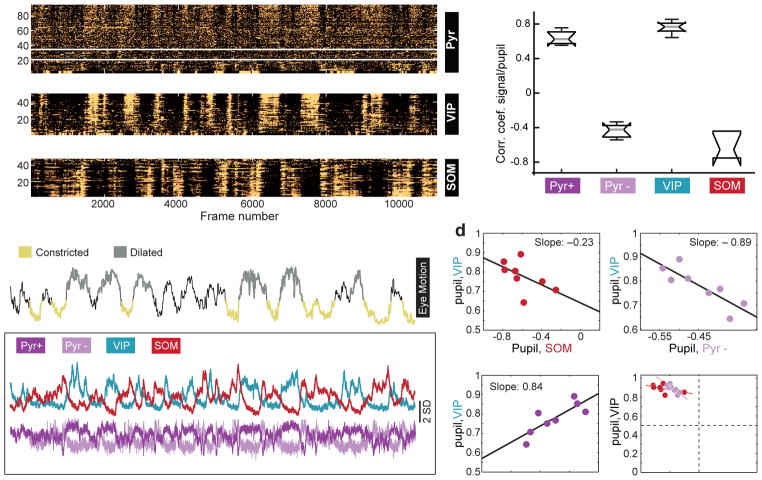
Figure 2. Co-variation of coefficients between VIP and Pyr cells across mice.
a) 3 panels showing z-scored time series responses of 95 excitatory neurons (top), 50 VIP cells (middle), and 48 SOM cells (bottom) imaged in the same mouse over 13 minutes. For the excitatory neurons, responses above and below the white lines are significantly positively and negatively correlated, respectively, with pupil size. Mice are triple transgenic SOM-Cre;VIP-Cre;Ai9.
b) Top trace: z-scored time series for the pupil diameter in the same mouse shown in panel a. Regions of the trace representing “constricted” or “dilated” pupil are color coded and represent regions that remained significantly above or below the mean for a duration of greater than 3 seconds. The traces in the lower boxed region show the mean response of the Pyr+ and Pyr– neurons, VIP and SOM cells in panel a.
c) Box plots of the distribution of correlation coefficients of mean responses for each group of cells to pupil. Whiskers define median, upper quartile, lower quartile, upper adjacent value, and lower adjacent value. n=747 pyramidal neurons, 357 VIP cells, 204 SOM cells, 4 mice, 8 fields of view.
d) Co-variation across animals of correlation coefficients among population activity and pupil size.
VIP-Pyr+: t-stat: 3.44; root mean squared error = 0.05; R-squared = 0.608, F-statistic vs. constant model: 11.9, p=0.0139; 6 degrees of freedom.
VIP-Pyr–: t-stat: −3.9; root mean squared error = 0.045; R-squared = 0.671; F-statistic vs. constant model: 15.3, p=0.0079; 6 degrees of freedom.
VIP-SOM: t-stat: −1.57; root mean squared error = 0.072; R-squared = 0.173; F-statistic vs. constant model: 2.47, p =0.167. Plots are of fields of view. Significance of the linear regression was determined using a 2-sided t-statistic with n-2 degrees of freedom.
We calculated the mean response for each of the 4 groups of neurons identified in these studies (VIP, SOM, Pyr+ and Pyr– cells; Fig. 2b) and calculated the correlation coefficient between these responses and pupil diameter and locomotion (Fig. 2c). These associations co-varied across mice, with the strongest association being between VIP and PYR- neurons (Fig. 2d). This means that if in a mouse the correlation between VIP and pupil size was higher than average (across our population), then the correlation between Pyr- and pupil size was correspondingly more negative. This relationship is suggestive of a strong, direct connection between VIP and Pyr- neurons.
We confirmed this connection using channelrhodopsin-assisted circuit mapping 13 (Fig 3a–e), and found that all pyramidal neurons receive direct VIP input, but that the strength of this input varies over a wide range (by almost 2 orders of magnitude; Fig. 3f). This heterogeneity it is consistent with the distribution of positive and negative correlations between pyramidal cell responses and behavioral state that we see, but it may also reflect differences between brain slices that we cannot control.
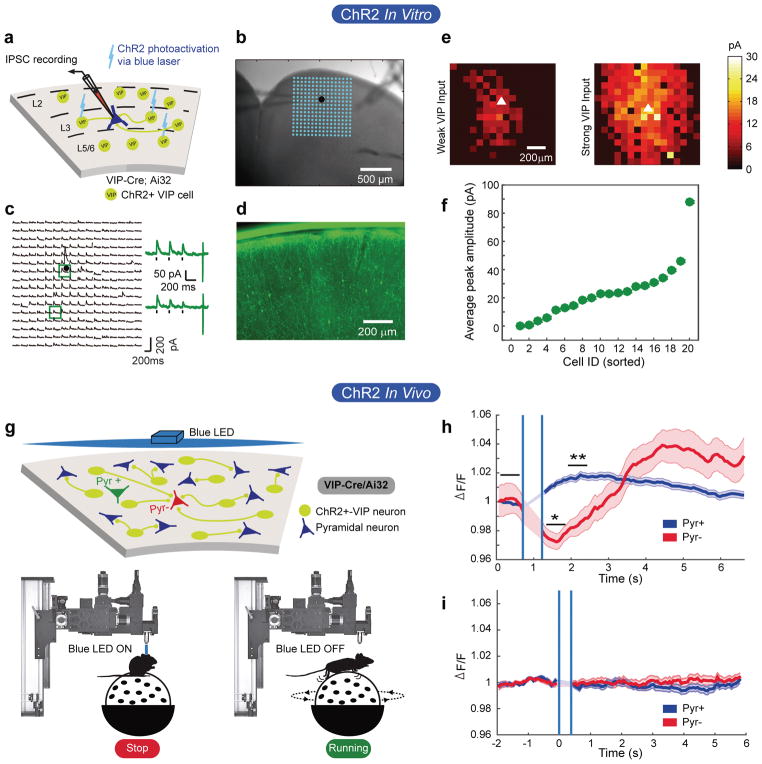
Figure 3. VIP cells act as a push-pull circuit in vivo.
a) Schematic of the approach used to map VIP cell-derived inhibitory post-synaptic currents in pyramidal neurons patched in layer 2/3 of FrA.
b) Brightfield image of a brain slice. The patch pipette can be seen entering from the left. The position of the patched cell is indicated by the black dot. The blue dots show the positions of each photostimulation site.
c) Inhibitory postsynaptic currents evoked from each photostimulation site. Responses evoked from stimulation at the two sites outlined by the blue squares are shown to the right.
d) Fluorescent image of section showing expression of YFP-tagged ChR2 in VIP interneurons. Scale bar is 200 micrometers.
e) Examples of a weak and strong heat map of VIP cell-derived inhibitory currents to two different pyramidal neurons.
f) Amplitude of the mean inhibitory current to each of 20 pyramidal neurons from all local VIP cells in the slice.
g) Schematic of experimental setup. Top: VIP cells are represented as yellow/green circles. Pyramidal cells as blue triangles. The green and red triangles represent pyramidal cells that are excited or inhibited during running, respectively. Bottom: Depiction of photostimulation through the objective lens. Stimulation was done when mice were quiescent.
h) Mean z-score of Pyr- responses to ChR2-VIP cell stimulation. Shaded areas define +/− 1 standard error of the responses. Vertical blue lines delineate the time when the LED was turned on and the PMT was gated off. A significant suppression of GCaMP6s response in the Pyr- cells occurs in ~575ms. P values result from Wilcoxon rank-sum test; 345 cells, 4 fields of view, 2 mice; Pyr+ = 6.14e-14; Pyr- = 0.024.
i) Same measure as in panel h, but in a mouse lacking ChR2 in VIP cells. 168 cells, 2 fields of view, 1 mouse.
In vivo, we found a significant suppression in the aggregate response of Pyr− neurons when VIP cells were optogenetically stimulated (Fig. 3g,h). In contrast, Pyr+ cells showed a significant increase in aggregate response (Fig. 3h), which peaked approximately 10 frames (645ms) after the dip seen in the Pyr− neurons. This delayed response is possibly due to the multi-synaptic, disinhibitory relay from VIP → SOM → Pyr 11, 14, 15. Light-activated responses were not observed in the absence of ChR2 (Fig. 3i).
These data identify a pull-push inhibitory circuit that is active during arousal. Pyramidal neurons likely receive both direct VIP cell-mediated inhibition and indirect VIP → SOM cell-mediated disinhibition. This inhibitory circuitry is an inversion of canonical excitatory push-pull circuitry 16, 17. An expected outcome from this circuitry is that variability in the net balance of inhibition and disinhibition generates a heterogeneous response of excitatory neurons, some of which are enhanced during arousal as others are suppressed. The mean balance between the strengths of direct inhibition and disinhibition appears to vary across cortical areas – in our measurements, the strength of direct VIP inhibition of pyramidal neurons is most pronounced in frontal cortex and weak in occipital cortex. The net effect of these two pathways on individual cells is expected to shift their operating point and, therefore, modulate the gain of pyramidal neurons during arousal.
Methods
Animals
All procedures were approved by UCLA’s Office of Animal Research Oversight (the Institutional Animal Care and Use Committee, IACUC) and UCI’s IACUC, and were in accord with guidelines set by the US National Institutes of Health. A total of 50 mice from Jackson Lab, both male (23) and female (27), aged P45–56, were used for in vivo imaging in this study as follow: 7 Viptm1(cre)Zjh/J (Stock#: 010908), 7 B6N.Cg-Ssttm2.1(cre)Zjh/J (Stock#: 018973), 27 Gad2tm2(cre)Zjh/J (Stock#: 010802), 4 Viptm1(cre)Zjh/J/Ssttm2.1(cre)Zjh/J and 2 (in vivo) + 4 (in vitro) Viptm1(cre)Zjh/J/ Ai32 (Stock#: 012569). All CRE lines were previously crossed with B6;129S6-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J (Stock#: 007905), to express td-Tomato in a CRE- dependent manner (except for the Vip-ChR2 mice). All lines were crossed into a C57 black 6 background for 7 generations or longer.
Mice were housed in groups of 2–3 per cage in a normal 12/12 light dark cycle. Animals were naïve subjects with no prior history of participation in research studies. Numbers of mice used, numbers of fields of view, and numbers of cells imaged are all provided in the appropriate figure legends.
Surgery
Carprofen (anti-inflamatory) was administered pre-operatively. Mice were then anesthetized with isoflurane (4–5% induction; 1.5–2% surgery). Core body temperature was maintained at 37.5C using a feedback heating system. Eyes were coated with a thin layer of ophthalmic ointment to prevent desiccation. Anesthetized mice were mounted in a stereotaxic apparatus. Blunt ear bars were placed in the external auditory meatus to immobilize the head. A portion of the scalp overlying the two hemispheres of the cortex (approximately 8mm by 6mm) was then removed to expose the underlying skull.
After the skull is exposed it was dried and covered by a thin layer of Vetbond. After the Vetbond dries (approximately 15 min) it provides a stable and solid surface to affix the aluminum bracket with dental acrylic. The bracket is then affixed to the skull and the margins sealed with Vetbond and dental acrylic to prevent infections.
Virus injection
A 2.5mm diameter region of skull overlying the FrA cortex (right hemisphere) was removed. Care was taken to leave the dura intact. Depending on the experiment, GCaMP6-slow, GCaMP6-fast or GCaMP6s-Floxed (UPenn Vector Core: AAV1.Syn.GCaMP6f.WPRE.SV40; #AV-1-PV2822, AAV1.Syn.GCaMP6s.WPRE.SV40 #AV-1-PV2824, AAV1.Syn.Flex.GCaMP6s.WPRE.SV40; #AV-1-PV2821) was expressed in cortical neurons using adeno-associated virus (AAV-GCaMP6 titer: ~13 genomes ml−1 aprox.). The virus was loaded into a glass micropipette and slowly inserted into the FrA using a micromanipulator. One injection site was made at 2.8mm from Bregma and 0.75mm from the midline. AAV-GCaMP6 was pressure injected using a PicoSpritzer III (Parker, Hollis, NH) (4 puffs at 15–20 pounds per square inch with a duration of 10 ms, each puff was separated by 4 s) starting at a depth of 350 microns below the pial surface and making injections every 10 microns moving up with the last injection made at 100 microns below the pial surface. The total volume injected across all depths was approximately 1μl. In some cases, GCaMP6s-Flex and GCaMPf were injected at the same time (0.5ul each). The injections were made by a computer program in control of the micro-manipulator and the PicoSpritzer.
A 1mm side square coverslip, previously cut with a diamond tip, was glued to a 2.5 mm diameter cover glass using an UV curing optical adhesive 18. UV light was used for gluing and sterilizing both coverslips and then they were placed directly on the dura and sealed at its edges with VetBond. When dry, the edges of the cover glass were further sealed with dental acrylic. At the end of the surgery, all exposed skull and wound margins were sealed with VetBond and dental acrylic. Mice were then removed from the stereotaxic apparatus, given a subcutaneous bolus of warm sterile saline, and allowed to recover on the heating pad. When fully alert they were placed back in their home cage.
Imaging
Once expression of GCaMP6 was observed in FrA, typically between 12–15 days after the injection, imaging sessions took place. Imaging was performed using a resonant scanning, two-photon microscope (Neurolabware, Los Angeles, CA) controlled by Scanbox acquisition software (Scanbox, Los Angeles, CA). The light source was a Coherent Chameleon Ultra II laser (Coherent Inc, Santa Clara, CA) running at 920nm. The objective was an x16 water immersion lens (Nikon, 0.8NA, 3mm working distance). The microscope frame rate was 15.6Hz (512 lines with a resonant mirror at 8kHz). Eye movements and pupil size were recorded via a Dalsa Genie M1280 camera (Teledyne Dalsa, Ontario, Canada) fitted with a 740nm long-pass filter. Images were captured at an average depth of 210 μm (90% of imaging fields within the range 125–300 microns). During imaging a substantial amount of light exits from the brain through the pupil. Thus, no additional illumination was required to image the pupil. Mice were head-fixed on a floating, spherical treadmill on which they had been trained to balance and run. All mice were trained on the spherical treadmill for 3 days, 20-minutes per day before experiments were started. Movement of the spherical treadmill was recorded via a Dalsa Genie M640 camera. Both locomotion and eye movement data were synchronized to the microscope frames.
We first summed images over a 30 sec. period to obtain a high signal-to-noise image of Cre-dependent tdTomato expression, using 1000nm excitation, to identify specific populations of inhibitory cells, depending on the experiment. We then changed the wavelength to 920nm and imaged GCaMP6(f/s) dynamics during 10–15 minutes per field. Mice were head fixed, but able to run during the entire imaging session.
Data processing
Motion stabilization
Calcium images were aligned to correct for motion artifacts in a two-step process. First, we aligned images rigidly in a recursive fashion to correct for slow drifts in the imaging plane. Pairs of neighboring images in time were aligned by finding the peak of their cross-correlation; then, pairs of averages of such pairs were aligned; and so on and so forth. In the second step, we aligned images non-rigidly to a reference mean image to correct for fast in-plane movements, which are frequently observed during grooming. We iteratively applied the Lucas-Kanade algorithm 19 to non-rigidly match a reference mean image, refining the estimate of this reference mean image after each alignment iteration.
Segmentation
Following motion stabilization, we used a Matlab graphical user interface (GUI) tool developed in our laboratory to manually define regions of interest corresponding to putative cell bodies. We used correlation and kurtosis images to identify cell candidates 20. The correlation image, corresponding to the average correlation of a pixel and its 8 neighbors across time, highlights regions of space which co-vary in time. The kurtosis image highlights regions in space with signals composed of large, infrequent deviations - putative spikes. These images were computed after subtracting a rank-2 factor from the calcium movies, corresponding to mean and linear temporal trends.
We used these images to visually identify approximately circular regions of space of an appropriate radius with high correlation, and high kurtosis. Clicking a seed pixel at the center of such a candidate patch allowed the definition of a region of interest by flood-filling an image corresponding to the correlation of the highlighted pixel and every other pixel in the image field 21. The interface then allowed the user to dynamically grow or shrink the region of interest to a desired size. Red and green channels were segmented separately, and the resultant masks were matched, enabling us to properly assign GCaMP signals to inhibitory or excitatory neurons appropriately.
Analysis
For data in figures 1 and 2, including supplementary figures 1 and 2, we determined the correlation coefficient between each cell’s activity and both locomotion and pupil diameter. For data in figure 3, we first calculated the correlation coefficient for each excitatory neuron’s activity and pupil diameter. Temporal responses of all cells that had significantly positive correlations coefficients were averaged together (Pyr+), and temporal responses of cells with significantly negative correlations were averaged (Pyr-). Mean time series were also derived for the VIP and SOM cells. These mean time series were then correlated with pupil diameter and their coefficients plotted in figure 3c. In figure 3d, for each mouse we plotted the coefficients for each cell type relative to the coefficients obtained for VIP cells. The slope, r-value, and p-value of linear regression lines of co-variance were measured.
Data in Figure 2
In this experiment, both SOM-expressing and VIP-expressing cells will also express tdTomato and GCaMP6s. Because we cannot distinguish SOM from VIP cells based on waveform, we assigned all tdTomato-expressing cells that had a positive correlation coefficient to pupil diameter as VIP cells, and all those with negative coefficients at SOM cells. From our data in Supplementary Figure 3, we know that of the 875 VIP cells that were imaged, 105 were negatively correlated with pupil diameter, and of the 417 SOM cells imaged, 70 were positively correlated with pupil diameter. That is, of the 1292 cells in the VIP/SOM pool, 175 (13.5%) would be incorrectly assigned using the strategy we employ here.
ChR2-assisted circuit mapping
Briefly, whole cell recordings were obtained from pyramidal neurons of layer 3 in FrA of VIP-Cre+/−/Ai32+/− mice at the age of ~7 weeks old. Spatial maps of VIP connectivity strength to each patched pyramidal neuron were derived by systematically stimulating ChR2-expressing VIP cells at a mapping grid of 256 different sites arranged in a 16x16 matrix spanning all cortical layers (Fig. 3). Spatially restricted optogenetic activation of ChR2-expressing VIP cells was accomplished using a 473 nm blue laser (30 mW, 2.5ms, laser spot diameter, ~50 μm). Voltage and current clamp controls were performed to normalize light stimulation protocols between slices and across animals. During optogenetic stimulation experiments, the ionotropic glutamate receptor antagonists (10 μM CNQX and 5 μM CCP) was added to the bath solution to block excitatory synaptic input (GABAergic transmission is unaffected) and avoid any potential dis-inhibition effects. Because interneurons can also be connected by electrical synapses, we blocked gap junctions using 100 μM carbenoxolone. Whole-cell voltage-clamp recordings were made from the recorded pyramidal neurons to measure photoactivation-evoked inhibitory post-synaptic currents at the empirically determined holding potential at +5 mV in voltage clamp mode with cesium-containing internal solution. Post hoc GFP staining (Abcam ab6556; 1:500 dilution) of the recorded slices verified ChR2-YFP expression in VIP neurons.
In vivo ChR2-VIP photostimulation with pyramidal cell imaging
VIP-Cre mice were crossed with the Ai32 line of mice housing a flexed ChR2 driven by a CAG promoter targeted to the Rosa26 locus (JAX Stock number: 012569). Offspring of these crosses expressed ChR2 in VIP interneurons and verified by genotyping. AAV-GCaMP6f was injected into frontal cortex of these mice as described above. Craniotomies were made as described above. Following strong GCaMP6f expression, mice were head fixed under the resonant scanning microscope and were imaged, head fixed and free to run to identify pyramidal neurons whose activities increased or decreased with running. A blue light emitting diode (ThorLabs M470L3) focused onto the objective back aperture was used to excite ChR2 in the imaging field during periods of behavioral quiescence (not running). The LED was controlled using custom Matlab software to generate 5V TTL signals. Each light pulse was 40ms duration. During the blue light flash, the PMTs were rapidly gated. For this experiment, we changed the PMT on our custom microscope to a Hamamatsu H11706-40, which has built in circuitry to rapidly gate the gain of the PMT within 400ns (rise and fall). These PMTs have a maximum shutter width of 10ms, therefore we triggered the PMT with 4 successive TTLs to obtain an effective open time of 40ms. Using this approach, we avoided light contamination from the LED in our GCaMP6 signals. GCaMP6f responses were recorded simultaneously. Movies of cell responses were motion corrected as described, cells were identified, and signals extracted. Because pyramidal cell firing is sparse, only a small fraction of the pyramidal neurons imaged were firing within the 40ms light flash. Therefore, we repeated the light flash 21 times at random intervals while imaging continuously. Cell responses were averaged across the 21 trials and across cells to produce the trace shown in Figure 3.
Statistics
Data collection and analysis was not blinded. No randomization was used to assign subject to the experimental groups. Experiments required mice of known genotype, making randomization impossible. No statistical methods were used to pre-determine sample sizes but our sample sizes are like those reported in previous publications (ref 18, 19, 20). Statistical tests are indicated in the figure legends with accompanying P values. Numbers of cells, numbers of fields of view, and numbers of mice are given in the appropriate figure legends.
Data availability
The data that support the findings of this study are available from the corresponding author upon request. Matlab scripts can be requested from the corresponding author.
Supplementary Material
Acknowledgments
This work was funded by R01 EY023871 (JTT), R01 EY018322 (DLR) and R01 EB022915 (DLR). PGJC was supported by postdoctoral Fellowship EX2009-0750 from the Spanish Ministry of Education, Culture and Sport and by postdoctoral contract from Junta de Andalucía (P12-CTS-2232).
Footnotes
Author contribuitions
PGJC collected and analyed the data in Figure 1&2 and all supplemenetary data figures. DLR analyzed the covariance data in Figure 2. ET, DLR, and JTT contributed to the in vivo photostimulation experiments in Figure 3. TI and XX contributed to the slice ChR2 mapping data in Figure 3.
Competing financial interests
None.
References
- 1.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGinley MJ, David SV, McCormick DA. Cortical Membrane Potential Signature of Optimal States for Sensory Signal Detection. Neuron. 2015;87:179–192. doi: 10.1016/j.neuron.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebitz RB, Platt ML. Neuronal activity in primate dorsal anterior cingulate cortex signals task conflict and predicts adjustments in pupil-linked arousal. Neuron. 2015;85:628–640. doi: 10.1016/j.neuron.2014.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zekveld AA, Heslenfeld DJ, Johnsrude IS, Versfeld NJ, Kramer SE. The eye as a window to the listening brain: neural correlates of pupil size as a measure of cognitive listening load. Neuroimage. 2014;101:76–86. doi: 10.1016/j.neuroimage.2014.06.069. [DOI] [PubMed] [Google Scholar]
- 5.Vinck M, Batista-Brito R, Knoblich U, Cardin JA. Arousal and locomotion make distinct contributions to cortical activity patterns and visual encoding. Neuron. 2015;86:740–754. doi: 10.1016/j.neuron.2015.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niell CM, Stryker MP. Modulation of visual responses by behavioral state in mouse visual cortex. Neuron. 2010;65:472–479. doi: 10.1016/j.neuron.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polack PO, Friedman J, Golshani P. Cellular mechanisms of brain state-dependent gain modulation in visual cortex. Nature neuroscience. 2013;16:1331–1339. doi: 10.1038/nn.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mineault PJ, Tring E, Trachtenberg JT, Ringach DL. Enhanced Spatial Resolution During Locomotion and Heightened Attention in Mouse Primary Visual Cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2016;36:6382–6392. doi: 10.1523/JNEUROSCI.0430-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Letzkus JJ, Wolff SB, Luthi A. Disinhibition, a Circuit Mechanism for Associative Learning and Memory. Neuron. 2015;88:264–276. doi: 10.1016/j.neuron.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 10.Pfeffer CK, Xue M, He M, Huang ZJ, Scanziani M. Inhibition of inhibition in visual cortex: the logic of connections between molecularly distinct interneurons. Nature neuroscience. 2013;16:1068–1076. doi: 10.1038/nn.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S, Kruglikov I, Huang ZJ, Fishell G, Rudy B. A disinhibitory circuit mediates motor integration in the somatosensory cortex. Nature neuroscience. 2013;16:1662–1670. doi: 10.1038/nn.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pi HJ, et al. Cortical interneurons that specialize in disinhibitory control. Nature. 2013;503:521–524. doi: 10.1038/nature12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nature neuroscience. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- 14.Karnani MM, et al. Opening Holes in the Blanket of Inhibition: Localized Lateral Disinhibition by VIP Interneurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2016;36:3471–3480. doi: 10.1523/JNEUROSCI.3646-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y, et al. A cortical circuit for gain control by behavioral state. Cell. 2014;156:1139–1152. doi: 10.1016/j.cell.2014.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson JS, Carandini M, Ferster D. Orientation tuning of input conductance, excitation, and inhibition in cat primary visual cortex. Journal of neurophysiology. 2000;84:909–926. doi: 10.1152/jn.2000.84.2.909. [DOI] [PubMed] [Google Scholar]
- 17.Douglas RJ, Martin KA. A functional microcircuit for cat visual cortex. The Journal of physiology. 1991;440:735–769. doi: 10.1113/jphysiol.1991.sp018733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber D, et al. Multiple dynamic representations in the motor cortex during sensorimotor learning. Nature. 2012;484:473–478. doi: 10.1038/nature11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenberg DS, Kerr JN. Automated correction of fast motion artifacts for two-photon imaging of awake animals. J Neurosci Methods. 2009;176:1–15. doi: 10.1016/j.jneumeth.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 20.Smith SL, Hausser M. Parallel processing of visual space by neighboring neurons in mouse visual cortex. Nature neuroscience. 2010;13:1144–1149. doi: 10.1038/nn.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozden I, Lee HM, Sullivan MR, Wang SS. Identification and clustering of event patterns from in vivo multiphoton optical recordings of neuronal ensembles. Journal of neurophysiology. 2008;100:495–503. doi: 10.1152/jn.01310.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. Matlab scripts can be requested from the corresponding author.