Abstract
Mantle cell lymphoma (MCL) is characterized by a clinically aggressive course with frequent relapse and poor survival. The p53 pathway is frequently dysregulated and p53 status predicts clinical outcome. In this report, we investigated whether modulation of p73 isoforms by diclofenac inhibits cell growth, induces apoptosis and/or cell cycle arrest in MCL relative to p53 status. Wild-type p53 [Granta-519 and JVM-2], mutant p53 [Jeko-1 and Mino-1] expressing cells, therapy resistant cell lines, and primary human cells isolated from MCL patients were used. Overexpression of pro-apoptotic TAp73 enhanced MCL cell apoptosis. Diclofenac induced a concentration- and duration-dependent increase in TAp73, cell cycle arrest, cell death, and inhibited MCL cell growth independent of p53 status. Diclofenac treatment was associated with increased activity of caspases 3, 7, and 8 and induction of p53 transcriptional target genes. These studies demonstrate the potential for diclofenac as novel therapeutic agent in MCL independent of p53 status.
Keywords: Apoptosis, cell cycle, diclofenac, mantle cell lymphoma, p73 isoforms
Introduction
Non-Hodgkin lymphoma (NHL) is a heterogeneous group of lymphoid neoplasms with distinct morphologic, immunologic, cytogenetic, and molecular features that are associated with a specific pathogenesis for each subtype.[1] Mantle cell lymphoma (MCL), a well-defined subtype of B-cell NHL, is characterized by a clinically aggressive course, a continuous pattern of relapse, and poor survival.[2] Intensive combination chemotherapy with stem cell transplantation has improved disease-free survival in MCL; however, the choice of a treatment for MCL patients remains complex and challenging due to the fact that the median age of MCL patients is about 60 years and these patients are not amenable to aggressive therapeutic approaches.[3,4] Therefore, novel and less toxic therapeutic agents targeting biological pathways relevant to MCL pathogenesis are warranted.
Molecularly, MCL is characterized by the hallmark t(11;14)(q13;q23) chromosomal translocation that juxtaposes the proto-oncogene CCND1, encoding the cell cycle regulatory protein cyclin D1, to the immunoglobulin heavy chain gene. Consequently, cyclin D1 which is not expressed in normal B cells is constitutively expressed in MCL cells, resulting in a deregulated cell cycle at the G1-S phase transition. This genetic alteration is considered the primary event in the pathogenesis of MCL, facilitating cell cycle progression of the cells as they accumulate secondary chromosomal alterations leading to genomic instability and progression of the disease.[5] This molecular cascade of events mandates a harmonizing deregulation of the p53 family signaling pathway; allowing the cells to further advance in the cell cycle and evade apoptosis despite having DNA damage. The aforementioned assumption is supported by reports showing that genes regulating the p53 pathway are frequently targeted by secondary genetic alterations in MCL patients, especially those with an aggressive cellular behavior and poor prognosis.[5,6] Interestingly, despite frequent p53 pathway alterations in MCL, mutations of the p53 gene are uncommon and occur mainly in clinically aggressive cases.[5,7] However, secondary chromosomal alterations of the 1p36 locus (the home for TP73; another functionally overlapping member of the p53 family) are frequent in MCL.[8,9] There is a high degree of homology between TP73 and TP53, which enables p73 to transactivate p53 target genes.[10–12] The TP73 gene locus encodes two types of isoforms due to alternate promoter usage and differential mRNA splicing. TAp73 isoforms (containing the transactivation domain) are tumor suppressive, whereas ΔNp73 isoforms (truncated and lacking the transactivation domain) are oncogenic by antagonizing both TAp73 and p53.[10–12] The balance between TAp73 and ΔNp73 isoforms and their harmony with other members of the TP53 family determines the net cellular responses.[10–12] The unique molecular pathogenesis of MCL with consistent alterations of the p53 family pathway, highlights the potential of targeting the p53 pathway as a therapeutic strategy in MCL.
Non-steroidal anti-inflammatory drugs (NSAIDs) are a structurally diverse group of drugs that are widely used for the treatment of pain and inflammation. More recently, in many cancer types, an anti-cancer effect has been shown for NSAIDs.[13] In contrast to the anti-inflammatory and analgesic effects that are COX-2-dependent, the anti-cancer effects are more complex, less well understood, and involve both COX-2-dependent and COX-2-independent mechanisms.[14] Studies in neuroblastoma have shown the ability of NSAIDs, particularly diclofenac and celecoxib, to increase activity of the p53 pathway. In two successive studies, the authors demonstrated the ability of diclofenac and celecoxib to inhibit growth and induce apoptosis in neuroblastoma cells, both in vitro and in vivo, through increased activity of wild-type p53,[15] and through increased activity of its structurally- and functionally-overlapping family member p73 in cell lines with mutant p53.[16] In the present study, we investigated whether genetic or pharmacological modulation of p73 isoforms modulates MCL cell survival, resistance to therapy, and p53 pathway activity. Our data demonstrates that TAp73 overexpression and modulation of p73 isoforms using Diclofenac (at nontoxic doses to normal cells) treatment on a panel of MCL cell lines, harboring wild-type or mutant p53, results in concentration- and duration-dependent growth inhibition, decreased cell cycle progression, and increased apoptosis. Diclofenac treatment was also associated with increased p53 pathway activity, independent of p53 status, and increased activity of its family member p73.
Materials and methods
Cell lines, culture conditions, plasmids, siRNA, and reagents
Four MCL cell lines including Granta-519 (ACC 342),[17] JVM-2 (ACC 18),[18] Jeko-1 (ACC 553),[19] and Mino-1 (ACC 687) [20] were obtained from DSMZ (Braunschweig, Germany). In addition, we used well characterized therapy-resistant Granta-519 cells that were established from metastasis to kidney (GRK), lung (GRR), or liver (GRL), and resistant to standard NHL therapy in a xenograft mouse model.[21,22] All the cell lines were authenticated at DSMZ (basic STR profiling) and UNMC (karyotypic analysis). Primary MCL cells were isolated from MCL patients in the leukemic phase. UNMC Institutional Review Board approval of the protocol and informed consents were obtained. Diclofenac was purchased from MP Biomedicals (Solon, OH). TAp73 over-expression was achieved using HA-p73α-pcDNA3 and pcDNA3 control vectors obtained from Addgene (Cambridge, MA). Three Trilencer-27 siRNA Duplexes for TP73 knockdown and negative control duplex were purchased from Origene (Rockville, MD). Lipofectamine LTX and plus reagent or Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA) were used for transfection with over-expression vectors or siRNA.
In vitro cell proliferation and cytotoxicity
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was used to determine the cytotoxicity and growth kinetics of the MCL cells. Briefly, 10 000 cells were incubated in a round-bottom 96-well plate in either media alone or media with diclofenac at different concentrations for various durations. Following treatment, cells were incubated with MTT reagent (MP Biomedicals) for 2 h. Cells were lysed with DMSO and optical density was measured using a microplate reader (BIO-TEK ELx-800) at 570 nm. For quantification of proliferation, BrdU incorporation was measured using Cell Proliferation ELISA, BrdU chemiluminescent immunoassay kit (Roche Applied Science, Indianapolis, IN). Ten thousand cells were incubated in a black 96-well plate with clear flat-bottom in either media alone or media with diclofenac at different concentrations for 48 h and then assessed following the manufacturer protocol. The luminescence of each sample was measured using a luminometer (SpectraMax GEMINI EM, CA).
Cell cycle analysis
Cells were fixed in cold 70% ethanol for 30 min, then washed twice with cold phosphate buffer solution (PBS) followed by addition of 1 U of DNase-free RNase to the cell suspension (106 cells/1 mL of PBS) for 30 min at 37 °C. Finally, 100 μL of propidium iodide (Roche Applied Science, Penzberg, Germany) was added and incubated for 30 min and analyzed by flow cytometer.
Cell death and apoptosis studies
Annexin-V-Fluos staining kit (Roche Applied Science) was used for detection and quantification of apoptosis. 1 × 106 cells were washed in PBS and then resuspended in 100 μM of Annexin-V-Fluos labeling solution and analyzed by flow cytometer and fluorescent microscopy. For morphologic evidence of cell death, cytospin preparations were labeled with Hema-3 (Fisher Scientific, Waltham, MA).
Caspase activity assay
For precise quantitative evaluation of the activity of both initiator (caspase-8) and effector (caspase-3/7) caspases, the caspase-Glo assay (Promega) was used according to the manufacturer’s instruction. The assay includes either luminogenic caspase-3/7 or caspase-8 substrates, which contain the tetrapeptide sequence DEVD for caspase 3/7 or LETD for caspase-8. If the cells have caspase 3/7 or 8 activity, caspase cleavage of the substrate and generation of a luminescent signal produced by thermostable luciferase (Ultra-Glo Recombinant Luciferase) will occur. Luminescence is proportional to the amount of caspase activity present and is used to quantitate the respective caspase activity in the test samples 1 × 104 control or diclofenac-treated cells were incubated in 25 μL of media in a white-walled 384 well plate. Then, 25 μL of Caspase-Glo 3/7 or 8 reagent (containing Caspase-Glo 3/7 or 8 buffer, luciferase, substrate, and MG-132 Inhibitor) was added to each well. Following incubation for 30 min at room temperature, the luminescence of each sample was measured using a luminometer (SpectraMax GEMINI EM). Sixty micro molar of the proteasome inhibitor MG-132, was added to avoid caspase-independent cleavage of the peptide substrate by the postglutamyl peptide hydrolytic activity (or caspase-like activity) of the proteasome.
Quantitative real-time PCR
Total RNA was isolated from samples using TRIzol reagent (Invitrogen, Carlsbad, CA) and quantified spectrophotometrically using the Ultrospec 2100 pro (Amersham Bioscience, Pittsburgh, PA). For real-time qRT-PCR analysis, first strand cDNA was generated using oligo (dT)18 (Fermentas, Glen Burnie, MD) and Superscript II RT (Invitrogen, Grand Island, NY). Two microliters of the resulting cDNA was used in real-time PCR reactions using the following primer sets: TAp73 Forward 5′-GCA CCA CGT TTGA GCA CCT CT-3′; Reverse 5′-GCA GAT TGA ACT GGG CCA TGA-3′; ΔNp73 Forward 5′-CAA ACG GCC CGC ATG TTC CC-3′; Reverse 5′-TGA ACT GGG CCG TGG CGAG-3′; β-Actin Forward 5′-TGA AGT GTG ACG TGG ACA TC-3′; and Reverse 5′-ACT CGT CAT ACT CCT GCT TG-3′. The primer set for TAp73 was designed to amplify all the isoforms containing the transactivation domain. For ΔNp73 the forward primer was designed to detect a sequence unique to the ΔNp73 transcript 5′ untranslated region of exon 3. The primers sets for p53 proapoptotic targets were: p21 forward primer: 5′-ATG AAA TTC ACC CCC TTT CC-3′ and reverse primer: 5′-AGG TGA GGG GAC TCC AAA GT-3′; BIM forward primer: 5′-TGG CAA AGC AAC CTT CTG ATG-3′ and reverse primer: 5′-CAG GCT GCA ATT GTC TAC CT-3′; PUMA, forward: 5′-GGA GGG TCC TGT ACA ATC TC-3′ and reverse: 5′-GCT ACA TGG TGC AGA GAA AG-3′; NOXA, forward: 5′-GCT GGA AGT CGA GTG TGC TA-3′ and reverse: 5′-CCT GAG CAG AAG AGT TTG GA-3′; and CD95 forward primer: 5′-AAT GGG GAT GAA CCA GAC-3′ and Reverse: 5′-ATC TTC CCC TCC ATC ATC-3′. Quantitative real-time PCR reactions were carried out using FastStart Sybr Green Master Mix (Roche, Indianapolis, IN) and a MyIQ iCycler (Bio-Rad, Hercules, CA). The ct value was normalized using β-actin for relative gene expression analysis. For calculation of the relative expression, ΔΔct was used.
Immunocytochemistry
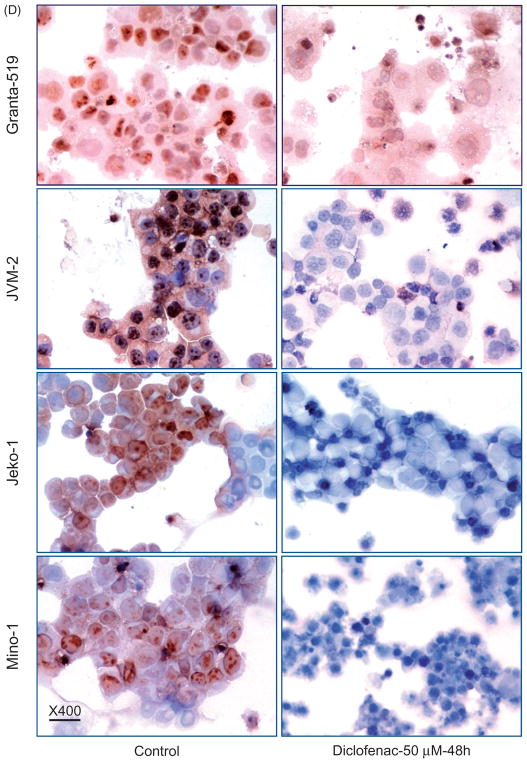
Immunocytochemical (ICC) analysis was performed on cytospin preparations of diclofenac-treated or control MCL cells. The following antibodies were diluted in antibody diluent (Pharmingen, San Diego, CA) in the following ratios: anti-Ki67 (H-300, Santa Cruz) (1:70) and anti-cleaved caspase-3 (ASP175 5A1E, Cell Signaling Technology, Danvers, MA) (1:200). Biotinylated secondary antibody either anti-mouse (BA-2000, Vector Laboratories, Burlingame, CA) or anti-rabbit (BA-1000 Vector Laboratories, CA) 1:500 in PBS were used.
Statistical analysis
All values in the figures are expressed as mean ± standard error of mean (SEM) from minimum three experiments with similar results. We used the Student t-test (two-tailed) for comparisons between treated and untreated cells. One way ANOVA test was used for comparisons between multiple groups followed by Tukey’s and Bonferoni tests for pair-wise comparison. SPSS software was utilized for performing statistical analysis (SPSS Inc., Chicago, IL).
Results
TAp73 modulation alters the behavior of MCL cells
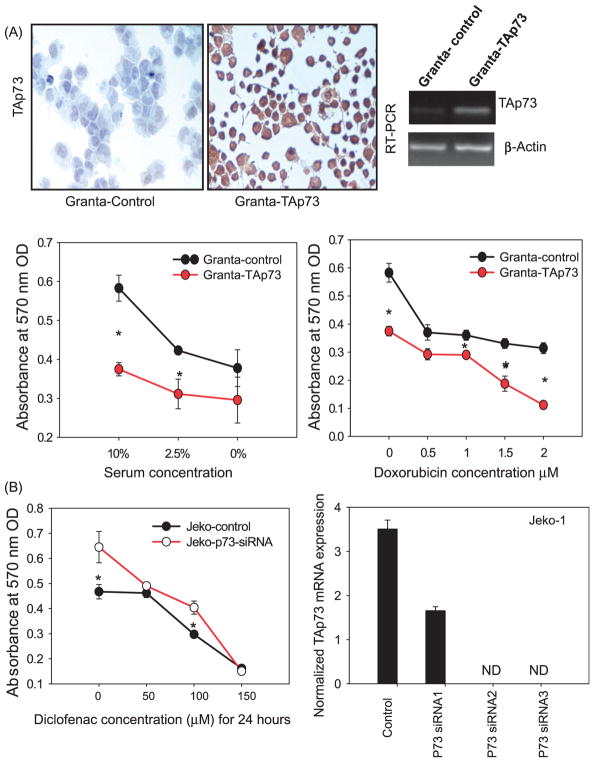
In order to determine whether p73 modulation can alter the behavior of MCL cells, Granta-519 cells were transfected with a TAp73 mammalian over-expression vector. TAp73-transfected Granta-519 cells showed lower basal growth and increased sensitivity to serum deprivation and the chemotherapeutic agent doxorubicin (Figure 1A). To elucidate whether p73 contributes to the diclofenac response, Jeko-1 cells (harboring mutant p53) were transfected with p73 siRNA. P73 siRNA-transfected cells showed a reduced response to diclofenac compared to control-vector transfected cells (Figure 1B).
Figure 1.
p73 modulation alters the behavior of MCL cells. (A) Granta-519 cells were transfected with a TAp73 mammalian expression vector or control vector, and the over-expression of TAp73 was confirmed using RT-PCR for TAp73 mRNA and immunocyto-chemistry for TAp73 protein (200×). The MTT assay was used to compare the basal growth of the cells with serum deprivation (2.5 or 0% serum) or with doxorubicin (0, 0.5, 1, 1.5, 2 μM) for 48 h. (B) Jeko-1 cells were transfected with p73 siRNA or control siRNA and the knockdown was confirmed by qRT-PCR for TAp73. After 48 h post-transfection, cells were treated with the indicated concentrations of diclofenac and the MTT assay was used to compare the growth of the cells.
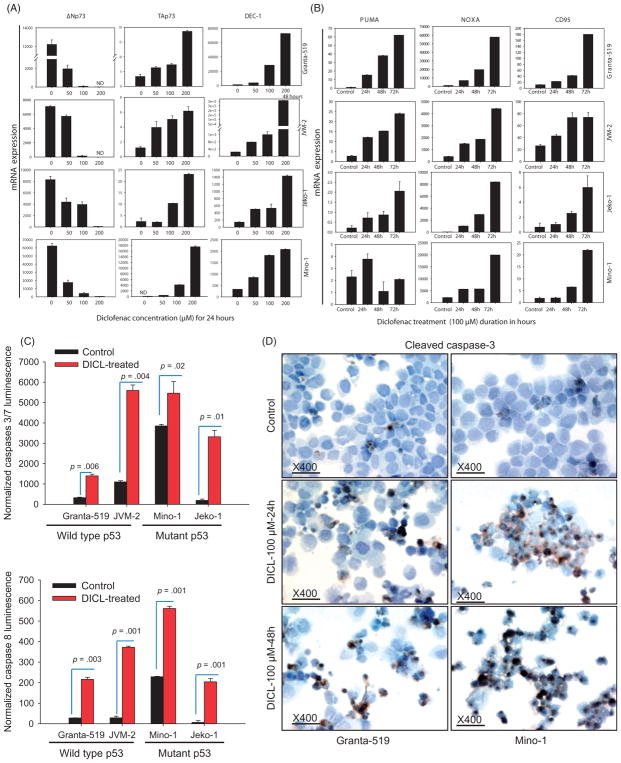
Diclofenac treatment enhances TAp73, decreases ΔNp73, and increases p53 family pro-apoptotic target expression
Real-time qRT-PCR was performed to compare mRNA expression of p73 transcripts (TAp73 and ΔNp73), DEC1, and three p53 family direct-transcriptional, pro-apoptotic targets (PUMA, NOXA, and BIM) between diclofenac-treated (100 μM for 24, 48, or 72 h) and control MCL cells. Diclofenac-treated MCL cells showed a duration-dependent increase of TAp73, the functionally active p73 isoforms and reduction of the transactivation-deficient functionally opposing ΔNp73 isoforms as compared to control untreated cells (Figure 2A).
Figure 2.
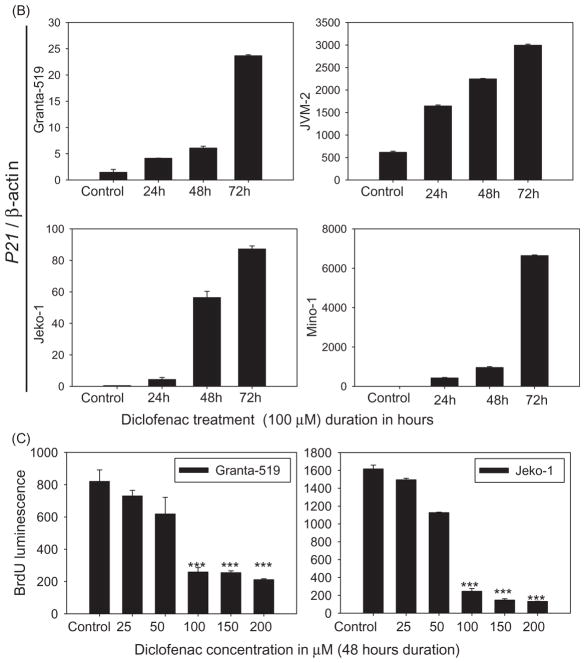
Diclofenac treatment of MCL cells induces TAp73 expression, p53 target genes and caspase cascade activation. (A and B) Diclofenac treatment of MCL cells enhances the expression of the p53 pro-apoptotic targets and differentially modulates the expression of p73 isoforms. MCL cell lines were incubated in media alone (control) or media with the indicated concentrations of diclofenac for the indicated times. Then, the mRNA expression of either: (A) TAp73, ΔNp73, DEC-1 or (B) PUMA, NOXA, or CD95 was analyzed by qRT-PCR and normalized to the housekeeping gene β-actin. The figure for DEC-1 expression in Granta was at 48 h of diclofenac treatment. (C) MCL cell lines were incubated in media alone (control) or media with 100 μM of diclofenac for 48 h. Then, Caspase Glo assay was performed and luminescence was measured after 30 min and plotted. (D) MCL cell lines were incubated in media alone (control) or media with 100 μM of diclofenac for 24 or 48 h, Cytospin preparations of the cells were then immunostained with an antibody against cleaved caspase 3 (the active form of the effector caspase 3) and representative images are shown.
Next we analyzed DEC-1 (Differentiated embryo-chondrocyte expressed gene 1) expression, which has been shown to regulate the p53-mediated cellular response.[23] DEC-1 expression was increased in diclofenac-treated cells compared to control cells (Figure 2A).
Next we examined whether modulation of p73 isoforms by diclofenac regulates p53 transcriptional targets in MCL cells with mutant p53 and wild type p73. NOXA expression was increased in a duration-dependent fashion following diclofenac treatment of the four MCL cell lines. PUMA was consistently increased over time in Granta-519, JVM-2, and Jeko-1 cells, but this trend was not evident in Mino-1 cells (Figure 2B). BIM showed a duration-dependent increase with treatment only in Granta-519 and JVM-2 cells, with no expression in either treated or control Jeko-1 and Mino-1 cells (data not shown). CD95 (Fas), is a pro-apoptotic and pro-necroptotic [20,21] transcriptional target of the p53 family.[22,24] A duration-dependent increase in the expression of CD95 was observed in all four diclofenac-treated (100 μM for 24, 48, or 72 h) cell lines over control (Figure 2B).
Diclofenac-induced cell death involves caspase activation
To delineate whether caspases are involved in diclofenac-induced cell death, we evaluated caspase 3, 7, and 8 activities using the Caspase Glo assay. Diclofenac treatment of MCL cells (100 μM for 24 h) resulted in significantly-increased activity of the effector caspases 3/7 in the four MCL cells used (Figure 2C). To investigate whether effector caspase 3 gets cleaved and activated, ICC staining for cleaved caspase 3 was performed. Diclofenac-treated MCL cells (100 μM for 24 or 48 h) showed increased cleaved caspase 3 immunostaining in all cell lines (Figure 2D) indicating activation of the caspase cascade in response to diclofenac treatment. To confirm upstream caspase activation, the activity of the initiator caspase 8 was evaluated by the Caspase Glo assay. Interestingly, diclofenac-treated MCL cells (100 μM for 24 h) exhibited increased activity of caspase 8 in all four cell lines (Figure 2C).
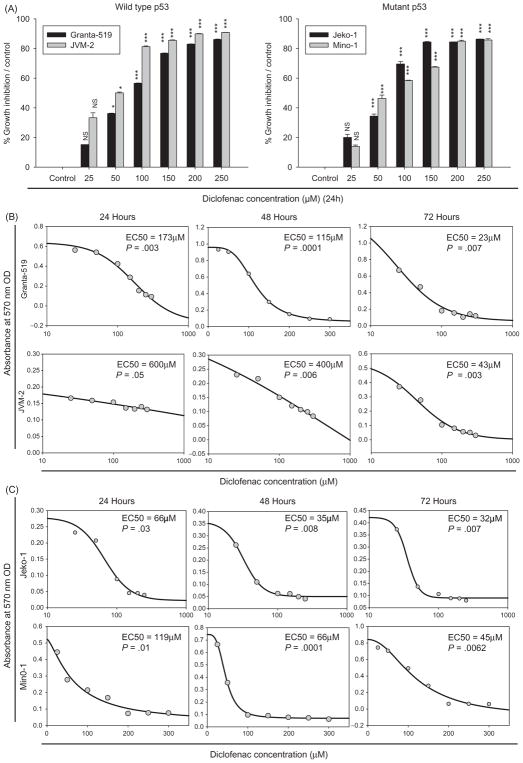
Diclofenac inhibits the growth of MCL cells irrespective of p53 status
To explore the response of MCL to diclofenac, cells with wild-type p53 [Granta-519 and JVM-2] and cells with mutant p53 [Jeko-1 and Mino-1] were used. Treatment of these cell lines with increasing doses of diclofenac (25, 50, 100, 150, 200, 250 μM) for 24 h resulted in concentration-dependent growth inhibition of MCL cells with wild-type p53 (Granta-519 and JVM-2) (Figure 3A), and those with mutant p53 (Jeko-1 and Mino-1) (Figure 3A) as measured with MTT assay. We evaluated the effect of the duration of diclofenac treatment on the cellular response at multiple time points: 24, 48, or 72 h. A duration-dependent response with reduction of the EC50 was observed (Figure 3B and C). To delineate whether the growth inhibitory effect was reversible, diclofenac treatment was removed at multiple time points (24, 48, or 72 h). A gradual duration-dependent recovery of the cells was observed following removal of diclofenac from the media (data not shown).
Figure 3.
Diclofenac inhibits the growth on MCL cells. (A) MCL cell lines were incubated in media alone (control) or media with increasing concentrations (25, 50, 100, 150, 200, 250 μM) of diclofenac for 24 h. The MTT assay was performed and the percentage growth inhibition of the treated cells relative to their respective control was calculated with the equation [100 − (treated/control) − 100]. NS = not significant, *=p<0.05, ***=p<0.001. (B) and (C) MCL cell lines were incubated in media alone (control) or media with increasing concentrations (25, 50, 100, 150, 200, 250 μM) of diclofenac for 24, 48, or 72 h. This is a representative of three experiments done in triplicate.
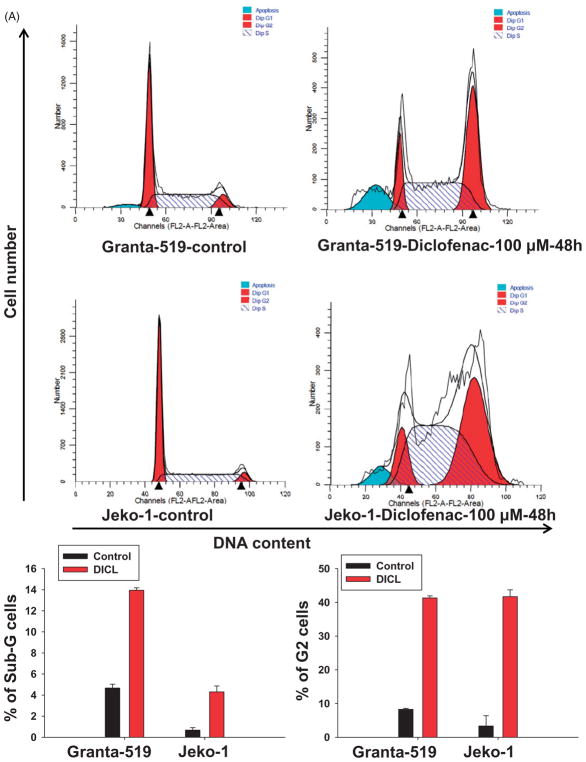
Diclofenac induces cell cycle arrest and inhibits proliferation in MCL cells
To study cell cycle progression, cell cycle analysis using propidium iodide was performed. Cell cycle analysis showed a marked increase in the G2/M sub-fraction of diclofenac-treated cells (100 μM for 48 h) as compared to control cells, indicating G2-M cell cycle arrest (Figure 4A). Furthermore, an increase in the sub-G1 fraction was also observed in diclofenac-treated cells indicating induction of apoptosis (Figure 4A). To test whether the cell cycle arrest was related to p53 pathway activation, the cell cycle regulator and pro-apoptotic molecule p21 (a well-established p53 family transcriptional target) was evaluated. An increase in p21 mRNA expression was observed in diclofenac-treated cells irrespective of p53 status (Figure 4B).
Figure 4.
Diclofenac treatment of MCL cells induces cell cycle arrest. (A) MCL cell lines were incubated in media alone (control) or media with 100 μM of diclofenac for 48 h. Cell cycle analysis was performed using propidium iodide followed by flow cytometric analysis. (B) MCL cell lines were incubated in media alone (control) or media with 100 μM of diclofenac for 24, 48, or 72 h, and then mRNA expression of p21 was analyzed by qRT-PCR and normalized to the housekeeping gene β-actin. This is a representative of three experiments done in triplicate. (C) Cells were incubated in either media alone or media with diclofenac at different concentrations for 48 h, and then BrdU incorporation was measured. (D) Diclofenac treatment inhibits proliferation of MCL cells. MCL cell lines were incubated in media alone (control) or media with 50 μM of diclofenac for 48 h. Cytospin preparations of the cells were then immunostained with an antibody against the proliferation marker Ki-67 and representative images are shown. This is a representative of two experiments done in duplicate.
For quantitation of cell proliferation, the chemiluminescence BrdU immunoassay was performed. Concentration-dependent reduction in cell proliferation was observed in diclofenac-treated MCL cells compared to control cells (p <0.001) (Figure 4C). To assess the effect of diclofenac treatment on proliferation, we immunostained cytospin preparations of diclofenac-treated and control cells for the proliferation marker Ki-67. A reduction in the frequency of Ki-67-positive cells was observed in diclofenac-treated MCL cells (50 μM for 48 h) (Figure 4D).
Diclofenac induces cell death in MCL cells
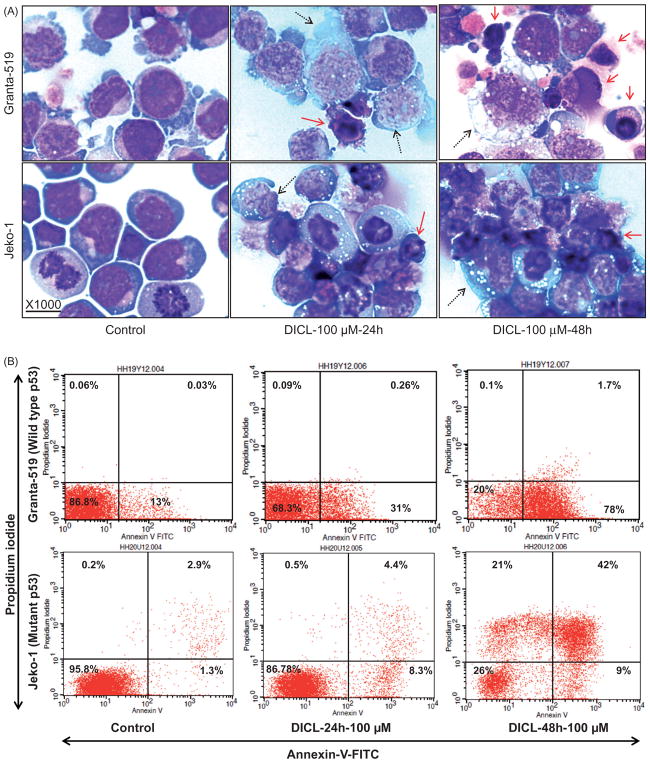
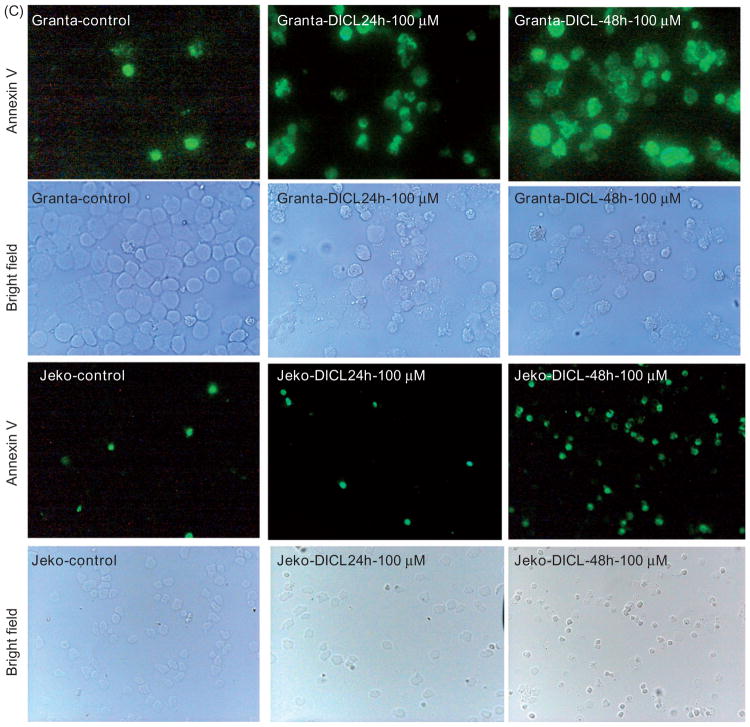
Cytospin preparations of diclofenac-treated MCL cells labeled with Hema-3 stain demonstrated morphologic evidence of cell death (Figure 5A). Unexpectedly, there was morphologic evidence of both apoptotic death, as indicated by nuclear condensation and fragmentation, as well as necroptotic (programed cell necrosis) cell death: as indicated by cellular swelling (Figure 5A). The Annexin V assay was performed to detect and quantify apoptosis and/or necrosis in diclofenac-treated (100 μM for 24 or 48 h) and control MCL cells. Diclofenac treatment resulted in a duration-dependent increase in the total fraction of cells showing evidence of cell death (positive for annexin and/or propidium iodide) reaching 70–80% with higher doses (100 μM for 24 or 48 h) (Figure 5B). There was an increase in both the apoptotic fraction (annexin positive and propidium iodide negative) and necrotic fraction (propidium iodide positive and annexin positive/negative) of MCL cells, confirming the morphologic evidence (Figure 5C).
Figure 5.
Diclofenac treatment of MCL cells induces cell death. (A) MCL cell lines were incubated in media alone (control) or media with 100 μM of diclofenac for 24 or 48 h. Then, cytospin preparations of the cells were stained with Hema-3 stain and representative images are shown. The solid arrows point to apoptotic cells and the dotted arrows indicate necroptosis. (B) and (C) MCL cell lines were incubated in media alone (control) or media with 100 μM of diclofenac for 24 or 48 h. Cells were labeled with Annexin-V-Fluorescein and propidium iodide followed by (B) flow cytometric analysis or (C) fluorescence microscopy and representative images are shown (at 400×).
Diclofenac treatment inhibits therapy-resistant MCL cell line model and MCL primary cells
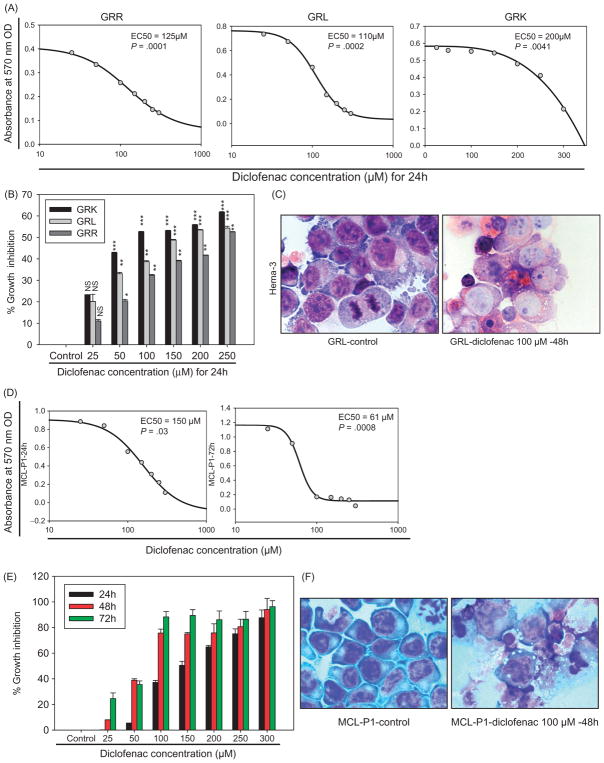
Therapy resistance is a major clinical problem for MCL patients. Therefore, we used therapy-resistant MCL cell line models (GRK, GRL, and GRR). Diclofenac treatment induced dose-dependent (25, 50, 100, 150, 200, 250 μM) and duration-dependent (24, 48, 72 h) growth inhibition of all three therapy-resistant cell lines (Figure 6A and B). Morphologically, Hema-3 staining showed evidence of cell death in diclofenac-treated therapy-resistant cells compared to control cells (Figure 6C). To validate the clinical relevance of our findings in MCL cell lines, the activity of diclofenac was also tested in primary human cells isolated from MCL patients. Diclofenac treatment also induced concentration- and duration-dependent inhibition of these cells, as indicated by the MTT assay (Figure 6D and E) associated with morphologic evidence of cell death (Figure 6F).
Figure 6.
Diclofenac inhibits the growth of resistant MCL cells and primary MCL cells. (A) and (D) The resistant MCL cell lines (GRK, GRL, GRR) or patient-derived primary MCL cells (MCL-P1) were incubated in media alone (control) or media with increasing doses (25, 50, 100, 150, 200, 250 μM) of diclofenac for the indicated durations. The MTT assay was done and SigmaPlot 11 (Systat Software, CA) was used to plot the standard curves and calculate the EC50. (B) and (E) GRK, GRL, GRR, or MCL-P1 were incubated in media alone (control) or media with increasing doses (25, 50, 100, 150, 200, 250 μM) of diclofenac for 24 h, and the percentage growth inhibition of the treated cells relative to their respective control was calculated with the equation [100 − (treated/control) × 100]. (C) and (F) MCL cells were incubated in media alone (control) or media with 100 μM of diclofenac for 24 or 48 h. Cytospin preparations of the cells were then stained with Hema-3 stain and representative images are shown. This is representative of three experiments done in triplicate.
Discussion
Treatment of MCL is challenging considering the constant relapse pattern, toxicity of the therapy, and the fact that more than half of the patients are above 60 years, a ‘watch and wait’ strategy is considered for those patients as well as asymptomatic relapsed patients.[24] Therefore, there is a clinical need for novel less toxic drugs that are biologically relevant for MCL. Clinical studies in MCL patients showed various alterations of the p53 upstream regulators consistently in MCL.[5,7,25] Furthermore, p53 mutations have been shown to be associated with aggressive histology, poor prognosis, and predicting clinical outcome.[6,26,27] Therefore, p53 pathway targeting is a promising therapeutic strategy for MCL. In this report, we demonstrate the potential for diclofenac as a novel therapeutic agent in MCL independent of p53 status through modulation of p73 expression.
The TP73 gene is a member of the TP53 family. As a result of its structural homology with p53, p73 can activate p53 target genes.[23,28] In this report, we observed that overexpression of TAp73 (the full length, transcriptionally-competent pro-apoptotic isoforms of p73) in Granta-519 cells reduced their survival and increased susceptibility to Doxorubicin. Next we analyzed whether Diclofenac modulates expression of the p73 isoforms TAp73 and ΔNp73. All the cell lines used in the study showed increased TAp73 and decreased ΔNp73 in diclofenac-treated cells. The ability of diclofenac to increase TAp73 is in agreement with an earlier report in neuroblastoma.[16] However, for the first time, we also observed reduction in the expression of ΔNp73. To verify the consequences of diclofenac treatment on the p53 pathway, we examined a panel of direct transcriptional targets of p53 that are imperative for cell death, including both the intrinsic apoptotic pathway (PUMA, NOXA, and BIM) and the extrinsic pathway (CD95). NOXA and CD95 were increased in all of the cell lines, Puma was increased in all of the cell lines except Mino-1 and Bim was increased only in the cell lines with wild type p53. Our explanation for these differences in the effects of diclofenac on p53 targets between cell lines is that TAp73 is reported to have a lower transactivation potential than p53 on target genes, and its transcriptional activity can differ between different targets.[29] Interestingly, mutant p53 is also reported to antagonize TAp73 on target genes.[25] Another modulator of p73 isoform expression might be DEC-1.[28] Our data demonstrate increased DEC-1 expression in diclofenac-treated cells compared to control cells.
Diclofenac is a NSAID that has been used as an anti-inflammatory and analgesic drug for a variety of conditions. Recently, diclofenac was shown to increase p53 pathway activity and inhibit the growth of neuroblastoma cells.[15,16] However, diclofenac has not been tested in MCL or any subtype of systemic NHL. The selective COX-2 inhibitor celecoxib has been shown to inhibit mycosis fungoides;[26] however, severe side effects have been reported with the use of COX-2 inhibitors and has necessitated some to be recalled.[27,30] Interestingly, in a screen for drug combinations that have synergistic cytotoxicity for melanoma cell lines, diclofenac showed the most robust results compared to 300 other drugs when combined with novel kinase inhibitors.[31] Furthermore, a recent study of chronic myeloid leukemia (CML) reported a unique synergistic effect for diclofenac on the imatinib response of tumor cells isolated from patients, as well as CML cell lines, and this effect was not observed with some of the other NSAIDs.[32] Together, the aforementioned data suggests a strong potential for diclofenac in cancer therapy.
Based on the need for novel, nontoxic agents, and the importance of the p73 and p53 pathways in MCL, we evaluated the effects of diclofenac on the behavior of MCL cells with wild type and mutated p53, including, cell cycle progression and apoptosis to define its relevance to therapy-resistant MCL. Our data demonstrates that diclofenac as a single agent induces concentration-dependent and duration-dependent growth inhibition of MCL cell lines (Granta-519, JVM-2, Jeko-1, and Mino-1), independent of p53 status, at clinically relevant concentrations (<0.2 mM).[33,34] Biologically, the growth inhibitory effect of diclofenac was associated with both cell cycle arrest at the G2/M phase and induction of apoptosis (intrinsic and extrinsic pathways). There was an increase in caspase activity (initiator caspase 8 and effector caspases 3/7), expression of p53 homolog p73, and expression of p53 transcriptional targets including p21, PUMA, NOXA, BIM, and CD95. Furthermore, diclofenac treatment inhibited the growth of therapy-resistant Granta subclones; GRK, GRL, and GRR which were associated with upregulation of p73. The growth inhibitory activity of diclofenac is in agreement with data from a recent study in cutaneous T-cell lymphoma [35] in which the authors reported superior cytotoxicity of diclofenac over other NSAIDs tested. Recent studies in neuroblastoma,[12,13] melanoma,[33,36] and squamous cell carcinoma of the skin [37] have also shown anticancer effect of diclofenac.
To understand the biological correlates of the suppressive effect of diclofenac on MCL cells, we analyzed cell cycle progression, proliferation, and apoptosis. Interestingly, diclofenac treatment was associated with G2/M arrest, as well as an increase in the sub-G1 fraction (cells with DNA content less than 2n) indicating cells undergoing apoptosis. Our observation of the G2/M phase as the main point of cell cycle arrest, rather than G1/S, with diclofenac treatment is a novel finding. In order to assess whether the effect of diclofenac on cell cycle progression is related to the p53 pathway, we analyzed p21 expression, a known p53 family direct transcriptional target that is reported to regulate cell cycle progression both at G1/S and G2/M.[38] We found a duration-dependent increase in p21 expression with diclofenac treatment, suggesting p53 pathway involvement. The absence of an effect on the G1/S transition is likely related to the constitutive expression of cyclin D1 (one of the main regulators of G1/S transition), thus masking the impact of p53 on that phase of the cell cycle. An earlier report found that if cells with DNA damage are not arrested at G1, the p53/p21 axis can arrest the cells at the G2/M phase.[39]
Our phenotypic and molecular analysis also found that diclofenac treatment of MCL cells induces apoptotic and necroptotic cell death. In several recent studies, a new form of necrosis known as programed cell necrosis (necroptosis) has been described (reviewed in [36]). Necroptosis occurs in a regulated manner and is mediated by specific signaling cascades, such as CD95 or TNF signaling, and is characterized morphologically by cellular swelling, early onset plasma membrane permeabilisation, and finally cellular rupture.[37,40] Mechanistically CD95 or TNFR can trigger either the extrinsic apoptotic pathway or necroptosis depending on the cellular context. In the presence of inhibitors of apoptosis such as cFLIP or cIAP (which inhibit caspases), necroptosis is favored and sometimes a combined phenotype occurs.[37,40,41] Also, a recent study demonstrated the ability of p53 to induce necrosis in MEF cells.[42] In our study, the finding of up-regulated CD95 expression in a duration-dependent manner following diclofenac treatment may explain the mixed cell death phenotype observed.
To confirm the mechanism of apoptosis at the molecular level, caspase Glo assay was used to evaluate the activity of both the initiator caspase 8 as well as the effector caspases 3/7. The activities of caspases 8, 3, and 7 showed a profound increase in diclofenac-treated Granta-519, JVM-2, Jeko-1 cells, and a modest increase in Mino-1 cells (especially caspases 3 and 7), as compared to control cells. However, immunostaining for the active form of caspase 3 (cleaved caspase 3) showed a marked increase in all the cell lines tested including Mino-1. This apparent contrast might be related to the short time of the caspase Glo assay (30 min incubation) as compared to the accumulation of caspase 3 over 48 h in the case of immunostaining for the active form, especially considering reported high levels of inhibitors of apoptosis and c-FLIP in MCL that can impede the activity of caspases.[43]
Given the fact that clinical relapse and therapy resistance are critical problems in MCL therapy, we next examined the activity of diclofenac treatment on established therapy-resistant sub-clones derived from the Granta-519 MCL cell line (GRK, GRL, GRR).[21,22] Diclofenac treatment showed activity on these resistant cells comparable to its effects on the parent cells, indicating the potential for diclofenac to kill cells resistant to the standard CHOP therapy.
In summary, our data demonstrates that diclofenac has dose-dependent and duration-dependent activity against MCL cells independent of p53 status, including experimental therapy-resistant models. Biologically, diclofenac treatment resulted in cell cycle arrest, mainly at the G2/M phase, and profound cell death (mainly apoptosis with findings suggesting also necroptosis). Molecularly, diclofenac treatment was associated with increased activity of caspases 8, 3, and 7, and increase of p53 pathway activity. The increased p53 pathway activity was suggested by a duration-dependent induction of the expression of a panel of p53 transcriptional targets including the cell cycle regulatory molecule p21 and the pro-apoptotic molecules, Puma, Noxa, Bim, and CD95. Importantly, diclofenac treatment was also associated with enhanced expression of the pro-apoptotic isoforms of the p73, TAp73. Given the relatively limited toxicity of diclofenac as compared to chemotherapeutic agents, our data suggests the potential for diclofenac as a novel therapeutic agent in MCL. In the present study, diclofenac demonstrated activity as a single agent in a spectrum of MCL cell line models, including therapy-resistant cells, which suggests that it could also be used as a novel therapy in MCL patients.
Footnotes
Potential conflict of interest: Disclosure forms provided by the authors are available with the full text of this article at http://dx.doi.org/10.3109/10428194.2016.1165814.
References
- 1.A clinical evaluation of the International Lymphoma Study Group classification of non-Hodgkin’s lymphoma. The Non-Hodgkin’s Lymphoma Classification Project. Blood. 1997;89:3909–3918. [PubMed] [Google Scholar]
- 2.Harris NL. Mantle cell lymphoma. J Clin Oncol. 1994;12:876–877. doi: 10.1200/JCO.1994.12.4.876. [DOI] [PubMed] [Google Scholar]
- 3.Martin P, Coleman M, Leonard JP. Progress in mantle-cell lymphoma. J Clin Oncol. 2009;27:481–483. doi: 10.1200/JCO.2008.19.5032. [DOI] [PubMed] [Google Scholar]
- 4.Williams ME, Bernstein SH, Jares P, et al. Recent advances in mantle cell lymphoma: report of the 2012 Mantle Cell Lymphoma Consortium Workshop. Leuk Lymphoma. 2013;54:1882–1890. doi: 10.3109/10428194.2013.771400. [DOI] [PubMed] [Google Scholar]
- 5.Jares P, Colomer D, Campo E. Molecular pathogenesis of mantle cell lymphoma. J Clin Invest. 2012;122:3416–3423. doi: 10.1172/JCI61272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greiner TC, Moynihan MJ, Chan WC, et al. p53 mutations in mantle cell lymphoma are associated with variant cytology and predict a poor prognosis. Blood. 1996;87:4302–4310. [PubMed] [Google Scholar]
- 7.Parekh S, Weniger MA, Wiestner A. New molecular targets in mantle cell lymphoma. Semin Cancer Biol. 2011;21:335–346. doi: 10.1016/j.semcancer.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dave BJ, Hess MM, Pickering DL, et al. Rearrangements of chromosome band 1p36 in non-Hodgkin’s lymphoma. Clin Cancer Res. 1999;5:1401–1409. [PubMed] [Google Scholar]
- 9.Espinet B, Salaverria I, Bea S, et al. Incidence and prognostic impact of secondary cytogenetic aberrations in a series of 145 patients with mantle cell lymphoma. Genes Chromosomes Cancer. 2010;49:439–451. doi: 10.1002/gcc.20754. [DOI] [PubMed] [Google Scholar]
- 10.Dominguez G, Garcia JM, Pena C, et al. DeltaTAp73 upregulation correlates with poor prognosis in human tumors: putative in vivo network involving p73 isoforms, p53, and E2F-1. J Clin Oncol. 2006;24:805–815. doi: 10.1200/JCO.2005.02.2350. [DOI] [PubMed] [Google Scholar]
- 11.Collavin L, Lunardi A, Del SG. p53-family proteins and their regulators: hubs and spokes in tumor suppression. Cell Death Differ. 2010;17:901–911. doi: 10.1038/cdd.2010.35. [DOI] [PubMed] [Google Scholar]
- 12.Bailey SG, Cragg MS, Townsend PA. Family friction as ΔNp73 antagonises p73 and p53. Int J Biochem Cell Biol. 2011;43:482–486. doi: 10.1016/j.biocel.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 13.Cha YI, DuBois RN. NSAIDs and cancer prevention: targets downstream of COX-2. Annu Rev Med. 2007;58:239–252. doi: 10.1146/annurev.med.57.121304.131253. [DOI] [PubMed] [Google Scholar]
- 14.Han JA, Kim JI, Ongusaha PP, et al. P53-mediated induction of Cox-2 counteracts p53- or genotoxic stress-induced apoptosis. EMBO J. 2002;21:5635–5644. doi: 10.1093/emboj/cdf591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau L, Hansford LM, Cheng LS, et al. Cyclooxygenase inhibitors modulate the p53/HDM2 pathway and enhance chemotherapy-induced apoptosis in neuroblastoma. Oncogene. 2007;26:1920–1931. doi: 10.1038/sj.onc.1209981. [DOI] [PubMed] [Google Scholar]
- 16.Lau LM, Wolter JK, Lau JT, et al. Cyclooxygenase inhibitors differentially modulate p73 isoforms in neuroblastoma. Oncogene. 2009;28:2024–2033. doi: 10.1038/onc.2009.59. [DOI] [PubMed] [Google Scholar]
- 17.Jadayel DM, Lukas J, Nacheva E, et al. Potential role for concurrent abnormalities of the cyclin D1, p16CDKN2 and p15CDKN2B genes in certain B cell non-Hodgkin’s lymphomas. Functional studies in a cell line (Granta 519) Leukemia. 1997;11:64–72. doi: 10.1038/sj.leu.2400555. [DOI] [PubMed] [Google Scholar]
- 18.Melo JV, Brito-Babapulle V, Foroni L, et al. Two new cell lines from B-prolymphocytic leukaemia: characterization by morphology, immunological markers, karyotype and Ig gene rearrangement. Int J Cancer. 1986;38:531–538. doi: 10.1002/ijc.2910380413. [DOI] [PubMed] [Google Scholar]
- 19.Jeon HJ, Kim CW, Yoshino T, et al. Establishment and characterization of a mantle cell lymphoma cell line. Br J Haematol. 1998;102:1323–1326. doi: 10.1046/j.1365-2141.1998.00911.x. [DOI] [PubMed] [Google Scholar]
- 20.Lai R, McDonnell TJ, O’Connor SL, et al. Establishment and characterization of a new mantle cell lymphoma cell line, Mino. Leuk Res. 2002;26:849–855. doi: 10.1016/s0145-2126(02)00013-9. [DOI] [PubMed] [Google Scholar]
- 21.Ahrens AK, Chaturvedi NK, Nordgren TM, et al. Establishment and characterization of therapy-resistant mantle cell lymphoma cell lines derived from different tissue sites. Leuk Lymphoma. 2012;53:2269–2278. doi: 10.3109/10428194.2012.691481. [DOI] [PubMed] [Google Scholar]
- 22.Hegde GV, Nordgren TM, Munger CM, et al. Novel therapy for therapy-resistant mantle cell lymphoma: multipronged approach with targeting of hedgehog signaling. Int J Cancer. 2012;131:2951–2960. doi: 10.1002/ijc.27602. [DOI] [PubMed] [Google Scholar]
- 23.Irwin M, Marin MC, Phillips AC, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;40738:648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 24.Vose JM. Mantle cell lymphoma: 2012 update on diagnosis, risk-stratification, and clinical management. Am J Hematol. 2012;87:604–609. doi: 10.1002/ajh.23176. [DOI] [PubMed] [Google Scholar]
- 25.Irwin MS, Kondo K, Marin MC, Jr, et al. Chemosensitivity linked to p73 function. Cancer Cell. 2003;3:403–410. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 26.Kopp KL, Dabelsteen S, Krejsgaard T, et al. COX-2 is a novel target in therapy of mycosis fungoides. Leukemia. 2010;24:2127–2129. doi: 10.1038/leu.2010.221. [DOI] [PubMed] [Google Scholar]
- 27.Bresalier RS, Sandler RS, Quan H, et al. Cardiovascular events associated with rofecoxib in a colorectal adenoma chemoprevention trial. N Engl J Med. 2005;352:1092–1102. doi: 10.1056/NEJMoa050493. [DOI] [PubMed] [Google Scholar]
- 28.Lunghi P, Costanzo A, Mazzera L, et al. The p53 family protein p73 provides new insights into cancer chemo-sensitivity and targeting. Clin Cancer Res. 2009;15:6495–6502. doi: 10.1158/1078-0432.CCR-09-1229. [DOI] [PubMed] [Google Scholar]
- 29.Luh LM, Kehrloesser S, Deutsch GB, et al. Analysis of the oligomeric state and transactivation potential of TAp73α. Cell Death Differ. 2013;20:1008–1016. doi: 10.1038/cdd.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nussmeier NA, Whelton AA, Brown MT, et al. Complications of the COX-2 inhibitors parecoxib and valdecoxib after cardiac surgery. N Engl J Med. 2005;352:1081–1091. doi: 10.1056/NEJMoa050330. [DOI] [PubMed] [Google Scholar]
- 31.Roller DG, Axelrod M, Capaldo BJ, et al. Synthetic lethal screening with small-molecule inhibitors provides a pathway to rational combination therapies for melanoma. Mol Cancer Ther. 2012;11:2505–2515. doi: 10.1158/1535-7163.MCT-12-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Hughes TP, Kok CH, et al. Contrasting effects of diclofenac and ibuprofen on active imatinib uptake into leukaemic cells. Br J Cancer. 2012;106:1772–1778. doi: 10.1038/bjc.2012.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chirasani SR, Leukel P, Gottfried E, et al. Diclofenac inhibits lactate formation and efficiently counteracts local immune suppression in a murine glioma model. Int J Cancer. 2013;132:843–853. doi: 10.1002/ijc.27712. [DOI] [PubMed] [Google Scholar]
- 34.Gottfried E, Lang SA, Renner K, et al. New aspects of an old drug-diclofenac targets MYC and glucose metabolism in tumor cells. PLoS One. 2013;8:e66987. doi: 10.1371/journal.pone.0066987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braun FK, Al-Yacoub N, Plotz M, et al. Nonsteroidal anti-inflammatory drugs induce apoptosis in cutaneous T-cell lymphoma cells and enhance their sensitivity for TNF-related apoptosis-inducing ligand. J Invest Dermatol. 2012;132:429–439. doi: 10.1038/jid.2011.316. [DOI] [PubMed] [Google Scholar]
- 36.Vandenabeele P, Galluzzi L, Vanden Berghe T, et al. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol. 2010;11:700–714. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- 37.Long JS, Ryan KM. New frontiers in promoting tumour cell death: targeting apoptosis, necroptosis and autophagy. Oncogene. 2012;31:5045–5060. doi: 10.1038/onc.2012.7. [DOI] [PubMed] [Google Scholar]
- 38.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 39.Gillis LD, Leidal AM, Hill R, et al. p21Cip1/WAF1 mediates cyclin B1 degradation in response to DNA damage. Cell Cycle. 2009;8:253–256. doi: 10.4161/cc.8.2.7550. [DOI] [PubMed] [Google Scholar]
- 40.Lu JV, Walsh CM. Programmed necrosis and autophagy in immune function. Immunol Rev. 2012;249:205–217. doi: 10.1111/j.1600-065X.2012.01147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silke J, Strasser A. The FLIP side of life. Sci Signal. 2013;6:e2. doi: 10.1126/scisignal.2003845. [DOI] [PubMed] [Google Scholar]
- 42.Vaseva AV, Marchenko ND, Ji K, et al. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell. 2012;149:1536–1548. doi: 10.1016/j.cell.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roue G, Perez-Galan P, Lopez-Guerra M, et al. Selective inhibition of IkappaB kinase sensitizes mantle cell lymphoma B cells to TRAIL by decreasing cellular FLIP level. J Immunol. 2007;178:1923–1930. doi: 10.4049/jimmunol.178.3.1923. [DOI] [PubMed] [Google Scholar]