Abstract
Deep learning, a relatively new branch of machine learning, has been investigated for use in a variety of biomedical applications. Deep learning algorithms have been used to analyze different physiological signals and gain a better understanding of human physiology for automated diagnosis of abnormal conditions. In this manuscript, we provide an overview of deep learning approaches with a focus on deep belief networks in electroencephalography applications. We investigate the state-of-the-art algorithms for deep belief networks and then cover the application of these algorithms and their performances in electroencephalographic applications. We covered various applications of electroencephalography in medicine, including emotion recognition, sleep stage classification, and seizure detection, in order to understand how deep learning algorithms could be modified to better suit the tasks desired. This review is intended to provide researchers with a broad overview of the currently existing deep belief network methodology for electroencephalography signals, as well as to highlight potential challenges for future research.
Keywords: Deep learning, machine learning, electroencephalography, classification
I. Introduction
Over the last decade, automated disease diagnosis has gained a lot of attention among artificial intelligence researchers [1]–[6]. The goal of this research is to increase the accuracy and the speed of disease diagnosis with the aid of automated computer analysis [3,7,8]. This is advantageous in different aspects of healthcare such as early diagnosis and decreasing medical expenses in areas that suffer from a lack of professional human resources [3,7,8]. Machine learning is a fast developing branch of artificial intelligence that helps machines to learn features from data and process it then to find patterns [9]. Machine learning techniques could also be used to analyze new datasets and make predictions about the nature of data sources based on old datasets [9,10]. Recently, these methods have been used to analyze physiological signals, such as electroencephalography (EEG), magnetoencephalography, electrocardiography, blood pressure, and heart rate [11]–[23]. However, the focus of our study is on the automated analysis of EEG signals.
EEG is a recording of the electrical activity in the brain [24,25]. EEG can be used as a diagnostic tool for several neurological disorders, as well as a practical tool to answer neuroscience questions in different research areas; such as psychology or rehabilitation [24,26]. The analysis of EEG signals can be challenging due to high levels of noise and high data dimensionality [27]–[29]. To deal with these problems, signal processing techniques such as filtering and dimensionality reduction methods have been introduced [12,18,28]–[31]. However, they demand careful execution so as not to remove important details from the original signal during processing stages. In addition, many physiological signals are non-stationary, which means that signal characteristics, such as frequency, change over time [15,27,32]–[35]. This makes it difficult to identify characteristics of the process underlying signal dynamics solely from patterns of observed time series [15,27,32,33]. There are some approaches, such as wavelet transform and time-varying autoregressive models, to help with the analysis of non-stationary signals like EEG [32,36,37]. Finally, the ultimate goal for automated analysis of EEG signals is for it to be considered as a practical clinical tool by producing a classifier with clinically-acceptable levels of accuracy. However, the efficiency of the algorithms regarding required execution time and memory is also an important factor, and the algorithm’s real-world practicality must also be considered [18,19]. Balancing these two conflicting requirements has been an ongoing demand in the field of EEG signal research.
In order to analyze EEG signals, one approach is to extract features from the signals in different domains [18,19,21], which are selected by data experts or by feature selection algorithms, such as principal components analysis (PCA) and independent components analysis (ICA) [38,39]. However, these methods are inherently limited by the choices made by the researcher, as only the features they manually select are considered. This method is also task specific, and changing the input or goals of the analysis will likely change what features are best suited for the task. Therefore, an automated feature selection and extraction from raw EEG data would be desirable to be less dependent on human expertise and more time-efficient [15,40].
Deep learning (DL) is an approach that can learn features purely from data [41]–[43], which is advantageous for two reasons. The first advantage is that this method intelligently learns features directly from the raw data through several layers in a hierarchical manner [42]–[45]. The high number of hidden layers means that deep learning takes into account higher-order features and relationships between those features, such that low-level information with simple concepts are learned through low layers and high-level information with complex concepts are learned at high layers [41]–[44]. The second advantage is that it can be applied to unlabeled data by using unsupervised methods [41,44,45], which makes it an applicable method for abundant unlabeled EEG data. Labeling EEG data requires time and expertise, whereas unsupervised methods are both time and cost efficient. DL is inspired by the inner workings of the human brain, which also exhibits a layered learning structure. Various DL architectures such as autoencoder [46]–[48], convolutional deep neural networks [22,48], and deep belief networks [11,14,18]–[20] have been applied to EEG data analysis where they have been shown to produce advanced results on different tasks.
In this paper, we review recent applications of Deep Belief Network (DBN) method for EEG signals. We identify various EEG applications in medicine and describe how DBN methods have been applied and modified based on the task desired. We begin by describing some of the DBN approaches in order to understand contributions made in the analysis of EEG signals. Using this understanding, we then explain the most recent applications of DBN to EEG signals.
II. DBN
In general, a Deep Neural Network (DNN) is superficially similar to a simple neural network. It contains an input layer and an output layer of ‘neurons’, separated by numerous layers of hidden units. However, there are differences in how these networks are trained [44,45]. Specifically, the DNN uses unsupervised learning techniques to adjust the weights between hidden layers, allowing the network to identify the best internal representation (features) of the inputs [45]. This property of DNN enables flexible, high-order modeling of the complex and non-linear relationship between the input and output of the network. The effectiveness of DNN in learning features and classification has been proven for different pattern recognition applications including speech, vision, and natural language processing [49]–[54]. These results have led to a new trend in research in automatic pattern recognition using DL techniques, although many areas lack the sufficient volume of research that would be necessary to make any conclusive declarations.
Despite some breakthroughs, attaining a proper training method for the DNN has been a major challenge [55,56]. DNNs have many hidden layers with large numbers of parameters that need to be trained. There are two steps for training DNNs. The first step is to randomly initialize the feature detection layers [55]. A cascade of generative models, including one visible input layer and one hidden layer, should be considered to initialize weights in the DNN [55]. These generative models are trained without considering discriminative information [43,55]. Finally, the whole DNN is discriminatively trained with the standard backpropagation algorithm [42,55]. Studies have shown that the standard gradient-based random initialization of network weights performs poorly with a DNN of more than two layers [42,55]. As the computational complexity and the large space parameters of a DNN with many hidden layers increases, they result in a lower speed of training [42]. Another problem associated with a large number, in addition to the low speed of training, is getting stuck in local minima which, in most cases, does not lead to desired results [43,55]. On the other hand, the machine learning literature has proposed semi-supervised algorithms in which an unsupervised pre-training procedure is used as an efficient regularizer to present more effective deep learning architectures [42,55]. In this method, the unsupervised pre-training initializes the parameters in a way such that the optimization process ends with a lower minima of the cost function. The first presented pre-training method is DBN [43,57]. Other pre-training methods such as Restricted Boltzmann Machine (RBM) [58] and Deep Boltzmann Machines (DBM) have also been presented.
RBMs can serve as a nuclear component of the DNN [44]. A Boltzmann Machine is a stochastic neural network where, in simplest terms, there are no discrete ‘layers’ and each neuron is bi-directionally connected to every other neuron [44,59,60]. A RBM morphs this structure into a more traditional form. It contains a dedicated input and output layers with unidirectional connections and no links between neurons of the same layer. A model of such a system is presented in Figure 1. This clear relation between input and output layers allows for much faster training of the network. A further advantage of the RBM is that it can easily be expanded by allowing the output layer of neurons from one RBM to serve as the input layer for another RBM. Cascading multiple RBMs in this manner produces a neural network with multiple hidden layers, commonly referred to as a DBN [43,44,55], as shown in Figure 2. While this superficially resembles a multi-layer feed-forward neural network, it differs in how the network is trained [42,44]. Specifically, each RBM is added to the network one layer at a time, and trained in an unsupervised manner before supervised learning and backpropagation are applied to the whole network [43,44,55]. In this method, a DBN is both able to self-select relevant features for analysis and is not subject to the impractically long convergence times of networks that utilize only back-propagation to modify weights between their many layers [60]. The unsupervised learning method commonly employed in this context is the contrastive divergence algorithm, which is summarized below.
Fig. 1.
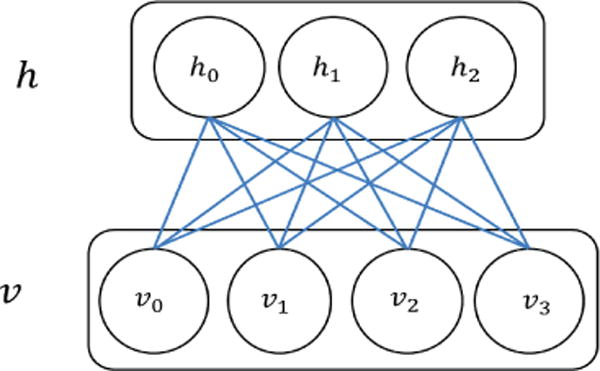
RBM: an undirected graphical model that includes one layer of hidden units (h) and one layer of visible units (v).
Fig. 2.
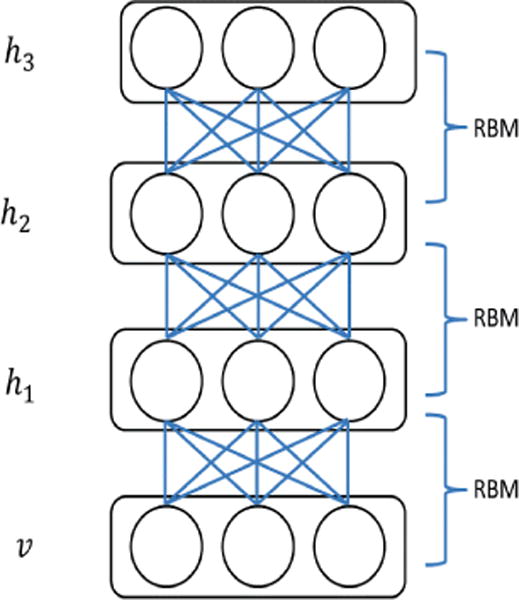
DBN: an example of DBN with one visible layer (v) and three hidden layers (h1, h2, h3).
A RBM has the general form of an energy function for a pair of visible and hidden vectors < v, h > with a matrix of weights W related to the connection between v and h as follows [61]:
| (1) |
where a and b are the bias weights for visible units and hidden units respectively. Probability distributions of v and h are constructed in terms of E:
| (2) |
where Z is a normalizing constant defined as:
| (3) |
Furthermore, the probability of a vector v equals the sum of the above equation over hidden units:
| (4) |
Differentiating a log-likelihood of training data with respect to W is computed as follows:
| (5) |
where 〈.〉data and 〈.〉model indicate expected values in the data or model distribution. The learning rules for weights of the network in the log-likelihood-based training data can be obtained as [61]:
| (6) |
where ε is the learning rate.
Since there is no connection between the neurons at either the hidden or visible layer, unbiased samples can be achieved from 〈vihj〉data. In addition, the hidden or visible unit activations are conditionally independent given visible or hidden units, respectively [61]. For instance, the conditional property of h given v is defined as:
| (7) |
where hj ∈ {0, 1} and the probability of hj = 1 is:
| (8) |
where σ is logistic function defined as:
| (9) |
Similarly, the conditional probability of vi = 1 is computed as:
| (10) |
In general, unbiased sampling from 〈vihj〉 is not straightforward, but it becomes applicable by first sampling a reconstruction of the visible units from hidden units and then using Gibbs sampling in multiple iterations [61]. By applying Gibbs sampling, all hidden units are updated in parallel using equation (8), and in following, visible units are updated using equation (10). Finally, the proper sampling from 〈vihj〉 could be achieved by computing the expected value of multiplying the updated values for hidden and visible units. Sigmoid equations (8) and (10) allow one to use the RBM weights to initialize feedforward neural networks with sigmoid hidden units.
It should be mentioned that there are several supervised and unsupervised algorithms used for the DL method, but this paper puts focus on only DBN, since it is a common algorithm used for EEG classification.
III. EEG
The general workflow of EEG application in medicine includes EEG data acquisition, preprocessing of EEG data by applying simple and/or advanced signal processing techniques, extracting features manually or automatically using DL methods, and finally applying classifiers for desired events or tasks. Figure 3 demonstrates the flowchart for this procedure. In the next subsections, three applications of DBNs for EEG data in medical applications will be described, including emotion recognition, seizure detection, and sleep stage classification.
Fig. 3.
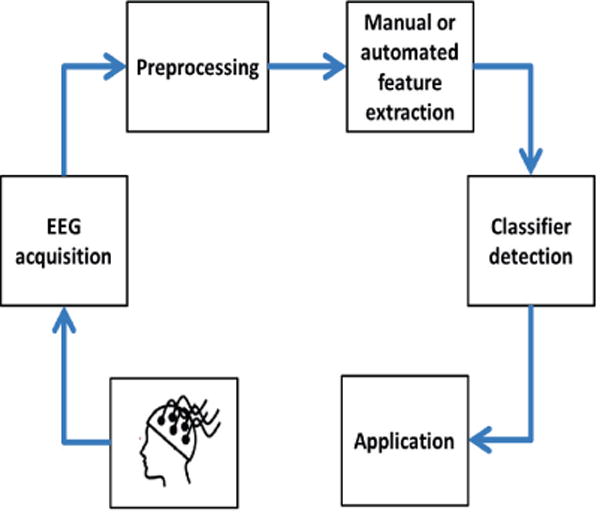
The general workflow of EEG application in medicine
A. Emotion recognition
Emotions are, arguably, one of the core aspects of human consciousness [62,63]. While such concepts are relatively simple to understand, being able to measure or quantify such high-level brain activity is an incredibly complex task that has yet to be solved in a meaningful fashion. However, there have been smaller scientific advances in this regard. Various commercial applications, such as monitors of attentiveness or enjoyment, have been proposed to improve the functionality of consumer devices [64]–[66]. Meanwhile, in the medical field, it has been proposed that monitoring a patient’s emotional state would be a great benefit to diagnosing or treating diseases [67,68]. As a result, efforts have been made to develop a technique to monitor these conditions in a person.
Humans express emotions, both voluntarily and involuntarily, in a variety of different ways including facial expressions, body gestures, voice, and physiological factors such as heart rate and blood pressure [69]–[74]. Therefore, there are several types of data sources such as speech, text, facial expression, body movement, and physiological measurement like EEG, finger temperature, skin conductance level, heart rate, and muscle activity [11,13,14,69]–[73]. There is a mass of emotion data for speech, text, and facial expression as these are details that can be easily gathered from devices used by people on a daily basis, such as cellphones and computers [75,76]. On the other hand, physiological signals can also be a good source of information since they could be collected continuously without participants interfering [77,78]. In addition, individuals can control factors like facial expression, text, and speech, but they cannot control physiological factors like heart rate or blood pressure [77,78]. However, recording physiological data is not convenient as it requires special sensors and equipment that may not be commonly available, or may exist only in the laboratory setting. Recently, several companies have provided small portable bio-sensors to assess human emotions based on different physiological indices of heart rate, skin conductance responses, skin temperature, respiration rate, and even EEG. Here, we can name two of these bio-sensors: ‘Affectiva’ by MIT Media Lab [79] and ‘Emotive Headset’ by the Emotive company [79,80], as shown in Figure 4. Yet, the extent of physiologically informative data is limited in comparison with other information sources for the assessment of human emotions, such as text sources.
Fig. 4.

Emotion measurement technology: (right picture) Affectiva by MIT’s Media Lab works based on facial cues or physiological responses; (left picture) Emotiv by Emotive Systems is an Australian electronics company developing brain-computer interfaces based on electroencephalography (EEG) technologies. We thank Emotiv Inc. and Affectiva for their permissions to use their images.
One source of data for emotion recognition that many studies have been acquired data from is the DEAP study [81]. In the DEAP study, EEG data (32 channels) and several peripheral physiological signals (8 channels), such as skin temperature, electrocardiogram, blood volume, electromyograms of Zygomaticus and Trapezius muscles, and electrooculogram (EOG) of 32 subjects were recorded while watching forty-minute long music videos clips. Participants then rated each video in terms of arousal, valence, like/dislike, dominance, and familiarity. In addition to the DEAP dataset, each researcher might design their own emotion recognition experiment. For instance, Zheng [13,14] used recorded EEG from subjects showing 12 clips to categorize emotions into positive and negative emotions.
The focus of this study is on emotion recognition based on EEG data. Features extracted from EEG signals can vary from simple statistical features such as mean and standard deviation to more complex features such as entropy rate and lempel-ziv complexity. For instance, some common extracted features from EEG signals for emotion recognition are computed in terms of frequency domain. As EEG has higher energy in low frequencies than in high frequencies, studies have shown that differential entropy would be a good choice to discriminate EEG patterns within low and high frequency energy [14].
The general theme of learning emotions is first feeding the algorithms tens of thousands examples of computed features from recorded EEG data in different emotions such as happiness, sadness, stress, etc., from individuals of different genders, cultures, ethnicities, and ages. Then, by using DL method, algorithms look for common characteristics of each emotion within the dataset, such as smiling as a characteristic of happiness. In this way, machines learn to recognize different emotions by using their characteristics and thus become able to recognize discrete emotions in unfamiliar EEG data sets.
Although there is a plenitude of features for each EEG data sample that can be used for emotion recognition, there is a lack of labeled EEG data for emotions as it is challenging to effectively label emotional experiments. This problem makes applying solely supervised learning inefficient for emotion recognition. Another problem is related to noisy EEG data. Here, ‘noisy’ refers to the effect of irrelevant EEG channels on recognition of emotion in EEG-based emotion recognition. In order to solve these two problems, Kang Li et al. [11] and Xiaoyi Li et al. [16,17] applied a DL architecture using DEAP dataset to learn emotions. They trained a two-layer DBN to extract high-level features and then applied a discriminative RBM upon the second hidden layer. In this way, by applying DBN to each of EEG channels, as shown in the Figure 5, low and latent features of EEG data were extracted to solve the problem of insufficient labeled data. Moreover, to deal with the noisy EEG data, the response of the zero-stimulus of each channel on the trained DBN is calculated to choose the critical channels for emotion recognition. In addition, to optimize the hyperparameters, Xiaoyi Li et al. [16] tried various combinations of parameters using training data to find the best combinations with the minimum recognition error. In one experiment, both the EEG (32 channels) dataset and peripheral physiological signals (8 channels) from DEAP dataset were fed into the DBN and its performance was compared to KNN, which demonstrated that the DBN is more capable of recognizing emotions. In another experiment, the DBN was applied only to EEG signals to compare the features extracted automatically using a DBN to those extracted with traditional manual extraction techniques. For DBN features, data from each EEG channel, 32 channels at 128 Hz for each subject, were fed into the DBN, while for the manual method, the PSD of EEG signals were computed. In order to compare the DBN features and PSD features, the SVM classifier with RBF kernel was implemented on both features. The outcome showed that feeding the raw EEG data into the DBN in order to learn effective features is as effective as manually selecting features for learning. Furthermore, Zheng et al. [14] applied DL models on differential entropies extracted from EEG signals to classify two emotions. He trained two DBN models with two hidden layers each and a DBN Hidden Markov Model (HMM). In doing so, he combined the DBN with HMM to form a static classifier with the potential of learning dynamic sequential patterns in human emotions. The results showed a slight improvement of accuracy in DBN models in comparison with KNN, SVM, and graph regularized Extreme Learning Machine (GELM) methods. In his next study [13], he added neutral to emotion categories and compared performance of the DBN model with KNN, LR, and SVM. DBN showed the best average accuracy of 86.08%. He also investigated the best channels and frequency bands for EEG-based emotion recognition and determined them to be the lateral temporal and prefrontal channels and beta and gamma bands, respectively [82]. The summary of all emotion recognition studies is presented in Table 1.
Fig. 5.
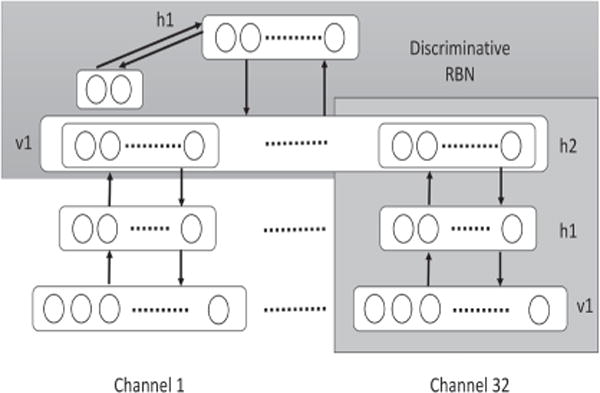
Architecture of unsupervised feature learning and classification that was used in [16] for emotion recognition.
TABLE 1.
Emotion recognition studies summary
| Author | Year | Data | DBN structure | Goal | Outcome | Reference |
|---|---|---|---|---|---|---|
| Zheng et al. | 2013 |
|
|
|
|
[14,83] |
| Li et al. | 2013 |
|
2 hidden layer DBN + critical channel selection the response of the zerostimulus |
|
|
[11] |
| Zheng et al | 2015 |
|
|
|
|
[13] |
| Li et al. | 2015 |
|
|
|
|
[16] |
| Li et al. | 2015 |
|
|
|
|
[17] |
B. Seizure detection
Epilepsy, or seizure disorder, is a common neurological disease, which affects approximately 50 million people globally including 3 million Americans [83,84]. Epilepsy is characterized by recurring seizures which are unpredictable interruptions of normal brain function [85]. Seizures can be induced by a number of different factors such as primary central nervous system (CNS) dysfunction, metabolic disorders, or a high fever [86]–[88]. Epilepsy may occur alongside cerebral palsy, cognitive impairments, developmental disabilities, and autism. There are several clinical assessment tools for epilepsy including CT, MRI, and EEG. The focus of this paper is on EEG data, which enables tracking the electrical activity of brain. Most of the time, various EEG rhythmic patterns can be seen in the beginning of seizures, as shown in Figure 6. EEG can be used for epilepsy as a diagnostic tool to support diagnosis, classify seizures, as well as a prognostic tool to adjust anti-epileptic treatment.
Fig. 6.
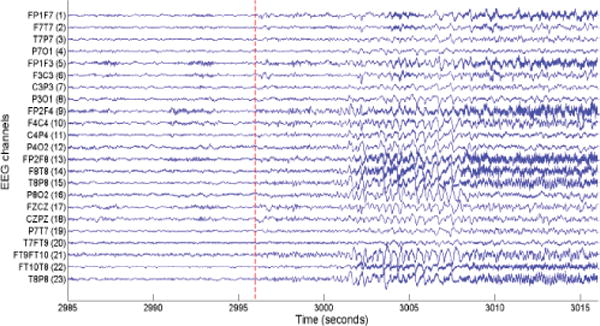
An example of a seizure pattern in the scalp EEG data of a patient in [15]. The dashed red line indicates the onset of seizure. We thank IEEE for providing a permission to use this figure, which was originally published in A. Supratak, L. Li, and Y. Guo, “Feature extraction with stacked autoencoders for epileptic seizure detection,” in Proc. 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC 2014), Chicago, IL, USA, Aug. 26–30, 2014, pp. 4184–4187.
Automated EEG-based seizure identification and classification is a challenging task as seizure patterns are very specific for each patient [15]. This is in addition to the complications brought by the inherently non-stationary nature of EEG signals [15]. So far, several classifiers have been applied to EEG data for seizure detection such as SVM and KNN [89,90], but the accuracies obtained from these classifiers are far from the high accuracy required for clinical use. DL has been proposed as an improved method for seizure detection in this context, as it is able to operate on raw EEG data inputs rather than performing only a narrow statistical interpretation of data as in the other techniques. Studies employing DL methods will be discussed later in this section.
Turner [18] and Page et al. [19] compared performance of different machine learning algorithms, including DBN, KNN, SVM, and logistic regression, for seizure detection using multi-channel EEG data. They emphasized the performance of algorithms, beyond the accuracy factor, including the computational complexity and memory requirements, given that the techniques will be implemented with wireless low-power embedded sensors. They extracted several features such as mean energy and average peak amplitude from EEG data of 23 channels at any given seconds. They then fed extracted features into both a DBN and several classifiers to compare the results. In the DBN case, they applied logistic regression as a classifier to the output of the DBN, as shown in Figure 7. The results showed that the logistic regression performed best with regard to complexity and accuracy when data from the same patient is used for both training and testing. However, in the situation where data from all patients except for one is used for training the model, and the excepted patient is then used as the test subject, the DBN showed the best results. However, adding a DBN stage to the system will increase both its memory and computational expense. Furthermore, Page et al. [19] justified the DBN model as a way to use multiple channels of data to determine the classification of one second of EEG, instead of using only one channel of EEG, by applying stacked denoising autoencoders on raw data before running the DBN. The summary of all seizure detection studies is presented in Table 2.
Fig. 7.
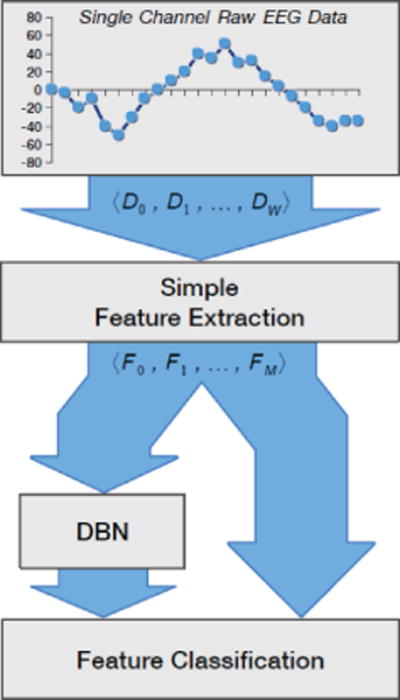
Flow diagram of the experiments for seizure detection in [18]. In this method, extracted features from raw EEG signals are fed directly into several classifiers and also into the DBN, and then logistic regression is used as a classifier. We thank for providing us with a permission to reproduce this figure, which was originally published in “Deep belief networks used on high resolution multichannel electroencephalography data for seizure detection,” by J. Turner, A. Page, T. Mohsenin, and T. Oates, in Proc. AAAI Spring Symposium Series, Palo Alto, California, USA, Mar. 24–26, Copyright ©2014, Association for the Advancement of Artificial Intelligence.
TABLE 2.
Seizure detection studies summary
| Author | Year | Data | DBN structure | Goal | Outcome | Reference |
|---|---|---|---|---|---|---|
|
2014 |
|
|
|
|
[18,19] |
C. Sleep stage classification
Sleep is needed for human survival, along with food, water, and oxygen [91]–[94]. Sleeping disorders have physiological consequences such as hypertension, diabetes, and depression [91,94,95]. Over 70 million Americans suffer from disorders of sleep and wakefulness [91]. To diagnose sleeping disorders, a sleep study called a polysomnographic (PSG) is used to evaluate patients’ sleeping [96,97]. A PSG usually includes all-night recording of EEG, electrooculogram (EOG) and electromyogram (EMG) at a hospital or at a sleep center. These recordings are then divided into short epochs of 20 to 30 seconds and scored by an expert based on one of the standard sets of criteria published by Rechtschaffen and Kales (R&K) [96] or the American Academy of Sleep Medicines (AASM) [97]. The R&K criteria consider two sleep stages which include non-rapid eye movement (NREM) and rapid eye movement (REM). NREM has four stages ranging from S1 to S4. However, because the characteristics of S3 and S4 are very similar to each other, AASM considers these two stages as one stage of slow wave sleep (SWS) [97]. The manual scoring of sleeping by trained experts is a time-consuming and demanding procedure. In addition, it is subjective and depends on the skill and experience of the scoring individuals. Therefore, developing a system of automated scoring would be beneficial, as it would require less time and would result in more unilaterally accurate scoring.
So far, many studies have developed algorithms for automated sleep scoring [21], such as the SVM and neural network approaches, which rely primarily on manual extraction of features from the signal. However, there is no general agreement between studies on which specific features are most applicable in the identification of a given sleeping disorder. Therefore, DL architecture has been recently considered as a new method to obtain latent information from PSG recordings. Langkvist et al. [20] and Zhang et al. [21] applied DBNs to EEG, EOG, and EMG signals in order to classify sleep stages. In both studies signals were preprocessed using notch filtering at 50 Hz to attenuate the power line disturbances, downsampling, and band-pass filtering. Langkvist et al. [20] conducted a 2-layer DBN with 200 hidden units in both layers and a HMM for validation of learned features. Their results showed that the DBN method increased the accuracy of sleeping classification by approximately 3% when compared with the handcrafted results. They also concluded that for multimodal signals such as PSG, it would be best to allocate separate DBNs for each of the signals and then to combine them using a secondary DBN at the top in order to achieve the highest accuracy. In this way, it is also possible to find out which signals provide better information regarding sleeping stages classification. On the other hand, Zhang et al. [21] applied the sparse version of DBN (SDBN) shown in Figure 8, as they believed that sparse models would be able to learn complex features from biological signals. They also used a voting principle based on classification entropy using combination of classifiers including SVM, KNN, and HMM. Their process attained a 91.31% accuracy. The summary of all sleep stage classification studies is presented in Table 3.
Fig. 8.
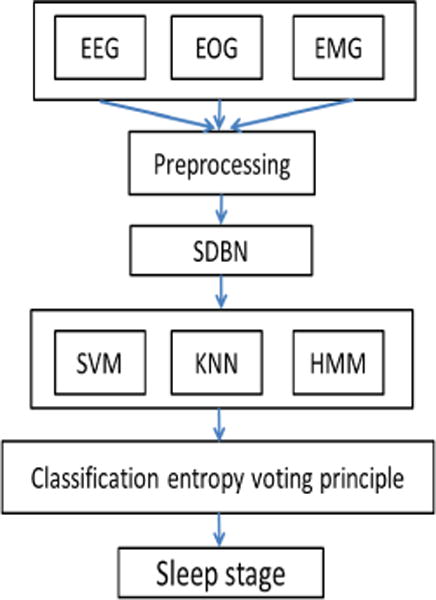
Flowchart of the multi-parameter sleep-staging method in [21]. Three signals of EEG, EOG, and EMG are fed into the sparse version of DBN (SDBN) to extract features automatically from input signals, after preprocessing. Then, several classifiers are applied to the output of SDBN. In the last step, a voting principle based on classification entropy is used to classify sleep stages.
TABLE 3.
Sleep classification studies summary
| Author | Year | Data | DBN structure | Goal | Outcome | Reference |
|---|---|---|---|---|---|---|
| Langkvist et al. | 2012 |
|
|
|
|
[20] |
| Zhang et al. | 2015 |
|
|
|
|
[21] |
IV. Discussion and future work
Experts have studied artificial neural networks for decades. The goal of this paper is not to show all approaches in this area, but to highlight one approach, deep learning (DL), that has demonstrated success in different applications, including automated classification of physiological signals. DL describes a neural network that has multiple hidden layers and can therefore create a much more effective inner representation of the data. This also allows it to identify relationships between higher-order features without the drawbacks encountered in other methods. Moreover, it is more flexible about which data are used as its input when compared with traditional machine learning methods, and is therefore better suited for unsupervised learning. Consequently, DL has the potential to make automatic analysis of physiological signals like EEG more accurate and less dependent on manual human analysis for either diagnosis or network training. However, there are several challenges to reach this point, as DL is a new approach with many open questions. In the following section, we will discuss some of the challenges to automated analysis of EEG data and how DL algorithms deal with them.
One of the challenges is dealing with multivariate data, such as multiple channels of EEG or a combination of multimodal physiological signals like EEG, skin temperature, and blood volume. Using multivariate data is advantageous as it can improve the accuracy of classification performance by interpreting multiple sources of data simultaneously. For instance, Page et al. [19] and Langkvist et al. [20] have applied DBNs on multi-channels signals from EEG and PSG (which includes EEG, EOG, and EMG) respectively, to classify seizures and sleep stages. The results of several studies have demonstrated good accuracy in classification using multimodal signals. The open question concerns what is the best optimized algorithm to combine these multi-modal information sources in order to achieve the most accurate result. In addition, it is critical to find out which physiological sources provide discriminative information regarding specific task classification. Langkvist et al. [20] have demonstrated that to reach a high accuracy for multimodal signals, it would be better to apply separate DBNs for each signal and then combine their outputs with a secondary DBN. In this way, it is possible to figure out which physiological signal provides the most discriminative information regarding specific task classification.
Another challenging concern is how to deal with an enormous amount of EEG data. For instance, a time-series sample of EEG data including 256 readings per second from 23 channels for several minutes, results in a large amount of data, which makes them difficult to deal with. One approach is to project the data to a smaller number of features, which appropriately describe the data. Based on the desired task for classification, various features might be extracted from EEG signals. This method relies on the data scientist’s expertise. In addition to the importance of choosing proper features, the number of extracted features is another critical point. Extracting many features from a small amount of data could lead to overfitting, whereby algorithms learn the training data instead of learning the dominant tendencies in the data. To overcome these problems, DL offers unsupervised algorithms to automatically select and extract features directly from data. In this way, the most relevant information can be extracted from the data, while the dependency on manual analysis is also decreased. As an example, Kang Li et al. [11] and Xiaoyi Li et al. [16] extracted hidden features from EEG data by applying DBN to each of EEG channels for emotion recognition. Results from different studies have demonstrated good performance for automated extraction of features using DL algorithms in regard to the classification accuracy.
The final goal of the automated analysis of EEG signals is to be implemented as a practical medical diagnostic tool in large-scale clinical settings. To reach this goal, it is necessary to boost the practicality of algorithms by improving both their accuracy and computational efficiency/complexity. Therefore, the complexity of DBN classifier algorithms is a critical point that needs to be addressed in subsequent studies. For instance, Turner et al. [18] and Page et al. [19] compared the complexity of DBN with other algorithms such as logistic regression, KNN, and SVM. They showed that computational complexity and memory were increased by DBN in comparison with logistic regression, although DBN provided higher accuracy. Therefore, there is still a need to investigate how to best improve the efficiency of algorithms to fill the gap between the academic and practical uses of DL algorithms.
We can conclude that challenges need to be addressed in future studies for one desired generalized DL method applicable to EEG classification that could satisfy three demanded conditions. First, it could handle multivariate EEG signals from different channels and advanced level multimodal physiological signals, such as EEG combined with heart rate or blood pressure measurements. Second, it could be generalized for any specific desired tasks such as sleep stage classification, seizure detection, and emotion recognition to save time and effort that would otherwise be needed to design a specific algorithm for each task. Lastly, it is critical to find an efficient algorithm that satisfies the time and memory requirements for practical usage of EEG classification in clinical settings.
Finally, it should be mentioned that there are several problems concerning the review of papers focusing on DBN methods and their applications to EEG signals. As the field is new, standards of analysis and experimentation have not been established. This makes it difficult to draw clear conclusions when comparing studies that might even be investigating identical conditions. Furthermore, these mathematically-focused contributions do not always control for physiological variables when obtaining data for classification. This issue is further exacerbated by the relatively a small amount of data that are typically used for these experiments, and a lack of sufficient details when reporting on how the data were obtained.
V. Conclusion
The development of DBN algorithms provides better data modeling and task classification for EEG data. In this paper, we reviewed studies that modified DBN algorithms in several EEG applications to reach a higher accuracy of feature extraction and classification processes. However, there is still a need for investigations to improve DBN algorithms in order to deal with challenges such as multimodal data, large datasets, and computational complexity issues.
Acknowledgments
Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute Of Child Health & Human Development of the National Institutes of Health under Award Number R01HD074819. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Faezeh Movahedi, Department of Electrical and Computer Engineering, Swanson School of Engineering, University of Pittsburgh, Pittsburgh, PA, USA.
James L. Coyle, Department of Communication Science and Disorders, School of Health and Rehabilitation Sciences, University of Pittsburgh, Pittsburgh, PA, USA
Ervin Sejdić, Department of Electrical and Computer Engineering, Swanson School of Engineering, University of Pittsburgh, Pittsburgh, PA, USA.
References
- 1.Sajda P. Machine learning for detection and diagnosis of disease. Annu Rev Biomed Eng. 2006;8:537–565. doi: 10.1146/annurev.bioeng.8.061505.095802. [DOI] [PubMed] [Google Scholar]
- 2.Orrù G, Pettersson-Yeo W, Marquand AF, Sartori G, Mechelli A. Using support vector machine to identify imaging biomarkers of neurological and psychiatric disease: A critical review. Neuroscience & Biobehavioral Rev. 2012;36(4):1140–1152. doi: 10.1016/j.neubiorev.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Klöppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR, Ashburner J, Frackowiak RS. Automatic classification of MR scans in alzheimer’s disease. Brain. 2008;131(3):681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alkım E, Gürbüz E, Kılıç E. A fast and adaptive automated disease diagnosis method with an innovative neural network model. Neural Netw. 2012;33:88–96. doi: 10.1016/j.neunet.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Übeyli ED. Implementing automated diagnostic systems for breast cancer detection. Expert Syst Appl. 2007;33(4):1054–1062. [Google Scholar]
- 6.Goldbaum MH, Sample PA, Chan K, Williams J, Lee T-W, Blumenthal E, Girkin CA, Zangwill LM, Bowd C, Sejnowski T, Weinreb RN. Comparing machine learning classifiers for diagnosing glaucoma from standard automated perimetry. Invest Ophthalmol Vis Sci. 2002;43(1):162–169. [PubMed] [Google Scholar]
- 7.Tsipouras MG, Exarchos TP, Fotiadis DI, Kotsia AP, Vakalis KV, Naka KK, Michalis LK. Automated diagnosis of coronary artery disease based on data mining and fuzzy modeling. IEEE Trans Inf Technol Biomedicine. 2008;12(4):447–458. doi: 10.1109/TITB.2007.907985. [DOI] [PubMed] [Google Scholar]
- 8.Lazarus R, Kleinman KP, Dashevsky I, DeMaria A, Platt R. Using automated medical records for rapid identification of illness syndromes (syndromic surveillance): The example of lower respiratory infection. BMC public health. 2001;1(1):1. doi: 10.1186/1471-2458-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fayyad U, Piatetsky-Shapiro G, Smyth P. From data mining to knowledge discovery in databases. AI Mag. 1996;17(3):37. [Google Scholar]
- 10.Witten IH, Frank E. Data mining: Practical machine learning tools and techniques Morgan Kaufmann. 2005 [Google Scholar]
- 11.Li K, Li X, Zhang Y, Zhang A. Affective state recognition from EEG with deep belief networks; Proc Int Conf IEEE on Bioinf Biomedicine; Shanghai, China. Dec 18–21; 2013. pp. 305–310. [Google Scholar]
- 12.Kim KH, Bang S, Kim S. Emotion recognition system using short-term monitoring of physiological signals. Med Biol Eng Comput. 2004;42(3):419–427. doi: 10.1007/BF02344719. [DOI] [PubMed] [Google Scholar]
- 13.Zheng WL, Guo HT, Lu BL. Revealing critical channels and frequency bands for emotion recognition from EEG with deep belief network; Proc Int Conf IEEE/EMBS Neural Eng; Montpellier, France. Apr 22–24; 2015. pp. 154–157. [Google Scholar]
- 14.Zheng WL, Zhu JY, Peng Y, Lu B-L. EEG-based emotion classification using deep belief networks; Proc Int Conf IEEE Multimedia and Expo; Chengdu, China. Jul 14–18; 2014. pp. 1–6. [Google Scholar]
- 15.Supratak A, Li L, Guo Y. Feature extraction with stacked autoencoders for epileptic seizure detection. Proc 36th Annu Int Conf IEEE Eng Med Biol Soc; Chicago, IL, USA. Aug. 26–30, 2014; pp. 4184–4187. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Zhang P, Song D, Yu G, Hou Y, Hu B. EEG based emotion identification using unsupervised deep feature learning. Proc 1st Int Workshop on Neuro-Physiological Methods in IR Research; Santiago, Chile. Aug. 13, 2015. [Google Scholar]
- 17.Li X, Zhang P, Song D, Hou Y. Recognizing emotions based on multimodal neurophysiological signals. A sponsored Suppl Sci : Adv Comput psychophysiology. 2015;350(6256):28–30. [Google Scholar]
- 18.Turner J, Page A, Mohsenin T, Oates T. Proc AAAI Spring Symposium Series. Palo Alto, California, USA: Mar 24–26, 2014. Deep belief networks used on high resolution multichannel electroencephalography data for seizure detection. [Google Scholar]
- 19.Page A, Turner J, Mohsenin T, Oates T. Comparing raw data and feature extraction for seizure detection with deep learning methods. Proc FLAIRS Conf; Florence, Italy. May 5–9, 2014. [Google Scholar]
- 20.Längkvist M, Karlsson L, Loutfi A. Sleep stage classification using unsupervised feature learning. Adv Artificial Neural Syst. 2012;2012:5. [Google Scholar]
- 21.Zhang J, Wu Y, Bai J, Chen F. Automatic sleep stage classification based on sparse deep belief net and combination of multiple classifiers. Trans Inst Meas Control. 2015 [Google Scholar]
- 22.Martinez HP, Bengio Y, Yannakakis GN. Learning deep physiological models of affect. IEEE Comput Intell Mag. 2013;8(2):20–33. [Google Scholar]
- 23.Martínez HP, Yannakakis GN. Deep multimodal fusion: Combining discrete events and continuous signals. Proc of the 16th Inte Confe Multimodal Interaction; Istanbul, Turkey. Nov 12–16, 2014; pp. 34–41. [Google Scholar]
- 24.Daly I, Pichiorri F, Faller J, Kaiser V, Kreilinger A, Scherer R, Müller-Putz G. What does clean EEG look like. Proc Conf proc IEEE Eng Med Biol Soc. 2012:3963–3966. doi: 10.1109/EMBC.2012.6346834. [DOI] [PubMed] [Google Scholar]
- 25.Binnie CD, Rowan AJ, Gutter T. A manual of electroencephalographic technology. (6th) 1982 CUP Archive. [Google Scholar]
- 26.Jestrović I, Coyle JL, Sejdić E. Decoding human swallowing via electroencephalography: A state-of-the-art review. J neural Eng. 2015;12(5):051001. doi: 10.1088/1741-2560/12/5/051001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Längkvist M, Karlsson L, Loutfi A. A review of unsupervised feature learning and deep learning for time-series modeling. Pattern Recogn Lett. 2014;42:11–24. [Google Scholar]
- 28.Brillinger DR. Time series: Data analysis and theory Siam. 2001;36 [Google Scholar]
- 29.Brockwell PJ, Davis RA. Time series: Theory and methods. Springer Science & Business Media; 2013. [Google Scholar]
- 30.Elger CE, Lehnertz K. Seizure prediction by non-linear time series analysis of brain electrical activity. European J Neuroscience. 1998;10(2):786–789. doi: 10.1046/j.1460-9568.1998.00090.x. [DOI] [PubMed] [Google Scholar]
- 31.Wagner J, Kim J, André E. From physiological signals to emotions: Implementing and comparing selected methods for feature extraction and classification. Proc Int Conf IEEE on Multimedia and Expo; Amsterdam, The Netherlands. July 6-8, 2005; pp. 940–943. [Google Scholar]
- 32.Krystal AD, Prado R, West M. New methods of time series analysis of non-stationary EEG data: Eigenstructure decompositions of time varying autoregressions. Clin Neurophysiol. 1999;110(12):2197–2206. doi: 10.1016/s1388-2457(99)00165-0. [DOI] [PubMed] [Google Scholar]
- 33.Ivanov PC, Amaral LAN, Goldberger AL, Havlin S, Rosenblum MG, Struzik ZR, Stanley HE. Multifractality in human heartbeat dynamics. Nature. 1999;399(6735):461–465. doi: 10.1038/20924. [DOI] [PubMed] [Google Scholar]
- 34.Sejdić E, Lipsitz LA. Necessity of noise in physiology and medicine. Comput methods and programs in biomedicine. 2013;111(2):459–470. doi: 10.1016/j.cmpb.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jestrović I, Coyle J, Sejdić E. The effects of increased fluid viscosity on stationary characteristics of eeg signal in healthy adults. Brain research. 2014;1589:45–53. doi: 10.1016/j.brainres.2014.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov PC, Rosenblum MG, Peng C, Mietus J, Havlin S, Stanley H, Goldberger AL. Scaling behaviour of heartbeat intervals obtained by wavelet-based time-series analysis. Nature. 1996;383(6598):323–327. doi: 10.1038/383323a0. [DOI] [PubMed] [Google Scholar]
- 37.Sejdić E, Djurović I, Jiang J. Time–frequency feature representation using energy concentration: An overview of recent advances. Digit Signal Process. 2009;19(1):153–183. [Google Scholar]
- 38.Subasi A, Gursoy MI. EEG signal classification using pca, ica, lda and support vector machines. Expert Syst Appl. 2010;37(12):8659–8666. [Google Scholar]
- 39.Polat K, Güneş S. Artificial immune recognition system with fuzzy resource allocation mechanism classifier, principal component analysis and fft method based new hybrid automated identification system for classification of EEG signals. Expert Syst Appl. 2008;34(3):2039–2048. [Google Scholar]
- 40.Guo L, Rivero D, Dorado J, Munteanu CR, Pazos A. Automatic feature extraction using genetic programming: An application to epileptic EEG classification. Expert Syst Appl. 2011;38(8):10425–10436. [Google Scholar]
- 41.Ngiam J, Khosla A, Kim M, Nam J, Lee H, Ng AY. Multimodal deep learning; Proc 28th Int Conf Mach Learn; Bellevue, Washington, USA. June 28 – July 2; 2011. pp. 689–696. [Google Scholar]
- 42.Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks; Proc Int Conf on artificial Intell and statistics; Sardinia, Italy. Mar 13-15, 2010; pp. 249–256. [Google Scholar]
- 43.Erhan D, Bengio Y, Courville A, Manzagol P-A, Vincent P, Bengio S. Why does unsupervised pre-training help deep learning? The J Mach Learn Research. 2010;11:625–660. [Google Scholar]
- 44.Le Roux N, Bengio Y. Representational power of restricted boltzmann machines and deep belief networks. Neural Comput. 2008;20(6):1631–1649. doi: 10.1162/neco.2008.04-07-510. [DOI] [PubMed] [Google Scholar]
- 45.Bengio Y. Learning deep architectures for AI. Foundations and trends® in Mach Learn. 2009;2(1):1–127. [Google Scholar]
- 46.Jirayucharoensak S, Pan-Ngum S, Israsena P. EEG-based emotion recognition using deep learning network with principal component based covariate shift adaptation. The Scientific World J. 2014;2014 doi: 10.1155/2014/627892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin Q, Ye S-q, Huang X-m, Li S-y, Zhang M-z, Xue Y, Chen W-S. Classification of epileptic EEG signals with stacked sparse autoencoder based on deep learning. International Conference on Intelligent Computing Springer. 2016:802–810. [Google Scholar]
- 48.Stober S. Using deep learning techniques to analyze and classify EEG recordings [Google Scholar]
- 49.Hinton G, Deng L, Yu D, Dahl GE, Mohamed Ar, Jaitly N, Senior A, Vanhoucke V, Nguyen P, Sainath TN, Kingsbury B. Deep neural networks for acoustic modeling in speech recognition: The shared views of four research groups. IEEE Signal Process Mag. 2012;29(6):82–97. [Google Scholar]
- 50.Dahl GE, Yu D, Deng L, Acero A. Context-dependent pre-trained deep neural networks for large-vocabulary speech recognition. Int Conf IEEE Acoustics, Speech and Signal Process. 2012 Mar 25–30;20(1):30–42. [Google Scholar]
- 51.Deng L, Hinton G, Kingsbury B. New types of deep neural network learning for speech recognition and related applications: An overview. Proc Int Conf IEEE Acoustics, Speech and Signal Process; Vancouver, BC, Canada. May 26-31, 2013; pp. 8599–8603. [Google Scholar]
- 52.Ciresan D, Meier U, Schmidhuber J. Multi-column deep neural networks for image classification. Proc IEEE Conf Comput Vision and Pattern Recognition; Providence, RI, USA. Jun 16-21, 2012; pp. 3642–3649. [Google Scholar]
- 53.Collobert R, Weston J. A unified architecture for natural language processing: Deep neural networks with multitask learning. Proc 25th Int Conf Mach Learn; Helsinki, Finland. July 5-9, 2008; pp. 160–167. [Google Scholar]
- 54.Bengio Y, Schwenk H, Senécal J-S, Morin F, Gauvain J-L. Neural probabilistic language models. Innovations in Mach Learn Springer. 2006:137–186. [Google Scholar]
- 55.Larochelle H, Bengio Y, Louradour J, Lamblin P. Exploring strategies for training deep neural networks. The J Mach Learn Research. 2009;10:1–40. [Google Scholar]
- 56.Nielsen M. Neural networks and deep learning. Determination Press; 2015. [Google Scholar]
- 57.Mohamed A, Hinton G, Penn G. Understanding how deep belief networks perform acoustic modeling. Int Conf IEEE on Acoustics, Speech and Signal Processing; Kyoto, Japan. Mar 25-30, 2012; pp. 4273–4276. [Google Scholar]
- 58.Salakhutdinov R, Hinton G. An efficient learning procedure for deep boltzmann machines. Neural Comput. 2012;24(8):1967–2006. doi: 10.1162/NECO_a_00311. [DOI] [PubMed] [Google Scholar]
- 59.Larochelle H, Bengio Y. Classification using discriminative restricted Boltzmann machines. Proc 25th Int Conf Mach Learn; Helsinki, Finland. July 5-9, 2008; pp. 536–543. [Google Scholar]
- 60.Hinton GE, Osindero S, Teh Y-W. A fast learning algorithm for deep belief nets. Neural Comput. 2006;18(7):1527–1554. doi: 10.1162/neco.2006.18.7.1527. [DOI] [PubMed] [Google Scholar]
- 61.Hinton G. A practical guide to training restricted Boltzmann machines. Momentum. 2010;9(1):926. [Google Scholar]
- 62.Schacter D, Gilbert DT, Wegner DM. Psychology. 2nd. New York: Worth; 2011. [Google Scholar]
- 63.Panksepp J. Affective consciousness: Core emotional feelings in animals and humans. Consciousness and cognition. 2005;14(1):30–80. doi: 10.1016/j.concog.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 64.Chen M, Zhang Y, Li Y, Mao S, Leung V. EMC: Emotion-aware mobile cloud computing in 5G. Network, IEEE. 2015;29(2):32–38. [Google Scholar]
- 65.Bahreini K, Nadolski R, Westera W. Towards multimodal emotion recognition in e-learning environments. Interactive Learn Environments. 2014:1–16. [Google Scholar]
- 66.Lisetti CL, Nasoz F. Using noninvasive wearable computers to recognize human emotions from physiological signals. EURASIP J Advances in Signal Process. 2004;2004(11):1–16. [Google Scholar]
- 67.Dickerson RF, Gorlin EI, Stankovic JA. Empath: A continuous remote emotional health monitoring system for depressive illness. Proc 2nd Conf Wireless Health; New York, USA. October 10 - 13, 2011; p. 5. [Google Scholar]
- 68.Hosseini HG, Krechowec Z. Facial expression analysis for estimating patient’s emotional states in RPMS. 26th Ann Int Conf IEEE Eng Med and Biol Soc1; San Francisco, California, USA. Sep 1-5, 2004; pp. 1517–1520. [DOI] [PubMed] [Google Scholar]
- 69.Tao J, Tan T. Affective computing: A review. Affective computing and intelligent interaction Springer. 2005:981–995. [Google Scholar]
- 70.Hudlicka E. To feel or not to feel: The role of affect in human–computer interaction. Int J human-computer Stud. 2003;59(1):1–32. [Google Scholar]
- 71.Koolagudi SG, Maity S, Kumar VA, Chakrabarti S, Rao KS. Contemporary Computing. Springer; 2009. IITKGP-SESC: Speech database for emotion analysis; pp. 485–492. [Google Scholar]
- 72.Koolagudi SG, Kumar N, Rao KS. Speech emotion recognition using segmental level prosodic analysis. Proc Int Conf Devices and Commun; Mesra, Ranchi, India. Feb 24-25, 2011; pp. 1–5. [Google Scholar]
- 73.Edwards J, Jackson HJ, Pattison PE. Emotion recognition via facial expression and affective prosody in schizophrenia: A methodological review. Clinical Psychology Review. 2002;22(6):789–832. doi: 10.1016/s0272-7358(02)00130-7. [DOI] [PubMed] [Google Scholar]
- 74.Larsen RJ, Fredrickson BL. Measurement issues in emotion research. Well-being: The foundations of hedonic psychology. 1999:40–60. [Google Scholar]
- 75.Lee K-K, Cho Y-H, Park K-S. Intell Comput Signal Process Pattern Recognition. Springer; 2006. Robust feature extraction for mobile–based speech emotion recognition system; pp. 470–477. [Google Scholar]
- 76.Gievska S, Koroveshovski K, Tagasovska N. Bimodal feature–based fusion for real–time emotion recognition in a mobile context. Proc Int Conf Affective Comput and Intelligent Interaction; Xi’an, China. Sep 21-24, 2015; pp. 401–407. [Google Scholar]
- 77.Kassam KS, Mendes WB. The effects of measuring emotion: Physiological reactions to emotional situations depend on whether someone is asking. PloS one. 2013;8(6):e64959. doi: 10.1371/journal.pone.0064959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.AlZoubi O, Mello SK, Calvo RA. Detecting naturalistic expressions of nonbasic affect using physiological signals. IEEE Trans Affective Comput. 2012;3(3):298–310. [Google Scholar]
- 79.Picard R. Affective media and wearables: Surprising findings. Proc ACM Int Conf Multimedia; Orlando, FL, USA. Nov 3-7, 2014; pp. 3–4. [Google Scholar]
- 80.Lan Z, Sourina O, Wang L, Liu Y. Real-time EEG-based emotion monitoring using stable features. The Visual Comput. 2015:1–12. [Google Scholar]
- 81.Koelstra S, Mühl C, Soleymani M, Lee J-S, Yazdani A, Ebrahimi T, Pun T, Nijholt A, Patras I. Deap: A database for emotion analysis; using physiological signals. IEEE Trans Affective Comput. 2012;3(1):18–31. [Google Scholar]
- 82.Zheng WL, Lu BL. Investigating critical frequency bands and channels for EEG-based emotion recognition with deep neural networks. IEEE Trans Auton Mental Develop. 2015;7(3):162–175. [Google Scholar]
- 83.Theodore WH, Spencer SS, Wiebe S, Langfitt JT, Ali A, Shafer PO, Berg AT, Vickrey BG. Epilepsy in north america: A report prepared under the auspices of the global campaign against epilepsy, the international bureau for epilepsy, the international league against epilepsy, and the world health organization. Epilepsia. 2006;47(10):1700–1722. doi: 10.1111/j.1528-1167.2006.00633.x. [DOI] [PubMed] [Google Scholar]
- 84.W. H. Organization. The global campaign against epilepsy: Out of the shadows. Geneva: WHO; 2000. [Google Scholar]
- 85.Fisher RS, Boas WvE, Blume W, Elger C, Genton P, Lee P, Engel J. Epileptic seizures and epilepsy: Definitions proposed by the international league against epilepsy (ILAE) and the international bureau for epilepsy (IBE) Epilepsia. 2005;46(4):470–472. doi: 10.1111/j.0013-9580.2005.66104.x. [DOI] [PubMed] [Google Scholar]
- 86.Devinsky O, Schein A, Najjar S. Epilepsy associated with systemic autoimmune disorders. Epilepsy Currents. 2013;13(2):62–68. doi: 10.5698/1535-7597-13.2.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sadleir LG, Scheffer IE. Febrile seizures. British Med J. 2007:307–311. doi: 10.1136/bmj.39087.691817.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Engel J. A proposed diagnostic scheme for people with epileptic seizures and with epilepsy: Report of the ILAE task force on classification and terminology. Epilepsia. 2001;42(6):796–803. doi: 10.1046/j.1528-1157.2001.10401.x. [DOI] [PubMed] [Google Scholar]
- 89.Asha S, Sudalaimani C, Devanand P, Thomas TE, Sudhamony S. Automated seizure detection from multichannel EEG signals using support vector machine and artificial neural networks. Proc Int Multi-Conf on Autom Comput Commun Control Compressed Sens; Kerala, India. Mar 22-23, 2013; pp. 558–563. [Google Scholar]
- 90.Shoeb AH, Guttag JV. Application of machine learning to epileptic seizure detection. Proc of the 27th Int Conf Mach Learn. 2010:975–982. [Google Scholar]
- 91.Altevogt BM, Colten HR. Sleep disorders and sleep deprivation: An unmet public health problem National. Academies Press; 2006. [PubMed] [Google Scholar]
- 92.Shephard RJ. Sleep, biorhythms and human performance. Sports Med. 1984;1(1):11–37. [Google Scholar]
- 93.Leach J. Survival psychology Palgrave Macmillan. 1994 [Google Scholar]
- 94.Roberts RE, Roberts CR, Duong HT. Sleepless in adolescence: Prospective data on sleep deprivation, health and functioning. J adolescence. 2009;32(5):1045–1057. doi: 10.1016/j.adolescence.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Banks S, Dinges DF. Behavioral and physiological consequences of sleep restriction. J Clin Sleep Med. 2007;3(5):519–528. [PMC free article] [PubMed] [Google Scholar]
- 96.Rechtschaffen A, Kales A. A manual of standardized terminology, techniques and scoring system for sleep stages of human subjects. US Public Health Service; 1968. [DOI] [PubMed] [Google Scholar]
- 97.Berry RB, Brooks R, Gamaldo CE, Hardling S, Marcus C, Vaughn B. The AASM manual for the scoring of sleep and associated events: Rules, terminology and technical specifications. American Academy of Sleep Medicine; 2012. [Google Scholar]


