Abstract
Many mammalian genes contain poorly spliced introns, resulting in nuclear detention of partially spliced transcripts, which may be exploited to modulate gene expression. In this issue of Cancer Cell, Braun et al. report that the arginine methyltransferase PRMT5 is critical for tumor cell proliferation by regulating numerous detained introns.
Expressed genes in mammalian genomes are known to undergo extensive alternative splicing (AS). A unique class of AS is intron retention (IR), which is more prevalent than previously thought based on analysis of a large number of cell types and tissues in mice and humans (Braunschweig et al., 2014). Interestingly, as illustrated in Figure 1A, IR can be further divided into two classes, one corresponding to those containing a premature termination codon (PTC), which elicits nonsense-mediated RNA decay (NMD) upon export to the cytoplasm, and the other corresponding to those retained in the nucleus, which are insensitive to NMD and collectively referred to as detained introns (DIs) (Boutz et al., 2015). DI-containing transcripts may be further spliced for nuclear export or degraded by the exosome (Fan et al., 2017). There must be specific cis-acting signals in nuclear-detained RNAs, which remain to be characterized.
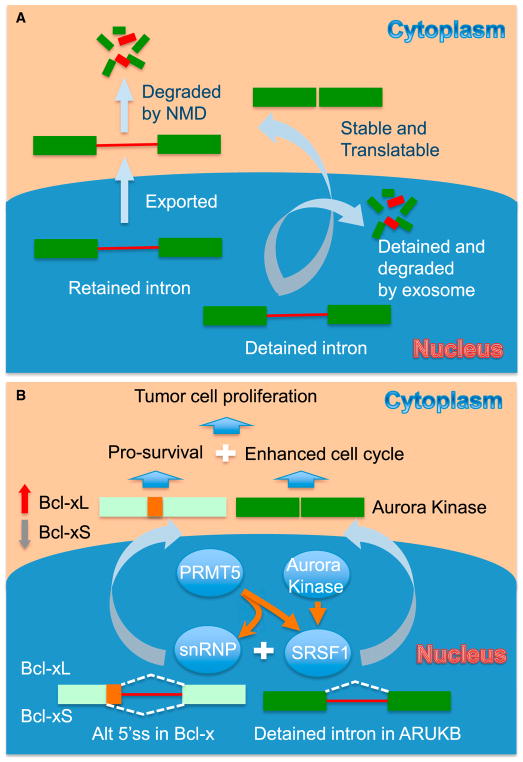
Figure 1. Exploring the Regulatory Window of Intron Retention for Cancer Therapeutics.
(A) Two types of intron retention. One type is retained introns that can be exported to the cytoplasm and then degraded by nonsense-mediated decay (NMD). The other type is detained introns that remain in the nucleus, where they are either further spliced to produce fully spliced mRNAs for export or degraded by the exosome. (B) PRMT5-regulated splicing important for tumor cell proliferation. PRMT5-mediated arginine methylation enhances the functions of the core splicing machinery (e.g., snRNP components) and splicing factors (e.g., SRSF1) to promote the removal of a detained intron in ARUKB and induce Bcl-x splicing. Efficient expression of ARUKB and the production of the pro-survival Bcl-xL isoform enable cancer cells to undergo more efficient cell cycle and less apoptosis. Inhibition of PRMT5 may not only reverse these processes, but also induce a self-enforcing loop in which increased intron retention represses ARUKB expression, which may somehow destabilize SRSF1 to cause further inhibition of ARUBK splicing and also induce a switch of Bcl-x splicing to generate the pro-apoptotic Bcl-xS isoform. These events may synergistically block tumor cell proliferation and increase tumor cell death, thus achieving potent anti-tumor effects.
Do retained introns result from splicing errors, thus representing noises in gene expression? Increasing evidence suggests that many retained introns are biologically relevant. From the evolution point of view, similar to alternative splice sites, splicing signals in retained introns are more conserved than those in constitutive introns (Boutz et al., 2015; Braunschweig et al., 2014). It has been suggested that those retained introns are involved in fine-tuning the transcriptome by eliminating physiologically irrelevant transcripts and/or preventing unwanted gene products during development and differentiation as a new mode of post-transcriptional regulation of gene expression (Braunschweig et al., 2014). On the other hand, cancer cells appear to also exploit those introns, particularly DIs, for their growth advantage, now revealed by the study in the current issue of Cancer Cell (Braun et al., 2017).
The study began with an unbiased screening of small hairpin RNAs (shRNAs) against various epigenetic regulators to identify genes important for growth of a grafted brain tumor cell line in the mouse. Using a well-studied KRAS gene as a positive control, candidate genes were identified based on depleted shRNAs, and the top hit was PRMT5, an arginine methyltransferase that has both histone and non-histone substrates. The requirement of PRMT5 for tumor growth was verified and extended to multiple brain tumor cell lines by using independent shRNAs through functional rescue of wild-type, but not catalytically dead enzyme, and with an existing small molecule chemical inhibitor EPZ015666. The broad anti-tumor effects of PRMT5 down-regulation paved the way for detailed mechanistic studies to understand how this arginine methyltransferase regulates gene expression via DIs.
The anti-tumor effect of EPZ015666 was further tested on a large set of tumor cell lines of diverse origin, many of which showed sensitivity while others were relatively resistant. After ruling out several potential candidate mechanisms, the authors used the existing RNA-seq data to identify gene signatures that are linked to responsiveness to EPZ015666. This comprehensive analysis landed on the ratio of expressions of a pair of genes, CLNS1A and RIOK1, with high CLNS1A-to-RIOK1 ratio correlated to EPZ015666 sensitivity. Importantly, CLNS1A and RIOK1 were mutually exclusive co-factors for PRMT5 to function in small nuclear ribonucleoprotein particle (snRNP) and ribosome biogenesis, respectively (Guderian et al., 2011); the positive correlation between high CLNSIA-to-RIKO1 expression ratio and EPZ015666 sensitivity thus suggested a role of PRMT5 in splicing control for its tumor-promoting effects. This elegant approach illustrates a general framework for using gene expression profiles to aid in pathway identification and dissection. Profiling PRMT5-dependent splicing then led to the discovery that inhibition of PRMT5 selectively and profoundly increased DIs. Among many of those DI-containing genes are critical cell-cycle regulators, such as AURKB, which is well known for its role in the regulation of mitosis.
The next challenge was to understand how PRMT5 modulates DIs. snRNP biogenesis appeared to be compromised, as evidenced by downregulation of Sm proteins SNRPB and SNRPD3 in EPZ015666-treated cells. Although the detailed mechanism still remains to be fully understood, especially with respect to potential defects in arginine methylation, the findings are connected to some critical information already in literature. Notably, a previous genome-wide small interfering RNA screen identified AURKA as a potent regulator of Bcl-x mRNA splicing (Moore et al., 2010). The Bcl-x mRNA is alternatively spliced via two competing 5′ splice sites to produce Bcl-xL, which is anti-apoptotic, and Bcl-xS, which is pro-apoptotic. Importantly, inhibition of either AURKA (with VX-680) or AURKB (with ZM447439) has been shown to induce Bcl-xS at the expense of Bcl-xL (Moore et al., 2010). Although not yet directly examined in the current study, the production of Bcl-xS (and perhaps other apoptosis regulators) might contribute to the tumor suppression phenotype in EPZ015666-treated cells.
Interestingly, Bcl-xS has been shown to be regulated by the SR protein SRSF1 (Paronetto et al., 2007), and, perhaps more than a coincidence, SRSF1 has been documented to be regulated by arginine methylation as well, the defect of which caused a shift of SRSF1 to the cytoplasm (Sinha et al., 2010). Furthermore, SRSF1 protein became downregulated in AURK inhibitor-treated cells (Moore et al., 2010). Taken together, these observations suggest a potential self-enforcing loop in which inhibition of PRMT5 may impair various splicing factors and regulators, such as snRNPs and SRSF1, leading to increased intron retention in AURKB (and likely other cell-cycle regulators); in addition, down-regulation of AURKB further diminishes the expression of SRSF1, thereby causing additional intron retention and other types of alternative splicing (Figure 1B). Induction of this loop (via inhibition of either PRMT5 or Aurora kinases) may thus render cancer cells less efficient in proliferation (e.g., impaired cell-cycle progression) and more vulnerable to apoptosis (e.g., via the induction of Bcl-xS). This loop may underlie, at least in part, both the oncogenic function of SRSF1 documented earlier (Karni et al., 2007) and the anti-cancer effect of PRIMT5 inhibition revealed in the current study (Braun et al., 2017).
Given that PRMT5 is an enzyme that can be targeted by small molecule inhibitors, the findings illustrate potential anti-cancer therapeutics by exploiting DIs. Notably, many mutations in splicing regulators have been linked to myelodysplastic syndromes (MDS), and small molecule inhibitors against the spliceosome, which likely affect many DIs, have been shown to be effective in treating MDS (Lee et al., 2016). These findings highlight the value of DIs in fine-tuning the transcriptome in disease cells without causing detrimental effects on healthy cells to achieve therapeutic effects. On this note, it might also be possible to decrease specific intron-retention events to increase protein production as a strategy to treat other types of diseases caused by inefficient splicing of certain DI-containing genes. These possibilities highlight DIs not as splicing noises but as gifts of evolution in biology.
References
- Boutz PL, Bhutkar A, Sharp PA. Genes Dev. 2015;29:63–80. doi: 10.1101/gad.247361.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun CJ, Stanciu M, Boutz PL, Patterson JC, Calligaris D, Higuchi F, Neupane R, Fenoglio S, Cahill DP, Wakimoto H, et al. Cancer Cell. 2017;32(this issue):411–426. doi: 10.1016/j.ccell.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, Frey B, Irimia M, Blencowe BJ. Genome Res. 2014;24:1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Kuai B, Wu G, Wu X, Chi B, Wang L, Wang K, Shi Z, Zhang H, Chen S, et al. EMBO J. 2017:e201696139. doi: 10.15252/embj.201696139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guderian G, Peter C, Wiesner J, Sickmann A, Schulze-Osthoff K, Fischer U, Grimmler M. J Biol Chem. 2011;286:1976–1986. doi: 10.1074/jbc.M110.148486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Dvinge H, Kim E, Cho H, Micol JB, Chung YR, Durham BH, Yoshimi A, Kim YJ, Thomas M, et al. Nat Med. 2016;22:672–678. doi: 10.1038/nm.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Wang Q, Kennedy CJ, Silver PA. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. J Cell Biol. 2007;176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R, Allemand E, Zhang Z, Karni R, Myers MP, Krainer AR. Mol Cell Biol. 2010;30:2762–2774. doi: 10.1128/MCB.01270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]