Abstract
The olfactory receptor (OR) gene families, which govern mammalian olfaction, have undergone extensive expansion and contraction through duplication and pseudogenization. Previous studies have shown that broadly defined environmental adaptations (e.g., terrestrial vs. aquatic) are correlated with the number of functional and non-functional OR genes retained. However, to date, no study has examined species-specific gene duplications in multiple phylogenetically divergent mammals to elucidate OR evolution and adaptation. Here, we identify the OR gene families driving adaptation to different ecological niches by mapping the fate of species-specific gene duplications in the OR repertoire of 94 diverse mammalian taxa, using molecular phylogenomic methods. We analyze >70,000 OR gene sequences mined from whole genomes, generated from novel amplicon sequencing data, and collated with data from previous studies, comprising one of the largest OR studies to date. For the first time, we demonstrate statistically significant patterns of OR species-specific gene duplications associated with the presence of a functioning vomeronasal organ. With respect to dietary niche, we uncover a novel link between a large number of duplications in OR family 5/8/9 and herbivory. Our results also highlight differences between social and solitary niches, indicating that a greater OR repertoire expansion may be associated with a solitary lifestyle. This study demonstrates the utility of species-specific duplications in elucidating gene family evolution, revealing how the OR repertoire has undergone expansion and contraction with respect to a number of ecological adaptations in mammals.
Keywords: olfactory receptors, mammalian evolution, ecological niche, adaptation, comparative genomics
Introduction
More than 5,400 species of extant mammals have been described to date, with a shared evolutionary history dating back at least 200 My (Luo 2007; Meredith et al. 2011; Dos Reis et al. 2014). Mammals have successfully exploited a wide range of ecological niches, and have varied habitats, diets, social structures, and rhythmic activity phases. These adaptations involve the accumulation of different biological characteristics (Peterson et al. 1999), involving changes in physiology, feeding ecology, and sensory perception. An example of this is diet, which has undergone numerous shifts to meet the energetic demands of novel environments (Luca et al. 2010). Adaptation to a new energy source requires changes at the genetic, metabolic and morphological levels. As feeding is multi-sensorial, dietary shifts also involve a number of changes to visual, olfactory and taste perception (Luca et al. 2010).
The Olfactory Receptor (OR) repertoire is the set of G-protein coupled receptor (GPCR) genes responsible for the perception of chemosensory information, found mainly in the cilia of the olfactory epithelium of the nasal passage in vertebrates. It is the largest multigene family in vertebrates, typically accounting for ∼4–5% of protein coding genes in mammals (Hayden et al. 2010). OR genes are intron-less and roughly 1 kb in size, with each mammalian species having an average of 1,259 genes (including both functional and non-functional genes; Hayden et al. 2010). The OR repertoire can be split into Class I receptors (binds water-borne odorants) and Class II receptors (binds mainly volatile odorants). These classes are split into four families (OR 51, OR52, OR55, and OR56) and nine OR gene families (OR 1/3/7, OR 2/13, OR 4, OR 5/8/9, OR 6, OR 10, OR 11, OR 12, and OR 14) respectively, with each family also having a range of smaller subfamilies. The OR gene repertoire follows a “birth and death” model of gene evolution, expanding through processes such as tandem gene duplication (Nei and Rooney 2005). ORs are connected to the main olfactory bulb (MOB) in the forebrain via OR neuron axons (Farbiszewski and Kranc 2013). The posterior region of the MOB also contains the accessory olfactory bulb, projecting to the vomeronasal organ (VNO). The VNO is involved in binding pheromones and contains its own multigene family receptors, the vomeronasal receptors (V1R, V2R, and formyl peptide receptors; Young et al. 2010), enabling an additional form of chemosensory perception.
The mammalian OR repertoire consists of functional and non-functional genes. A number of these genes may have been present in the most recent common ancestor of a target species and its closest living sister taxon, possibly with the same or similar functionality. However, with adaptation to new or changing environments and gene duplications giving rise to new ORs, the fixation or loss of novel genes can be specific to an individual species and/or that species’ ecological niche. Previous research has focused on total number of OR genes in an attempt to characterize how the distribution of genes across OR gene families differs as a function of ecological niche adaptation. In 2010, Hayden et al., using normalized total gene counts, showed that mammals can be categorized into different ecological habitats based on the distribution of OR genes per gene family, highlighting differences between aquatic, volant, and terrestrial mammalian OR repertoires. A further association between the evolution of OR family 1/3/7 with frugivory among bat species has also been demonstrated (Hayden et al. 2014). Similar associations between niche and OR gene families have also been documented in birds (Khan et al. 2015). Yet total counts cannot enable a full understanding of how the OR repertoire has evolved. To truly elucidate the role of OR gene families in mammalian evolution, paralogous OR genes born through duplication events specific to a target species must be uncovered, and the subsequent fate of these OR genes (e.g., retained, lost, or gained new function) must be assessed. Retention of functionality in the new gene after duplication suggests either a critical role of the original gene or “neofunctionalization”, thus a novel role for that gene, perhaps in some form of environmental adaptation.
Here, we explored which OR gene families are potentially driving adaptation to different ecological niches by uncovering the fate of paralogous OR genes resulting from species-specific gene duplications. We identified these genes that have been retained in the genomes of 94 diverse mammalian taxa using molecular phylogenomic methods. We analyzed >70,000 OR gene sequences to investigate if such OR gene duplication and subsequent fate of the daughter OR correlates with the presence or absence of a functional VNO, dietary niche, habitat, sociality, and rhythmic activity phase. These data were mined from publicly available whole-genome sequences, generated from de novo OR amplicon next generation sequencing (NGS) data (14 species) and collated with OR genes from previous studies. This data set represents one of the largest mammalian OR studies to date, in terms of taxonomic and genic representation. Our results show that OR gene expansion correlates with the presence versus absence of a functioning VNO, emphasizing the link between odorant and pheromone chemosensory detection mechanisms. We show that there are statistically significant patterns of gene duplications with respect to OR gene families and dietary niche. Specifically, OR gene family 5/8/9 shows a large number of gene duplications and retention of function in postduplication paralogs across a wide range of mammalian species with an herbivorous diet. We find significant differences in the number of OR gene duplication events across ecological niches with respect to habitat and sociality but show that rhythmic activity phase (time when a species is most active) does not correlate with OR repertoire expansion. Our results shed light on how the olfactory gene repertoire has undergone expansion and contraction with respect to a variety of ecological niche adaptations in mammals and how the analysis of species-specific gene duplications can be used to elucidate the evolution of gene families.
Results
Number of Contigs Recovered from 454-NGS Amplicon Sequencing
A total of 744,432 reads were generated for 14 new species using 454 NGS amplicon data (supplementary table S1, Supplementary Material online). Mean read length was 736 bp, with a modal length of 744 bp. The number of contigs generated using the de Bruijn graph assembly methods SOAPdenovo, ABySS, and our clustering method using CD-HIT 454 are displayed in supplementary table S2, Supplementary Material online. Contigs from each different assembly method were compared to determine the optimum method of gene reconstruction given the read data. In the cluster method, a number of various clustering identities were tested. Ultimately, 97% identity was determined as the optimal threshold based on comparisons between reference genomes (dog, cat, little brown bat, and greater horseshoe bat) and read data, for the clustering assembly method. The number of ORs per family, the average number of reads per cluster, the interquartile range across clusters and the maximum cluster sizes are displayed in supplementary tables S2 and S3. The assembly using the SOAPdenovo program recovered the most genes for dog (528 ORs), while clustering recovered the highest number of genes for the cat, little brown bat and greater horseshoe bat (521, 378, and 218, respectively, supplementary table S2, Supplementary Material online).
NGS Amplicon Comparisons
Each species showed on average 99% sequence identity to their respective target gene, except for contigs generated through clustering in the little brown bat (supplementary table S3, Supplementary Material online). The ABySS assembly had the largest average sequence lengths at 595, 617, 616, and 599 bp for dog, cat, little brown bat and greater horseshoe bat, respectively (supplementary table S3, Supplementary Material online). SOAPdenovo had the lowest number of redundant sequences for the cat, little brown bat and greater horseshoe bat at 12%, 14%, and 13%, respectively (supplementary table S3, Supplementary Material online). The distribution of assembled ORs for the cat, little brown bat and greater horseshoe bat were not significantly different from their respective genomic distributions for all methods of contig assembly (supplementary table S3, Supplementary Material online). For the dog data, the number of OR genes represented by the sequenced amplicons in OR gene family 6 appeared underrepresented, considering the size of this gene family (118 ORs) mined from the C. familiaris genome (Aloni et al. 2006), compared to the number of contigs assembled (27, 36, and 21 ORs; supplementary table S2 and fig. S1, Supplementary Material online). The underrepresentation of this family has previously been observed in the dog (Hayden et al. 2010). This is perhaps due to the high number of diverse OR genes in OR family 6 causing problems with amplification using degenerate primers, or problems in the primer design. Comparisons to full repertoires conducted both with and without OR family 6 (supplementary table S3, Supplementary Material online) showed no significant differences. Based on different metrics used to compare assembly versus cluster methods, it was determined that SOAPdenovo was the optimal method to reconstruct the 454 OR amplicon sequence data. The final number of contigs generated for each species using SOAPdenovo is displayed in supplementary table S1, Supplementary Material online (5,114 contigs in total) and were used for further analyses. The species with the smallest number of ORs assembled using NGS methods was Megaptera novaeangliae (humpback whale) at 192 ORs. This low number reflects the small OR repertoire size previously documented in cetaceans (Hayden et al. 2010).
Analysis of OR Repertoires from WGS-Based Genome Assemblies
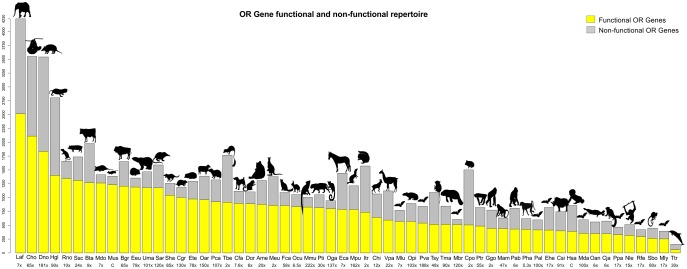
Of the 58 mammals with whole genome sequence (WGS; “Genomic”) based data, the species with the highest and lowest number of OR genes were Loxodonta africana (African elephant) and Tursiops truncatus (common bottlenose dolphin) with 4,230 and 156 ORs, respectively (supplementary table S4, Supplementary Material online). Using fully sequenced genomes, an average of 1,213 ORs were found per mammal. The number of OR genes that we considered putatively functional (coding sequence of 650 bp or more with no in-frame stop codons) ranged from 58 (dolphin) to 2,514 (elephant; supplementary table S4, Supplementary Material online), with an average of 794 functional ORs (65% of repertoire) and 419 (35%) ORs considered non-functional per mammal (coding sequence <650 bp or contained in frame stop codon). The full repertoire of functional and nonfunctional ORs in the “Genomic” data set is displayed in figure 1. The total number of genes for each mammalian OR repertoire differs slightly from previous studies such as Niimura and Nei (2007), Hayden et al. (2010), and Niimura et al. (2014), most likely as a consequence of using different methods, workflows and genome assemblies to mine and annotate the OR data. Such discrepancies have also been observed in OR gene studies in the class Aves (Lu et al. 2016). A total of 9,108 OR genes were added from an additional 36 mammals sequenced from OR amplicons (NGS data set; supplementary table S4, Supplementary Material online). The species with the most and fewest assembled ORs are Ursus arctos (brown bear; Illumina) and Globicephala sp. (pilot whale, Sanger sequencing) at 496 ORs and 42 ORs, respectively.
Fig. 1.
The full OR repertoire of 58 mammalian genomes. The sequenced genomes of 58 ecologically and phylogenetically diverse mammals were mined for OR gene sequences. The number of functional ORs (yellow bars) and non-functional ORs (grey bars) in the full OR gene repertoire varies between different species. Species are ranked based on the size of their functional repertoire. The coverage of the genomes from which these data were mined, as noted in their assembly builds online, are also included with “C” representing “complete” for human and mouse.
OR Species-Specific Gene Duplication Events
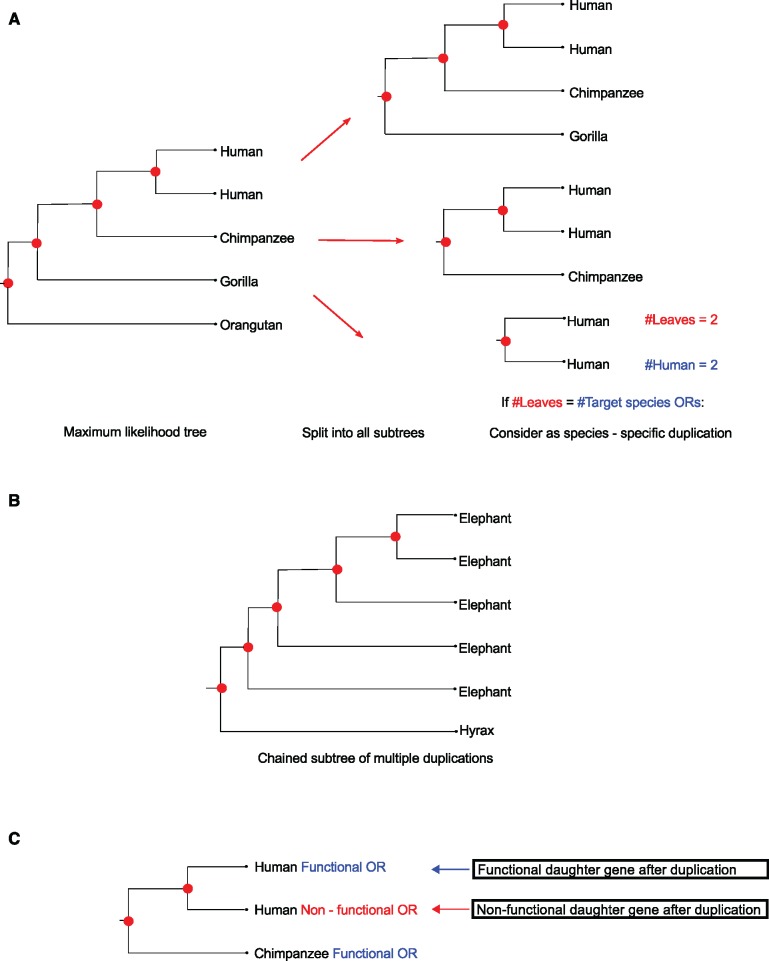
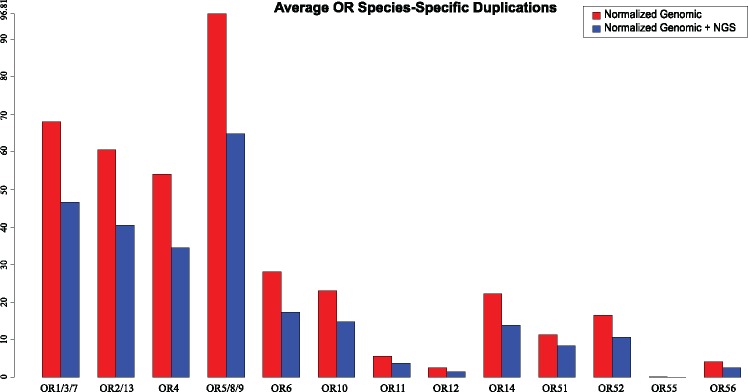
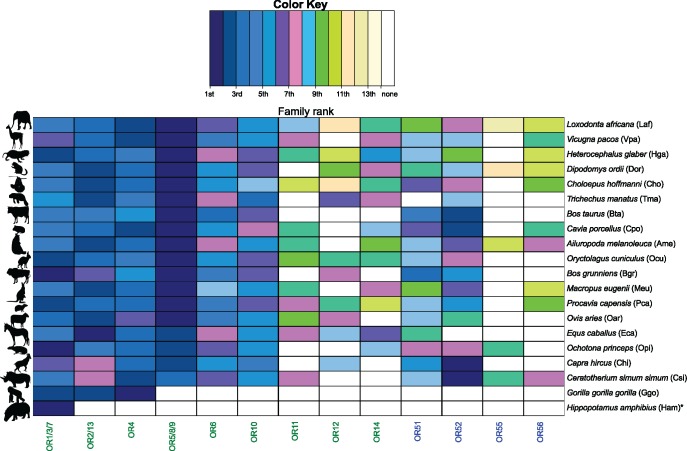
For the “Genomic” data set, the number of species-specific duplication (SSD) events was determined for all OR gene families using a tree parsing methodology (fig. 2). A total of 22,826 duplications across 58 mammals were detected. These SSD events have given rise to 31,595 paralogous OR genes, 18,907 of which remain functional (postduplication functional) while 12,688 have since lost their function (postduplication non-functional) through pseudogenisation (table 1). OR gene families 5/8/9, 1/3/7, 2/13, and 4 have on average the largest number of SSDs (table 1 and fig. 3) and the most functional and non-functional ORs after duplication (table 1). When both daughter ORs have retained function after a duplication event, this may suggest subfunctionalization or neofunctionalization, and differ across taxa (fig. 4). Family 55 had the smallest average number of SSDs suggesting very few gene births, or alternatively, that duplication events have occurred but subsequent loss of function and degradation has made them undetectable. The subsequent fates of OR genes after a duplication for the “Genomic” data are displayed in supplementary figure S2, Supplementary Material online.
Fig. 2.
Method for identifying gene “birth” and “death” events. (a) Evolutionary relationships between ORs are established based on a maximum likelihood tree (JTT + Γ + F). Receptors born through gene duplication appear as paralogs. (b) Chained subtrees composed entirely of paralogs for one species are counted as multiple duplications (four duplication events in this example). (c) ORs undergoing duplication may retain function or become pseudogenes (death).
Table 1.
The Number of Species-Specific Duplication Events, Paralogous ORs Retaining Function and Paralogous ORs Having Lost Function After Duplication, in Addition to the Average Per OR Family for Each Data Set Are Displayed.
| Total Duplications | Mean | Total Functional | Mean | Total non-functional | Mean | |
|---|---|---|---|---|---|---|
| Genomic data (N = 58 mammals) | ||||||
| OR 1/3/7 | 3,948 | 68.07 | 2,680 | 44.66 | 2,444 | 40.73 |
| OR 2/13 | 3,518 | 60.55 | 2,820 | 47 | 1,991 | 33.18 |
| OR 4 | 3,138 | 54.10 | 2,505 | 41.75 | 1,967 | 32.78 |
| OR 5/8/9 | 5,616 | 96.81 | 4,716 | 78.6 | 959 | 49.31 |
| OR6 | 1,630 | 28.10 | 1,479 | 24.65 | 861 | 14.35 |
| OR 10 | 1,336 | 23.03 | 1,415 | 23.58 | 539 | 8.98 |
| OR 11 | 330 | 5.69 | 316 | 5.26 | 185 | 3.08 |
| OR 12 | 149 | 2.57 | 128 | 2.13 | 74 | 1.23 |
| OR 14 | 1,295 | 22.32 | 825 | 13.75 | 37 | 12.28 |
| OR 51 | 659 | 11.36 | 709 | 11.81 | 341 | 5.68 |
| OR 52 | 958 | 16.52 | 1,052 | 17.53 | 472 | 7.86 |
| OR 55 | 11 | 0.19 | 11 | 0.18 | 9 | 0.15 |
| OR 56 | 239 | 4.12 | 251 | 4.18 | 109 | 1.81 |
| Total | 22,826 | 393.55 | 18,907 | 315.11 | 12,688 | 211.46 |
| Genomic + NGS data (N = 94 mammals) | ||||||
| OR 1/3/7 | 4,383 | 47.13 | 3,045 | 31.71 | 2,723 | 28.36 |
| OR 2/13 | 3,809 | 40.96 | 3,110 | 32.39 | 2,175 | 22.65 |
| OR 4 | 3,238 | 34.82 | 609 | 27.17 | 1,992 | 20.75 |
| OR 5/8/9 | 6,096 | 65.55 | 5,250 | 54.68 | 3,187 | 33.19 |
| OR6 | 1,635 | 17.58 | 1,502 | 15.64 | 853 | 8.88 |
| OR 10 | 1,393 | 14.98 | 1,449 | 15.09 | 599 | 6.23 |
| OR 11 | 352 | 3.78 | 345 | 3.59 | 192 | 2 |
| OR 12 | 148 | 1.59 | 124 | 1.29 | 80 | 0.83 |
| OR 14 | 1,307 | 14.05 | 37 | 8.71 | 737 | 7.67 |
| OR 51 | 790 | 8.49 | 834 | 8.68 | 414 | 4.31 |
| OR 52 | 1,003 | 10.78 | 1,108 | 11.54 | 499 | 5.19 |
| OR 55 | 10 | 0.11 | 10 | 0.10 | 8 | 0.08 |
| OR 56 | 244 | 2.62 | 255 | 2.65 | 118 | 1.22 |
| Total | 24,408 | 262.45 | 20,478 | 213.31 | 13,577 | 141.42 |
Note.—The nomenclature as described in Hayden et al. (2010) is used to classify and describe OR families and clusters.
Fig. 3.
Average number of gene duplication events for the “Genomic” and “Genomic + NGS” data sets. The number of species-specific gene duplication (SSD) events were counted for each OR family based on a gene tree established using maximum likelihood.
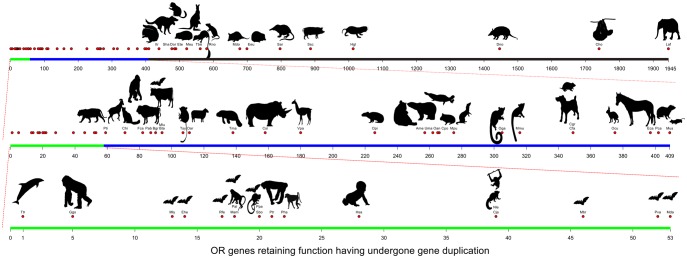
Fig. 4.
The number of functional ORs after species-specific duplications (SSDs) in “Genomic” data. The 58 mammalian species that are in the “Genomic” data are ranked based on the number of OR paralogs born through duplication that are putatively functional. The top line (black, blue, and green) shows the highest ranked species. As not all mammals can be represented on a single line, the different numbers of functional ORs after duplication are expanded upon for visualization via the second and third line.
With the inclusion of the NGS data set (supplementary table S5, Supplementary Material online), the total number of SSDs was 24,408 leading to a total of 34,055 ORs from duplication for the “Genomic + NGS” data set. Of these, 20,478 are functional while 13,577 have since lost their function (table 1 and supplementary tables S5 and S6, Supplementary Material online). The OR families with the highest average number of SSDs were 5/8/9, 1/3/7, 2/13, and 4 (table 1 and fig. 3). Family 55 still had the smallest number of SSDs when the NGS data was added. The overall average duplications for the “Genomic + NGS” data set was systematically lower than the “Genomic” data, due to the addition 36 species representing a subset of OR genes affecting internal arrangement of the OR gene trees, and was therefore used to investigate robustness of duplication rates observed relative to the “Genomic” data set. Only one species, Aonyx cinerea (oriental small-clawed otter) did not show any identifiable SSDs. The varying number of node types (based on fate of the daughter ORs) for the NGS data were expressed as a percentage of the total duplication nodes, as they did not represent the full OR subgenome (supplementary fig. S3, Supplementary Material online).
Ecological Niche and Species-Specific Duplications
The number of subfamilies within each OR gene family showing retention of functionality in genes born through SSD ranged from 1 (OR Family 55) to 78 (OR family 5/8/9) subfamilies, suggesting OR family diversification (table 2). Nucleotide and inferred translated amino acid sequence identity ranged from an average of 89% to 94.51% and 86% to 91.98% across all OR genes after an SSD, respectively (table 2), highlighting differences between sister ORs, indicating sequence diversification. The average number of OR gene SSDs in each niche type in the “Genomic” data is displayed in table 3 and supplementary table S7, Supplementary Material online.
Table 2.
The Number of Different Subfamilies Showing Species-Specific Duplication Events for Each or Gene Family Is Displayed, Including the Maximum Number of Subfamilies in a Single Taxon and the Mean Percentage Identity between Putative Functional Postduplication ORs within These Subfamilies.
| OR Family | Total Subfamilies Represented | Max Subfamilies in a Single Taxon Showing Gene Duplication | IQR of Subfamilies Showing Duplications Across Taxa | Mean Percentage Identity (DNA) (%) | Mean Percentage Identity (Protein) (%) |
|---|---|---|---|---|---|
| 1/3/7 | 44 | 24 | 8.75 | 92.5 | 88.69 |
| 2/13 | 73 | 50 | 17.75 | 93.59 | 90.81 |
| 4 | 51 | 40 | 15.75 | 93.53 | 90.64 |
| 5/8/9 | 78 | 52 | 21 | 93.05 | 89.69 |
| 6 | 28 | 17 | 6.75 | 94.01 | 91.43 |
| 10 | 40 | 18 | 8.75 | 93.71 | 91.13 |
| 11 | 9 | 8 | 3 | 93.62 | 90.57 |
| 12 | 2 | 2 | 1 | 93.49 | 90.25 |
| 14 | 7 | 6 | 3 | 92.47 | 88.18 |
| 51 | 22 | 19 | 5 | 93.69 | 91.32 |
| 52 | 28 | 20 | 10 | 93.96 | 91.64 |
| 55 | 1 | 1 | 0 | 89.45 | 86 |
| 56 | 7 | 6 | 2 | 94.51 | 91.98 |
| Total | 389 | 258 | 92.75 | 93.32 | 90.23 |
Table 3.
The Total and Average Number of a Species-Specific Duplication Events per Niche Is Displayed for the “Genomic” Data.
| Niche | Average % of Total OR Repertoire that is Functional | Total Duplications | Average Duplications | Average % OR Repertoire from Gene Duplications | |
|---|---|---|---|---|---|
| Diet | Carnivore (n = 8) | 68.54 | 1410 | 176.25 | 27.47 |
| Frugivore (n = 5) | 59.91 | 271 | 54.2 | 12.84 | |
| Insectivore (n = 12) | 70.26 | 5861 | 488.42 | 38.85 | |
| Herbivore (n = 19) | 60.96 | 11,200 | 589.47 | 42.25 | |
| Omnivore (n = 14) | 70.89 | 4084 | 291.71 | 31.88 | |
| Sociality | Social (n = 36) | 65.57 | 9350 | 259.72 | 25.91 |
| Solitary (n = 22) | 67.33 | 13,476 | 612.55 | 48.47 | |
| Rhythmic activity phase | Crepuscular (n = 3) | 70.46 | 842 | 280.67 | 39.11 |
| Diurnal (n = 29) | 60.07 | 12,201 | 420.72 | 32.91 | |
| Nocturnal (n = 26) | 72.63 | 9783 | 376.27 | 35.67 | |
| Habitat | Volant (n = 9) | 67.52 | 359 | 39.88 | 11.37 |
| Terrestrial (n = 46) | 67.11 | 21,937 | 476.89 | 38.95 | |
| Aquatic (n = 2) | 42.2 | 160 | 80 | 13.13 | |
| Vomeronasal organ | VNO Present (n = 41) | 68.33 | 22,071 | 525.5 | 43.31 |
| VNO Absent (n = 15) | 60.16 | 755 | 47.19 | 11.24 | |
Note.—The average percentage of the entire OR repertoire that is functional in mammals per niche is displayed, as is the percentage of each repertoire that has come from gene duplication events after diverging from a common ancestor.
When OR families were ranked from most to least functional ORs born through SSD, family 5/8/9 in herbivores had the highest quantity in the majority of species (fig. 5). Family 1/3/7 showed some retention of OR genes after duplication in frugivores and insectivores (specifically in bats), however, no family specific functional retention was apparent in carnivores or omnivores (supplementary figs. S4–S7, Supplementary Material online). Fisher’s exact test did not detect any significant differences in the distributions of SSD events across OR gene families as estimated based on: 1) “Genomic” and (2) “Genomic + NGS” for the 58 species present in both. This suggests that inclusion of the additional 36 species does not significantly affect the estimation of SSDs across the entire repertoire, indicating robustness of these data.
Fig. 5.
Herbivore OR families ranked by functional OR genes after a duplication event. OR gene families for each herbivorous species from the “Genomic” data were ranked by the number of OR genes born through species-specific gene duplication (SSD) that have retained function. Herbivores from the “Genomic + NGS” data (delimited by a black star) are also included.
Phylogenetic Generalized Least Squares Analyses
Using Phylogenetic Generalized Least Squares (PGLS) analyses, significant differences were found for OR gene families across multiple niches with respect to the number of SSDs detected and the functional status of their daughter OR genes (table 4). Only OR families and ecological niches showing significant differences in both data sets (“Genomic”, “Genomic + NGS”) are reported here (see supplementary tables S8–S10, Supplementary Material online for OR gene families specific to each data set). For dietary niche, family 5/8/9 showed significant differences for herbivory compared with all other diets with respect to number of SSDs and functional/non-functional daughter ORs. Family 1/3/7 showed a significant difference in the number of SSDs in insectivores and in the number of functional daughter ORs for habitat. Family 5/8/9 had significantly more SSDs and functional daughters in taxa with a functional VNO. Rhythmic activity phase and sociality showed significant differences in the number of non-functional daughters for OR families 4 and 11, respectively (more pseudogenization in diurnal and crepuscular mammals versus nocturnal for OR family 4 and for solitary mammals in OR family 11).
Table 4.
OR Families Were Compared Across Ecological Niches Using PGLS to Identify Families Showing Significantly Differences with Respect To Species-Specific Duplication Events, Paralogous ORs Retaining Function and Paralogous ORs Having Lost Function After Duplication.
| Family | Niche | Data Set | PGLS P Value | Estimated λ | Significant Niches |
|---|---|---|---|---|---|
| Duplications | |||||
| OR 1/3/7 | Diet | Genomic | P = 0.011 | 0.28 | Insectivory |
| Genomic+NGS | P = 0.005 | 0 | |||
| OR 5/8/9 | Diet | Genomic | P = 0.004 | 0.43 | Herbivory |
| Genomic+NGS | P = 0.005 | 0.13 | |||
| VNO | Genomic | P = 0.000 | 0.28 | VNO | |
| Genomic+NGS | P = 0.002 | 0.30 | |||
| OR 52 | Habitat | Genomic | P = 0.009 | 0 | Aquatic |
| Genomic+NGS | P = 0.034 | ||||
| Genomic | P = 0.0003 | Terrestrial | |||
| Genomic+NGS | P = 0.02 | ||||
| Postduplication functional | |||||
| OR 1/3/7 | Habitat | Genomic | P = 0.0009 | 0 | Aquatic |
| Genomic+NGS | P = 0.009 | ||||
| Genomic | P = 0 | Terrestrial | |||
| Genomic+NGS | P = 0 | ||||
| OR 5/8/9 | Diet | Genomic | P = 0.029 | 0.23 | Herbivory |
| Genomic+NGS | P = 0.014 | 0.29 | |||
| VNO | Genomic | P = 0.0004 | 0.07 | VNO | |
| Genomic+NGS | P = 0.001 | 0.33 | |||
| OR 52 | Habitat | Genomic | P = 0.023 | 0 | Aquatic |
| Genomic+NGS | P = 0.044 | ||||
| Postduplication non-functional | |||||
| OR 4 | Rhythmic Activity Phase | Genomic | P = 0.02 | 0 | Diurnal |
| Genomic+NGS | P = 0.01 | ||||
| Genomic | P = 0.001 | Nocturnal | |||
| Genomic+NGS | P = 0.002 | ||||
| OR 5/8/9 | Diet | Genomic | P = 0.001 | 0.55 | Herbivory |
| Genomic+NGS | P = 0.002 | 0.03 | |||
| VNO | Genomic | P = 0.001 | 0.59 | VNO | |
| Genomic+NGS | P = 0.02 | 0.29 | |||
| OR 11 | Diet | Genomic | P = 0.02 | 0 | Frugivory |
| Genomic+NGS | P = 0.008 | ||||
| Sociality | Genomic | P = 0.02 | 0.15 | Sociality | |
| Genomic+NGS | P = 0.01 | 0 | |||
| OR 56 | Diet | Genomic | P = 0.001 | 0 | Herbivory |
| Genomic+NGS | P = 0.01 | ||||
Note.—OR gene families significant in both “Genomic” and “Genomic + NGS” data sets are displayed (P < 0.05, λ<0.9).
Poisson Modeling of Ecological Niche Data
Results for the AICc model selection for each data set (SSDs; functional daughters; non-functional daughters) are given in supplementary table S11, Supplementary Material online. With the exception of non-functional OR daughters, the best supported model in all data sets modeled SSD rates as a function of the presence or absence of a functional VNO, with a posterior probability of 1.00 (supplementary table S11, Supplementary Material online), with individual gene families having separate rates of duplication. In all cases, the estimated species-specific rates were substantially higher for taxa with a functional VNO (supplementary table S11, Supplementary Material online). For non-functional daughters, dietary niche separated by individual OR gene families, was the most supported model. Dietary niche, with all diets as independent categories, ranked second for the rate of SSDs in both data sets.
Discussion
Gene duplication through processes such as tandem gene duplication, segmental duplication or whole-genome duplication is an important means by which novel diversifying genotypes and phenotypes can arise (Cotton 2008; Chang and Duda 2012). Expansion or reduction of gene families in this way can be random or a consequence of selective forces. However, demonstrating evidence of selection on expanding gene families can be a difficult task (Harris and Hofmann 2015).
The elephant has the largest OR repertoire among mammals (Hayden et al. 2010; Niimura et al. 2014), with African elephants being able to distinguish between family members using olfaction (Bates et al. 2008) and Asian elephants being able to distinguish between enantiomer odorant pairs (Rizvanovic et al. 2013). This exceptional olfactory capability is reflected in the number of detectable species-specific duplications (SSD) observed in the African elephant genome and its subsequent ORs that have putatively retained function. In contrast, the dolphin has the smallest number of SSDs, reflecting the diminished role of olfaction after the transition from land to water in the evolutionary history of cetaceans (Springer and Gatesy 2017). To explore how gene expansion has occurred in the OR gene family, with respect to ecological niche adaptation, we investigated SSD events and the fate of the genes such events give rise to in 94 mammals (58 sequenced genomes and 36 from NGS amplicon sequence data). These data included species representatives from all superordinal groups.
OR Gene Evolution in Dietary Niche
Our findings show that in herbivores, OR gene family 5/8/9 had the highest number of SSDs and retained more functional OR genes born through duplication than in nonherbivores. These SSD events were found in a diverse number of subfamilies within the OR 5/8/9 gene family, suggesting diversification through duplication. The sampled mammalian herbivores span clades that are distantly related: Laurasiatheria (Cetartiodactyla, Perissodactyla, and Carnivora), Euarchontoglires (Rodentia), Afrotheria (Paenungulata), Xenarthra (Pilosa), and Metatheria (Diprodontia) (Meredith et al. 2011), with herbivory evolving independently from omnivorous or carnivorous diets (Price et al. 2012), yet showing convergent functional expansion of OR 5/8/9. For the giant panda (Ailuropoda melanoleuca), an herbivorous mammal that still maintains the genetic requirements for a carnivorous diet (Li et al. 2009), family 5/8/9 had slightly more functional ORs after SSDs than any other OR family highlighting a potential role in the evolution of herbivory. In humans and mice, family 5/8/9 is linked to binding odorants whose “descriptive aroma” has been associated with oils and plants in addition to fruity or floral aromas (coumarin, β-ionone, menthol, prenyl acetate, heptanone, acetophenone, eugenol, vanillin; Dunkel et al. 2014) suggesting its importance in a herbivorous diet. However, family 5/8/9 can bind odorants that are not related to plant matter (nonanoic acid; Dunkel et al. 2014), highlighting the need for further research into the role of receptors and diet.
For frugivores, OR family 1/3/7 shows a higher number of SSDs in bats compared to others species and families. Frugivores sampled in this study come from the orders Chiroptera and Primates. Hayden et al. (2014) established an association between frugivory and functional ORs in family 1/3/7 in bats, and this is corroborated by the SSD data observed here. We also note high numbers of SSDs for family 1/3/7 in the orangutan and white-cheeked gibbon, whose diets are highly frugivorous (Mitani 1989; Kissling et al. 2014; Muchlinski and Deane 2014). The ancestor to all modern bats is inferred to have been insectivorous (Schondube et al. 2001) but dietary shifts to frugivory occurred, independently, in Phyllostomidae and in Pteropodidae (Hayden et al. 2014). We find that OR 1/3/7 also shows significant duplication rates associated with insectivory across a wide range of mammalian orders, with mammalian dietary shifts potentially converging on the same gene family. In humans, family 1/3/7 ORs have been shown to bind to different molecules that are associated with fruity or floral aromas, similar to OR 5/8/9 (citronellal, benzyl acetate, heptanal, γ-decalactone, lilial, ethyl-2-methyl proponoate; Dunkel et al. 2014). Future deorphaning of OR and odorant molecules may allow a more comprehensive study of mammalian olfactory space, and the role of diet in olfactory evolution. Duplication and retention of function in both frugivorous and insectivorous species across major placental supraordinal clades suggests family 1/3/7, like family 5/8/9, has undergone multiple expansions in phylogenetically diverse mammals with respect to dietary niche.
The Evolution of ORs and the VNO
The Vomeronasal or Jacobson organ (VNO) is used to detect pheromones: water-soluble chemical signals that trigger a social response in a number of mammals (Zhang and Webb 2003). When looking at whole genome and NGS amplicon data, significant differences between the number of putatively functional ORs born through SSD suggests a link exists between the expansion of the OR repertoire and the presence/absence of a functional VNO. This pattern was observed across all statistical analyses and was the best-supported niche to explain observed duplication events. The VNO has previously shown the ability to express OR genes (Keller and Vosshall 2008), and it has been demonstrated that pheromones can stimulate olfactory neurons, which suggests a complex interaction between the two sensory modes. A transcriptome analysis of the olfactory system in mice (Ibarra-Soria et al. 2014) highlighted at least 17 OR genes expressed on the VNO. The OR gene with the highest gene expression in the mouse VNO, olfr124, was one of a pair of OR genes born through a SSD event that we have identified in mice. Additionally, olfr533, olfr741, and olfr536 were also found to be expressed in the mouse VNO, and are postduplication functional OR genes in mice.
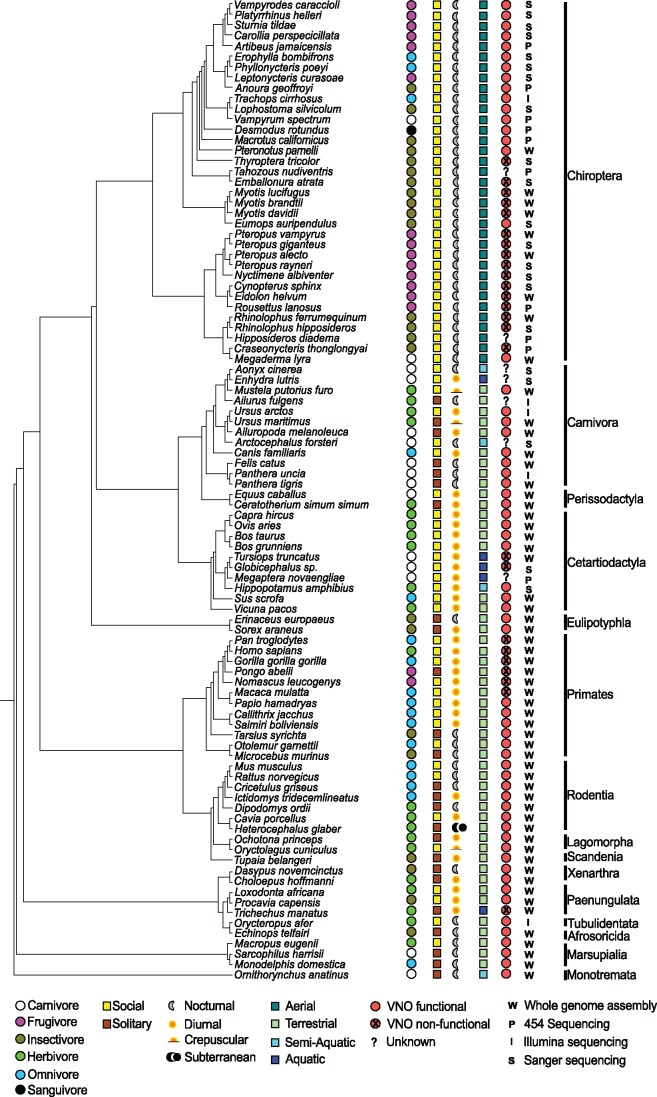
Our results indicate that the OR repertoire has not evolved to compensate for the complete loss of a VNO, but rather is augmented by the presence of a functioning one in extant taxa. Such a relationship may explain the highly developed OR and VNO systems of the Asian elephant (Rizvanovic et al. 2013) and the fact that a number of mammals without a functional VNO (16 bat species, six primates, and two cetaceans across both data sets; fig. 6) demonstrate a stronger reliance on alternative means of sensory perception, namely echolocation, and vision. Further studies into the differential expression of VNO receptor genes in olfactory epithelia, the expression of OR genes in VNO tissue and their potential correlations with OR subgenome evolution may help elucidate the patterns observed here.
Fig. 6.
Ecological niches of 94 mammals. The ecological niche data for 94 mammals used in this study are displayed.
OR Gene Evolution in Social versus Solitary Mammals
Solitary species have on average twice the number of SSDs compared with social species, with no family in particular showing significant differences to others. This suggests that individual OR gene families may have expanded at similar high rates with respect to sociality, with broader repertoires being adaptive to a solitary lifestyle. It is possible that mammals living in large groups may rely less on olfaction and the detection of odorant molecules as a whole, due to the greater protection and cooperation in finding resources conferred by a social group membership (Chesser et al. 1993; Silk 2007). In this case, vocalization or display may be the dominant form of communication. Solitary mammals may need to be more vigilant with respect to prey, predators and mating and as such, an expanded OR repertoire could potentially allow a finer tuned sense of olfaction. An exception to this observation is the African elephant, which shows more OR gene duplication events than any other taxon, despite predominantly living in social groups. (Wittemyer et al. 2005). Interestingly, the OR gene families that are associated with diet (1/3/7 and 5/8/9) do not show a significant difference between social and solitary taxa, but also suggests that the increases in SSD events in solitary mammals are contained largely in OR subfamilies associated with surveying the physical environment around the individual.
OR Evolution and Rhythmic Activity Phase
Given the importance of light in maintaining circadian rhythm (Challet 2007), activity phase in mammals has previously been linked to adaptations in visual perception. Eye morphology has previously shown to be a discriminator between nocturnal and diurnal species (Schmitz and Motani 2010), and a loss of function in opsin color vision genes has been reported in nocturnal mammals (Zhao et al. 2009; see section below). In reduced light, olfaction likely plays an important role in hunting, identifying potential threats and communication with conspecifics for both nocturnal and crepuscular species. Bats have maintained the ancestral mammalian OR repertoire with little loss of function associated with gain of echolocation suggesting an important role for olfaction in bat survival, potentially driven by their nocturnality (Hayden et al. 2010). Odorant detection has previously been used to elicit some circadian response in mice (Granados-Fuentes et al. 2006) and nocturnal mammals contain more intact vomeronasal type 1 receptor (V1R) genes for pheromone perception compared to diurnal taxa (Wang et al. 2010). While nocturnal mammals showed a lower number of non-functional daughters after SSD in OR family 4 compared with crepuscular and diurnal species, the lack of an observed significant correlation between activity phase and OR gene family expansion through duplication in our data is surprising. It is possible that selective forces are acting on the preservation of OR gene function in nocturnal mammals after duplication, similar to that of the VNO (Wang et al. 2010). This could be due to different taxa occupying the same ecological niches but at different times. As rhythmic activity phase is affected by a number of internal and external cues, including geographical distribution (Bennie et al. 2014), a more focused, in-depth analysis of rhythmic activity phase and its regulation with respect to chemosensory perception is needed to truly elucidate any patterns of olfaction and activity phase.
The Evolution of OR Genes and Habitat
In a previous study carried out by Hayden et al. (2010), differences in the number of functional ORs between terrestrial and volant mammals suggested a correlation between the evolution of the OR repertoire and adaptation to various habitat types. However, when accounting for the percentage of the OR repertoire attributable to SSD, terrestrial mammal OR repertoires have undergone almost three times as much OR gene duplication as volant or aquatic mammals. This reflects previous conclusions regarding OR repertoire composition and ecological adaptation (Hayden et al. 2010). Terrestrial mammals occupy a variety of different niches, which may be associated with such high rates of gene duplications. The expansion of OR gene families for terrestrial adaptation can also be observed in the high rate of duplication in Class I OR genes (which typically bind water odorants) compared to aquatic mammals, suggesting a potential novel usage or cooptimization for this class of gene families in adaptation to a nonaquatic environment, as previously observed in the dog (Olender et al. 2004). The relatively small number of OR genes in aquatic mammals would imply relaxed selective pressures on olfactory mechanisms for an aquatic lifestyle. Nonetheless, we observed some duplication of OR genes in aquatic taxa, with a higher rate in the manatee compared to cetacean species, suggesting some putatively retained OR functionality in an aquatic environment.
Conclusion
Our findings indicate that the evolution and diversification of the mammalian OR repertoire through SSD (large expansions within a specific OR family, or of the OR repertoire as a whole) and the retention of function in duplicated OR genes is associated with adaptations to a range of ecological niches. We have identified specific OR families that appear to be associated with adaptation to different dietary niches (herbivory: 5/8/9, insectivory and frugivory: 1/3/7). Furthermore, our findings suggest that the OR repertoire and the VNO are intimately linked, with a functional VNO associated with rapid expansion of the OR gene repertoire. In addition, solitary mammals have undergone nearly twice as many SSD events as social mammals. However, there were no significant differences observed between rhythmic activity phases with respect to OR gene evolution through SSD. The wide range of different terrestrial ecological niches to which mammals have successfully adapted is reflected in the fact that three times as many OR genes are born through gene duplication in terrestrial mammals compared to volant or aquatic mammals.
In this study, we have shown the utility of using phylogenetically independent SSDs as a means of elucidating how environmental niche adaptation is associated with the evolution of sensory perception. As amplicon sequencing and whole genome sequencing become cheaper and faster, future studies will allow the possibility of further exploring these results. Higher quality genomes will allow further exploration of the associations between OR gene evolution and ecological niche adaptation, possibly uncovering more functional genes or resolving any instances of incorrectly identified pseudogenes. The Genome 10 K sequencing project (G10K; Haussler et al. 2009), which aims to sequence the full genomes of >10,000 vertebrates, includes a large set of mammalian taxa. Such a large set of genomic data will play a huge role in the investigation and understanding of mammalian sensory evolution, shedding more light on the relationship between sensory evolution and niche adaptation. Additionally, the modeling of 3D structures of ORs, and deorphaning of receptor and ligand may also help further explore associations between the role of OR repertoire and ecological niche.
Materials and Methods
454. Sequencing and Assembly
A representative subset of OR genes from 14 species, ten of which are new to this study, (supplementary table S1, Supplementary Material online) were amplified using primer pairs GPC1 and GPC2, (degenerate primers designed to amplify a representative subsample of mammalian OR genes), using protocols described by Hayden et al. (2010), modified from Gilad et al. (2004). PCRs were performed in a total volume of 25 µl containing: 2.5 µl 10× PCR Buffer, 2 mM MgCl2, 1 U Platinum Taq (Invitrogen Corporation), 0.2 µM dNTP’s, 0.2 µM of each primer, and approximately 10 ng of DNA. Conditions for initial PCR were modified from Gilad et al. (2004). A first step of denaturation for 10 min at 94°C followed by 35 cycles of denaturation for 15 s, annealing for 30 s at a temperature gradient of 38–50°C, and an extension for 1 min at 72°C, with a final extension for 10 min at 72°C. PCR products were visualized on a 1% agarose gel using SYBR Safe DNA gel stain (Invitrogen Corporation). Samples were quantified using Qubit® 3.0 Fluorometer (Life Technologies). The two PCR products per species were pooled then purified and concentrated using Millipore Centrifugal filter units. Samples were sequenced on the Roche 454 FLX+ Titanium sequencer using Lib-L sequencing, with an estimated average amplicon size of 700–750 bp.
Sequence Assembly
The OR gene amplicons were sequenced and assembled for all 14 species (supplementary table S1, Supplementary Material online). For each read of the 14 species, tailing nucleotides with a Q-score of ≤30 were removed using the FastX toolkit (Pearson et al. 1997). Two de Bruijn graph based assemblers, SOAPdenovo (Luo et al. 2012), and ABySS (Simpson et al. 2009) were used to assemble the ORs contained in the read data, collapsing multiple amplicons into their target genes for each species. Such assembly algorithms use a parameter k to control the assembly of reads into contiguous sequences (contigs). Each read was split into smaller fragments of length “k”, termed k-mers. Low values for k increase the chances of successful overlaps, and are more sensitive regarding the assembly of fragments compared to larger values. Larger k values however reduce the risk of incorrect overlaps, and are thus more specific. The relatively large average read lengths (700 bp) in these read data allowed a wide range of k-values to be investigated. Two independent k ranges were tested: a low k-value range (20–127) using SOAPdenovo, which allows a maximum k of 127, and high k-value ranges using ABySS. As the number of assembled contigs dropped dramatically for higher k values, a range of k-values between 200 and 500 was applied.
Assembly parameters and approaches were tested using four species with previously sequenced genomes: Canis familiaris (dog), Felis catus (domestic cat), Myotis lucifugus (little brown bat), and Rhinolophus ferrumequinum (greater horseshoe bat) to ensure that the NGS data generated was a true representation of the OR subgenome. OR sequences were mined from these genomes using the Olfactory Receptor Assigner (ORA; Hayden et al. 2010) and TBLASTX (Altschul et al. 1990) using previously annotated ORs and served as a benchmark for comparing assembly methods. ORA uses profile Hidden Markov Models (HMMs), based on alignments of mammalian ORs, to scan through a given contig or sequence and identify putative OR genes present. It was used to assign each assembled OR to the correct gene family (as defined by Hayden et al. 2010).
Sequence Clustering
As each read represented a potential amplified OR gene sequence, a clustering method was applied in addition to de Bruijn graph assemblers to collapse amplicon data. Reads for the four test species were sorted into clusters using the CD-HIT 454 clustering package (Fu et al. 2012). With an error rate of 1% per read for 454 sequencing (Glenn 2011), at least seven erroneous bases per read were expected (based on a mean read length of 736 bp), hence clusters were created based on ≥97% sequence identity. This allowed reads to join clusters despite potential sequence errors and undetermined nucleotide positions. Consensus sequences were then made for each cluster. Undetermined nucleotide positions represented by “N” in consensus sequences may lead to multiple sequences representing the same gene. To account for this, all consensus sequences showing ≥98% sequence identity were merged together, considered as one gene and assigned to their appropriate OR family.
Clustering versus Assembly
To determine which method of sequence reconstruction (assembly vs. clustering) produced the best representation of the OR repertoire, cluster-consensus and assembled OR sequences were mapped to the genomes of the four test species using BLASTN (Altschul et al. 1990). The clustering and assembly methods were compared based on:
Number of unique ORs present in the data;
Data redundancy (number of unique ORs vs. number of contigs);
Comparison of gene distributions between assembled and genomic ORs;
Average percentage identity between sequencing and genomic data;
Average length of sequenced genes.
The distributions of generated ORs and genomic ORs for each species were compared in R using both Pearson’s chi-square test and Fisher’s exact test.
In Silico Whole Genome Assembly OR Data
Olfactory Receptor gene sequences were mined from 58 different sequenced mammalian genomes, covering 17 mammalian orders (supplementary table S12, Supplementary Material online and fig. 1). Genes were detected using TBLASTX (Altschul et al. 1990) in addition to gene mining using the ORA (Hayden et al. 2010), as above. While OR annotations of a number of genomes in this study have already been characterized (Glusman et al. 2001; Olender et al. 2004; Lee et al. 2013), we nonetheless applied our mining methods to the raw genome assemblies to ensure a ubiquitous application of the same threshold parameters (such as determining functionality) and methods of data collection across all taxa. Due to this, and differences in genome assembly versions in different studies, we expect to see potential differences in the size of the OR repertoires we have mined compared to previously characterized repertoires (Niimura and Nei 2007; Hayden et al. 2010; Niimura et al. 2014). As some of the genomes used in this study have not yet been annotated, it is possible that a number of identical redundant contigs were present in the whole genome assemblies, either due to different alleles or multiple copies of the same contiguous sequence, leading to an overestimation of the number of ORs per genome. Such potential duplicate sequences were identified through searching for genes with three or fewer nucleotide differences compared to others (≥99% sequence identity), and removed. ORs were grouped into clusters according to their families based on Hayden et al. 2010 (OR 1/3/7, OR 2/13, OR 4, OR 5/8/9, OR 6, OR 10, OR 11, OR 12, OR 14, OR 51, OR 52, OR 55, OR 56), were further assigned to a subfamily based on homology to reference OR genes, for example, OR51A1 indicates OR gene family 51, gene 1 of subfamily A, (Glusman et al. 2000) and translated into amino acid sequences. OR genes were considered “putatively functional” (referred to throughout simply as “functional”) if they were >650 bp, having the potential to encode the 7-transmembrane GPCR protein structure, and did not contain an in-frame stop codon, as in Hayden et al. (2010) and Khan et al. (2015), rather than functionality inferred from expression studies. This length threshold allowed us to identify and include functional ORs from fragmented whole genome assemblies or genomes with long, unresolved sequence regions that did not contain a complete ORF. Such an approach allowed an upper estimate of the functional OR repertoire size for species with lower quality genome assemblies, but differs slightly from more conservative methods such as Montague et al. (2014). An OR gene was treated as non-functional if it contained an in-frame stop codon, insertion or deletion frame-shift mutation. Our computational methods detected functional OR genes, OR genes with a premature stop codon, and truncated or degraded pseudogenes. However, given that relaxed selection after a pseudogenization event can change a gene to the point that it can no longer be detected using sequence homology, we will always underestimate their occurrence with these methods (Balasubramanian et al. 2009). Therefore, our counts of OR pseudogenes represent a minimum number of detectable pseudogenes within the genome.
The genomic data set, consisting of ORs from 58 fully sequenced mammals, was termed “Genomic” and contained 70,369 OR genes. These OR genes showed on average 95.91% nucleotide identity when mapped to a subset of annotated RefSeq genes (mean of 98.81% with pseudogenes excluded), confirming they represented true OR sequences.
Combined Genomic and NGS Data
A second data set containing 94 taxa, termed “Genomic + NGS”, was considered. This data set consisted of OR data from the 58 fully sequenced mammalian genomes coupled with the assembled 454 gene data from the ten species (had no genome sequenced) described above (3,338 OR sequences). In addition, using the GPC1 and GPC2 primer pairs, similar representative subsets of OR genes were sequenced in five additional mammalian species (Ailurus fulgens [red panda], Orycteropus afer [aardvark], Panthera uncia [snow leopard], Trachops cirrohsus [fringe-lipped bat], and Ursus arctos [brown bear]; 2,933 total contigs) using the Illumina sequencing platform, laboratory protocols, and bioinformatic pipelines described by Hughes et al. (2013). These genes, as well as OR genes from 21 species amplified in previous studies (Hayden et al. 2010, 2014; Hughes et al. 2013), were added to the “Genomic + NGS” data set. This resulted in 10,536 OR sequences for 36 taxa (10,454 generated, five Illumina generated, all new to this study; 21 from Hayden et al. (2010) using Sanger sequencing). OR sequences were classified and converted to amino acid sequences as described above. A length of 80 amino acids (240 bp) was used as an operational minimum cutoff in the NGS data to differentiate between short read/assembly fragments and definitive OR sequences (Hughes et al. 2013), allowing for the identification of putative degraded pseudogenes. This reduced the NGS data down to 9,083 sequences, equating to 79,452 ORs in the “Genomic + NGS” data set. As the amplified data represent only a subset rather than the full OR repertoire, analyses were performed with and without the additional 36 taxa (i.e., “Genomic” data set), to investigate potential biases in our results. This allowed us test the robustness of our gene data with respect to additional taxa, niches and sequences.
Tree Building
The two data sets, “Genomic” and “Genomic + NGS” contained 58 and 94 mammalian species, respectively. OR families were aligned individually using the full distance matrix for guide tree calculation in Clustal Omega (Sievers et al. 2011). Functional and non-functional OR genes were included in each alignment. As both data sets contained 13 OR families, a total of 26 protein alignments were generated. Each alignment was then used to generate phylogenetic trees using RAxML-LITE (Stamatakis et al. 2012), with the Jones–Taylor–Thornton model of sequence evolution, gamma model of rate heterogeneity, and observed amino acid frequencies (JTT+Γ+F). This model was considered the best-fit model of sequence evolution using ProtTest 3 (Darriba et al. 2011) on small alignments (OR families 10, 11, 12, 14, 51, 55, and 56) and PartitionFinder (Lanfear et al. 2012) on the larger ones (OR families 1/3/7, 2/13, 4, 5/8/9, 6, 52).
Counting OR Gene Duplication Events
We counted species-specific OR gene duplication (SSD) events, whereby OR genes have undergone duplication leading to two or more paralogous genes (daughters) in a single species alone, that are retained in the genome. To do this, we parsed gene trees for each OR family to identify nodes indicating a SSD or “birth” event. A node was considered to represent an SSD if it contained two or more leaf nodes, all from the same species (fig. 2a). Chained sub trees consisting of a single target species were counted as multiple SSD events (fig. 2b). Under this method of counting duplications, we cannot rule out the possibility of a duplication event in which one daughter gene retains function whilst the other degrades beyond recognition in the genome, and therefore cannot be documented as a duplication event. Although we expect the frequency of such events in extant taxa to be low, the estimates here refer to SSDs resulting in paralogous OR genes (daughters) that are retained and detectable in the genome, and therefore represent minimum levels of observable OR duplication. Additionally, this method will not recover shared ancestral duplication events.
The R package “ape” was used to decompose each gene tree into all possible subtrees. A perl script was used to parse these trees, highlighting instances of SSD while relaying specific information about each gene. We identified three types of duplication nodes: duplication events where all daughter paralogous genes are functional, duplication events where all daughter genes have since lost their function and duplication events showing a combination of both.
With respect to non-functional OR genes that are one of a duplicated pair, it is unknown if function was lost soon after duplication or at a later point in the evolutionary history of that species. There also exists the possibility that a stop codon is a consequence of sequencing error, however the majority of genome assemblies had a read coverage between 5× and 222×, reducing the likelihood of an incorrect pseudogene assignment. The number of SSDs, the number of OR functional and non functional daughter genes per species (fig. 2c) were counted, with tables generated for each data type. To investigate if duplication was diversifying the OR repertoire, the number of different subfamilies within each OR gene family showing SSD events was determined.
Comparison of Mammalian Niches
To investigate the potential link between OR gene duplication and ecological adaptation, we considered the following niches: dietary (carnivore, frugivore, insectivore, herbivore, or omnivore), functional VNO(present/absent, see supplementary table S12, Supplementary Material online), sociality (social and solitary), rhythmic activity phase (crepuscular, nocturnal, and diurnal), and habitat (volant, terrestrial, and aquatic). These niches imply many different ecological modalities, environmental conditions, and sensory perception repertoires. Species were assigned to each niche according to data from the literature (supplementary table S12, Supplementary Material online and fig. 6). Data was normalized to allow comparisons across niches and comparisons between genes mined from whole genomes and NGS data. The distribution of SSDs detected in each family was normalized by dividing the number of observed SSDs per family by the total SSDs observed for that species (supplementary table S13, Supplementary Material online, see worked example), and this normalization was also applied to the counts of functional and non-functional daughters. The percentage of the OR repertoire that has undergone SSD in a given taxon was determined as the ratio of total SSD daughters to the total number of OR genes (functional and non-functional) in the full repertoire (e.g., see supplementary table S13, Supplementary Material online). This was used to investigate if the OR repertoire expansion differs between various niche types. It was only estimated from the “Genomic” data set, as this data set was considered a “complete” catalogue of all OR genes present in the whole genome assemblies for species included.
Statistical Comparisons of Species-Specific OR Duplication Data
We analyzed the pattern of OR gene evolution relative to niche adaptation using several statistical comparisons. First, OR families within each species were ranked from highest to lowest based on the number of OR SSD functional daughters. To investigate if the addition of taxa from the NGS data set affected observed rates of OR gene family evolution significantly, we compared the distributions of SSDs in the 58 “Genomic” species with the combined “Genomic” and additional 36 NGS species, using Fisher’s exact test.
We analyzed mammalian OR gene duplication data relative to the phylogenetic tree using two complimentary techniques. First, we performed a PGLS (Kamilar and Cooper 2013). Second, we fitted a Poisson model to the SSD data, comparing rates of duplication across ecological niches.
Phylogenetic generalized least squares is a general linear model, which accounts for phylogenetic history by allowing correlations between predictor and response variables while controlling for the nonindependence due to phylogeny. Analysis of the data in this manner considers OR SSDs explicitly in the context of the phylogenetic tree. PGLS can accommodate different models of character evolution and branch scaling parameters (Garland and Ives 2000). Here, we used Pagel’s λ (Pagel 1999) as a scaling factor, estimated using maximum likelihood. The λ value can range from 0 (no phylogenetic signal) to 1 (trait data matched a Brownian model of evolution). For this study, the trait data consisted of the total number of SSD functional and non-functional daughter ORs across taxa with known niche data. We used a composite mammalian phylogeny based on Meredith et al. (2011) and Foley et al. (2016) for interordinal relationships, with species relationships added based on Ruedi et al. (2013), Almeida et al. (2014), and Hayden et al. (2014; supplementary table S12, Supplementary Material online). We estimated branch lengths using the method devised by Grafen (1989), implemented in the “ape” package in R (Paradis et al. 2004). We used the R packages “ape” (Paradis et al. 2004), “geiger” (Harmon et al. 2008), “nlme” (Pinheiro et al. 2015), and “phytools” (Revell 2012) to read phylogenetic trees and run the PGLS. Instances of λ > 0.90 were not considered evidence of ecological adaptation.
We modeled the rate of SSDs detected in each Class II OR gene family using a fitted Poisson model (Edwards 1992; Burnham and Anderson 2002; supplementary table S11, Supplementary Material online). This model relates the rate of SSDs to different ecological factors. Analysis of the data in this manner considers OR gene duplications in the context of different classes or partitions of the ecological variables. As such, rates of SSDs are analyzed with respect to these variables in a manner similar to “treatment classes” (Finarelli and Goswami 2013, Goswami and Finarelli 2016; Vartia et al. 2016). Taking the sociality variable as an example, we compared rates of SSDs across the two sociality “treatments”: social versus solitary. It should be noted that this model is being used to identify the relative strengths of association between ecological variables and the SSD counts observed in mammalian species, not to construct predictive model for the purpose of estimation of SSD counts.
We considered a total of 23 models. Duplication rate estimates were modeled for each variable as: 1) a single rate for all gene families for each ecological niche category, 2) as a distinct estimated rate for each individual OR gene family, and 3) as a distinct rate for each of the two major groups apparent in the data: (OR1/3/7, OR2/13, OR4, and OR5/8/9) versus (OR6, OR10, OR11, OR12, and OR14). Taking sociality again for the purpose of example, we modeled a single rate for all gene families for social and for solitary taxa (two rate parameters), distinct rates for social and solitary taxa for each of the gene families (18 rate parameters), and rates for social and solitary taxa for each of the two major OR family groups (four rate parameters). We employed two coding schemes for dietary niche: a five-state condition with carnivory, frugivory, insectivory, herbivory, and omnivory, and a three-state partition of “animalivorous” (carnivory and insectivory), “herbivory” (frugivorous and herbivory), and omnivorous. Model selection was carried out using the Akaike Information Criterion (Akaike 1973; Burnham and Anderson 2002) using the finite sample corrected AIC (AICc; Hurvich and Tsai 1989) converting AICc scores to posterior probabilities across the set of examined models. This model selection procedure was performed using the same set of models for SSDs; functional daughters; non-functional daughters in both data sets (“Genomic” and “Genomic + NGS”).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This project was funded: by the Irish Research Council (IRC) Graduate Research Education Programme (GREP) awarded to G.H., E.C.T., and D.H.; a Science Foundation Ireland PIYRA 06/YI3/B932; a Short Term Travel Fellowship 06/Y13/B932STTF08 awarded to E.C.T. Currently, E.C.T. is funded by a European Research Council Starting grant ERC-2012-StG311000. W.J.M. acknowledges funding from NSF (EF0629849). We thank Michael Bekaert for ORA and Nicole Foley and all of Batlab for their input. Animal silhouettes used in figures were downloaded from phylopic.org under Creative Commons Public Domain Dedication (CC0 1.0) and Creative Commons Attribution 3.0 Unported, unaltered images (CC BY 3.0, Sarah Werning, T. Michael Keesey, after Mauricio Antón). We would like to thank Amanda Melin and our other anonymous reviewers for taking the time to read our work and providing constructive suggestions.
References
- Akaike H. 1973. Information theory as an extension of the maximum likelihood principle In: Petrov BN, Casaki F, editors. Second international symposium on information theory. Budapest: Akademiai Kiado, p. 267–281. [Google Scholar]
- Almeida FC, Giannini NP, Simmons NB, Helgen KM.. 2014. Each flying fox on its own branch: a phylogenetic tree for Pteropus and related genera (Chiroptera: Pteropodidae). Mol Phylogenet Evol. 77:83–95. [DOI] [PubMed] [Google Scholar]
- Aloni R, Olender T, Lancet D.. 2006. Ancient genomic architecture for mammalian olfactory receptor clusters. Genome Biol. 7(10): R88.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3): 403–410. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Zheng D, Liu Y-J, Fang G, Frankish A, Carriero N, Robilotto R, Cayting P, Gerstein M.. 2009. Comparative analysis of processed ribosomal protein pseudogenes in four mammalian genomes. Genome Biol. 10(1): R2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates LA, Sayialel KN, Njiraini NW, Poole JH, Moss CJ, Byrne RW.. 2008. African elephants have expectations about the locations of out-of-sight family members. Biol Lett. 4(1): 34–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennie JJ, Duffy JP, Inger R, Gaston KJ.. 2014. Biogeography of time partitioning in mammals. Proc Natl Acad Sci U S A. 111 (38): 13727–13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR.. 2002. Model selection and multimodel inference: a practical information–theoretic approach. New York: Springer. [Google Scholar]
- Challet E. 2007. Minireview: entrainment of the suprachiasmatic clockwork in diurnal and nocturnal mammals. Endocrinology 148 (12): 5648–5655. [DOI] [PubMed] [Google Scholar]
- Chang D, Duda TF Jr.. 2012. Extensive and continuous duplication facilitates rapid evolution and diversification of gene families. Mol Biol Evol. 29(8): 2019–2029. [DOI] [PubMed] [Google Scholar]
- Chesser RK, Sugg DW, Rhodes OE Jr, Novak JM, Smith MH.. 1993. Evolution of mammalian social structure. Acta Theoriol. 38:163–174. [Google Scholar]
- Cotton JA. 2008. The impact of gene duplication on human genome evolutionEncyclopedia of life sciences. Chichester: John Wiley and Sons. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27(8): 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Reis M, Donoghue PCJ, Yang Z.. 2014. Neither phylogenomic nor paleontological data support a paleogene origin of placental mammals. Biol Lett. 10 (1): 20131003.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkel A, Steinhaus M, Kotthoff M, Nowak B, Krautwurst D, Schieberle P, Hofmann T.. 2014. Nature’s chemical signatures in human olfaction: a foodborne perspective for future biotechnology. Angew Chem. 53 (28): 7124–7143. [DOI] [PubMed] [Google Scholar]
- Edwards AWF. 1992. Likelihood: expanded edition. Baltimore: The Johns Hopkins University Press. [Google Scholar]
- Farbiszewski R, Kranc R.. 2013. Olfactory receptors and the mechanism of odor perception. Pol Ann Med. 20(1): 51–55. [Google Scholar]
- Finarelli JA, Goswami A.. 2013. Potential pitfalls of reconstructing deep time evolutionary history with only extant data, a case study using the Canidae (Mammalia, Carnivora). Evolution 67 (12): 3678–3685. [DOI] [PubMed] [Google Scholar]
- Foley NM, Springer MS, Teeling EC.. 2016. Mammal madness: is the mammal tree of life not yet resolved?. Philos Trans R Soc Lond B Biol Sci. 371:20150140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W.. 2012. CD-HIT: accelerated for clustering the next generation sequencing data. Bioinformatics 28(23): 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T Jr, Ives AR.. 2000. Using the past to predict the present: confidence intervals for regression equations in phylogenetic comparative methods. Am Nat. 155(3): 346–364. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Wiebe V, Przeworski M, Lancet D, Paabo S.. 2004. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2(1): E5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn T. 2011. Field guide to next generation DNA sequencers. Mol Ecol Res. 11 (5): 759–769. [DOI] [PubMed] [Google Scholar]
- Glusman G, Bahar A, Sharon D, Pilpel Y, White J, Lancet D.. 2000. The olfactory receptor gene superfamily: data mining, classification, and nomenclature. Mamm Genome 11 (11): 1016–1023. [DOI] [PubMed] [Google Scholar]
- Glusman G, Yanai I, Rubin I, Lancet D.. 2001. The complete human olfactory subgenome. Genome Res. 11(5): 685–702. [DOI] [PubMed] [Google Scholar]
- Goswami A, Finarelli JA.. 2016. EMMLi: a maximum likelihood approach to the analysis of modularity. Evolution 70 (7): 1622–1637. [DOI] [PubMed] [Google Scholar]
- Grafen A. 1989. The phylogenetic regression. Philos Trans R Soc Lond B Biol Sci. 326(1233): 119–157. [DOI] [PubMed] [Google Scholar]
- Granados-Fuentes D, Tseng A, Herzog ED.. 2006. A circadian clock in the olfactory bulb controls olfactory responsivity. J Neurosci. 26 (47): 12219–12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W.. 2008. GEIGER: investigating evolutionary radiations. Bioinformatics 24 (1): 129–131. [DOI] [PubMed] [Google Scholar]
- Harris RM, Hofmann HA.. 2015. Seeing is believing: dynamic evolution of gene families. Proc Natl Acad Sci U S A. 112 (5): 1252–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussler D, O’Brien SJ, Ryder O, Barker FK, Clamp M, Crawford AJ, Hanner R, Hanotte O, Johnson WE, McGuire JA.. 2009. Genome 10K: a proposal to obtain whole-genome sequence for 10,000 vertebrate species. J Hered. 100 (6): 659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden S, Bekaert M, Crider TA, Mariani S, Murphy WJ, Teeling EC.. 2010. Ecological adaptation determined functional mammalian olfactory subgenomes. Genome Res. 20(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden S, Bekaert M, Goodbla A, Murphy WJ, Dávalos LM, Teeling EC.. 2014. A cluster of olfactory receptor genes linked to frugivory in bats. Mol Biol Evol. 31 (4): 917–927. [DOI] [PubMed] [Google Scholar]
- Hughes GM, Gang L, Murphy WJ, Higgins DG, Teeling EC.. 2013. Using illumina next generation sequencing technologies to sequence multigene families in de novo Species. Mol Ecol Res. 13(3): 510–533. [DOI] [PubMed] [Google Scholar]
- Hurvich CM, Tsai C–L.. 1989. Regression and time series model selection in small samples. Biometrika 76 (2): 297–307. [Google Scholar]
- Ibarra-Soria X, Levitin MO, Saraiva LR, Logan DW.. 2014. The olfactory transcriptome of mice. PLoS Genet. 10(9): e1004593.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamilar JM, Cooper N.. 2013. Phylogenetic signal in primate behaviour, ecology and life history. Philos Trans R Soc Lond B Biol Sci. 368(1618): 20120341.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Vosshall LB.. 2008. Better smelling through genetics: mammalian odor perception. Curr Opin Neurobiol, 18 (4): 364–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I, Yang Z, Maldonado E, Li C, Zhang G, Gilbert MTP, Jarvis ED, O’Brien SJ, Johnson WE, Antunes A.. 2015. Olfactory receptor subgenomes linked with broad ecological adaptations in Sauropsida. Mol Biol Evol. 32 (11): 2832–2843. [DOI] [PubMed] [Google Scholar]
- Kissling WD, Dalby L, Fløjgaard C, Lenoir J, Sandel B, Sandom C, Trøjelsgaard K, Svenning J-C.. 2014. Establishing macroecological trait datasets: digitization, extrapolation, and validation of diet preferences in terrestrial mammals worldwide. Ecol. Evol. 4 (14): 2913–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, Guindon S.. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29 (6): 1695–1701. [DOI] [PubMed] [Google Scholar]
- Lee K, Nguyen DT, Choi M, Cha S-Y, Kim J-H, Dadi H, Seo HG, Seo K, Chun T, Park C.. 2013. Analysis of cattle olfactory subgenome: the first detailed study on the characteristics of the complete olfactory receptor repertoire of a ruminant. BMC Genomics 14(1): 596.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, et al. . 2009. The sequence and de novo assembly of the giant panda. Nature 463:311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Wang K, Lei F, Yu D, Zhao H.. 2016. Penguins reduced olfactory receptor genes common to other waterbirds. Sci Rep. 6:31671.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca F, Perry GH, Di Rienzo A.. 2010. Evolutionary adaptations to dietary changes. Annu Rev Nutr. 30:291–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Liu B, Yinlong X, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. . 2012. SOAPdenovo2: an empiricaly improved memory-efficient short-read de novo assembler. GigaScience 1(1):. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z-X. 2007. Transformation and diversification in early mammal evolution. Nature 450(7172): 1011–1019. [DOI] [PubMed] [Google Scholar]
- Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simao TLL, Stadler T, et al. . 2011. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334(6055): 521–524. [DOI] [PubMed] [Google Scholar]
- Mitani JC. 1989. Orangutan activity budgets: monthly variations and the effects of body size, partruition and sociality. Am J Primatol. 18:87–100. [DOI] [PubMed] [Google Scholar]
- Montague MJ, Li G, Gandolfi B, Khan R, Aken BL, Searle SMJ, Minx P, Hillier LW, Koboldt DC, Davis BW, et al. . 2014. Comparative analysis of the domesticated cat genome reveals genetic signatures underlying feline biology and domestication. Proc Natl Acad Sci U S A. 111 (48): 17230–17235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchlinski MN, Deane AS.. 2014. The interpretive power of infraorbital foramen area in making dietary inferences in extant apes. Anat Rec. 297(8): 1377–1384. [DOI] [PubMed] [Google Scholar]
- Nei M, Rooney AP.. 2005. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 39:121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M.. 2007. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS One 2 (8): e708–e710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Matsui A, Touhara K.. 2014. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 24(9): 1485–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olender T, Fuchs T, Linhart C, Shamir R, Adams M, Kalush F, Khen M, Lancet D.. 2004. The canine olfactory subgenome. Genomics 83 (3): 361–372. [DOI] [PubMed] [Google Scholar]
- Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401(6756): 877–884. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K.. 2004. APE: analysis of phylogenetics and evolution in R language. Bioinformatics 20(2): 289–290. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Wood T, Zhang Z, Miller W.. 1997. Comparison of DNA sequences with protein sequences. Genomics 46 (1): 24–36. [DOI] [PubMed] [Google Scholar]
- Peterson AT, Soberon J, Sanchez-Cordero V.. 1999. Conservation of ecological niches in evolutionary time. Science 285:1265–1267. [DOI] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D.. R Core Team. 2015. Nlme: linear and nonlinear mixed effects models [Internet]. R package version 3.1–128. Available from: https://cran.r-project.org/web/packages/nlme/nlme.pdf. [Google Scholar]
- Price SA, Hopkins SSB, Smith KK, Roth VL.. 2012. Tempo of trophic evolution and its impact on mammalian diversification. Proc Natl Acad Sci U S A. 109 (18): 7008–7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ. 2012. Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3 (2): 217–223. [Google Scholar]
- Rizvanovic A, Amundin M, Laska M.. 2013. Olfactory discrimination ability of Asian elephants (Elephas maximus) for structurally related odorants. Chem Senses 38(2): 107–118. [DOI] [PubMed] [Google Scholar]
- Ruedi M, Stadelmann B, Gager Y, Douzery EJP, Francis CM, Lin L-K, Guillén-Servent A, Cibois A.. 2013. Molecular phylogenetic reconstructions identify east Asia as the cradle for the evolution of the cosmopolitan genus myotis (Mammalia, Chiroptera). Mol Phylogenet Evol. 69(3): 437–449. [DOI] [PubMed] [Google Scholar]
- Schmitz L, Motani R.. 2010. Morphological differences between the eyeballs of nocturnal and diurnal amniotes revisited from optical perspectives of visual environments. Vision Res. 50(10): 936–946. [DOI] [PubMed] [Google Scholar]
- Schondube JE, Herrera–M LG, Del Rio C.. 2001. Diet and the evolution of digestion and renal function in phyllostomid bats. Zoology 104(1): 59–73. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. . 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol Syst Biol. 7:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk JB. 2007. The adaptive value of sociality in mammalian groups. Philos Trans R Soc Lond B Biol Sci. 362(1480): 539–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I.. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19 (6): 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer MS, Gatesy J.. 2017. Inactivation of the olfactory marker protein (OMP) gene in river dolphins and other odontocete cetaceans. Mol Phylogenet Evol. 109:375–387. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Abere AJ, Goll C, Smith SA, Berger SA, Izquierdo-Carrasco F.. 2012. RaxML-Light: a tool for computing terabyte phylogenies. Bioinformatics 28 (15): 2064–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartia S, Villanueva-Cañas JL, Finarelli J, Farrell ED, Collins PC, Hughes GM, Carlsson JEL, Gauthier DT, McGinnity P, Cross TF, et al. . 2016. A novel method of microsatellite genotyping-by-sequencing using individual combinatorial barcoding. R Soc Open Sci. 3(1): 150565.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittemyer G, Douglas-Hamilton I, Getz WM.. 2005. The socioecology of elephants: analysis of the processes creating multitiered social structures. Anim Behav. 69 (6): 1357–1371. [Google Scholar]
- Young JM, Massa HF, Hsu L, Trask BJ.. 2010. Extreme variability among mammalian V1R gene families. Genome Res. 20 (1): 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Webb DM.. 2003. Evolutionary deterioration of the vomeronasal pheromone signalling transduction pathway in catarrhine primates. Proc Natl Acad Sci U S A. 100 (14): 8337–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Rossiter SJ, Teeling EC, Li C, Cotton JA, Zhang S.. 2009. The evolution of color vision in nocturnal mammals. Proc Natl Acad Sci U S A. 106 (22): 8980–8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Xu D, Zhang S, Zhang J.. 2011. Widespread losses of vomeronasal signal transduction in bats. Mol Biol Evol. 28(1): 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, Hanrahan F, Pertea G, Van Tassell CP, Sonstegard TS, et al. . 2009. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 10(4): R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.