Abstract
The Molecular Evolutionary Genetics Analysis (Mega) software implements many analytical methods and tools for phylogenomics and phylomedicine. Here, we report a transformation of Mega to enable cross-platform use on Microsoft Windows and Linux operating systems. Mega X does not require virtualization or emulation software and provides a uniform user experience across platforms. Mega X has additionally been upgraded to use multiple computing cores for many molecular evolutionary analyses. Mega X is available in two interfaces (graphical and command line) and can be downloaded from www.megasoftware.net free of charge.
Keywords: software, evolution, Linux
Introduction
Over the past decade, genome sequencing has become an efficient and potent means to investigate a broad array of biological systems, from large-scale studies of biological diversity to tracking the evolution and origin of pathogenic microbes. The numerous steps required to glean interpretable and actionable results from raw sequence data invariably require comparative analysis of molecular sequences to discover functional and adaptive genome differences. Molecular Evolutionary Genetics Analysis (Mega) software provides tools to conduct such analyses. Mega includes a large repertoire of programs for assembling sequence alignments, inferring evolutionary trees, estimating genetic distances and diversities, inferring ancestral sequences, computing timetrees, and testing selection (Kumar et al. 2016). Over the last 25 years, Mega’s use in evolutionary analysis has been cited in over one hundred thousand studies in diverse biological fields.
Mega across Computing Platforms (Mega X)
Mega was first developed for MS DOS in the early 1990s (Kumar et al. 1994) and then upgraded for use in MS Windows eight times, including MEGA 1 to MEGA 6 and MEGA-CC and MEGA-MD (Kumar et al. 2001, 2016). Some of the Mega releases have been packaged for Linux systems using the WINE compatibility layer for POSIX-compliant operating systems and the Wineskin tool (built on WINE) for macOS systems. These versions have been downloaded over 200,000 times. But the ad hoc Windows-emulation solution is sluggish and relatively unstable when compared with the performance in MS Windows. Emulators cannot be used effectively for the latest 64-bit version of Mega that is built to handle memory-intensive analyses of large contemporary data sets (Kumar et al. 2016), so a more comprehensive solution is required for users of alternate platforms.
Therefore, Mega has been transformed into a cross-platform version that runs natively on Linux and Microsoft Windows. This advancement eliminates the Windows-only limitation of Mega, which has become particularly acute due to the increasing use of Linux in biological research. This transformation also paves the way for development of a MEGA X version for macOS in the near future.
Cross-Platform Graphical User Interface (GUI) for Mega X
The GUI has been programmed by refactoring Mega’s source code to be compatible with the Lazarus integrated development environment (Lazarus Team) and the Free Pascal compiler. Extensive reprogramming of visual components was needed to compile cross-platform Mega, including the Alignment Explorer, Sequence Data Explorer, and Tree Explorer. Additionally, all the internal control code that used the MS Windows system application programming interface (API) was advanced to work across platforms.
The reprogramming was accompanied with visual modernization of many parts of the GUI (fig. 1). To provide a uniform user experience across computing platforms, many user interface elements have been implemented using the Chromium Embedded Framework (CEF), which is an open source framework based on the Google Chromium web browser core. With this framework, we have implemented GUI elements using HTML and JavaScript in Mega X desktop GUI (fig. 2). As a result, Mega X has the same look-and-feel in Windows and Linux. We are currently developing Mega X for macOS, which will share this user interface.
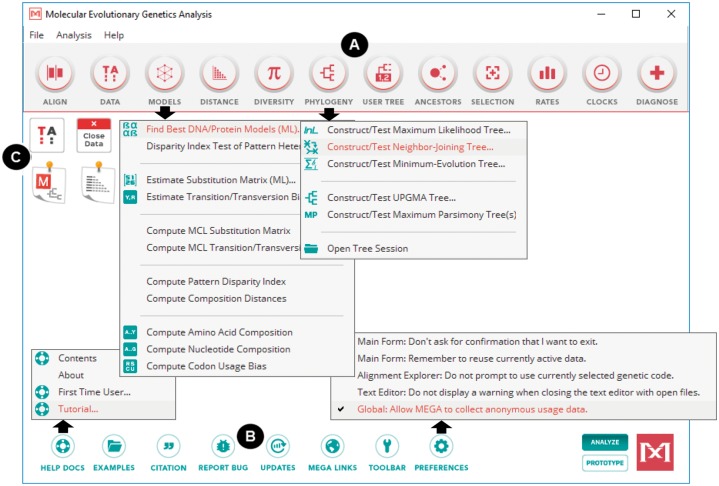
Fig. 1.
The Mega X main form has a modernized look-and-feel, but it maintains the familiar structure of the previous versions of Mega. (A) The top toolbar organizes the large number of analyses available in Mega X into logical groups accessed via drop down menus. (B) The bottom toolbar provides access to utility functions such as the help system, example input data files and application preferences. (C) Icons on the main form provide convenient access to input data and results explorer windows. Mega X can now be used in two modes: one for data analysis (ANALYZE mode) and another for prototyping (PROTOTYPE mode).
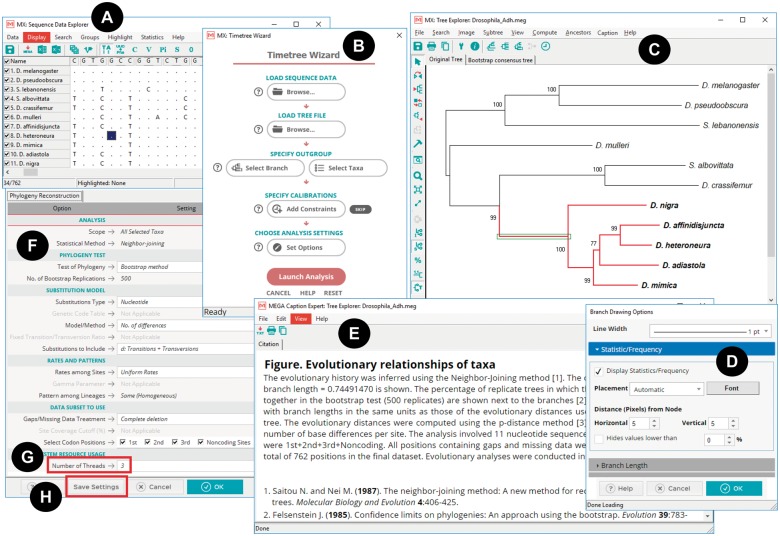
Fig. 2.
The redesigned GUI elements in Mega X include explorer windows such as the (A) Sequence Data Explorer, (B) Timetree Wizard, and (C) Tree Explorer. These use the native widget set of the target operating system. (D) Options dialogs and (E) Caption Expert were developed using HTML and JavaScript and are displayed via Mega’s integrated web browser that is built on the Chromium Embedded Framework. (F) The redesigned “Analysis Preferences” dialog box for selecting options for data analysis, including (G) the number of threads to employ and (H) a “Save Settings” button for generating an “.mao” file for use with MEGA-CC for high throughput analysis.
Parallelization of Computational Core
We have also improved Mega’s computational core by increasing the number of methods that can utilize multiple processors and cores on the same machine. This will greatly reduce the run time for computationally demanding inferences. Users can specify the maximum number of cores to deploy in the Analysis Preferences dialog box (fig. 2G). By default, Mega X uses all available cores, minus one, such that no >80% of RAM is consumed. Computational methods benefitting from calculation parallelization include those for selecting the best model for nucleotide or amino acid substitutions and bootstrap tests of molecular phylogenies employed with distance-based and parsimony-based methods. Users can already employ multiple cores during the heuristic search for the Maximum Likelihood (ML) tree in Mega.
Distributions
On Microsoft Windows, Mega X is intended to be used on 64-bit systems having version 10 and later. It will, however, work smoothly on previous versions of 64-bit Windows. We also provide a 32-bit version of Mega X for Windows for those with 32-bit computers. On Linux platforms, Mega X has been optimized and tested for use on Debian/Ubuntu and RedHat/CentOS based distributions. Because users may customize their Linux environments easily, we recommend use of Mega X for Linux in tested desktop environments. Specifically, we have tested Mega X on Ubuntu 16.04.4 (with Unity desktop), Mint 18.1 (with Cinnamon desktop), and CentOS 7 (with GNOME desktop).
Mega X now generates a configuration file specifying analysis options for user-desired analysis in Mega CC, which many researchers use for high throughput and iterative analysis in Windows, Linux and macOS environments (Kumar et al. 2012). The “Save Settings” button provides this functionality (fig. 2H). In fact, Mega X can now be used in the prototype mode (see fig. 1, lower right corner), if desired, which obviates the need to distribute and maintain a separate application for generating the configuration file for running Mega CC, so Mega-Proto is no longer required or supported.
In conclusion, the cross-platform Mega X will benefit many existing users and enable many others to consider utilizing Mega X in their research and teaching endeavors. The consistency of user experience across platforms should be particularly useful in facilitating collaborative analyses within and among research groups as well as in teaching a diverse group of students. In the near future, we will complete the development of a macOS version to make Mega X available for all major operating systems.
Acknowledgments
We thank our laboratory members and many beta testers for providing invaluable feedback and bug reports. This work was supported in part by research grants from National Institutes of Health (R01GM126567-01), National Science Foundation (ABI 1661218) to S.K., and Japan Society for the Promotion of Science (JSPS) grants-in-aid for scientific research (DB5) to K.T.
References
- Kumar S, Stecher G, Peterson D, Tamura K.. 2012. MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics 28(20):2685–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M.. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17(12):1244–1245. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M.. 1994. MEGA: molecular Evolutionary Genetics Analysis software for microcomputers. Comput Appl Biosci 10(2):189–191. [DOI] [PubMed] [Google Scholar]
- Lazarus Team. Lazarus Homepage. Available from: https://www.lazarus-ide.org/.