Abstract
The human body represents a striking example of ciliary diversification. Extending from the surface of most cells, cilia accomplish an astonishingly diverse set of tasks. Predictably, mutations in ciliary genes cause a wide range of human diseases such as male infertility or blindness. In C. elegans sensory cilia, this functional diversity appears to be traceable to the differential regulation of the kinesin-2-powered intraflagellar transport (IFT) machinery. Here, we reconstituted the first functional, multi-component IFT complex that is deployed in the sensory cilia of C. elegans. Our bottom-up approach revealed the molecular basis of specific motor recruitment to the IFT trains. We identified the key component that incorporates homodimeric kinesin-2 into its physiologically relevant context which in turn allosterically activates the motor for efficient transport. These results lay the groundwork for a molecular delineation of IFT regulation that eluded understanding since its ground-breaking discovery more than two decades ago.
Having been neglected for decades, the biology of cilium underwent a renaissance after the realization that ciliogenesis is central to development and disease 1–5 Virtually all cilia are built by the remarkably conserved IFT machinery with a few intriguing exceptions where the cilium is built in the cytoplasm in an IFT-independent manner5–8. Numerous in vivo studies unmasked the universal hallmarks of IFT in diverse model systems. The IFT trains display continuous movement either towards the ciliary tip (kinesin-2-dependent) or the ciliary base (dynein-2-dependent) without apparent reversals in between5,7,9–11. Once a kinesin-2-powered IFT train arrives at the ciliary tip, it undergoes an enigmatic remodeling that deactivates the kinesin-2 motor and restructures the train for dynein-2-dependent transport back to the base5,7,9–11. It is long known that C. elegans employs two different kinesin motors, a heterotrimeric kinesin-2 called kinesin-II and a homodimeric kinesin-2 called OSM-3, to build its functionally distinct cilia12. Loss of the homodimeric OSM-3 function invariably leads to the loss of the distal portion of the canonical cilium and to deficient osmotic avoidance in the so-called rod-shaped or “canonical” cilia while the loss of both, heterotrimeric and homodimeric kinesin-2 motors, leads to loss of entire axonemes12. In the wing-shaped AWC cilia, on the other hand, loss of the heterotrimeric kinesin-2 motor leads to deficient chemotaxis suggesting that the kinesin-dependent trafficking of cilia-specific components define the functional identity of the respective cilia13,14. In contrast to the heterotrimeric kinesin-2, the precise role of the homodimeric kinesin-2 in other organisms is much less understood7. For instance, the homodimeric kinesin-2 moves along the mammalian cilia, if it also functions as an IFT motor, however, has not yet been demonstrated15,16.
Despite its essential role in ciliogenesis and its emerging role in ciliary diversity13,14,17–19, mechanisms of IFT train assembly, motor recruitment, and the timely activation and deactivation of the oppositely directed kinesin-2 and dynein-2 motors remained mysterious. To lay the foundation towards a comprehensive molecular understanding of IFT, here we employed a bottom-up approach to dissect the OSM-3-dependent IFT in C. elegans sensory cilia. The OSM-3 function was previously linked to the so-called IFT-B complex20. Indeed, the loss of function of many IFT-B subunits in corresponding mutants have been proposed to interfere with OSM-3 function in vivo many of which were shown to be part of the so-called IFT-B core complex (ref 20–24; Extended Data Fig. 1). Previous work with green algae C. reinhardtii was instrumental to the detailed characterisation of the IFT components. The entire IFT-B complex consisting of the so-called peripheral and core complexes has been reconstituted from 15 recombinantly expressed subunits25. Specifically, nine core components (IFT-74, IFT-81, IFT-27, IFT-25, IFT-22, IFT-52, IFT-46, IFT-88, IFT-70) formed a stable complex, several of which were capable of autonomously forming sub-complexes in vitro26–29. Guided by these previous findings, here, we concentrated on the IFT-B core complex and started our bottom-up approach by first assembling the presumptive sub-complexes using recombinantly expressed wild-type, full-length subunits from C. elegans. Next, we systematically probed the interaction of the sub-complexes as well as their individual subunits with the OSM-3 motor using multifaceted techniques. To visualize the motor activity in functional transport assays, we used a previously described, constitutively active OSM-3G444E motor that we C-terminally Halo-tagged for fluorophore labeling (ref. 30; OSM-3G444EHalo hereafter).
Building the first IFT-kinesin-2 complex
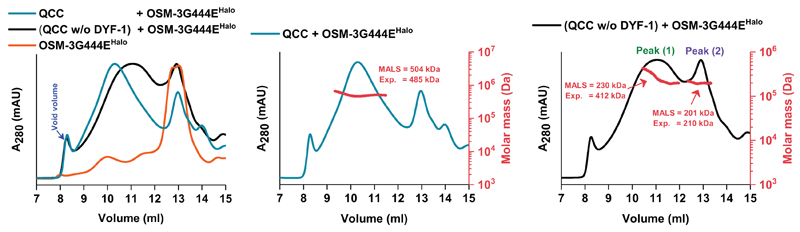
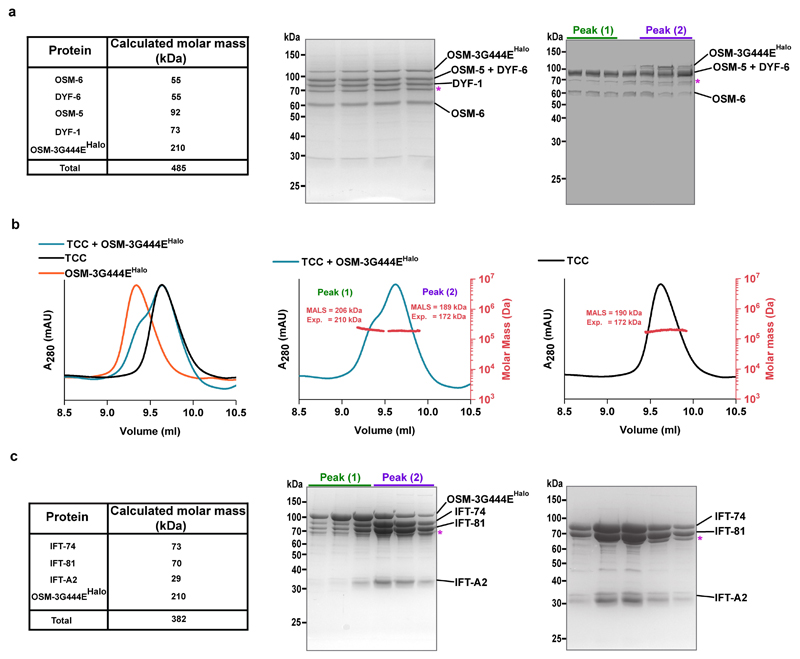
To monitor complex formation, we made use of size-exclusion chromatography (SEC) that was coupled to multiple-angle light scattering (MALS) analysis to gauge the molecular weight of the presumptive complexes. We first assessed whether the Tripartite IFT-74/IFT-81/IFT-22(IFTA-2) Core Complex (TCC hereafter, blue subunits in Extended Data Fig.1), and the Quadripartite IFT-52(OSM-6)/IFT-46(DYF-6)/IFT-88(OSM-5)/IFT-70(DYF-1) Core Complex (QCC hereafter, magenta subunits in Extended Data Fig. 1) are formed stably or if additional factors were necessary (i.e. IFT-27 and IFT-25 subunits that are present in the IFT-B core complex in C. reinhardtii but not in C. elegans; ref. 29). Whereas both complexes were formed, only the QCC incorporated the motor protein (Fig. 1 and Extended Data Fig. 2a) but not the TCC (Extended Data Fig. 2b and c). Importantly, removal of the IFT-70(DYF-1) subunit from the QCC (QCC w/o DYF-1 hereafter), disrupted the complex formation between the motor and its IFT-B complex suggesting a IFT-70(DYF-1)-specific recruitment of the motor (Fig. 1, middle vs. right panels). Consistent with this notion, the motor was found under the elution peak with QCC and was absent under QCC w/o DYF-1 when IFT-70(DYF-1) subunit was removed (Extended Data Fig. 2a, middle vs. right panels).
Figure 1. IFT-70(DYF-1) is key for the incorporation of the OSM-3 motor into the QCC.
Overlay of the elution profiles of the (QCC w/o DYF-1 + OSM-3G444EHalo) and (QCC + OSM-3G444EHalo) along with the OSM-3G444EHalo motor show the IFT-70(DYF-1)-dependent shift of the respective elution peaks upon motor incorporation into the QCC (left panel). The MALS analyses of the respective complexes are shown in the middle and right panels. The molar mass determined from the MALS fit and the expected mass of the complexes are shown in red (the right axis and insets). Consistent results were obtained from three independent experiments.
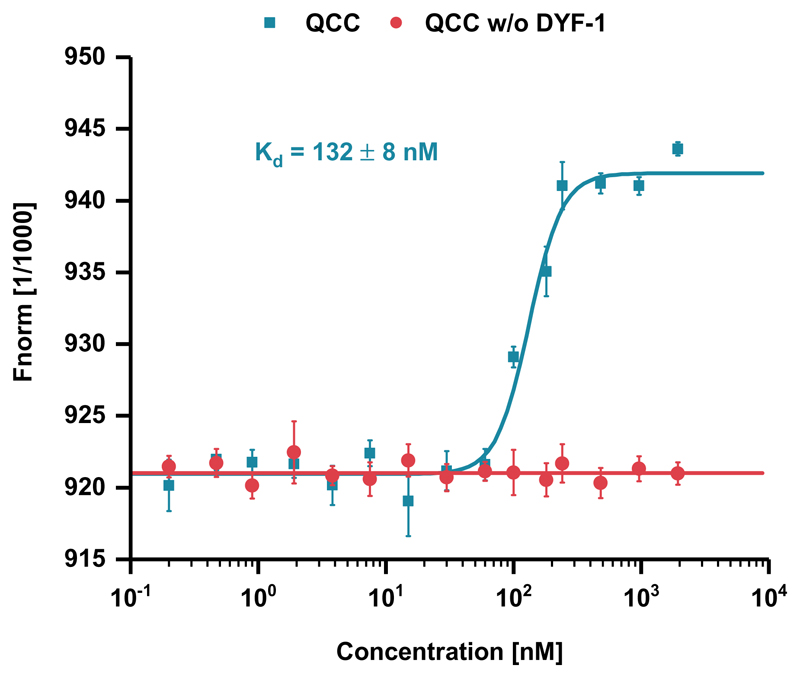
To provide independent biochemical evidence for the IFT-70(DYF-1)-mediated interaction of the motor with the QCC, we turned to in-solution protein-binding assays. By MicroScale Thermophoresis (MST), we observed robust, sub-micromolar binding of the motor protein to the QCC, whereas removal of the IFT-70(DYF-1) subunit abolished this interaction (Fig. 2). Taken together, our solution-based SEC-MALS and MST analyses both argued for a specific, IFT-70(DYF-1)-mediated recruitment of the OSM-3 motor to the IFT-B complex.
Figure 2. IFT-70(DYF-1)-mediated binding of OSM-3G444EHalo motor to the QCC measured by MST.
The fluorophore-labeled motor was titrated with both the QCC and QCC w/o DYF-1, respectively. Data points presented as means of n=3 independent experiments ± SD (error bars; source data is provided online).
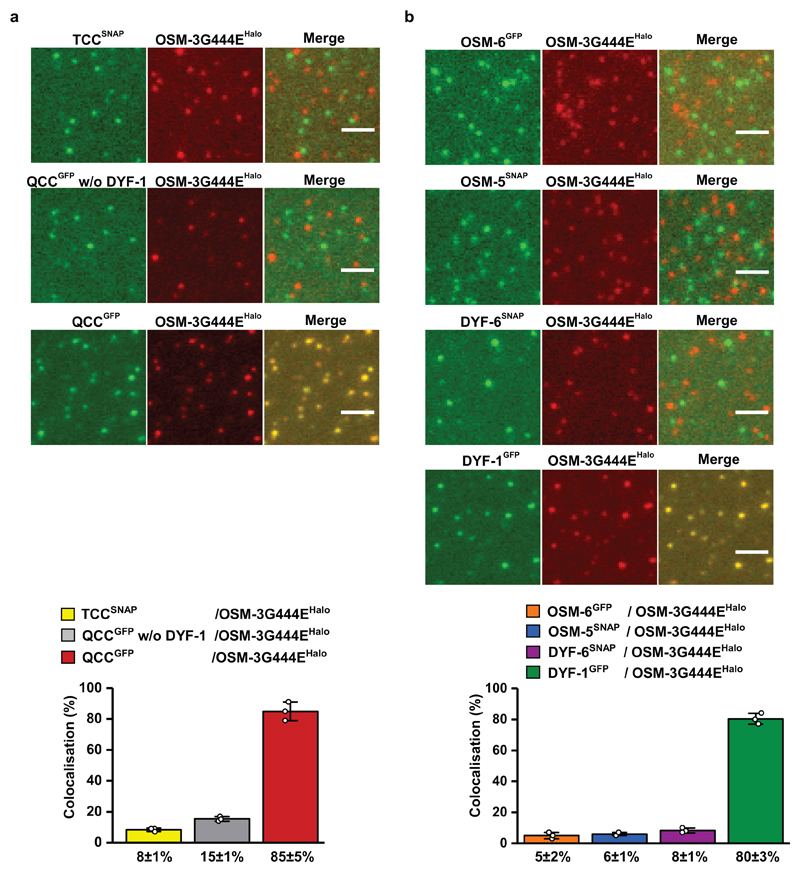
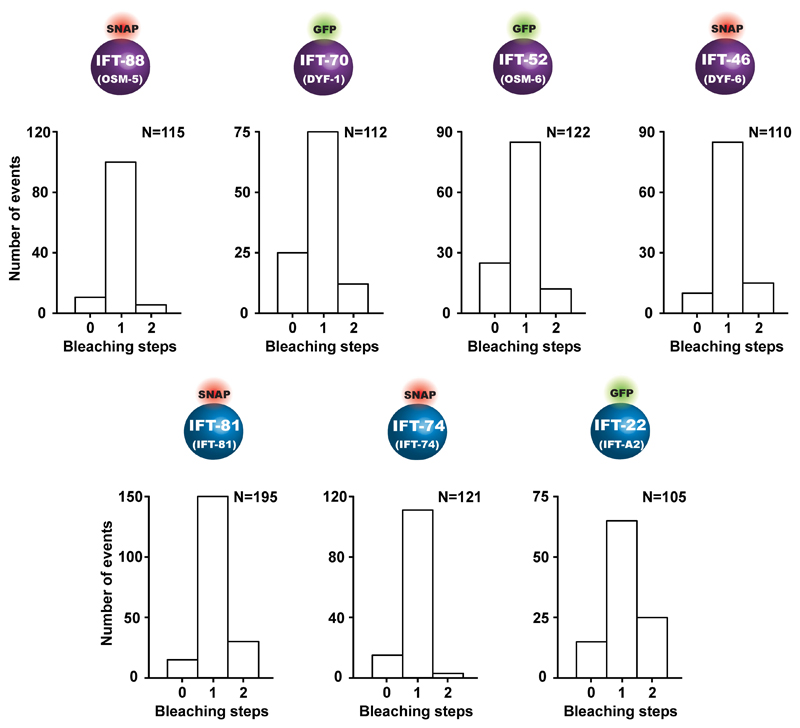
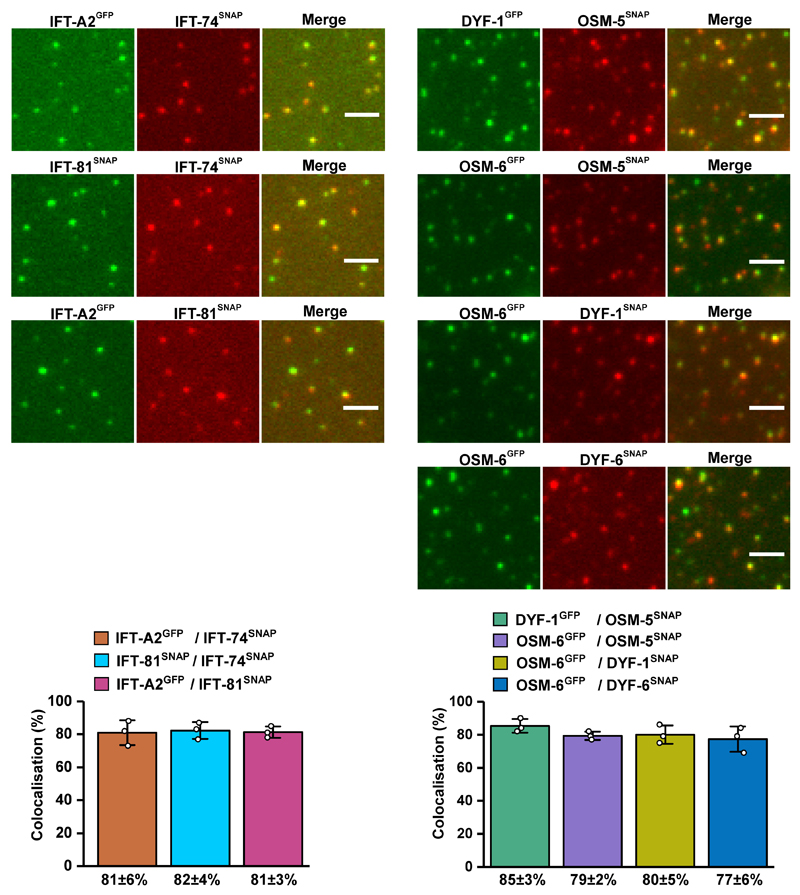
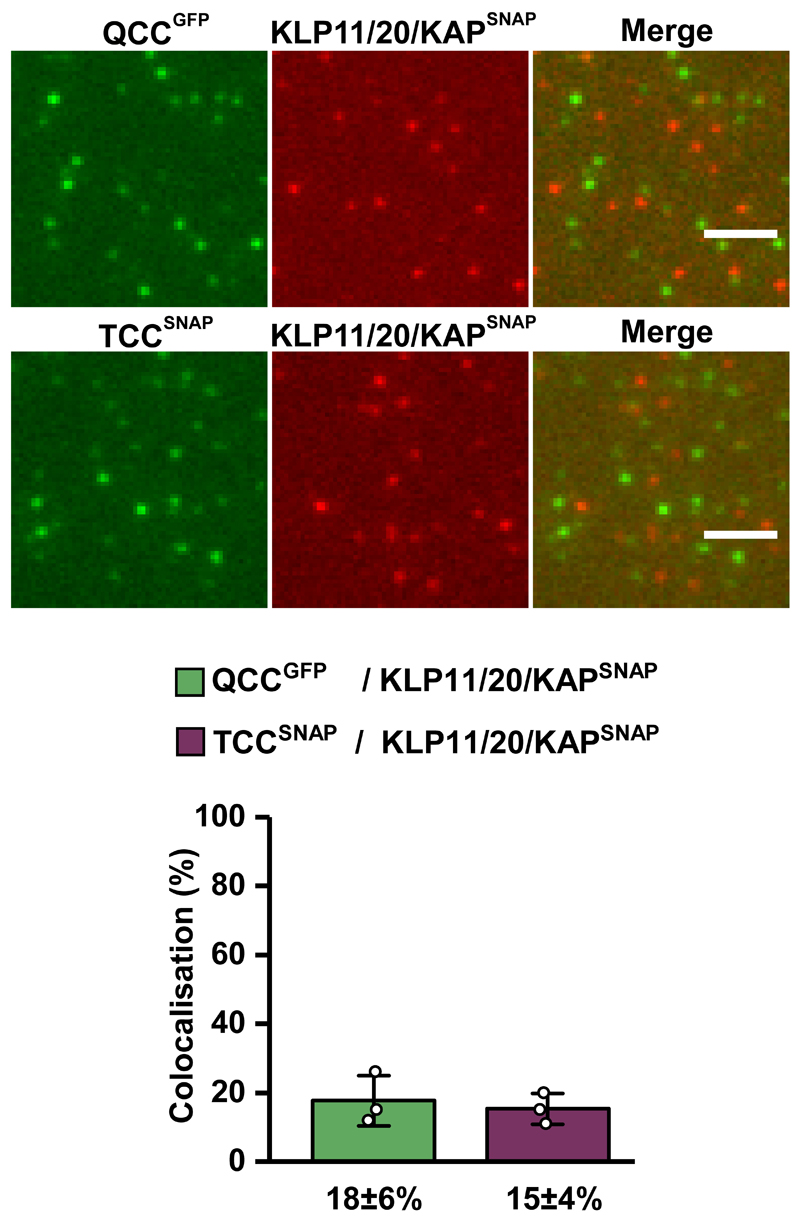
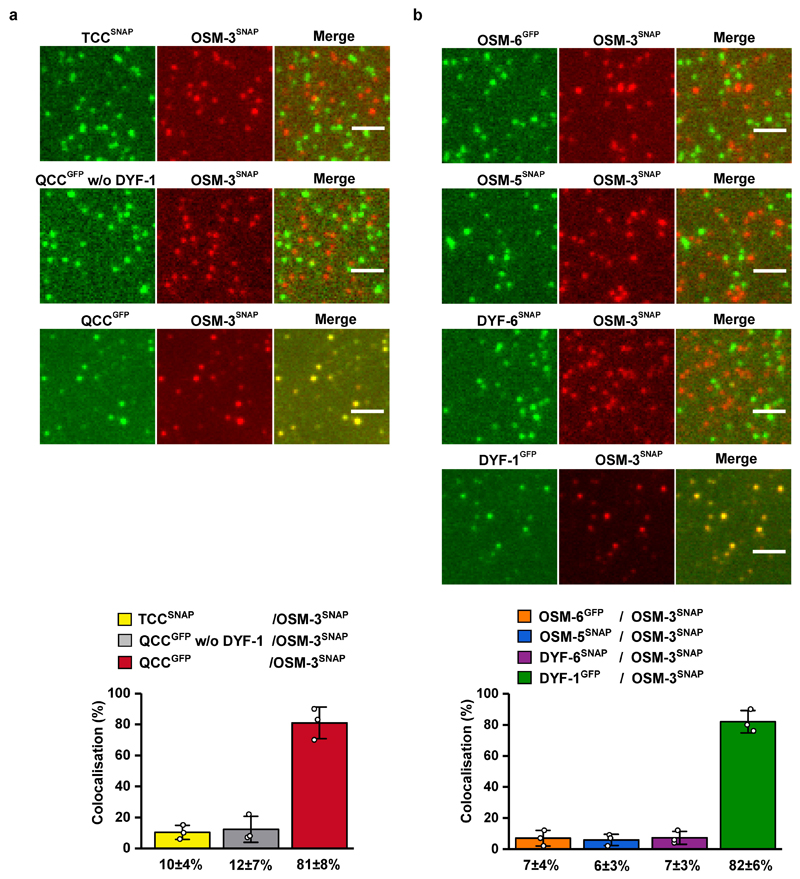
To further corroborate the key role of the IFT-70(DYF-1) subunit in OSM-3 recruitment, we next turned to fluorescence-based microscopy assays. To this end, we functionalised the respective IFT-B subunits with different fluorescent tags and photobleaching assays demonstrated that we detected non-aggregated, single subunits after functionalisation (Extended Data Fig. 3). Next, we determined the pairwise colocalisation efficiencies by differentially labeling the respective subunits. This exclusion process demonstrated that the functionalised subunits retained their capability to incorporate into a stable complex under assay conditions (Extended Data Fig. 4). To probe which of these sub-complexes displays efficient colocalisation with the motor, we differentially labeled the OSM-3G444EHalo and the respective sub-complexes. Consistent with our previous assays that argued for a IFT-70(DYF-1)-dependent interaction between the motor and its IFT-B complex (Figs. 1 and 2), OSM-3G444EHalo failed to colocalise efficiently with both, the TCC and QCC that lacked the IFT-70(DYF-1), but displayed significant colocalisation with the QCC which contained the IFT-70(DYF-1) subunit (Fig. 3a). Finally, the efficient colocalisation of the IFT-70(DYF-1) subunit with the OSM-3G444EHalo, but not with IFT-52(OSM-6), IFT-88(OSM-5) or IFT-46(DYF-6) subunits provided direct evidence for the IFT-70(DYF-1) to be the key subunit to mediate the interaction between the motor and the IFT-B core complex (Fig. 3b). In contrast, the heterotrimeric KLP11/20/KAP motor which cooperates with OSM-3 to build the rod-shaped sensory cilium in C. elegans, failed to colocalise with the IFT-B complex (Extended Data Fig. 5). Previous in vivo experiments in fact suggested the KLP11/20/KAP motor to associate with the IFT-A complex20.
Figure 3. Colocalisation efficiency of the OSM-3G444EHalo motor with its IFT-B complex is solely dependent on the IFT-70(DYF-1) subunit.
(a) Neither TCC nor QCC w/o DYF-1 sub-complexes displayed efficient colocalisation with the OSM-3G444EHalo motor. In the presence of the IFT-70(DYF-1) subunit on the other hand, the QCC complex efficiently colocalised with OSM-3G444EHalo (85±5%). (b) Consistently, OSM-3G444EHalo failed to interact with the individual IFT-52(OSM-6), IFT-88(OSM-5), and IFT-46(DYF-6) subunits but showed robust colocalisation (80±3%) with the IFT-70(DYF-1) subunit. IFT-81 and IFT-52(OSM-6) were fluorescently labeled with SNAP- and GFP-tags, respectively. Values presented as means of n=3 independent experiments ± SD; source data are provided online. Scale bar: 3 µm.
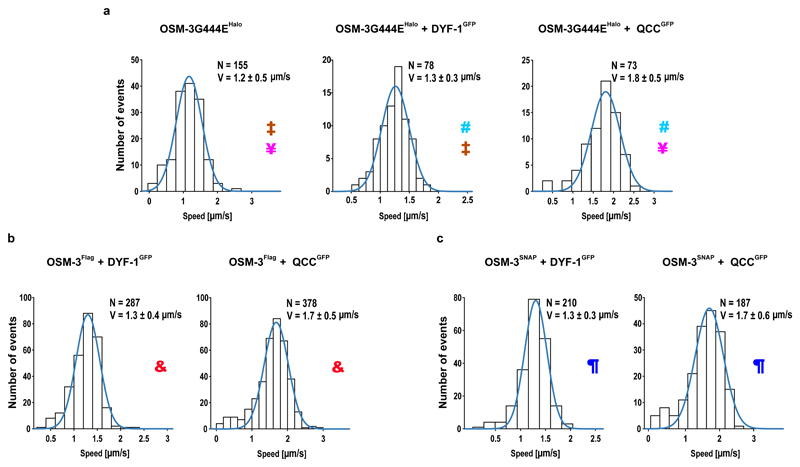
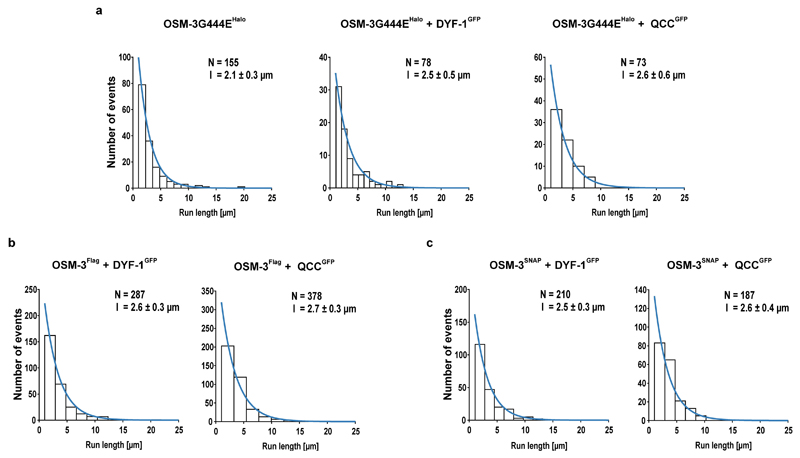
Functional transport assays arguably provide the most direct evidence for the specific recruitment and activation of the OSM-3 motor by the IFT-70(DYF-1) subunit. To this end, we tracked the differentially fluorophore labeled OSM-3G444EHalo motor and its IFT-B complex in a TIRF microscope to assess if our reconstituted OSM-3/IFT-B complex is capable of directional movement in vitro. Indeed, most of the respective fluorophore signals from the motor and its complex were colocalised and moved directionally on surface-attached microtubules (Fig. 4a and Supplementary Video 1). Again consistent with our previous assays, removal of IFT-70(DYF-1) terminated the interaction of the motor with QCC and the OSM-3G444EHalo motor moved alone along microtubules (Supplementary Video 1). Intriguingly, incorporation of the motor into the QCC significantly enhanced the velocity of the OSM-3G444EHalo, whereas the velocities of the motor alone or in the presence of the IFT-70(DYF-1) subunit were indistinguishable (Fig. 4a). The processivity of the respective motors, in contrast, was independent of the IFT-70(DYF-1) subunit or the QCC (Extended Data Fig. 6a).
Figure 4. IFT-70(DYF-1)-dependent incorporation into the QCC fully activates OSM-3 in vitro.
(a) In the absence (top, left panel) and presence (top, middle panel) of IFT-70(DYF-1), the velocity of OSM-3G444EHalo is indistinguishable but incorporation into the QCC (top, right panel) significantly increases the motor’s velocity. p-values (from a two-tailed t-test assuming unequal variances): ‡0.013; ¥2.5*10-18; #3.4*10-15. (b and c) Consistently, QCC fully activates the OSM-3Flag and OSM-3SNAP but not the IFT-70(DYF-1) subunit alone. p-values: &9.6*10-25; ¶1.8*10-12. N= number of events obtained from three different flow chambers from n=3 independent experiments (velocities fit to a Gaussian distribution ± SD).
The point mutation in the stalk of the OSM-3G444EHalo construct hampers the auto-inhibitory folding which in turn results in an activated motor in vitro30. We thus next asked if the IFT-70(DYF-1) subunit can also activate the auto-inhibited OSM-3 motor in our reconstitution assays. To this end, we designed a motor with a wild type stalk that lacked the G444E mutation. We introduced an N-terminal Flag and a SNAP-tag for affinity purification and fluorophore labeling of the motor (OSM-3SNAP hereafter), respectively. In this construct the entire C-terminus that followed the catalytic heads remained unmodified. As demonstrated with the OSM-3G444EHalo (Fig. 3), the OSM-3SNAP motor with unmodified C-terminus displayed efficient colocalisation with the QCC in an IFT-70(DYF-1)-dependent manner but not with TCC (Extended Data Fig. 7a) and interacted with the IFT-70(DYF-1) subunit but not with IFT-52(OSM-6), IFT-88(OSM-5), IFT-46(DYF-6) subunits (Extended Data Fig. 7b).
However, the auto-regulation in kinesin motors is achieved by folding of the distal tail domain at the C-terminus onto the N-terminal head domains30. To ensure that the N-terminal SNAP-tag does not impede the auto-inhibitory folding of the motor per se, we additionally designed a construct that was solely Flag-tagged at its C-terminus for affinity purification (OSM-3Flag hereafter). Due to the lack of fluorescence information, this motor cannot be followed directly in the functional transport assays, however, it represents the wild type OSM-3 as close as possible. Both motors containing the wild type stalk were recruited and activated in an IFT-70(DYF-1)-specific manner (Figs. 4b and c, Supplementary Videos 2 and 3). In fact, the IFT-70(DYF-1)-dependent activation was much more obvious with the OSM-3SNAP motor that lacked the G444E mutation. As expected from an auto-inhibited kinesin, and in stark contrast to the OSM-3G444EHalo motor, OSM-3SNAP with its wild type stalk was barely capable of directional movement and displayed two-dimensional diffusion instead (compare Supplementary Video 1, top left to Supplementary Video 2, top left). The presence of IFT-70(DYF-1) or the QCC initiated robust directional movement of the auto-inhibited OSM-3SNAP motor (Supplementary Video 2). Lastly, as seen with the OSM-3G444EHalo motor (Extended Data Fig. 6a), the processivity of OSM-3SNAP and OSM-3Flag motors remained unmodified by the presence of IFT-70(DYF-1) (Extended Data Fig. 6b and c).
Taken together, our functional transport assays consistently showed that the presence of IFT-70(DYF-1) is essential to recruit the OSM-3 motor to the IFT-B complex. Our results further revealed that the presence of the IFT-70(DYF-1) subunit alone is not sufficient to fully unleash the motor’s activity. Instead, it is the IFT-70(DYF-1)-mediated incorporation of the OSM-3 into the IFT-B complex that allosterically activates the motor. Importantly, the transport rates extracted from our in vitro reconstitution assays were consistent with rates extracted from in vivo studies12,20,31. The physiological relevance of our reconstitution is underscored in particular by previous in vivo findings in the C. elegans sensory cilium. In the absence of dyf-1 function, OSM-3 not only appeared to detach from the IFT trains, it also failed to move directionally but diffused along the cilium instead (see Supplementary Video 2, top left; ref. 20), which now can be understood by the direct role of the IFT-70(DYF-1) subunit in specific motor recruitment and activation as unmasked in this study. In fact, these results now represent a functional reciprocity which is particularly remarkable given the daunting complexity of the IFT machinery. Collectively, our work directly unmasked for the first time the molecular mechanism of how an IFT motor is recruited to its IFT train and demonstrated the power of bottom-up approaches to delineate the underlying mechanisms of highly convoluted processes such as the ciliogenesis.
Methods
DNA and protein constructs
All DNA constructs used in this study were commercially synthesized (GenScript) based on the available sequence information from the WormBase (http://www.wormbase.org) and cloned into the pFastBac1 vector according to the manufacturer’s instructions (Thermo Fisher). In addition, the pFastBac1 vector was functionalised with Halo, SNAP, or GFP genes for C-terminal tagging of several constructs as described in the manuscript. Three different OSM-3 constructs were designed for this study (see SI for detailed information about DNA and protein sequences).
Protein expression, purification, and fluorescent labeling
The Baculovirus Expression System (Thermo Fisher) was used to express all proteins in the insect cells (Sf9) according to the manufacturer’s instructions.
For protein purification Sf9 cells at concentration of 2 x 106 cells/ml were infected with the respective viruses. After 65h of incubation at 28°C, the cells were harvested by centrifugation for 15 min at 2,600 g. Cell pellets were lysed in PIPES buffer (50 mM PIPES, pH 6.9, 300 mM potassium acetate, 1 mM MgCl2, 1mM DTT, 0.5% Triton X-100, 10% glycerine, 0.1mM ATP, Complete Protease Inhibitor Cocktail (Roche). Lysed cells were pelleted by centrifugation for 10 min at 40,000 g. The supernatant was incubated with 60 µl of ANTI-Flag M2 Affinity Gel (Sigma) for 90 min. The Flag-resin was washed three times with 1 ml of wash buffer 1 (80 mM PIPES, pH 6.9, 500 mM potassium acetate, 1mM MgCl2, 1 mM DTT, 5 µM ATP, 10% glycerine, 0.1% Tween-20, 1 mM EGTA) and three times with 1 ml of wash buffer 2 (80 mM PIPES, pH 6.9, 200 mM potassium acetate, 1mM MgCl2, 1 mM DTT, 0.1 mM ATP, 10% glycerine, 0.1% Tween-20, 1 mM EGTA). The Flag-resin was incubated in a rotator for 30 min at room temperature with 150 µl wash buffer 2 containing either 1 mM HaloTag® Alexa Fluor®660 ligand (for labeling OSM-3G444EHalo) or 1 mM SNAP-Surface® Alexa Fluor® 647 or 488 (for labeling OSM-3SNAP and the corresponding IFT-B subunits). The Flag-resin was washed three times with 1 ml of wash buffer 2 and eluted with 70µl of elution buffer (wash buffer 2 containing 0.5 mg/ml of 1× Flag Peptide (Sigma)) for 60 min at 4°C32,33.
Size-exclusion chromatography coupled to multi-angle light scattering (SEC-MALS)
SEC-MALS analyses were used to determine the absolute molar masses of the protein complexes. The TSKgel G4000SWXL (Tosoh Biosciences) and the Superose 6 Increase 10/300 GL columns (GE Healthcare) were calibrated with at least 2 column volume of the gel filtration buffer (25mM PIPES, pH 7.0, 200 mM NaCl, 1mM MgCl2, 1mM EGTA, 1mM DTT). The columns were inline with a variable UV-absorbance detector (Agilent 1260 Infinity series) and a DAWN8+ MALS detector (Wyatt Technology). Molar masses were calculated with ASTRA 6 software (Wyatt Technology) with the dn/dc value set to 0.185 ml/g. Bovine serum albumin (BSA) was used as a calibration standard.
Microscale thermophoresis (MST) assays
OSM-3G444EHalo interaction with either the QCC or the QCC w/o DYF-1 was measured using MST with a Monolith NT.115 (NanoTemper Technologies GmbH). The fluorophore-labeled OSM-3G444EHalo at 10 nM was titrated with the respective (unlabeled) binding partner whose concentration varied between 0.2 nM and 2 µM in the MST buffer (50 mM tris-HCl (pH 7.6), 150 mM NaCl, 1 mM MgCl2, 0.1% Tween 20, 1 mM EGTA, 1 mM DTT, 10 % glycerine). Standard coated capillaries were used in all measurements. Kd values were determined using NanoTemper Analysis software.
Photobleaching experiments and single-molecule assays
Photobleaching experiments and single-molecule assays were conducted as described previously32,33. Briefly, after purifying the proteins as described above the respective motor was mixed with its corresponding IFT-B proteins in equimolar ratio and incubated overnight at 4°C in a rotator. To track the movement of the motor alone or in the presence of IFT-B proteins, microtubules were attached to the surface of a flow chamber (coated with 1 mg/ml biotinylated BSA and 1 mg/ml streptavidin (Sigma)) and the fluorescently labeled proteins were diluted to the desired concentration in BRB80 motility buffer (80 mM Pipes, pH 6.9, 10 mM ATP, 0.145 mg/ml glucose oxidase (Sigma), 0.0485 mg/ml catalase (Sigma), 20% glucose) and flowed into the chamber. Movement of the respective fluorescent signals was recorded with a cycle time of 235 ms with an objective-type Leica DMI6000 B TIRF microscope (Leica, Germany), equipped with a plan objective lens (100×, N.A. 1.47 Oil), and a back-illuminated Andor U897 EMCCD camera (Andor, UK). Excitation was achieved with the help of solid-state laser at 488 and 635 nm wavelength, and frames were recorded and analysed with AF 6000 software (Leica, Germany). The velocities and run lengths were analysed with custom-written programs using the MATLAB software (Mathworks Inc.). Spots were selected automatically according to their brightness compared with the mean brightness in each frame. The position of the spots was determined with subpixel accuracy using a radial center approach. Runs were considered processive with a minimal run length of 1 µm. The run length data were fit to a truncated (χ0 = 1µm) single-exponential distribution. Parts of the distance over time data were considered for speed calculation in a linear fit of at least six frames resulted in a r2-value > 95%. Landing events were counted from the data obtained by the tracking algorithm described above. Events that started after the first frame and showed a displacement from the binding position of at least 2 µm while associated with the microtubule were counted (n). As no processive movement was observed for the OSM-3Snap, runs were not selected for unidirectionality. Images of the microtubule positions were obtained from maximum intensity projections of the single molecule movies and their length (l) were measured using ImageJ. Considering the total length of a movie (t), the landing rate for a movie was calculated as e = n/l/t. This value was then corrected for dilution (c) of the motor (0.5) over the motor plus adapter (1) e_corr = e/c. The mean over three independent purifications of this value was calculated alongside the standard deviation and reported above. For the colocalised movements, runs were compared pairwise from the two respective channels. Weighted penalties resulted from the mean distances of tracked positions (pixel, factor 1/3) and the difference of the starting time (frames, factor 1). Runs with a penalty below 10 were considered and parameters are averages of the single runs.
Colocalisation assays
Proteins were mixed in equimolar concentrations, incubated overnight at 4°C and flowed into chambers as described above for single-molecule assays. Fluorescence was detected by TIRF illumination. The colocalised images were analysed using custom-written routines in MATLAB software (MathWorks).
Extended Data
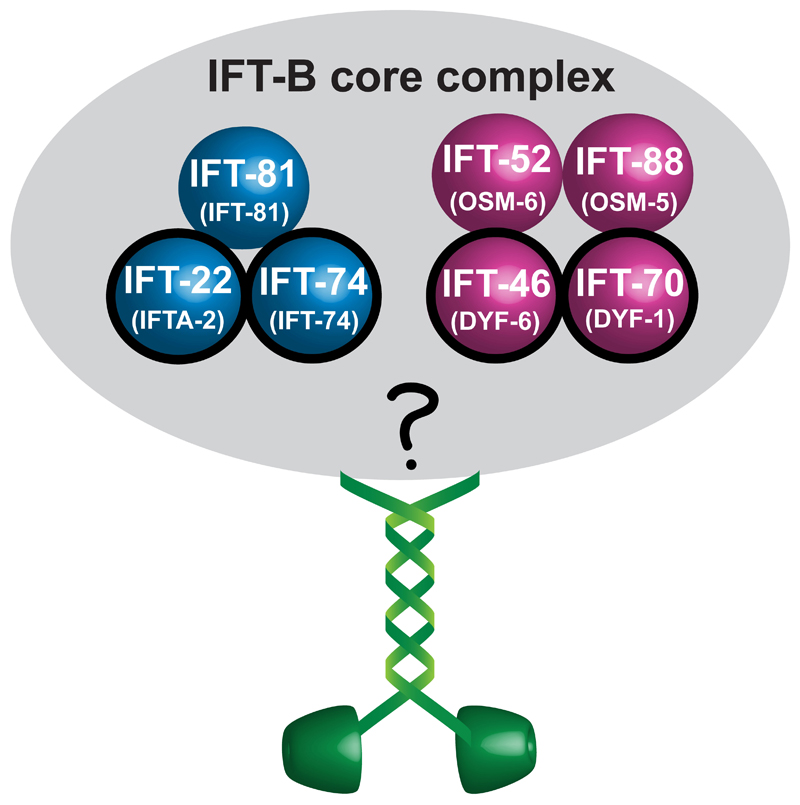
Extended Data Fig. 1. Overview of recombinant constructs and their proposed assembly in vitro.
Schematic representation of the presumptive IFT-B core complex from C. elegans (corresponding nomenclature of the subunits in C. elegans is shown in brackets). Subunits known to autonomously form sub-complexes in C. reinhardtii are colour-coded. The OSM-3 motor is shown in green. Subunits of the IFT-B core complex that are proposed to interfere with OSM-3 function in vivo are highlighted with black circles.
Extended Data Fig. 2. Interaction of OSM-3G444EHalo with QCC and TCC.
(a) Calculated molar mass of QCC subunits and the OSM-3G444EHalo motor along with their expected sum (left panel). The SDS-PAGE analyses of the respective elution peaks in Figure 1 (middle and right panels) show that OSM-3G444EHalo co-elutes with the complex in the presence (middle panel) but does not co-elute with the complex in the absence (right panel) of the IFT-70(DYF-1) subunit (TSKgelG4000SWXL column). (b) Overlay of the elution profiles of the TCC with and without the OSM-3G444EHalo motor and OSM-3G444EHalo alone (left panel). Note that the left shoulder of the (TCC + OSM-3G444EHalo) complex overlaps with the elution profile of the OSM-3G444E Halo motor, and the right shoulder with the TCC. Consistently, the molar masses determined for the (TCC + OSM-3G444EHalo) under peak 1 correspond to the OSM-3G444EHalo motor and peak 2 to the TCC, respectively (middle vs. right panels). (c) Left panel summarises the calculated molar mass of the TCC subunits and the OSM-3G444EHalo along with their expected sum. Middle and right panels show the SDS-PAGE analyses of the respective elution peaks shown above in (b). Consistent results obtained with three independent experiments. The identities of all subunits were confirmed by LC-MS/MS analysis. *Hsp70 protein.
Extended Data Fig. 3. Overview of the IFT-B subunits functionalised with C-terminal tags and their photobleaching properties.
The respective subunits were functionalised either with a GFP- or with a SNAP-tag for fluorescence labeling. All subunits displayed mostly single-step photobleaching consistent with non-aggregated, single subunits after functionalisation. N represents the number of events obtained from three different slides from three independent experiments.
Extended Data Fig. 4. Pairwise colocalisation efficiency of IFT-B subunits.
Pairs of differentially labeled subunits of the IFT-B sub-complexes were incubated and analysed for their colocalisation efficiency. The columns (bottom panels) represent the percentage of colocalised spots in the corresponding colocalised images (top panels). All assayed combinations of the labeled subunits displayed significant colocalisation efficiencies demonstrating that C-terminal functionalisation of the subunits does not interfere with their complex formation capabilities. Values presented as means of n=3 independent experiments ± SD; source data are provided online. Scale bar: 3µm. C. elegans nomenclature was used in the figure due to space limitations.
Extended Data Fig. 5. Colocalisation of the heterotrimeric KLP11/20/KAP motor with the IFT-B complex.
Neither QCC (top panel) nor TCC (bottom panel) of the IFT-B complex displayed efficient colocalisation with the KLP11/20/KAP motor. The IFT-81 subunit of TCC was fluorescently labeled via SNAP-tag and the IFT-52(OSM-6) subunit of QCC was GFP-tagged, respectively. Values presented as means of n=3 independent experiments ± SD; source data are provided online. Scale bar: 3 µm.
Extended Data Fig. 6. The IFT-70(DYF-1)-dependent activation does not alter the processivity of the OSM-3 motor.
(a) OSM-3G444EHalo, (b) OSM-3Flag, and (c) OSM-3SNAP motors display similar processivity that is independent of the presence of the IFT-70(DYF-1) subunit or the QCC. N represents the number of events obtained from three different flow chambers from n=3 independent experiments. Run length was fit to a single exponential ± confidence interval.
Extended Data Fig. 7. OSM-3SNAP motor containing the wild type stalk colocalised with QCC in an IFT-70(DYF-1)-dependent manner.
(a) Neither TCC nor QCC w/o DYF-1 displayed efficient colocalisation with the OSM-3SNAP motor. In the presence of the IFT-70(DYF-1) subunit on the other hand, the QCC efficiently colocalised with OSM-3SNAP (81±8%). (b) Consistently, OSM-3SNAP showed robust colocalisation (82±6%) with the IFT-70(DYF-1) subunit but not with IFT-52(OSM-6), IFT-88(OSM-5), and IFT-46(DYF-6) subunits. IFT-81 from TCC was fluorescently labeled with SNAP-tag and IFT-52(OSM-6) from QCC and QCC w/o DYF-1 were GFP-tagged, respectively. Data presented as means of n=3 independent experiments ± SD; source data are provided online. Scale bar: 3 µm. C. elegans nomenclature was used in the figure due to space limitations.
Supplementary Material
Acknowledgements
We thank Günther Woehlke (Technische Universität München), Erwin J. G. Peterman (Vrije Universiteit Amsterdam), Esben Lorentzen (Aarhus University), Michael Taschner (University of Lausanne) and Felix Müller-Planitz (Ludwig-Maximilians-Universität München) for fruitful discussions throughout this work and Thi-Hieu Ho (Technische Universität München) for excellent technical assistance. We are grateful to Angela Oberhofer (Technische Universität München) and Felix Müller-Planitz (Ludwig-Maximilians-Universität München) for critically reading the manuscript. This work was supported by European Research Council Grant 335623 (to Z.Ö.). We apologize to our colleagues whose work could not be cited due to space limitations.
Footnotes
Author contributions
M.A.A.M. and Z.Ö. designed the experiments. M.A.A.M. performed the experiments and analysed the data. W.L.S. wrote all the customized MATLAB routines. Z.Ö. and M.A.A.M. wrote the manuscript.
Reprints and permissions information is available at www.nature.com/reprints.
Competing financial interests
The authors declare no competing financial interests.
Data Availability
All data that support the findings of this study are available within the paper and its Supplementary Information including uncropped SDS-PAGE analyses of purified complexes, the detailed sequences of the motor constructs used in the study, source data for Figs. 2 and 3, ED Figs. 4, 5, and 7, and from the corresponding author upon reasonable request).
Code Availability
Custom-written codes used in this study are available from the corresponding author upon reasonable request.
References
- 1.Sharma N, Berbari NF, Yoder BK. Ciliary dysfunction in developmental abnormalities and diseases. Curr Top Dev Biol. 2008;85:371–427. doi: 10.1016/S0070-2153(08)00813-2. [DOI] [PubMed] [Google Scholar]
- 2.Pazour GJ, Rosenbaum JL. Intraflagellar transport and cilia-dependent diseases. Trends Cell Biol. 2002;12:551–555. doi: 10.1016/s0962-8924(02)02410-8. [DOI] [PubMed] [Google Scholar]
- 3.Marshall WF. The cell biological basis of ciliary disease. J Cell Biol. 2008;180:17–21. doi: 10.1083/jcb.200710085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- 6.Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prevo B, Scholey JM, Peterman EJG. Intraflagellar transport: mechanisms of motor action, cooperation, and cargo delivery. FEBS J. 2017;284:2905–2931. doi: 10.1111/febs.14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avidor-Reiss T, Leroux MR. Shared and Distinct Mechanisms of Compartmentalized and Cytosolic Ciliogenesis. Curr Biol. 2015;25:R1143–1150. doi: 10.1016/j.cub.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholey JM. Intraflagellar transport. Annu Rev Cell Dev Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- 10.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 11.Taschner M, Lorentzen E. The Intraflagellar Transport Machinery. Cold Spring Harb Perspect Biol. 2016;8:a028092. doi: 10.1101/cshperspect.a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snow JJ, et al. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- 13.Mukhopadhyay S, et al. Distinct IFT mechanisms contribute to the generation of ciliary structural diversity in C. elegans. EMBO J. 2007;26:2966–2980. doi: 10.1038/sj.emboj.7601717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans JE, et al. Functional modulation of IFT kinesins extends the sensory repertoire of ciliated neurons in Caenorhabditis elegans. J Cell Biol. 2006;172:663–669. doi: 10.1083/jcb.200509115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkins PM, et al. Ciliary targeting of olfactory CNG channels requires the CNGB1b subunit and the kinesin-2 motor protein, KIF17. Curr Biol. 2006;16:1211–1216. doi: 10.1016/j.cub.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 16.Leaf A, Von Zastrow M. Dopamine receptors reveal an essential role of IFT-B, KIF17, and Rab23 in delivering specific receptors to primary cilia. Elife. 2015;4:e06996. doi: 10.7554/eLife.06996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silverman MA, Leroux MR. Intraflagellar transport and the generation of dynamic, structurally and functionally diverse cilia. Trends Cell Biol. 2009;19:306–316. doi: 10.1016/j.tcb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Bae YK, Barr MM. Sensory roles of neuronal cilia: cilia development, morphogenesis, and function in C. elegans. Front Biosci. 2008;13:5959–5974. doi: 10.2741/3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bae YK, et al. General and cell-type specific mechanisms target TRPP2/PKD-2 to cilia. Development. 2006;133:3859–3870. doi: 10.1242/dev.02555. [DOI] [PubMed] [Google Scholar]
- 20.Ou G, Blacque OE, Snow JJ, Leroux MR, Scholey JM. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- 21.Hao L, et al. Intraflagellar transport delivers tubulin isotypes to sensory cilium middle and distal segments. Nat Cell Biol. 2011;13:790–798. doi: 10.1038/ncb2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ou G, et al. Sensory ciliogenesis in Caenorhabditis elegans: assignment of IFT components into distinct modules based on transport and phenotypic profiles. Mol Biol Cell. 2007;18:1554–1569. doi: 10.1091/mbc.E06-09-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burghoorn J, et al. Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of Caenorhabditis elegans. Proc Natl Acad Sci USA. 2007;104:7157–7162. doi: 10.1073/pnas.0606974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masyukova SV, et al. A Screen for Modifiers of Cilia Phenotypes Reveals Novel MKS Alleles and Uncovers a Specific Genetic Interaction between osm-3 and nphp-4. PLoS Genet. 2016;12:e1005841. doi: 10.1371/journal.pgen.1005841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taschner M, et al. Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J. 2016;35:773–790. doi: 10.15252/embj.201593164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taschner M, Bhogaraju S, Lorentzen E. Architecture and function of IFT complex proteins in ciliogenesis. Differentiation. 2012;83:S12–22. doi: 10.1016/j.diff.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piperno G, Mead K. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc Natl Acad Sci USA. 1997;94:4457–4462. doi: 10.1073/pnas.94.9.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cole DG, et al. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taschner M, Bhogaraju S, Vetter M, Morawetz M, Lorentzen E. Biochemical mapping of interactions within the intraflagellar transport (IFT) B core complex: IFT52 binds directly to four other IFT-B subunits. J Biol Chem. 2011;286:26344–26352. doi: 10.1074/jbc.M111.254920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imanishi M, Endres NF, Gennerich A, Vale RD. Autoinhibition regulates the motility of the C. elegans intraflagellar transport motor OSM-3. J Cell Biol. 2006;174:931–937. doi: 10.1083/jcb.200605179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prevo B, Mangeol P, Oswald F, Scholey JM, Peterman EJ. Functional differentiation of cooperating kinesin-2 motors orchestrates cargo import and transport in C. elegans cilia. Nat Cell Biol. 2015;17:1536–1545. doi: 10.1038/ncb3263. [DOI] [PubMed] [Google Scholar]
- 32.Oberhofer A, et al. Myosin Va's adaptor protein melanophilin enforces track selection on the microtubule and actin networks in vitro. Proc Natl Acad Sci USA. 2017;114:E4714–E4723. doi: 10.1073/pnas.1619473114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stepp WL, Merck G, Mueller-Planitz F, Okten Z. Kinesin-2 motors adapt their stepping behavior for processive transport on axonemes and microtubules. EMBO Rep. 2017;18:1947–1956. doi: 10.15252/embr.201744097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.