Abstract
Introduction
We have comprehensively described the expression profiles of mitochondrial DNA and nuclear DNA genes that encode subunits of the respiratory oxidative phosphorylation (OXPHOS) complexes (I–V) in the hippocampus from young controls, age matched, mild cognitively impaired (MCI), and Alzheimer’s disease (AD) subjects.
Methods
Hippocampal tissues from 44 non-AD controls (NC), 10 amnestic MCI, and 18 AD cases were analyzed on Affymetrix Hg-U133 plus 2.0 arrays.
Results
The microarray data revealed significant down regulation in OXPHOS genes in AD, particularly those encoded in the nucleus. In contrast, there was up regulation of the same gene(s) in MCI subjects compared to AD and ND cases. No significant differences were observed in mtDNA genes identified in the array between AD, ND, and MCI subjects except one mt-ND6.
Discussion
Our findings suggest that restoration of the expression of nuclear-encoded OXPHOS genes in aging could be a viable strategy for blunting AD progression.
Keywords: Aging, Alzheimer’s disease, Mild cognitively impaired (MCI), Mitochondria, Oxidative phosphorylation-related genes expression, Microarray, Postmortem brains
1. Introduction
Alzheimer’s disease (AD) is a progressive, degenerative brain disease and the most common cause of dementia [1,2]. AD is characterized by a gradual decline in cognitive function and the presence of pathologic inclusions including amyloid β (Aβ) plaques and neurofibrillary tangles [1]. The disease is preceded by a pre-symptomatic stage that can last for years during which time the clinical symptoms are undetectable. An intermediate stage between normal aging and AD has been identified as mild cognitive impairment. This disease state is characterized by problems with memory, language acquisition and processing, critical thinking, and judgment problems that are greater than normal age-related changes [2].
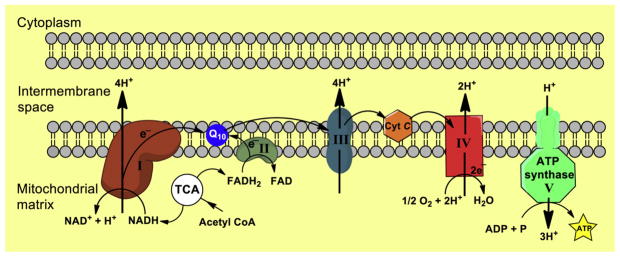
Bioenergetic failure and mitochondrial dysfunction in AD are well documented [2–10]. Several studies using multiple preclinical in vitro and in vivo AD models have demonstrated a decline in mitochondrial function before the development of AD pathology [2–8]. Mitochondria are key organelles that regulate a multitude of metabolic and signaling pathways including programmed cell death [11–15]. The primary function of mitochondria is to produce ATP through the process of oxidative phosphorylation (OXPHOS), which is regulated through four respiratory multi-subunit enzyme complexes (complexes I–IV) and ATP synthase (complex V), all located in the inner mitochondrial membrane (Fig. 1) [16–18]. Mammalian mitochondrial DNA (mtDNA) is a maternally inherited, double-stranded circular genome of approximately 16.6 kb [19–21]. It contains 37 genes encoding 13 protein subunits of enzymes involved in oxidative phosphorylation, two ribosomal RNAs, and 22 transfer RNAs necessary for translation of these proteins. The remaining subunits are encoded by the nuclear genome, synthesized in the cytosol, and subsequently imported into mitochondria through protein translocation machineries of the outer and inner membranes [19–21].
Fig. 1.
Mitochondrial oxidative phosphorylation system (OXPHOS) shown: The five respiratory chain complexes with the corresponding electron transport chain numbers (I–V). In the bottom part, these are indicated by the subunits of each respiratory chain complex gene numbers, which were selected by microarray analysis. Among the ~80 polypeptides constituting the electron transport chain, 13 are encoded by mtDNA, and the rest are all encoded by nDNA, synthesized in the cytosol, and translocated to the mitochondria. TCA, trichloroacetic acid; Cyt C, cytochrome C; ADP, adenosine diphosphate; ATP, adenosine triphosphate.
A critical role of mitochondrial dysfunction has been hypothesized in both aging and neurodegenerative diseases [2–10]. Numerous studies use animal models based on genetic mutations found in rare early onset familial AD cases that represent <1% of AD patients [1]. Recently, it has been demonstrated that mitochondrial bioenergetic deficits precede AD pathology in the female triple transgenic mouse model of AD (3xTgAD) [4]. Converging lines of evidence indicate that mitochondria are direct targets of Aβ [22–24], and that Aβ is directly responsible for impaired electron transport chain function [22–28]. Accumulation of Aβ in neurons is believed to be an essential step leading to Aβ-mediated mitochondrial dysfunction and contributes to energy failure, neuronal apoptosis, and production of reactive oxygen species (ROS) in AD brain tissue [25–29]. Mitochondrial dysfunction and oxidative damage occur early in the course of disease, before the onset of significant plaque pathology, and act causally in disease pathogenesis [4,30]. Normal aging and AD are both marked by prominent defects in brain metabolism and increased oxidative stress. Although inheritance of certain susceptibility genes increases the risk of the disease, aging is the most prominent risk factor for the non-Mendelian sporadic AD which affects most patients diagnosed for dementia over the age of 60 years [31]. Several studies have provided evidence of neuronal metabolic impairments at the transcript and protein level in AD brain, which was ascribed to the down regulation of mitochondrial-associated genes; in particular, oxidative phosphorylation genes [27,32–37]. Following this line of thought, we present here a microarray-based study that focuses specifically on the role of OXPHOS-related genes in aging and in AD.
It is clear from the literature that the etiology of AD is not completely understood; hence, the use of a wide array of animal and cellular models to address the pathological process of the early onset, dominantly inherited familial form of this disease. Although these model system(s) do feature particular aspects of the disease process, no model has yet fully recapitulated the human disease. There is much evidence that late onset AD overlaps with normal aging in many clinical and pathologic features [38–43]. Therefore, the availability of human postmortem brain tissue is of great importance for biological studies on human brain aging and the progression to disease. These unique tissue specimens make it possible to investigate potential gene expression alterations before, during, and after the onset of AD. In this study, we compared expression profiles of mitochondrial genes (e.g., OXPHOS) from the hippocampus from clinically and pathologically confirmed AD cases, young controls, age-matched controls, and MCI cases.
2. Methods
Frozen unfixed hippocampus was obtained from 44 non-AD controls (age, 20–99 years), 10 amnestic mild cognitively impaired cases, and 18 sporadic AD cases (age, 74–95 years; Table 1) from seven nationally recognized brain banks. Inclusion criteria can be found in Berchtold et al. [38]. Total RNA was extracted from heterogeneous population across hippocampal subfields as described previously [38], and RNA quality was assessed using the Agilent Bio-Analyzer (Agilent Technologies, Palo Alto, CA). Average RIN for the samples was 8.29 (SD, 0.775; range, 6.7–9.6), with 93% of the cases (67 of 72) having RIN > 7. There were no significant differences in RNA integrity across groups [39]. Seventy-two hippocampal tissue samples were individually hybridized to Affymetrix HgU133 plus 2.0 arrays.
Table 1.
Description of subjects studied
| Group# ()*S | Sex ()*S | Braak ()*S | Age ()*S |
|---|---|---|---|
| Young control | Male | unknown | 20 |
| Young control | Male | unknown | 20 |
| Young control | Male | unknown | 20 |
| Young control | Male | unknown | 22 |
| Young control | Female | unknown | 26 |
| Young control | Male | unknown | 28 |
| Young control | Female | unknown | 30 |
| Young control | Male | unknown | 33 |
| Young control | Female | unknown | 34 |
| Young control | Female | unknown | 37 |
| Young control | Male | unknown | 42 |
| Young control | Female | unknown | 44 |
| Young control | Male | unknown | 45 |
| Young control | Female | unknown | 45 |
| Young control | Male | unknown | 45 |
| Young control | Female | unknown | 47 |
| Young control | Female | unknown | 48 |
| Young control | Male | unknown | 52 |
| Age match control | Female | 0 | 64 |
| Age match control | Male | 2.5 | 69 |
| Age match control | Male | 1 | 69 |
| Age match control | Male | 1 | 69 |
| Age match control | Female | 1 | 70 |
| Age match control | Female | 2 | 74 |
| Age match control | Female | 2 | 74 |
| Age match control | Female | 0 | 74 |
| Age match control | Male | 2 | 75 |
| Age match control | Male | 2 | 80 |
| Age match control | Male | 3 | 82 |
| Age match control | Female | 3 | 82 |
| Age match control | Female | 2 | 82 |
| Age match control | Male | 2 | 83 |
| Age match control | Female | 2 | 83 |
| Age match control | Male | 2 | 84 |
| Age match control | Male | 2 | 85 |
| Age match control | Male | 1 | 86 |
| Age match control | Female | 3 | 87 |
| Age match control | Male | unknown | 91 |
| Age match control | Female | unknown | 91 |
| Age match control | Female | 2 | 91 |
| Age match control | Male | 1 | 95 |
| Age match control | Female | 4 | 96 |
| Age match control | Male | 2 | 97 |
| Age match control | Female | unknown | 99 |
| MCI | Female | 4 | 74 |
| MCI | Male | 4 | 75 |
| MCI | Female | 1.5 | 83 |
| MCI | Male | 2 | 85 |
| MCI | Male | 3 | 86 |
| MCI | Female | 3 | 88 |
| MCI | Female | 3 | 88 |
| MCI | Male | 3 | 88 |
| MCI | Male | 4 | 89 |
| MCI | Female | 4 | 89 |
| AD | Female | 6 | 60 |
| AD | Female | 6 | 74 |
| AD | Male | 5 | 76 |
| AD | Female | 5 | 76 |
| AD | Male | 3 | 76 |
| AD | Male | 6 | 79 |
| AD | Female | 6 | 79 |
| AD | Male | unknown | 80 |
| AD | Male | 4 | 85 |
| AD | Male | 3 | 85 |
| AD | Male | 4 | 86 |
| AD | Male | 5 | 87 |
| AD | Female | 6 | 90 |
| AD | Female | 4 | 90 |
| AD | Female | 3 | 90 |
| AD | Female | 5.5 | 91 |
| AD | Male | 6 | 94 |
| AD | Male | 4 | 94 |
Abbreviations: MCI, mild cognitively impaired; AD, Alzheimer’s disease.
Samples were randomized across batch runs, and no batch effects were detected. Guanine-cytosine robust multi-assay analysis (GC-RMA) expression values were calculated, followed by per-chip and per-gene normalization, log-transformation of the geometric mean of expression using Gene-Spring 7.3.1 software (Agilent Technologies, Palo Alto, CA). The data underwent the following normalization steps: (1) per chip: normalize to 50th percentile, (2) per gene: normalize to median. Genes were not normalized to “housekeeping” genes. Eighty-six mitochondrial genes, 41 of 44 from complex I, four of four from complex II, eight of ten from complex III, 16 of 19 from complex IV, and 17 of 19 from complex V, were investigated for statistical significance (P < .01) in aging comparing young (n = 18, age 20–59 years, mean age 35.4 ± 10.9 years) versus aged controls (n = 26, age 69–99 years, mean age 82 ± 9.8 years; Table 1). AD/MCI-related gene expression changes were based on the comparison of aged controls (n = 26; age, 69–99 years; mean age, 82 ± 9.8 years), versus mild cognitive impairment (n = 10, mean age 84.5 ± 5.6 years) versus AD (n = 18, mean age 83 ± 8.7 years). A complete list of genes and descriptions can be found in Supplementary Table 1.
We preferentially selected those probe sets that were most specific, such that they were annotated with the smallest number of Ensembl gene IDs. After applying this criterion, if there remained multiple probe sets for any one gene, we excluded those probe sets expected to hybridize with targets in a nonspecific fashion (i.e., those with “_x_” in the Affymetrix identifier), which was the case for seven of the OX-PHOS gene. If there still remained multiple probe sets for a given gene, we took the mean of both probe sets.
2.1. Statistical analysis
Analysis between individuals, groups, and disease status were performed. All genes that did not meet the 50% present call threshold were removed by gene-spring G 7.3.1 Expression Analysis software. A two-tailed paired t test, assuming equal variance (using multiple testing corrections, by Benjamin and Hochberg False Discovery Rate) was applied to locate genes that were significant (P = .05 or less) in differentiating expression between AD, ND, and MCI cases. The results presented here are, of course, subject to the limitations of microarray-based approaches. These include false discovery resulting from multiple statistical tests of significance, although this source of error was limited by the use of two different analytical methods to validate the gene lists, as well as conventional validation by RT-PCR. Taqman primers (Thermo Fisher Scientific) were designed to six randomly chosen mitochondrial transcripts MT-ND1 (Hs02596873_s1), NDUFA6 (Hs00899690_m1), NDUFC2 (Hs01072843_m1), UQCRC1 (Hs00429571_g1), ATP5D (Hs00961521_m1), and MT-ATP6 (Hs02596862_g1). All six transcripts showed the same pattern of expression identified in the array.
3. Results
3.1. Aging and AD-related changes in gene expression levels of OXPHOS-related genes from hippocampal autopsy tissue
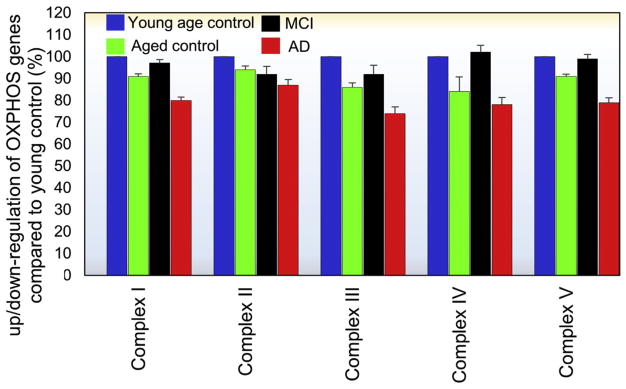
Aging is the most salient risk factor for cognitive decline in the elderly and the predominant risk factor for late-onset of AD. Thus, in this study, an aging cohort was included for a better understanding of age-related effects on the expression of OXPHOS-related genes. Young controls (20–59 years) and age-matched controls (69–99 years) were included as shown in the demographic details (Table 1). Our microarray data revealed age-related alterations of the OXPHOS-related genes which encode mitochondrial respiratory chain complexes I, II, III, IV, and V. Ninety-five percent of the identified genes were down regulated in aging and 100 percent of genes in AD, particularly those which were nuclear encoded (Fig. 2). These data indicate that aging may be a contributing factor for the decline in OXPHOS-related gene observed in AD. This is in agreement with the previous finding that aging resulted in down-regulation of genes in the respiratory chain complexes I, III, IV, and V in the brains of aged C57/BL6 mice [38]. In the context of aging, our results (Fig. 2), a general trend can be observed in complexes I, III, IV, and V; as age increases the mean expression of OXPHOS-related genes decrease, a phenomenon which is exacerbated in AD. Correspondingly, in these same complexes (I, III, IV, and V), MCI cases demonstrate an unequivocal increase in these same OXPHOS-related genes, suggesting a compensatory-like mechanism at the early stages of disease. An exception to this general trend is observed in complex II where only a modest difference can be observed across groups. To determine whether a significant correlation was present between Braak stage and a decrease in OXPHOS-related transcripts in AD vs. aged controls, we ran a Pearson’s correlation test. Accordingly, a significant (P <.001, r = .78) correlation was observed between Braak stage and a decrease in OXPHOS-related transcripts, indicating that disease-related pathology may account, to some extent, for the decrease in OXPHOS-related transcripts in AD.
Fig. 2.
Mean expression of oxidative phosphorylation system (OXPHOS)–related genes within complexes I–V in the hippocampus compared to young controls (20–52 years). As a function of age, a general trend can be observed in complexes I, III, IV, and V; as one’s age increases, the mean expression of OXPHOS-related genes decreases, a phenomenon which is exacerbated in AD. Correspondingly, in these same complexes (I, III, IV, and V), MCI cases demonstrate an unequivocal increase in these same OXPHOS-related genes, suggesting a compensatory-like mechanism at the early stages of disease. An exception to this general trend is observed in complex II where only a modest difference can be observed across groups. Error bars generated using standard error of the mean. MCI, mild cognitively impaired; AD, Alzheimer’s disease.
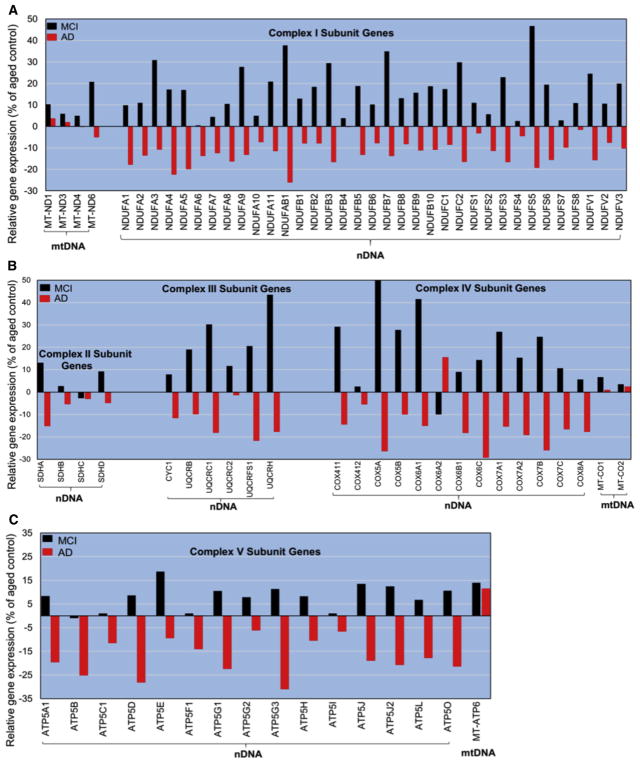
Relative gene expression profiles of OXPHOS-related genes from hippocampal tissue of young, MCI, AD, and age-matched controls were subdivided into two groups: nuclear-encoded genes and mitochondrial-encoded genes (Fig. 3). Seventy of the 73 nuclear-encoded genes and six of the seven mitochondrial-encoded genes identified in the Affymetrix array showed a robust correspondence in the overall direction in the expression levels (e.g., up/down regulation) within groups (Fig. 3A–C). As such, in complex I (3a), AD cases showed significantly less expression in all 37 OXPHOS-related genes, compared to control, where MCI cases showed a significant increase in all 37 genes. In contrast, MT-ND6 gene (see Fig. 3A) was the only mitochondrial-encoded transcript identified in the array that was significantly upregulated in MCI and down regulated in AD, reflecting the same trend as nuclear-encoded OXPHOS genes. Interestingly, all other OXPHOS mitochondrial-encoded genes identified proved to be equivalent to control and appear to be unaffected in the disease progression. Similar findings were observed in complexes II–IV (Fig. 3B) and complex V (Fig. 3C). This finding suggests that, in AD, aging is also an important contributing factor in disease progression, consistent with previous reports [39–43]. The proportion of significantly expressed OXPHOS-related genes in the hippocampal sampled tissue for the different groups in this study is shown in Supplementary Table 2. The three nuclear-encoded and one mitochondrial-encoded transcript not included in the analysis were removed because they were found to hybridize to gene targets in a nonspecific fashion (i.e., those with “_x_” in the Affymetrix identifier).
Fig. 3.
Relative gene expression profiles of oxidative phosphorylation system (OXPHOS)–related genes from hippocampal tissue are shown in, and grouped by, OXPHOS complexes, compared to age-matched controls (62–99 years). Up regulation/down regulation of OXPHOS-related gene(s) within complex I relative to aged controls (A). Up regulation/down regulation of OXPHOS-related gene(s) within complex II–IV relative to aged controls (B). Up regulation/down regulation of OXPHOS-related gene(s) within complex V relative to aged controls (C). The nDNA-encoded OXPHOS-related genes profile was significantly reduced in AD compared to CTL for all complexes, where MCI cases show a significant increase in all 37 genes (a significant P value of <.05 (two-tailed student t test), graphs a, b, and c). Mitochondrial-encoded transcript showed no significant difference from control for all complexes with one exception (MT-ND6). MCI, mild cognitively impaired; Alzheimer’s disease; MT-ND1, mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 1; MT-ND3, NADH dehydrogenase, subunit 3 (complex I); MT-ND4, Homo sapiens DC24 mRNA, complete cds.; MT-ND6, NADH dehydrogenase, subunit 6 (complex I); NDUFA, NADH dehydrogenase (ubiquinone) 1 alpha subcomplex; NDUFAB1, NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex; NDUFB, NADH dehydrogenase (ubiquinone) 1 beta subcomplex; NDUFC, NADH dehydrogenase (ubiquinone) 1, subcomplex unknown; NDUFS, NADH dehydrogenase (ubiquinone) Fe-S protein; NDUFV, NADH dehydrogenase (ubiquinone) flavoprotein; SDH, succinate dehydrogenase; CYC1, cytochrome c-1; UQCR, ubiquinol-cytochrome c reductase; COX, cytochrome c oxidase; MTC, mitochondrial electron transport chain; ATP, adenosine triphosphate.
4. Discussion
Evidence has accumulated over decades that mitochondria play a role in the metabolic deficits of AD. Indeed, studies using the same data set revealed a global decrease in many mitochondrial transcripts in AD [39]. A recent PubMed search of “Mitochondria and Alzheimer’s” yielded 1908 hits. Among the significant proponents of the “mitochondrial cascade hypothesis” have been RH Swerdlow [44] and MF Beal [45]. More recent work has shown that mitochondrial dysfunction is, in fact, an early event in AD that is detectable in “preclinical” AD CSF [46] and in imaging data [47]. Recently, a nonamyloid driven hypothesis known as the “inverse Warburg effect” has drawn significant attention in the field. The theory behind the inverse Warburg effect is that AD is the result of a cascade of three events—mitochondrial dysregulation, metabolic reprogramming (the Inverse Warburg effect), and natural selection [48,49]. Although these papers among others make valid arguments in proving there hypothesis, none are absolute. The one absolute finding among all these articles are that mitochondria are affected in the progression of AD, and here, we provide evidence of mitochondrial gene expression changes as a function of age and complement some studies and argue against others.
Herein, we present microarray data in brain that demonstrate significant down regulation of nuclear-encoded mitochondrial genes within respiratory chain complexes in aging, and in AD, but not in mitochondrial-encoded genes. Additionally, we show that these same genes are over-expressed in amnestic MCI cases; which is suggestive of a compensatory-like mechanism in these subjects. Differential expression of OXPHOS-related transcripts could have deleterious effects on ATP production, providing further insight into preclinical strategies for therapy.
4.1. Early AD is related to selected cellular phenomena
Deficits in several cellular events have been shown to occur early in the course of AD cellular pathology. These include metabolic deficits [7,50], altered synaptic structure and function [51], altered phosphorylation and configuration of tau [52], and alterations in the endocytic/autophagy system [53]. Relatively recently, protein misfolding has received more attention [54,55]. In addition, we have recently shown that intracellular redistribution of the key epigenetic molecule, H3K4me3, is also an early event in intracellular AD pathology, indicating that epigenetic regulation of gene expression may be an early event in AD, capable of modulating expression of a wide variety of the molecules attributed to early AD, notably synapses [56].
4.2. Prospective causal mechanism for the decrease in nuclear-encoded OXPHOS genes in AD
We have previously proposed a model of AD pathophysiology in which oligomeric Aβ disrupts the exchange of molecules between the cell nucleus and the cytoplasm [57]. Disruption of this exchange then jeopardizes critical functions of the cell leading to selected phenotypes of AD [58]. The present data showing the effect of AD on expression of nuclear-encoded but not mitochondrial-encoded OXPHOS genes identifies another example of nucleocytoplasmic dysfunction in AD. Because nuclear-encoded mitochondrial proteins require transport molecules for delivery, and mitochondrial-encoded proteins do not, this may implicate the exchange process from the cell nucleus to the mitochondria in AD.
Although other OXPHOS expression changes and causal mechanism have been reported [36], we believe our data set is more comprehensive and addresses the need to analyze the entire mitochondrial transcriptome. Although our data contradicts work by Chandrasekaran et al. [36], we stress the importance of sample size and more importantly brain regions. It was reported that a 50%–60% reduction in mRNA levels of nDNA-encoded subunit IV of COX (COX411) and β-subunit of the F0F1-ATP synthase (ATP5B) in the mid temporal cortex in AD compared to control brains. A single mt-DNA-encoded gene was used in their study (mt-ND1) and showed a similar reduction (50%) in mRNA levels in AD brain tissue when compared to control. It is very difficult to compare the two analysis when only two nDNA-OXPHOS-encoded genes and one mt-DNA-encoded gene were used in their study. Whereas, in our study, 98% of OXPHOS encoded genes and 65% of mt-DNA genes were used for analysis. The cause for the discrepancy in the mt-DNA-encoded gene is unclear but may reflect the different brain regions studied, the small sample size, and the different method used. We know for example that the hippocampus is particularly vulnerable in AD [59], it may be that the amount of pathology is directly correlated to mitochondrial gene expression levels, which we found in our study as a strong correlate (P < .001, r = 0.78). We expect, however, that other, particularly less-affected brain regions would yield different results.
4.3. Compensatory mechanisms are initiated in mild cognitively impaired subjects
Compensatory mechanisms are not new to the field of aging or age-related diseases [33,40,60,61]. It has been postulated that compensatory mechanisms like the one we show (e.g., up regulation of mitochondrial genes in MCI) are largely the product of survival and protection from disease progression. These processes are innately activated due to various brain insults (e.g., oligomeric amyloid β [OAβ], inflammation, ROS production, among others).
Severe metabolic deficit has been shown to be a prominent feature of AD in human brain [44,62], as well as in vitro [63] and animal models [4,64]. In vitro [65], mouse model [6,7] and human studies [5] reveal that Aβ and OAβ are both toxic to mitochondria, suggesting that Aβ may be responsible for the metabolic deficit observed in AD. Although they do not represent the exclusive source of cellular ATP, mitochondria are the primary source of ATP in brain cells and provide the energy required to perform metabolic processes from cell signaling to calcium homeostasis [66]. Deficiencies in energy metabolism have been shown to be one of the earliest events in AD pathobiology [2,33]. Although a number pathogenic pathways have the ability to affect mitochondrial function, the amyloid precursor protein and Aβ are at the forefront [67]. Progressive accumulation of Aβ in the mitochondria of AD neurons is believed to be an essential step leading to Aβ-mediated mitochondrial dysfunction, and several studies provide substantial evidence that mitochondria contain Aβ in AD [21–23,28,68], and that Aβ is directly responsible for impaired respiratory chain function [69–71]. It is interesting that, although complex I inhibition is believed to result from direct interaction with Aβ, impairment of complex IV function results primarily from ROS production at complex I [63]. The recent emergence of a putatively more toxic form of Aβ, oligomeric Aβ (OAβ), has prompted several groups to evaluate mitochondrial function in the presence of OAβ. Collectively, they have found that OAβ permeated mitochondrial membranes [71], increased reactive oxygen species levels [72], and impaired ATP production [73]. These data reinforce the toxicity of OAβ on mitochondrial function.
4.4. Potential significance of the only affected mitochondrially encoded transcript identified in the Affymetrix array
NADH: ubiquinone oxidoreductase core subunit 6 (mt-ND6) was the only of the seven identified mitochondrially encoded transcripts affected in AD. The reason for this alteration in mt-ND6 (5% decrease in AD) is unclear but quite intriguing. It is important to note that the mt-ND6 is the only protein-encoding gene present in the L strand of mtDNA, whereas the rest of the 13 protein-encoding mitochondrial genes all reside in the H strand. Furthermore, the mtDNA-encoded subunits are transcribed in a polycistronic fashion and are regulated by the same promoter (excluding mt-ND6) [74]. Recently, it has been found that the myocyte enhancer factor 2 (MEF2) family member (nuclear transcription factor), exclusively regulates expression of the mt-ND6 gene without significant effects on several other protein-encoding mtDNA genes [75]. It is important to note that the full biological significance of this added layer of regulation of mt-ND6 gene expression by MEF2D requires further exploration.
Although select papers make a distinction between the amyloid hypothesis and hypotheses emphasizing bioenergetics (e.g. Demetrius et al., 2015), we believe that our data presented here and in other papers references suggest a blending of the two classes of hypotheses in which an age-related decline in bioenergetics is exacerbated by OAβ. Our data in this and other articles suggest a model in which actions of OAβ lead to early events in the cascade of AD pathologies which directly impedes mitochondrial function. Thus, these findings suggest that restoration of mitochondrial function could blunt disease progression, and that a focused molecular strategy may be applicable to a range of neurodegenerative diseases, notably AD. A novel type of catalytic antioxidant is suggested for possible therapeutic application, and its mechanism(s) for blunting neurodegeneration and cell death should be explored. As such, we have recently described the ability of novel co-enzyme Q10 analogs to reverse the effect of OAβ-induced mitochondrial dysfunction [76]. We show that treatment with these novel compounds also leads to restoration of mitochondrial function as observed in nonaffected middle aged or younger individuals. Although enhancing ATP production would likely be beneficial to the aging population in general, other significant factors such as mitochondria half-life and abundance and changes in protein expression may or may not result in functional alterations at the protein level. Detailed analysis of compensatory mechanisms as shown here might be a starting point for further treatment and diagnostic strategies to combat AD.
Supplementary Material
RESEARCH IN CONTEXT.
Systemic review: Alzheimer’s disease (AD) is a complex disease with a prolonged trajectory of etiopathogenic changes in brain bioenergetics decades before the onset of measureable cognitive deficits.
Interpretation: Increasing evidence suggests that alterations in energy metabolism are among the earliest events that occur in AD. Previous studies, including our own, have identified expression changes in OX-PHOS genes in AD, but our detailed expression array study identifies deficits in nuclear not mitochondrial-encoded transcripts.
Future directions: To elucidate the specific mechanism(s) by which OAβ interferes with the transport of nuclear-derived mitochondrial mRNAs, and to demonstrate that these mechanisms are necessary and sufficient to explain the mitochondrial dysfunction observed in aging and AD.
Acknowledgments
The authors declare no competing financial or conflict of interests. We are grateful to the Banner Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona for the provision of human brain samples. This work was supported by NIRG-15-321390 and ADHS16-104646 FY2015-16 to D.M. Supported by RO1 to C.C. and 1R01AGO36400 to P.D.C.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jalz.2016.09.003.
References
- 1.Alzheimer’s Association. 2015 Alzheimer’s disease facts and figures. Alzheimers Dement. 2015;11:332–84. doi: 10.1016/j.jalz.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Hauptmann S, Scherping I, Dröse S, Brandt U, Schultz KL, Jendrach M, et al. Mitochondrial dysfunction: an early event in Alzheimer pathology accumulates with age in AD transgenic mice. Neurobiol Aging. 2009;30:1574–86. doi: 10.1016/j.neurobiolaging.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Du H, Guo L, Yan S, Sosunov AA, McKhann GM, Yan SS. Early deficits in synaptic mitochondria in an Alzheimer’s disease mouse model. Proc Natl Acad Sci U S A. 2010;107:18670–5. doi: 10.1073/pnas.1006586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao J, Irwin RW, Zhao L, Nilsen J, Hamilton RT, Brinton RD. Mitochondrial bioenergetic deficit precedes Alzheimer’s pathology in female mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2009;106:14670–5. doi: 10.1073/pnas.0903563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy PH, Tripathi R, Troung Q, Tirumala K, Reddy TP, Anekonda V, et al. Abnormal mitochondrial dynamics and synaptic degeneration as early events in Alzheimer’s disease: implications to mitochondria-targeted antioxidant therapeutics. Biochim Biophys Acta. 2012;1822:639–49. doi: 10.1016/j.bbadis.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie H, Guan J, Borrelli LA, Xu J, Serrano-Pozo A, Bacskai BJ. Mitochondrial alterations near amyloid plaques in an Alzheimer’s disease mouse model. J Neurosci. 2013;33:17042–51. doi: 10.1523/JNEUROSCI.1836-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leuner K, Hauptmann S, Abdel-Kader R, Scherping I, Keil U, Strosznajder JB, et al. Mitochondrial dysfunction: the first domino in brain aging and Alzheimer’s disease? Antioxid Redox Signal. 2007;9:1659–75. doi: 10.1089/ars.2007.1763. [DOI] [PubMed] [Google Scholar]
- 8.Pedrós I, Petrov D, Allgaier M, Sureda F, Barroso E, Beas-Zarate C, et al. Early alterations in energy metabolism in the hippocampus of APPswe/PS1dE9 mouse model of Alzheimer’s disease. Biochim Biophys Acta. 2014;1842:1556–66. doi: 10.1016/j.bbadis.2014.05.025. [DOI] [PubMed] [Google Scholar]
- 9.Calkins MJ, Manczak M, Mao P, Shirendeb U, Reddy PH. Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of Alzheimer’s disease. Hum Mol Genet. 2011;20:4515–29. doi: 10.1093/hmg/ddr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parihar MS, Brewer GJ. Mitoenergetic failure in Alzheimer’s disease. Am J Physiol Cell Physiol. 2007;292:C8–23. doi: 10.1152/ajpcell.00232.2006. [DOI] [PubMed] [Google Scholar]
- 11.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 12.Graier WF, Frieden M, Malli R. Mitochondria and Ca2+ signaling: old guest, new functions. Pflugers Arch. 2007;455:375–96. doi: 10.1007/s00424-007-0296-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bras M, Queenan B, Susin SA. Programmed cell death via mitochondria different modes of dying. Biochemistry (Mosc) 2005;70:231–9. doi: 10.1007/s10541-005-0105-4. [DOI] [PubMed] [Google Scholar]
- 14.Pradelli LA, Beneteau M, Ricci JE. Mitochondrial control of caspase-dependent and -independent cell death. Cell Mol Life Sci. 2010;67:1589–97. doi: 10.1007/s00018-010-0285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henze K, Martin W. Evolutionary biology: essence of mitochondria. Nature. 2003;426:127–8. doi: 10.1038/426127a. [DOI] [PubMed] [Google Scholar]
- 16.Saraste MW. Oxidative phosphorylation at the fin de siècle. Science. 1999;283:1488–93. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–8. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 18.Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 2003;112:481–90. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 19.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–65. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 20.Attardi G, Chomyn A, Montoya J, Ojala D. Identification and mapping of human mitochondrial genes. Cytogenet Cell Genet. 1982;32:85–98. doi: 10.1159/000131689. [DOI] [PubMed] [Google Scholar]
- 21.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–8. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 22.Manczak M, Anekonda TS, Henson E, Park BS, Quinn J, Reddy PH. Mitochondria are a direct site of A beta accumulation in Alzheimer’s disease neurons: implications for free radical generation and oxidative damage in disease progression. Hum Mol Genet. 2006;15:1437–49. doi: 10.1093/hmg/ddl066. [DOI] [PubMed] [Google Scholar]
- 23.Cha MY, Han SH, Son SM, Hong HS, Choi YJ, Byun J, et al. Mitochondria-specific accumulation of amyloid beta induces mitochondrial dysfunction leading to apoptotic cell death. PLoS One. 2012;7:e34929. doi: 10.1371/journal.pone.0034929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhein V, Song X, Wiesner A, Ittner LM, Baysang G, Meier F, et al. Amyloid-beta and tau synergistically impair the oxidative phosphorylation system in triple transgenic Alzheimer’s disease mice. Proc Natl Acad Sci U S A. 2009;106:20057–62. doi: 10.1073/pnas.0905529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker WD, Parks JK, Filley CM, Kleinschmidt-DeMasters BK. Electron transport chain defects in Alzheimer’s disease brain. Neurology. 1994;44:1090–6. doi: 10.1212/wnl.44.6.1090. [DOI] [PubMed] [Google Scholar]
- 26.Rhein V, Baysang G, Rao S, Meier F, Bonert A, Müller-Spahn F, et al. Amyloid-beta leads to impaired cellular respiration, energy production and mitochondrial electron chain complex activities in human neuroblastoma cells. Cell Mol Neurobiol. 2009;29:1063–71. doi: 10.1007/s10571-009-9398-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandrasekaran K, Hatanpaa K, Brady DR, Rapoport SI. Evidence for physiological down-regulation of brain oxidative phosphorylation in Alzheimer’s disease. Exp Neurol. 1996;142:80–8. doi: 10.1006/exnr.1996.0180. [DOI] [PubMed] [Google Scholar]
- 28.Caspersen C, Wang N, Yao J, Sosunov A, Chen X, Lustbader JW, et al. Mitochondrial Abeta: a potential focal point for neuronal metabolic dysfunction in Alzheimer’s disease. FASEB J. 2005;19:2040–1. doi: 10.1096/fj.05-3735fje. [DOI] [PubMed] [Google Scholar]
- 29.Nunomura A, Perry G, Aliev G, Hirai K, Takeda A, Balraj EK, et al. Oxidative damage is the earliest event in Alzheimer disease. J Neuropathol Exp Neurol. 2001;60:759–67. doi: 10.1093/jnen/60.8.759. [DOI] [PubMed] [Google Scholar]
- 30.Pratico D, Uryu K, Leight S, Trojanoswki JQ, Lee VM. Increased lipid peroxidation precedes amyloid plaque formation in an animal model of Alzheimer amyloidosis. J Neurosci. 2001;21:4183–7. doi: 10.1523/JNEUROSCI.21-12-04183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin MT, Simon DK, Ahn CH, Kim LM, Beal MF. High aggregate burden of somatic mtDNA point mutation in aging and Alzheimer’s disease brain. Hum Mol Genet. 2002;11:133–45. doi: 10.1093/hmg/11.2.133. [DOI] [PubMed] [Google Scholar]
- 32.Liang WS, Reiman EM, Valla J, Dunckley T, Beach TG, Grover A, et al. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci U S A. 2008;105:4441–6. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manczak M, Park BS, Jung Y, Reddy PH. Differential expression of oxidative phosphorylation genes in patients with Alzheimer’s disease: implications for early mitochondrial dysfunction and oxidative damage. Neuromolecular Med. 2004;5:147–62. doi: 10.1385/NMM:5:2:147. [DOI] [PubMed] [Google Scholar]
- 34.Chandrasekaran K, Giordano T, Brady DR, Stoll J, Martin LJ, Rapoport SI. Impairment in mitochondrial cytochrome oxidase gene expression in Alzheimer disease. Brain Res Mol Brain Res. 1994;24:336–40. doi: 10.1016/0169-328x(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 35.Chandrasekaran K, Hatanpää K, Brady DR, Stoll J, Rapoport SI. Downregulation of oxidative phosphorylation in Alzheimer disease: loss of cytochrome oxidase subunit mRNA in the hippocampus and entorhinal cortex. Brain Res. 1996;796:13–9. doi: 10.1016/s0006-8993(98)00248-0. [DOI] [PubMed] [Google Scholar]
- 36.Chandrasekaran K, Hatanpää K, Rapoport SI, Brady DR. Decreased expression of nuclear and mitochondrial DNA-encoded genes of oxidative phosphorylation in association neocortex in Alzheimer disease. Brain Res Mol Brain Res. 1997;44:99–104. doi: 10.1016/s0169-328x(96)00191-x. [DOI] [PubMed] [Google Scholar]
- 37.Rice AC, Ladd AC, Bennett JP. Postmortem Alzheimer’s disease hippocampi show oxidative phosphorylation gene expression opposite that of isolated pyramidal neurons. J Alzheimers Dis. 2015;45:1051–9. doi: 10.3233/JAD-142937. [DOI] [PubMed] [Google Scholar]
- 38.Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A. 2008;105:15605–10. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berchtold N, Sabbagh M, Beach T, Kim R, Cribbs D, Cotman C. Brain gene expression patterns differentiate mild cognitive impairment from normal aged and Alzheimer’s disease. Neurobiol Aging. 2014;35:1961–72. doi: 10.1016/j.neurobiolaging.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manczak M, Jung Y, Park BS, Partovi D, Reddy PH. Time-course of mitochondrial gene expressions in mice brains: implications for mitochondrial dysfunction, oxidative damage, and cytochrome c in aging. J Neurochem. 2005;92:494–504. doi: 10.1111/j.1471-4159.2004.02884.x. [DOI] [PubMed] [Google Scholar]
- 41.Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–7. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 42.Blalock EM, Chen KC, Sharrow K, Herman JP, Porter NM, Foster TC, et al. Gene microarrays in hippocampal aging: statistical profiling identifies novel processes correlated with cognitive impairment. J Neurosci. 2003;23:3807–19. doi: 10.1523/JNEUROSCI.23-09-03807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fraser HB, Khaitovich P, Plotkin JB, Paabo S, Eisen MB. Aging and gene expression in the primate brain. PLoS Biol. 2005;3:e274. doi: 10.1371/journal.pbio.0030274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swerdlow RH, Burns JM, Khan SM. The Alzheimer’s disease mitochondrial cascade hypothesis. J Alzheimers Dis. 2010;20:S265–79. doi: 10.3233/JAD-2010-100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johri A, Calingasan NY, Hennessey TM, Sharma A, Yang L, Wille E, et al. Pharmacologic activation of mitochondrial biogenesis exerts widespread beneficial effects in a transgenic mouse model of Huntington’s disease. Hum Mol Genet. 2012;21:1124–37. doi: 10.1093/hmg/ddr541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Podlesniy P, Figueiro-Silva J, Llado A, Antonell A, Sanchez-Valle R, Alcolea D, et al. Low cerebrospinal fluid concentration of mitochondrial DNA in preclinical Alzheimer disease. Ann Neurol. 2013;74:655–68. doi: 10.1002/ana.23955. [DOI] [PubMed] [Google Scholar]
- 47.Dowling NM, Johnson SC, Gleason CE, Jagust WJ. The mediational effects of FDG hypometabolism on the association between cerebrospinal fluid biomarkers and neurocognitive function. Neuroimage. 2015;105:357–68. doi: 10.1016/j.neuroimage.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demetrius LA, Magistretti PJ, Pellerin L. Alzheimer’s disease: the amyloid hypothesis and the Inverse Warburg effect. Front Physiol. 2015;5:522. doi: 10.3389/fphys.2014.00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grimm A, Friedland K, Eckert A. Mitochondrial dysfunction: the missing link between aging and sporadic Alzheimer’s disease. Biogerontology. 2016;17:281–96. doi: 10.1007/s10522-015-9618-4. [DOI] [PubMed] [Google Scholar]
- 50.Minoshima S, Giordani B, Berent S, Frey KA, Foster NL, Kuhl DE. Metabolic reduction in the posterior cingulate cortex in very early Alzheimer’s disease. Ann Neurol. 1997;42:85–94. doi: 10.1002/ana.410420114. [DOI] [PubMed] [Google Scholar]
- 51.Masliah E, Mallory M, Alford M, DeTeresa R, Hansen LA, McKeel DW, et al. Altered expression of synaptic proteins occurs early during progression of Alzheimer’s disease. Neurology. 2001;56:127–9. doi: 10.1212/wnl.56.1.127. [DOI] [PubMed] [Google Scholar]
- 52.Weaver CL, Espinoza M, Kress Y, Davies P. Conformational change as one of the earliest alterations of tau in Alzheimer’s disease. Neurobiol Aging. 2000;21:719–27. doi: 10.1016/s0197-4580(00)00157-3. [DOI] [PubMed] [Google Scholar]
- 53.Nixon RA, Cataldo AM, Mathews PM. The endosomal-lysosomal system of neurons in Alzheimer’s disease pathogenesis: A review. Neurochem Res. 2000;25:1161–72. doi: 10.1023/a:1007675508413. [DOI] [PubMed] [Google Scholar]
- 54.Frost B, Diamond MI. Prion-like mechanisms in neurodegenerative diseases. Nat Rev Neurosci. 2010;11:155–9. doi: 10.1038/nrn2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tai HC, Wang BY, Serrano-Pozo A, Frosch MP, Spires-Jones TL, Hyman BT. Frequent and symmetric deposition of misfolded tau oligomers within presynaptic and postsynaptic terminals in Alzheimer’s disease. Acta Neuropathol Commun. 2014;2:146. doi: 10.1186/s40478-014-0146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheung G, Jupp OJ, Cousin MA. Activity-dependent bulk endocytosis and clathrin-dependent endocytosis replenish specific synaptic vesicle pools in central nerve terminals. J Neurosci. 2010;30:8151–61. doi: 10.1523/JNEUROSCI.0293-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mastroeni D, Delvaux E, Nolz J, Tan Y, Grover A, Oddo S, et al. Aberrant intracellular localization of H3k4me3 demonstrates an early epigenetic phenomenon in Alzheimer’s disease. Neurobiol Aging. 2015;36:3121–9. doi: 10.1016/j.neurobiolaging.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Currais A, Hortobágyi T, Soriano S. The neuronal cell cycle as a mechanism of pathogenesis in Alzheimer’s disease. Aging. 2009;1:363–71. doi: 10.18632/aging.100045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mu Y, Gage FH. Adult hippocampal neurogenesis and its role in Alzheimer’s disease. Mol Neurodegener. 2011;6:85. doi: 10.1186/1750-1326-6-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ashraf A, Fan Z, Brooks DJ, Edison P. Cortical hypermetabolism in MCI subjects: A compensatory mechanism? Eur J Nucl Med Mol Imaging. 2015;42:447–58. doi: 10.1007/s00259-014-2919-z. [DOI] [PubMed] [Google Scholar]
- 61.Saura CA, Parra-Damas A, Enriquez-Barreto L. Gene expression parallels synaptic excitability and plasticity changes in Alzheimer’s disease. Front Cell Neurosci. 2015;9:318. doi: 10.3389/fncel.2015.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Del Sole A, Clerici F, Chiti A, Lecchi M, Mariani C, Maggiore L, et al. Individual cerebral metabolic deficits in Alzheimer’s disease and amnestic mild cognitive impairment: an FDG PET study. Eur J Nucl Med Mol Imaging. 2008;35:1357–66. doi: 10.1007/s00259-008-0773-6. [DOI] [PubMed] [Google Scholar]
- 63.Bobba A, Petragallo VA, Marra E, Atlante A. Alzheimer’s proteins, oxidative stress, and mitochondrial dysfunction interplay in a neuronal model of Alzheimer’s disease. Int J Alzheimers Dis. 2010 doi: 10.4061/2010/621870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cavallucci V, Berretta N, Nobili A, Nisticò R, Mercuri N, D’Amelio M. Calcineurin inhibition rescues early synaptic plasticity deficits in a mouse model of Alzheimer’s disease. Neuromolecular Med. 2013;15:541–8. doi: 10.1007/s12017-013-8241-2. [DOI] [PubMed] [Google Scholar]
- 65.Ma T, Trinh MA, Wexler AJ, Bourbon C, Gatti E, Pierre P, et al. Suppression of eIF2a kinases alleviates Alzheimer’s disease-related plasticity and memory deficits. Nat Neurosci. 2013;16:1299–305. doi: 10.1038/nn.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gleichmann M, Mattson M. Neuronal calcium homeostasis and dysregulation. Antioxid Redox Signal. 2011;14:1261–73. doi: 10.1089/ars.2010.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin MT, Beal MF. Alzheimer’s APP mangles mitochondria. Nat Med. 2006;12:1241–3. doi: 10.1038/nm1106-1241. [DOI] [PubMed] [Google Scholar]
- 68.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–95. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 69.Cardoso SM, Santos S, Swerdlow RH, Oliveira CR. Functional mitochondria are required for amyloid beta-mediated neurotoxicity. FA-SEB J. 2001;15:1439–41. doi: 10.1096/fj.00-0561fje. [DOI] [PubMed] [Google Scholar]
- 70.Veereshwarayya V, Kumar P, Rosen KM, Mestril R, Querfurth HW. Differential effects of mitochondrial heat shock protein 60 and related molecular chaperones to prevent intracellular beta-amyloid-induced inhibition of complex IV and limit apoptosis. J Biol Chem. 2006;281:29468–78. doi: 10.1074/jbc.M602533200. [DOI] [PubMed] [Google Scholar]
- 71.Camilleri A, Zarb C, Caruana M, Ostermeier U, Ghio S, Högenc T, et al. Mitochondrial membrane permeabilisation by amyloid aggregates and protection by polyphenols. Biochim Biophys Acta. 2013;1828:2532–43. doi: 10.1016/j.bbamem.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 72.Sanz-Blasco S, Valero RA, Rodriguez-Crespo I, Villalobos C, Nunez L. Mitochondrial Ca2+ overload underlies Abeta oligomers neurotoxicity providing an unexpected mechanism of neuroprotection by NSAIDs. PLoS One. 2008;3:e2718. doi: 10.1371/journal.pone.0002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moreira PI, Carvalho C, Zhu X, Smith MA, Perry G. Mitochondrial dysfunction is a trigger of Alzheimer’s disease pathophysiology. Biochim Biophys Acta. 2010;1802:2–10. doi: 10.1016/j.bbadis.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 74.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–99. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 75.She H, Yang Q, Shepherd K, Smith Y, Miller G, Testa C, et al. Direct regulation of complex I by mitochondrial MEF2D is disrupted in a mouse model of Parkinson disease and in human patients. J Clin Invest. 2011;121:930–40. doi: 10.1172/JCI43871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mastroeni D, Khdour OM, Arce PM, Hecht SM, Coleman PD. Novel antioxidants protect mitochondria from the effects of oligomeric amyloid Beta and contribute to the maintenance of epigenome function. ACS Chem Neurosci. 2015;15:588–98. doi: 10.1021/cn500323q. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.