Summary
‘Photoarsenotrophy’, the use of arsenite as an electron donor for anoxygenic photosynthesis, is thought to be an ancient form of phototrophy along with the photosynthetic oxidation of Fe(II), H2S, H2 and . Photoarsenotrophy was recently identified from Paoha Island's (Mono Lake, CA) arsenic-rich hot springs. The genomes of several photoarsenotrophs revealed a gene cluster, arxB2AB1CD, where arxA is predicted to encode for the sole arsenite oxidase. The role of arxA in photosynthetic arsenite oxidation was confirmed by disrupting the gene in a representative photoarsenotrophic bacterium, resulting in the loss of light-dependent arsenite oxidation. In situ evidence of active photoarsenotrophic microbes was supported by arxA mRNA detection for the first time, in red-pigmented microbial mats within the hot springs of Paoha Island. This work expands on the genetics for photosynthesis coupled to new electron donors and elaborates on known mechanisms for arsenic metabolism, thereby highlighting the complexities of arsenic biogeochemical cycling.
Introduction
Despite its toxicity arsenic can be used by certain organisms in a favourable way to generate cellular energy (Oremland, 2003; Saltikov, 2011; Amend et al., 2014). Oremland et al. (2009). More specifically, this has been defined as ‘arsenotrophy’ or the coupling of arsenic reduction or oxidation to cellular energy production (Oremland et al., 2009). The interconversion of arsenate and arsenite also has environmental consequences that are known to affect arsenic contamination of water (Harvey, 2002; Kocar et al., 2008; Fendorf et al., 2010). Arsenotrophic microbes oxidize/reduce two major forms of arsenic oxyanions: arsenite [As(III)] and arsenate [As(V)] (Islam et al., 2004; Oremland and Stolz, 2005; Dhar et al., 2011; Stuckey et al., 2015). The latter arsenical is typically less toxic, more stable under oxidizing conditions, and favours sequestration within minerals (O'Day et al., 2004; Kocar and Fendorf, 2009). However, the onset of reducing conditions stimulates microbial arsenate reduction to arsenite. Relative to arsenate, arsenite is more toxic, has a greater partitioning into the aqueous phase, and is more readily transported within anoxic groundwater (Masscheleyn and Delaune, 1991; Kocar et al., 2008). South Asian countries have the most serious arsenic groundwater problems that are manifested by high prevalence of arsenic-related illnesses such as arsenicosis and cancer (Nickson et al., 1998; Acharyya et al., 1999; Smith et al., 2000). The underlying microbial genes responsible for microbial reactions relevant to public health likely developed on the early Earth when the prevailing environmental conditions were reducing (Sessions et al., 2009). During this time, arsenic may have been more abundant in aquatic environments because of increased volcanic activity, which would have brought arsenite and other chalcophilic compounds to the Earth's surface (Oremland et al., 2009). This would have provided the necessary niche that allowed the evolution of autotrophic pathways for arsenite oxidation. Finally, recent discoveries where both arsenic and microbial organic signatures found in a Mars (Wallis and Wickramasinghe, 2015) meteoroid and within stromatolites (Sforna et al., 2014) from 2.7 b.y.a., raises the question on how arsenite may have played a role in arsenotrophy as life evolved within arsenic rich environments.
Most of the well-known pathways of arsenotrophy have been determined in chemotrophic microbes. However, arsenotrophic reactions now include a light-dependent pathway involving anoxygenic photosynthesis with arsenite as an electron donor (Budinoff and Hollibaugh, 2008; Kulp et al., 2008; Hoeft et al., 2010), or ‘photoarsenotrophy’. Kulp et al. (2008) first described the occurrence of photosynthetic arsenite oxidation in red-pigmented biofilms found within the hot springs of Paoha Island Mono Lake, CA, an arsenic-rich, hypersaline, alkaline lake. Moreover, the first photoarsenotroph, Ectothiorhodospira sp. str. PHS-1, was isolated from the Paoha Island hot spring milieu. The genome of PHS-1 lacked evidence for an aioA-type arsenite oxidase, which at the time was the prevailing consensus pathway for arsenite oxidation (Lett et al., 2011). Instead, Zargar et al. (2010) revealed a potentially ‘new’ genetic pathway for arsenite oxidation based on a recently identified anaerobic arsenite oxidase gene, arxA, in a related chemoautotroph, Alkalilimnicola ehrlichii str. MLHE-1. The predicted ArxA enzyme was shown to form a new clade within the DMSO reductase family of oxidoreductases (Zargar et al., 2012), which also contains a separate clade for the AioA enzyme. In the absence of a genetic system in a photoarsenotroph, circumstantial evidence suggested that the arxA-like gene of PHS-1 encodes for a molybdenum-containing arsenite oxidase. Furthermore arsenite exposure resulted in the induction of the arxA gene in PHS-1. However, the function of ArxA in PHS-1 could not be ascertained because of difficulties in establishing a genetic system in this organism. Ultimately, the ArxA phylogenetic analysis and molecular genetic studies with the chemotrophic, MLHE-1 (Zargar et al., 2010) bacterium suggested that ArxA is the most plausible pathway for photoarsenotrophy in PHS-1.
The ecological distribution and environmental impact of photoarsenotrophs to the arsenic cycle remains unknown. Currently our awareness of the distribution of photoarsenotrophy is limited to the environment where PHS-1 was isolated. Molecular detection of arxA from other extreme environments such as Yellowstone, WY, and other hot spring environments around the Mono Lake, CA (Zargar et al., 2012) area, suggest the possibility of anaerobic arsenite oxidation. These extreme, arsenic-rich environments are analogous to the conditions of early Earth (Kempe and Kazmierczak, 1997) that may have favoured the evolution of anoxygenic photosynthetic pathways that are present in green and purple (non-) sulfur bacteria. In order to comprehend the evolutionary trajectory of these metabolisms and to better appreciate the past, present, and future impact of photoarsenotrophs on arsenic geochemistry, additional work is needed to confirm that arxA is responsible for arsenite oxidation in photoarsenotrophs.
The purpose of this report is to present the first genetic evidence that arxA is the main arsenite oxidase in a photoarsenotroph. We demonstrated that the arxA gene is also expressed in the Paoha Island red-mat biofilms where PHS-1 was first isolated. These results not only validate the function of the arx gene pathway but also further expand on the complexities of the arsenic biogeochemical cycle by adding ‘light’ as a considering factor. Investigating the geochemical impact of photoarsenotrophy will depend on further understanding the regulation and kinetic parameters of arx genes and protein products, and developing molecular tools to track the genetic signatures of arx-containing microorganisms. Thus, this work lays the critical groundwork for these future studies.
Results and discussion
Molecular mechanism for photoarsenotrophy
Our current understanding of the molecular mechanism for anoxygenic photosynthesis coupled to arsenite oxidation is based on previous genetic confirmation that the arx gene cluster in Alkalilimnicola ehrlichii str. MLHE-1 is required for oxidation and anaerobic growth on arsenite coupled to nitrate reduction (Zargar et al., 2010). The identification of arx gene homologs in the PHS-1 genome (Zargar et al., 2010; 2012) and the absence of an aio-type gene cluster further supports the hypothesis that photosynthetic arsenite oxidation is conferred by arx genes. In anoxygenic photosynthesis electrons are required in order to generate a proton motive force, which in turn can drive numerous biological reactions in the cell such as producing ATP and NAD(P)H that are used for anabolic metabolisms. Due to thermodynamic constraints reduced substrates such as sulfide, nitrite (Griffin et al., 2007), iron(II) (Widdel et al., 1993) and arsenite (Budinoff and Hollibaugh, 2008) may be oxidized via cyclic or non-cyclic electron flow within anoxygenic photosynthetic bacteria. During cyclic electron flow, light energizes electrons within the photosynthetic reaction centre (PRC) followed by electron transport through quinone intermediates to cytochrome bc1, which generates a proton motive force. The cycle is completed when bc1 transfers its electrons to a periplasmic c-type cytochrome (e.g., c2) and ultimately back to the PRC for another cycle (Lavergne et al., 2009). In non-cyclic electron flow, reduced quinols (e.g., ubiquinol/ubiquinone, +70 to +112 mV) can be used to drive reverse electron transport, which uses the energy from the proton motive force to generate reducing power [e.g., NAD(P)H] to fuel carbon dioxide assimilation and biosynthesis. Based on the redox potential of +154 mV (Budinoff and Hollibaugh, 2008), electrons from arsenite could be transferred to either a c2-like cytochrome (+325 mV) (Cammack et al., 1981) similar to the predicted cyclic electron transfer for photosynthetic Fe(II) oxidation (Ehrenreich and Widdel, 1994) or the quinone pool (ubiquinone, +70 to +112 mV). Although it is not known how arsenite oxidation is coupled to photosynthesis, we hypothesize that arsenite is oxidized to arsenate by ArxA followed by electron transfer to Fe-S containing subunits (either ArxB2 and/or ArxB1). The predicted protein sequences for ArxB2 and ArxB1 share low amino identities/similarities (14.4%/22.8%) however they both have predicted amino acid motifs for 4Fe-4S clusters (based on pfam analysis http://pfam.xfam.org). Their function with respect to the electron transport chain for photosynthesis or in chemoautotrophs remains unknown. Electrons could then either be transferred to c2 (or similar c-type cytochromes) and back to the PRC or to the quinone pool mediated by ArxC, a predicted membrane protein homologous to the polysulfide reductase NrfD-like membrane protein. The latter is known to transfer electrons from the quinone pool during polysulfide reduction (Jormakka et al., 2008). Nonetheless the generation of NAD(P)H is dependent on electrons from arsenite and coupled to the proton motive force, generated by cytochrome bc1, which together ultimately generate reducing power, NAD(P)H, and ATP for biological reactions within the cell.
To begin testing the photosynthetic reaction mechanism for arsenite oxidation, we first addressed the hypothesis that arxA (or genes within the arxB2AB1CD cluster) is required for photosynthetic arsenite oxidation. Because no genetic systems were available for a photoarsenotrophic microbe, our first task was to identify a suitable strain for genetic system development. PHS-1 was determined to be intractable for this purpose because of difficulties in reliably culturing the strain on solid media. We then isolated another photoarsenotroph, Ectothiorhodospira sp. str. BSL-9 from Big Soda Lake (NV), an arsenic-rich hypersaline alkaline crater lake similar to Mono Lake (Cloern et al., 1983; Zehr et al., 1987). BSL-9 was facile to work with relative to PHS-1 and could be grown under less “extreme” conditions such as pH 8 and 20 g/L salt. Although BSL-9 was unable to grow in the presence of oxygen, the strain was aerotolerant facilitating the preparation of conjugation reactions and streak plates on the bench prior to incubation under anaerobic conditions. Genetic system development was also aided by completing the BSL-9 genome. Analysis of the genome indicated an arx gene cluster with 100% synteny to that found in PHS-1 and MLHE-1 genomes. The BSL-9 ArxA was homologous to other ArxA sequences from Ectothiorhodospira sp. str. PHS-1 (75% similarities), Alkalilimnicola ehrlichii str. MLHE-1 (86% similarities), and other sequences found in NCBI database such as Halorhodospira halophila str. SL-1 (78% similarities). As previously described for other PHS-1 and MLHE-1 ArxA sequences, the BSL-9 ArxA clustered within these sequences and formed its own distinct clade within the DMSO reductase family of molybdenum containing enzymes (Fig. 1). The genomic and phylogenetic evidence strongly supported that the arxB2AB1CD gene cluster of BSL-9 encoded for an anaerobic arsenite oxidase, arxA (ECTOBSL9_2837, locus tag number) and that it was the best candidate gene for further genetic manipulation.
Fig. 1.
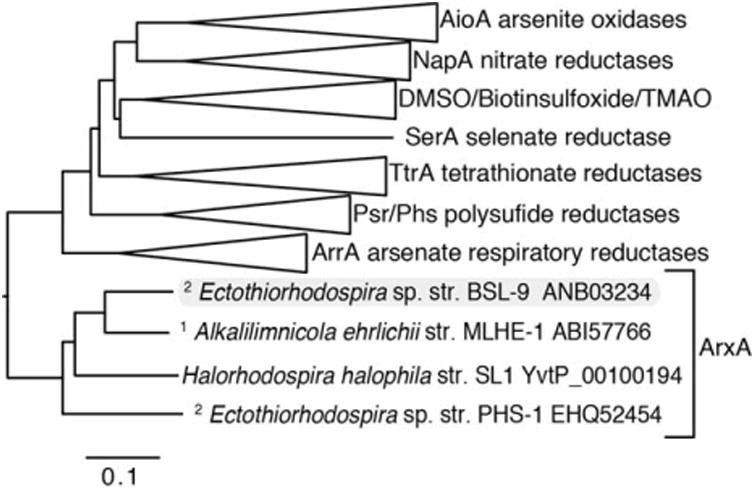
ArxA forms a clade within the DMSO reductase family of molybdenum containing oxidoreductases. Phylogenetic analysis indicates that ArxA clusters more closely to the arsenate reductase, ArrA than the arsenite oxidase, AioA. However ArxA is distinct from other known DMSO oxidoreductase family enzymes. The ArxA clade contains chemoautorophic1 and photoautotrophic2 arsenite oxidizers. The Genbank accession numbers are indicated for the ArxA containing taxa. The scale bar indicates changes per position within the multisequence amino acid alignment.
We were interested in determining if expression of arxA in BSL-9 was regulated by arsenite. In our experiments, we used custom-built infrared (IR) LED (850 nm peak centred between 800 and 900 nm) arrays as the primary light source because this was originally used for enrichment cultures to eliminate the growth of oxygenic photoautotrophs, such as cyanobacteria, which absorb light in the visible range spectrum. Oxygen produced by cyanobacteria and algae inhibited the growth of anoxygenic phototrophs in our enrichment cultures. In purple sulfur bacteria the absorbance spectra depends on the bacterial species, light, and growth conditions but most typically have peak absorbance within the near-infrared (∼ 800–900 nm) (Overmann, 2008; Robert, 2009). BSL-9 cells grown under IR had comparable absorbance peaks near 800, 860 and 900 nm. To demonstrate the regulation and induction of arxA only in the presence of arsenite (Fig. 2, bar graph), BSL-9 cultures were grown on acetate (Fig. 2, solid lines) under infrared (IR) lights to mid-log phase. We then extracted total RNA from BSL-9 cultures spiked with 300 μM arsenite and compared with BSL-9 cultures lacking arsenite over time. After 2 hours of the arsenite spike, arxA expression increased ∼ 20-fold relative to cultures without arsenite (Fig. 2, bar graph). Peak arxA expression (40-fold) was detected after 12 hours following the arsenite spike. This result is consistent with previous reports that showed arsenite also induced arxA expression in PHS-1 (Zargar et al., 2012) and MLHE-1 (Zargar et al., 2010). The increased expression of arxA by the addition of arsenite is evident of arxA being regulated under different growth conditions. The aio-type arsenite oxidase is also regulated in response to arsenite (Kashyap et al., 2006; Sardiwal et al., 2010), which is mediated by a two-component sensor histidine kinase (AioS) and response regulator (AioR). Genes for an analogous regulatory system, arxXSR (ArxX, periplasmic binding protein; ArxS, sensor histidine kinase; and ArxR, response regulator), are present immediately upstream and in the reverse direction of the arxB2AB1CD gene clusters in BSL-9 (Fig. 3A), PHS-1 and MLHE-1. This gene cluster, arxXSR, is of great interest to future studies regarding the regulation of arxA in both chemoautotrophic and photoautotrophic organisms. Together with the ArxA phylogenetic analysis, the conservation of arxXSR and arxB2AB1CD gene clusters within the chemoautotroph, MLHE-1 and photoautotrophs, PHS-1, SL-1 and now BSL-9, paved the way for developing a genetic system in BSL-9 to confirm the function of arx genes in photoarsenotrophy.
Fig. 2.
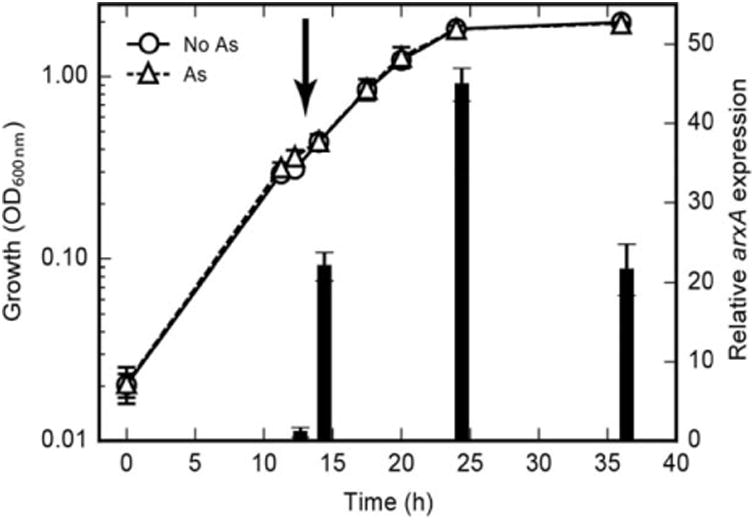
arxA transcription induced by arsenite in Ectothiorhodospira sp. str. BSL-9. The wild-type BSL-9 strain was grown photosynthetically with acetate (solid lines) and monitored over time for arxA expression (indicated by the bars). After addition of arsenite (300 μM) at 12 hours (bold vertical arrow) arxA gene expression increased only in samples receiving the arsenite spike (dashed lines). Relative arxA gene expression at each time point was determined using the ΔΔCt method with the corresponding “no arsenic” condition as the control condition and 16S rRNA gene as the reference gene. Both growth and gene expression data points and error bars represent the averages and standard deviation from triplicate cultures.
Fig. 3.
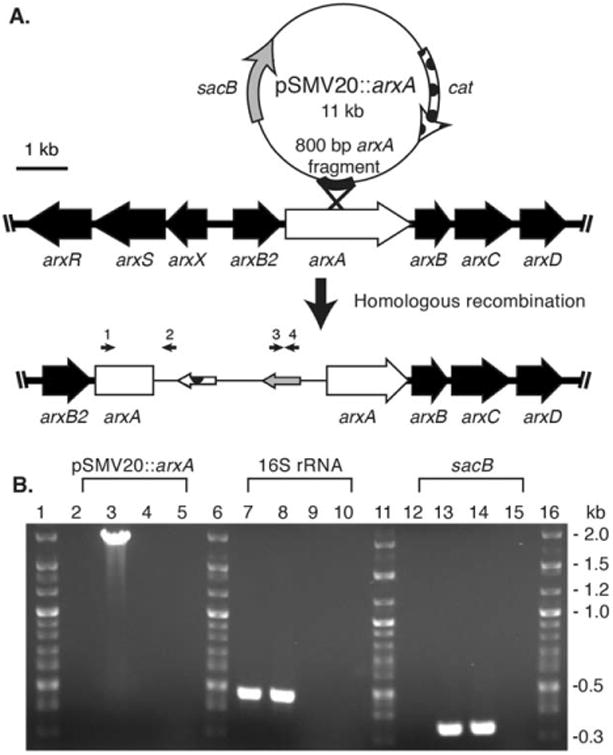
Development and verification of arxA disruption mutation in Ectothiorhodospira sp. str. BSL-9. (A) The pSMV20 vector was inserted within arxA via homologous recombination. (B) PCR analysis for DNA from wild-type BLS-9 (2, 7, 12), ARXA1 (3, 8, 13), empty pSMV20 vector (4, 9, 14), and water (5, 10, 15). The following PCR primers were used: (i) arxA (Ext_arxA_F, arrow 1) gene and vector-specific (M13_R, arrow 2) (lanes 2-5), (ii) 16S rRNA (0341_16SV3V4-F and 0785_16SV3V4-R) gene (lanes 7-10) and (iii) sacB (sacB_F and sacB_R, arrows 3 and 4) (lanes 12-15). PCRs were analysed by agarose (1%) gel electrophoresis.
To begin elucidating the genetics of photoarsenotrophy we developed a genetic system to test if arxA was required for photosynthetic arsenite oxidation. Mutagenesis often requires the use of selectable markers, typically encoded by antibiotic resistance genes. Our analyses indicated that BSL-9 was resistant to kanamycin, ampicillin, gentamycin, but sensitive to chloramphenicol (Cm). A Cm-resistant broad host range plasmid was used to develop a conjugation method for introducing foreign DNA into BSL-9 wild-type strains from an Escherichia coli WM3064 donor strain (Saltikov and Newman, 2003), yielding ∼ 103–104 CmR CFU ml−1. We then took a gene disruption approach to ‘knock out’ the function of arxA gene in BSL-9, using a mutagenesis plasmid, pSMV20 (Zargar et al., 2010), which contains the chloramphenicol acetyltransferase resistance encoding cat gene. An 800 base pair internal fragment of arxA was cloned into the mutagenesis plasmid (pSMV20). The new plasmid (pSMV20∷arxA) was introduced into BSL-9 wild-type by conjugation with an E. coli donor strain. The partial arxA DNA fragment in pSMV20 allowed the disruption of arxA within the BSL-9 genome by a single homologous recombination event (Fig. 3A). PCR was utilized to verify that the BLS-9 exconjugants integrated pSMV20∷arxA within the arxA gene. Figure 3B shows positive detection for the recombination event within the arxA gene indicated by a ∼ 2 kb PCR product in Cm resistant BSL-9 colonies using the primer set Ext_arxA_Forward and M13_Reverse primer, which binds upstream of the arxA disruption and within pSMV20 DNA, respectively. As expected, only DNA from BSL-9 Cm resistant colonies produced a 2 kb PCR product (Fig. 3B, lane 3). Furthermore, Fig. 3B shows positive detection for sacB gene, which is present in pSMV20 constructs and yields a 200 bp product (lane 14). Detection of sacB was observed only in genomic DNA of a BSL-9 Cm resistance isolates (lane 13) and no detection in BSL-9 wild-type genomic DNA (lane 12) was observed. The data in Fig. 3B confirms that the BSL-9 Cm resistant exconjugant colonies contain a genomic plasmid insertion mutation within arxA, potentially disrupting the function of the arsenite oxidase. The BSL-9 arxA insertion mutant is referred to as ARXA1.
We performed various physiological growth curve experiments with different electron donors to test for specificity of arxA disruption to arsenite. The ARXA1 mutant grew similarly to the wild-type under photoautotrophic (thiosulfate) and photoheterotrophic (acetate) conditions (Supporting Information Fig. S1). Photoheterotrophic growth was also observed with sucrose, lactate, pyruvate, succinate, malate, propionate and photoautotrophic growth with sulfide as electron donors (data not shown). Photoautotrophic growth on arsenite only occurred in BSL-9 wildtype cells with an intact arxA gene (Fig. 4A). Moreover, ARXA1 strain was only deficient when grown with arsenite as an electron donor (Fig. 4A). Background arsenate carryover is observed initially when adding arsenite. However there was no indication of arsenite oxidation in ARXA1 even though there was an increase of 33 μM of arsenite and 30 μM of arsenate over time (Fig. 4C). More evidently, arsenite oxidation was observed in BSL-9 wild-type cells with the conversion of ∼ 400 μM arsenite to ∼ 400 μM arsenate (Fig. 4B). Together the arsenite growth inhibition (Fig. 4A) and no arsenite oxidation (Fig. 4C) in the ARXA1 mutant strongly support the hypothesis that arxA is the sole arsenite oxidase within the arxB2AB1CD gene cluster. Thus far, our genetic model development system establishes a successful way to disrupt gene functions in Ectothiorhodospira sp. str. BSL-9.
Fig. 4.
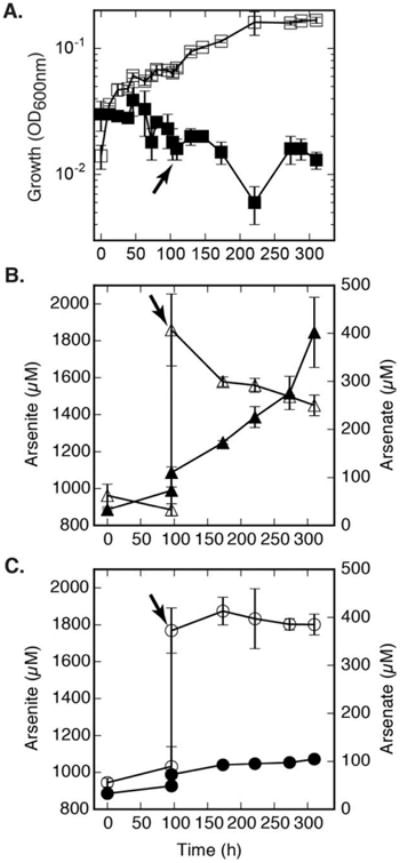
Effects of ARXA1 mutant on anoxygenic photosynthetic arsenite oxidation. (A) Growth of ARXA1 (arxA mutant) and BSL-9 wild-type strains were compared under arsenite (600 μM) conditions (■,□). All samples received an additional 1000 μM arsenite spike at the 96th hour, indicated by the arrow. Growth was detected only in BLS-9 wild-type (□) and absent in BSL-9 ARXA1 mutant ■. (B) Arsenite (Δ) oxidation to arsenate (▲) in BSL-9 wild-type cells. (C) Arsenite (○) to arsenate (●) oxidation inhibition in ARXA1 cells. No clear indication of arsenic transformations in ARXA1 mutant. Data points and error bars in represent the averages and standard deviation of triplicate cultures, respectively.
Photosynthetic bacteria are unique in evolving diverse strategies to capture light in order to generate biomass. The uniqueness of photoarsenotrophy is how arsenite oxidation can be driven by light energy. We were interested in addressing if arsenite oxidation was light dependent. Figure 5A shows arsenite oxidation in the presence of light in cell suspension experiments (∼ 108 cells ml−1) by utilizing BSL-9 and ARXA1 mutant. When cell suspensions were initiated in the dark and then shifted to the light, as shown in Fig. 5B, arsenite oxidation was not observed until the suspensions were shifted into the light. Arsenite oxidation promptly occurred in BSL-9 but not in ARXA1. The cell suspension experiments in Fig. 5 confirmed that arsenite oxidation is coupled to light in BSL-9 and dependent on the arxAB1CD gene cluster. These findings highlight arxAB1CD as the essential genes to carry out photoarsenotrophy.
Fig. 5.
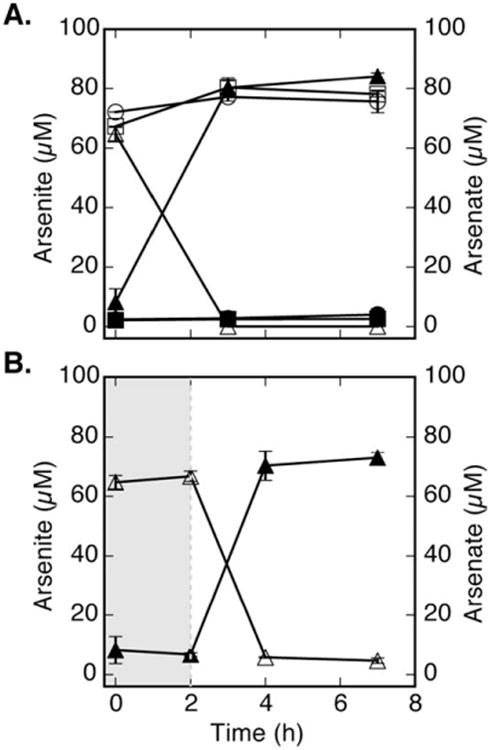
Light-dependent and growth-independent arsenite oxidation. Cell suspension samples were spiked with approximately 75 μM arsenite and monitored for arsenic speciation. (A) Growth independent arsenite oxidation (○) to arsenate (●) was inhibited in ARXA1 (arxA mutant). However, wild-type BSL-9 was able to oxidize arsenite (Δ) to arsenate (▲) within 3 hours. No abiotic arsenic speciation was observed (■/□). (B) Light dependent arsenite oxidation (Δ) to arsenate (▲) was observed only after BSL-9 wild-type cells were shifted to IR light conditions (t = 2 hours). Data points and error bars in represent the averages and standard deviation of triplicate samples, respectively.
We then investigated the polar effects of the pSMV20∷arxA plasmid insertion mutation in ARXA1 to determine if transcription of the downstream arxBCD genes were affected by the mutation. To investigate polar effects of ARXA1 mutant, we first observed evidence for the induction of the arxB2AB1CD gene cluster by the addition of arsenite in the BSL-9 wild-type strain (Supporting Information Fig. S2A, lanes 2, 4, 6, 8, 10 and 12). Arsenite (300 μM) was added to BSL-9 wild-type and ARXA1 mutant cultures grown initially on acetate. The expression of the arx gene cluster was determined by reverse transcription-polymerase chain reaction (RT-PCR). For the BSL-9 wild-type strain each gene, arxB2AB1CD, was expressed when induced with arsenite (Supporting Information Fig. S2A, lanes 2, 4, 6, 8, 10 and 12). However, only arxB2 and arxA upstream of the mutation were expressed in the ARXA1 strain (Supporting Information Fig. S2A, lanes 3 and 4), and little to no RT-PCR products were observed for arxA downstream of the mutation, arxB1 and arxC (Supporting Information Fig. S2A, lanes 7, 9 and 11). RT-PCR without reverse transcriptase enzyme lacked detectable PCR products. These results support the conclusion that the arxA insertion mutation caused polar effects on the transcription of the downstream genes. Although we cannot conclusively state that arxA is responsible for photosynthetic arsenite oxidation our data strongly suggests that the arx gene cluster is responsible for photoarsenotrphy in BSL-9. Future work will be needed to generate gene deletions. However, this has been challenging because of difficulties optimizing sucrose counter selection using sacB as a sensitivity marker.
Environmental impact of photoarsenotrophy
Lastly, we were interested in determining if arxA transcription could be detected in an arsenic rich environment where previous photoarsenotrophs have been identified. The expression of the arxA gene in the environment would signal potential arsenite oxidation activity in situ. We therefore sampled the red biofilms within the Paoha Island hot spring microbial mats and analysed the total RNA for the presence of arxA mRNA by RT-PCR. As previously reported in 2008, arsenite oxidation was confirmed geochemically in microcosm red mat slurries containing artificial hot spring media. The presence of the arxA DNA was also detected within the red mat material (Kulp et al., 2008). Our analysis of the total RNA extracts from these same hot spring biofilms demonstrated in situ arxA expression (Fig. 6A) and indirectly arsenite oxidase activity. As a control we also detected 16S rRNA gene expression within the environmental samples (Fig. 6B). Both arxA and 16S rRNA gene DNA and cDNA PCR products from the red mats were cloned and sequenced. All the environmental arxA cDNA sequences were nearly 100% identical to the arxA of PHS-1. The 16S rRNA gene sequencing data also confirmed that the red mat was dominated by a PHS-1-like bacterium, suggesting that photoarsenotrophy is active in this extreme environment and likely carried out by Ectothiorhodospria species similar to PHS-1. Our microbial ecology data when considered in the context of Kulp et al. (2008), provides additional evidence that ArxA is active within the environment and likely impacting arsenic cycling in the Paoha Island hot springs.
Fig. 6.
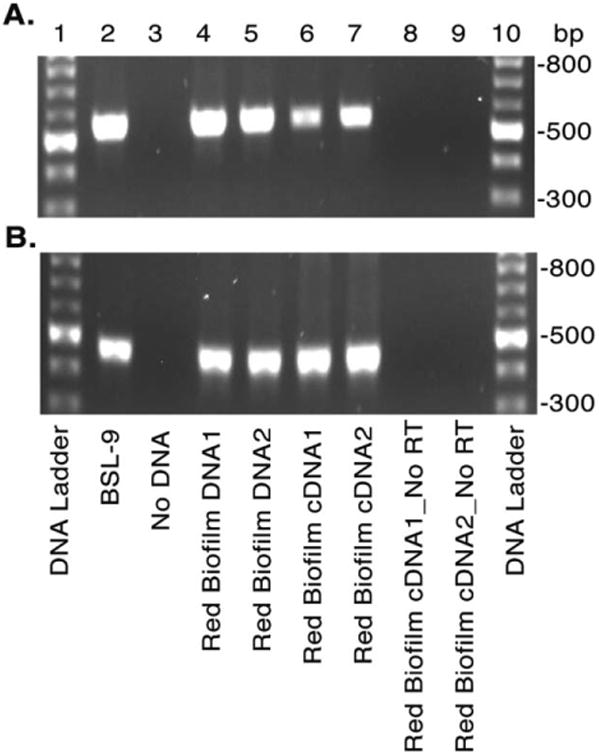
Evidence for environmental photoarsenotrophy activity determined by arxA gene expression in situ. Red biofilm-like microbial mats within hot springs of Paoha Island Mono Lake, CA were collected and the RNA and DNA extracts analysed for arxA (arxA_1824_deg_F and arxA_2380_deg_R primers) (A) and 16S rRNA (B) by PCR and RT-PCR respectively. Agarose gel electrophoresis of the PCRs and RT-PCRs are shown in A and B. For both panels lanes correspond to: DNA ladder (1, 10), BSL-9 genomic DNA (2), water negative control (3), environmental DNA (4–5), and cDNA of environmental RNA samples with (6–7) and without (8–9) reverse transcriptase.
In conclusion, we provided genetic evidence that supports arxB2AB1CD gene cluster as essential genes to the photoarsenotrophy mechanism and potentially ArxA as the sole arsenite oxidase in Ectothiorhodospira sp. strain BSL-9. These findings raise new questions about the biochemical mechanism, such as how the pathway is regulated, and what are the potential environmental impacts of ArxA-dependent arsenite fuelled anoxygenic photosynthesis. For the latter question, we cannot rule out the possibility that AioA could be a contributor to photoarsenotrophy. Chloroflexus is an abundant phototrophic organism in many hot spring microbial mat environments and Engel et al. (2013) identified Chloroflexus-like AioA sequences in microbial mat samples from El Tatio Geyser Field in Chile. Consequently, this opens up the possibility for an AioAdependent pathway for photoarsenotrophy. In terms of biogeochemical cycling of arsenic, the environmental impacts of photosynthetic arsenite oxidation needs further investigation.
Results from this study for the first time demonstrate that the arx gene cluster is responsible for photosynthetic arsenite oxidation. The implications of our microbial ecology survey further builds upon the case that photoarsenotrophs containing arxA may be impacting the arsenic geochemical cycle as demonstrated by the in situ detection of arxA mRNA in Paoha Island hot springs of Mono Lake containing ∼ 6 mM sulfide (Kulp et al., 2008). Moreover a broader distribution of arxA-like sequences occurs in the environment as observed in non-extreme environmental metagenomic studies (Supporting Information Table S3). It may be plausible that photoarsenotrophs are present within diverse euphotic environment such as freshwater aqueous, sediment, subsurface groundwater and marine sediment microbial communities as indicated by the presence of arxA-like metagenomic sequences (Supporting Information Table S3). Detection of arx like sequences in groundwater microbial communities may indicate arsenite oxidation coupled to nitrate by chemotroautotrophs (Supporting Information Table S3), which is tremendously relevant in regions such as Bangladesh and other south East Asia countries that suffer from groundwater arsenic contamination. This work is foundational to future studies regarding the potential contribution of photoarsenotrophs in diverse non-extreme environments. Results from this work also have implications for arsenic cycling on the early Earth. Sforna et al. (2014) provided evidence for the occurrence of microbial arsenic metabolism and cycling nearly 2.7 billion years ago. This predates the rise of oxygen. During this time, photoarsenotrophy may have been a key metabolism to providing arsenate as a by-product, which would be used as a terminal electron acceptor for arsenate respiring microbes. This would provide a mechanism for photoarsenotrophy to co-exist with anaerobic arsenate-respiring microbes, as currently occurs in the Paoha Island biofilms (Hoeft et al., 2010). Finally photosynthetic microorganisms containing arxA capable of oxidizing arsenite in anoxic environments are potential candidate metabolisms capable of surviving beyond the Earth, on or within other planet(oid)s such as Mars and Europa. The recent finding of arsenic minerals within the Tissint Martian meteorite raises the possibility of a primitive microbial arsenic metabolism having once occurred on Mars (Wallis and Wickramasinghe, 2015).
Material and methods
Sampling site description
Ectothiorhodospira sp. strain BSL-9 was isolated from Big Soda Lake (BSL), a meromictic hypersaline alkaline crater lake containing approximately 25 μM arsenic located in western Nevada (39°31′N118°52′W) (Cloern et al., 1983; Zehr et al., 1987). Red colored biofilm-like microbial mat samples were collected from Paoha Island hot springs, located (37°59.633 N, 119°01.376 W) in Mono Lake, CA. Paoha Island hot pools are alkaline anoxic hypersaline environments containing ∼ 100 μM arsenic and ∼ 6 μM sulfide (Kulp et al., 2008).
Media composition and growth conditions for photoarsenotrophs
Strain BSL-9 was grown in Basal Salt Media (BSM) with the following composition (g/L): KH2PO4 (0.24), K2HPO4 (0.30), (NH4)2SO4 (0.23), MgSO4 (0.12), NaCl (20), yeast extract (0.25), SL-10 mineral mix (5 mL/L), Wolfe's vitamin mix (10 mL/L), [2 g/L] Vitamin B-12 (100 μL). Electron donors were added as needed. After autoclaving the basal medium, 1 and 10 mL of filter sterilized stock solutions of NaCO3 (300 g/L) and NaHCO2 (600g/L) were added respectively. The media was cooled in anaerobic chamber containing 95% nitrogen, 5% hydrogen mixed gas. Typically the pH of the media was 8.0–8.3. Agar (15 g/L) was added to solidify the media when needed. BSL-9 physiology experiments for various growth experiments are typically initiated from frozen (−80°C) 20% glycerol stock cultures by the following method: using a sterile wooden stick, some ice from the glycerol stock is streak plated onto BSM/acetate (20 mM) agar plates in the bench top. Agar plates are then transferred to an anaerobic chamber and incubated at 358C for 3 days. Single colony forming units (CFU's) are observed within 3 days with continuous illumination of IR (850 nm) LED lights. For liquid cultures, a CFU is picked with a sterile loop and inoculated into an anaerobic Balch tube containing 20 mM acetate BSM media and grown for 2 days at 35°C under IR LED lights. To test other electron donors, cultures grown under acetate were harvested from turbid cultures by centrifugation in the anaerobic chamber. Cell pellets were washed three times in 1× PBS in order to re-suspended to a final optical density (OD) at 600 nanometers (nm) of 0.6 and then inoculated at 1:50 dilution into anaerobic media containing electron donors of choice, all prepared in anaerobic Balch tubes with sterile butyl rubber stoppers. Growth was followed over time using a spectrophotometer (Spectronic 20D×). All growth experiments where done in triplicates unless otherwise stated as similarly reported (Zargar et al., 2012).
Environmental sample collection for RNA extractions
An environmental sample collection trip was conducted in October 2015 to collect red coloured biofilm material from Paoha Island Mono Lake, CA. The red-coloured biofilm-like coatings on the rocks were collected from the Paoha Island hot pools by scraping the material off with a sterile metal spatula. Replicate microbial mat samples were added to RNase/DNase free 1.5 mL microcentrifuge tubes and immediately flash frozen in liquid nitrogen on site. The samples were transported back to the laboratory the next day and stored at −80°C. Total RNA was extracted 2 weeks later and analysed by RT-PCR for 16S rRNA and arxA gene expression.
DNA, RNA extractions and cDNA synthesis
In triplicates, environmental microbial genomic DNA and total RNA transcripts were extracted by Qiagen RNeasy Mini kit and Qiagen DNA extraction kit according to the manufacturer's instructions. After RNA extractions, 500 ng of RNA was then treated with RQ1 DNase (Promega) in order to degrade any potential DNA carryover. RNA treated samples were then followed by reverse transcriptase reactions, synthesized by the Applied Biosystems TaqMan kit (Part No. N808-0234) in order to generate cDNA. The extracted DNA and RNA concentrations were quantified by using a NanoDrop spectrophotometer at absorbance 260 nm. All samples were store at −20°C until performing further analysis.
Amplification and detection of genes through PCR
PCR was performed in order to amplify and detect genes of interest in environmental, BSL-9 and ARXA1 mutant DNA and environmental cDNA samples. DNA Sequencing was performed by Sequetech (San Jose, CA). The arxA-1824-deg-F and arxA-2380-deg-R primer set was used as a positive control to detect the presence of arxA gene, which yields approximately 500 base pair fragments. As a positive control for bacteria DNA, 16S rRNA gene was amplified with the primer set 0341_16SV3V4-F and 0785_16SV3V4-R. In order to generate the BSL-9 ARXA1 mutant an 800 base pair (bp) BLS-9 arxA homology region was amplified with X-arxA_Int —F1 and X-arxA_int_R1 primers and cloned into pSMV20. To verify plasmid insertion within arxA in BSL-9 chloramphenicol resistant colonies two primer sets were used. First the arxA_Ext_F and M13_R primer set was used which bind outside arxA and within the pSMV20 DNA, and yields 2 kb DNA fragments. Secondly a partial DNA sequence of sacB which encodes for the enzyme levansucrase and is only found in pSMV20 plasmids was amplified with sacB_FOR and sacB_REV primer set, generating 200 bp DNA products. Amplification of sacB was observed in positive control pSMV20 and in genomic DNA of BSL-9 ARXA1 mutant. BSL-9 arxB2AB1CD gene cluster was amplified with the following primer sets, arxB2_F and arxB2_R, arxA_UpS_F and arxA_UpS_R, arxA_DwnS_F and arxA_DwnS_R, arxB1_F and arxB1_R, arxC_F and arxC_R, arxD_F and arxD_R. A thermocycler profile was set to 28 cycles of denaturation, annealing, and extension for all PCR reactions. The primer annealing temperature and the extension time varied due to the primer set composition and the product size being amplified. Typically PCR parameters were set to 95°C for 5 min to denature DNA, [28 cycles of 95°C for 30 s to denaturation DNA, primer-specific annealing temperature for 30 s, extension at 72°C and a final extension of 72°C for 5 min. Supporting Information Table S1 shows specific annealing and extension PCR parameters for specific primer sets. Typically PCR reactions contained a final concentration of the following constituents: genomic DNA 10–50 ng] or [10 ug] of plasmid DNA, [0.4 uM] for each primer, 1× PCR Master Mix from Promega containing, buffers, Taq DNA polymerase, dNTP's and Mg2+.
Strains, plasmids and primer sets
Strains and plasmids used in this study are listed in Supporting Information Table S1. Primer composition, annealing and extension times are listed in Supporting Information Table S2.
Isolation of Ectothiorhodospira sp. str. BSL-9
Strain BSL-9 was isolated from swamp sediments samples collected in Big Soda Lake, Nevada. Basal salt media containing 2 mM arsenic and trace amounts of yeast (0.01 g/L) was used to set-up enrichment cultures. Enrichment cultures were streaked on BSM agar plates containing 2 mM arsenite in order to isolate single colonies. Purple like single colony forming units (CFU's) were re-streaked in order to acquire a pure culture. Big Soda Lake candidate isolates 1–14 were obtained; however, BSL-9 was selected since it grew well in BSM liquid cultures and in solid agar plates. The BLS-9 genome was sequenced using PacBio technology by UC Davis genome sequencing centre. The genome assembly was done by Pac-Bio HGAP_v2 assembly pipeline within approximate 300× coverage. The annotation was done through NCBI Public Genome Annotation Pipeline service. The Ectothiorhodospira sp. str. BSL-9 can be accessed from NCBI BioProject: PRJNA232800 or using the GeneBank accession number: CP011994.
arxA mutagenesis plasmid (pSMV20∷arxA) construct
In order to disrupt arxA in BSL-9 wild-type, by single homologous recombination, pSMV20∷arxA was used. To construct pSMV20∷arxA primer Int_arxA_F and Int_arx_R primers were used to amplified approximately 800 base pair homology region from BSL-9 arxA wild-type genomic DNA. The arxA homology sequence was ligated to pSMV20 (Zargar et al., 2010) by Gibson Assembly Kit (Gibson et al., 2009) according to the manufacture protocol. The Gibson Assembly reaction was then transformed into E. coli DH5 alpha λpir cell line (Saltikov and Newman, 2003). Cells were recovered in LB for an hour at 37°C and then plated on chloramphenicol 25 μg/ml agar plates. CFU's were then picked from chloramphenicol agar plate and grown in chloramphenicol LB liquid media in order to extract plasmid DNA and screen for the presence of pSMV20∷arxA plasmid. Plasmid DNA was extracted and screened with M13 F/R primer set for 800 bp fragments. The arxA homology sequence was approximately 800 bp, which was obtained and verified through sequencing. Plasmid (pSMV20∷arxA) was then transformed into E. coli WM3064 (Saltikov and Newman, 2003) mating strain in order to carry out conjugation reactions with BSL-9 wild-type strain. Diaminopimelic acid (DAP) was added to growth media since E. coli WM3064 is an auxotroph DAP mutant strain. BSL-9 exconjugants were then screened with primer sacB_FOR and sacB_REV and Ext_arxA_F and M13_R primer sets in order to detect pSMV20∷arxA genomic insertion within arxA gene in chloramphenicol resistant colonies.
Generation of BSL-9 arxA gene disruption mutant
To perform mating reactions E. coli strain WM3064 cells containing pSMV20∷arxA were grown overnight at 37°C in Luria Broth (LB) containing DAP and BSL-9 wild-type strain was grown in BSM for 2 days at 35°C under infrared (IR) lights. About 1 ml of E. coli WM3064 containing pSMV20∷arxA and BSL-9 Wild-type strain liquid cultures were combined and spun down at 10,000 RPM for 1 min. The supernatant was removed (∼ 2 ml), and the combined cells were suspended in 1 ml of LB containing DAP. About 60 μl of the combined cells were spot plated on the centre of an LB plate containing acetate and nitrate with approximately pH 8.2. Reactions were incubated for 6 hours at room temperature under IR lights in an anaerobic chamber. After 6 hours the mating reaction was collected with a sterile metal loop and re-suspended in 200 μl BSM liquid media containing 5 μg/ml chloramphenicol. The mixture was then plated on BSM agar plates containing 5 μg/ml chloramphenicol. After a week, BSL-9 chloramphenicol resistant colonies were picked and inoculated into BSM media containing chloramphenicol. Two days thereafter, BSL-9 chloramphenicol resistance cells were harvested by centrifugation and used to extract genomic DNA for analysis of genomic plasmid integration.
arxA mRNA quantification by qRT-PCR
BSL-9 wild-type cells were grown in BSM medium with 10 mM acetate to stationary phase then diluted to OD600nm of 0.6. A 1:100 dilution was made into 6 Balch tubes containing BSM media with 10 mM acetate. After cells reached an OD600nm of 0.2, 1 mM arsenite was spiked into each triplicate culture, while three other replicates did not receive arsenite. The tubes were sampled before the arsenite spike and 2, 12 and 24 hours post arsenite addition. Cells were processed for RNA purification and cDNA synthesis as described elsewhere (Saltikov et al., 2005). Briefly 500 ng of DNase treated RNA was conveted to cDNA using Applied Biosystems TaqMan kit (Part No. N808-0234). To quantify arxA gene expression relative to the 16S rRNA gene a probe-based real-time PCR assay was used. The IDT PrimeTime qPCR primer probe sets for arxA were (arxA: Probe 50′-/56-FAM/CCATCGCCA/ZEN/ATGGCAAGCTGTG/3IABkFQ/-30′; Primer 1, 5′-CAAGGTCACCGCCATCTAC-3′; Primer 2, 5′-GTCCGGGTCATACAGGAAATAG-3′), and 16S rRNA gene were (Probe 5′-/56-FAM/CGGTGTAGC/ZEN/GGTGAAATGCGTAGA/3IABkFQ/-3′; Primer 1, 5′-GCATGGCTAGAGTTTGGTAGAG-3′; Primer 2, 5′-CGCACCTCAGTGTCAGTTT-3′). Real-time PCR reactions consisted of 20 μl volume per reaction; 10 μl of 2×Taq Mix (Promega), 1 μl of 20× probe-primer, 5 μl of H2O and 4 μl of cDNA (diluted 1:4 in H2O). BSL-9 wild-type DNA was used as a positive control for generating standard curves in order to quantify arxA relative expression. The Biorad Real-Time PCR machine was used with the following thermal profile: 95°C for 5 min, 40 cycles of 95°C/30 s, 55°C/30 s and 72°C/30 s. The quantification analysis of arxA expression was done using the ΔΔCq method (Livak and Schmittgen, 2001). The ΔCq values were calculated using Cq values of arxA and 16S rRNA gene. The no arsenite condition was used as a reference for the ΔΔCq calculation and the fold change corresponded to 2−ΔΔCq
Cell suspension arsenite oxidation assay
To test for growth independent arsenite oxidation, high-density cell suspension assays were examined. BSL-9 wild-type and BSL-9 arxA mutant were grown in BSM media containing 10 mM acetate. Cells were washed and transfer to BSM media lacking yeast and acetate but containing approximately 100 μM arsenite. Each growth condition was done in triplicates with an initial OD600nm of 0.5 (∼ 4.0 × 108 cells/ml). Anaerobic samples were stored either in the dark or under IR lights at 35°C. In order to analyse and quantify arsenite and arsenate, 500 μl of each sample was taken out over time, every hour and incubated at 4°C. Arsenic analysis was done routinely two days after sampling.
Analytical techniques
Arsenic speciation was analysed and quantified by High Performance Liquid Chromatography (HPLC) coupled to an Inductively Coupled Plasma-Mass Spectroscopy (ICP-MS). Elemental detection was carried out by Thermo X-Series2 ICP-MS equipment. The isocratic mobile phase contained 30 mM phosphoric acid, which was also used in the autosampler flush. The flow rate was 1 min/ml with 100 μl sample injection loop. An anion exchange column (Hamilton PRP-X-100, particle size 10 mm, size 4.1 × 50 mm) was used with a run time of 3 min per sample. The column temperature was ambient. All samples were diluted in 30 mM phosphoric acid mobile phase containing germanium as an internal control.
Phylogenetic analysis
The phylogenetic analysis of ArxA sequence of Ectothiorhodospira sp. str. BSL-9 and other DMSO reductase family molybdenum-containing oxidoreductases was done as previously described (Oremland, 2003).
Supplementary Material
Fig. S1. Phototrophic growth curve analysis of BSL-9 wildtype and ARXA1 (arxA mutant) with acetate and thiosulfate. Strains were grown under photoautotrophic conditions in the presence of thiosulfate (diamonds) and photoheterotrophic conditions in the presence of acetate (circles) and compared with samples lacking electron donors as a negative control (triangles). No general growth defects were observed in BSL-9 wild-type and ARAX1 when providing thiosulfate (diamonds) or acetate (circles) as electron donors. No growth was observed in the absence of an electron donor (triangles) relative to acetate or thiosulfate. Data points and error bars in corresponding figures represent the averages and standard deviation of replicate samples.
Fig. S2. Transcription analysis of arx genes in BSL-9 wildtype and BSL-9 arxA mutant strain (ARXA1). To investigate the polar effects and impact of arxA gene insertion on downstream genes such as arxB1CD, we analyzed activity of the arxB2AB1CD gene cluster. BSL-9 ARXA1 and wildtype cells were grown (in triplicates) photosynthetically on 20 mM acetate to early exponential phase (OD600nm 0.2) and induced with 300 μM arsenite for 2 hours. Panels (A) and (B) are agarose gel electrophoresis (1%) images of the RT-PCR analyses for a single representative culture: BSL-9 wild-type (lane 2, 4, 6, 8, 10, 12, 14) and ARXA1 mutants (lane 3, 5, 7, 9, 11, 13, 15). The other two replicates showed identical results. cDNAs were generated with (A) and without (B) reverse transcriptase and analyzed for the presence of arxB2 (lane 2-3), a region flanking the arxA before (lane 4-5) and after (lane 6-7) the plasmid insertion mutation, arxB1 (8-9), arxC (10-11), arxD genes (lanes 12-13), 16S rRNA gene as a positive control for bacterial mRNA expression (lanes 14-15), and No DNA samples as a control for contamination. Genes were down regulated in BSL-9 ARXA1 mutant (lanes 7, 9, 11, 13) relative to BSL-9 wild-type cells (lanes 6, 8, 10, 12). Panel (C) shows PCR results using genomic DNA for BSL-9 wild-type and BSL-9 ARXA1 mutant as a positive control for gene amplification Detection was observed for all genes in the arxB2AB1CD gene cluster (same primer sets were used in RT-PCR).
Table S1. Bacterial strains and plasmids used in this study.
Table S2. Primers and probes PCR parameters.
Table S3. Metagenomic arxA detection in non-extreme environments. The BSL-9 arxA gene was used in a BLAST search of the JGI metagenome data sets. The sequences that were similar where then BLAST searched in NCBI to verify their similarties to BSL-9 wild-type strain and other arxA-like sequences. The data reported here are the identities/similarties to the BSL-9 arxA gene (BLASTn) and predict ArxA protein sequence (BLASTx).
See Excel sheet: Table_3S_arxA_metagenomes.xlsx
See Fasta File: Table_3S_arxA_metagenomes.fna
Acknowledgments
This work was supported by the National Science Foundation (EAR-1349366) award to CWS; UCSC RMI fellowship (NIHNHGRI 1R25HG006836-01A1) and UCSC IMSD fellowship (NIH/HIGMS 5R25GM058903-14) to Jaime Hernandez-Maldonado; UCSC PREP fellowship (NIH 5R25GM104552) to Benjamin Sedillo-Sanchez. We thank Professor Dianne Newman for constructive advise on sampling red biofilm-like mats in Paoha Island; John Stolz for discussions on earlier versions of the manuscript. We thank Rob Franks (UC Santa Cruz) for tremendous help in developing a rapid automated HPLC-ICP-MS arsenic analysis method. We thank the Sierra Nevada Aquatic Research Laboratory for facilitating fieldwork.
Footnotes
Author contributions: J. Hernandez-Maldonado performed genetic system development, mutant generation, physiology experiments, arsenic analysis, and manuscript writing. B. Sanchez-Sedillo performed qRT-PCR and B. Stoneburner performed cloning and environmental sequencing of arxA PCR amplicons. All authors helped with field sampling on Paoha Island Mono Lake, California. A. Boren (Conrad) isolated Ectothiorhodospira sp. str. BSL-9. M. Rosen provided access to USGS research vessel on Mono Lake and Big Soda Lake. C.W Saltikov contributed to manuscript writing process and served as PI on the project. R.S. Oremland contributed to manuscript writing, field sampling logistics, and funding USGS Menlo Park personnel in support of field sampling.
Competing financial interests: The author declares no competing financial interests.
Supporting information: Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
References
- Acharyya SK, Chakraborty P, Lahiri S, Raymahashay BC, Guha S, Bhowmik A. Arsenic poisoning in the Ganges Delta. Nature. 1999;401:545–546. doi: 10.1038/44052. [DOI] [PubMed] [Google Scholar]
- Amend JP, Saltikov C, Lu GS, Hernandez J. Microbial arsenic metabolism and reaction energetics. Rev Miner Geochem. 2014;79:391–433. [Google Scholar]
- Budinoff CR, Hollibaugh JT. Arsenite-dependent photoautotrophy by an Ectothiorhodospira-dominated consortium. ISME J. 2008;2:340–343. doi: 10.1038/ismej.2007.115. [DOI] [PubMed] [Google Scholar]
- Cammack R, Rao KK, Hall DO. Metalloproteins in the evolution of photosynthesis. Biosystems. 1981;14:57–80. doi: 10.1016/0303-2647(81)90022-8. [DOI] [PubMed] [Google Scholar]
- Cloern JE, Cole BE, Oremland RS. Autotrophic processes in meromictic Big Soda Lake, Nevada. Limnol Oceanogr. 1983;28:1049–1061. [Google Scholar]
- Dhar RK, Zheng Y, Saltikov CW, Radloff KA, Mailloux BJ, Ahmed KM, van Geen A. Microbes enhance mobility of arsenic in Pleistocene aquifer sand from Bangladesh. Environ Sci Technol. 2011;45:2648–2654. doi: 10.1021/es1022015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich A, Widdel F. Anaerobic oxidation of ferrous iron by purple bacteria, a new type of phototrophic metabolism. Appl Environ Microbiol. 1994;60:4517–4526. doi: 10.1128/aem.60.12.4517-4526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel AS, Johnson LR, Porter ML. Arsenite Oxidase gene diversity among Chloroflexi and Proteobacteria from El Tatio Geyser Field, Chile. FEMS Microbiol Ecol. 2013;83:745–756. doi: 10.1111/1574-6941.12030. [DOI] [PubMed] [Google Scholar]
- Fendorf S, Michael HA, van Geen A. Spatial and temporal variations of groundwater arsenic in South and Southeast Asia. Science. 2010;328:1123–1127. doi: 10.1126/science.1172974. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Meth. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Griffin BM, Schott J, Schink B. Nitrite, an electron donor for anoxygenic photosynthesis. Science. 2007;316:1870. doi: 10.1126/science.1139478. [DOI] [PubMed] [Google Scholar]
- Harvey CF. Groundwater flow in the Ganges Delta. Science. 2002;296:1563. doi: 10.1126/science.296.5573.1563a. [DOI] [PubMed] [Google Scholar]
- Hoeft SE, Kulp TR, Han S, Lanoil B, Oremland RS. Coupled arsenotrophy in a hot spring photosynthetic biofilm at Mono Lake, California. Appl Environ Microbiol. 2010;76:4633–4639. doi: 10.1128/AEM.00545-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam FS, Gault AG, Boothman C, Polya DA, Charnock JM, Chatterjee D, Lloyd JR. Role of metal-reducing bacteria in arsenic release from Bengal Delta sediments. Nature. 2004;430:68–71. doi: 10.1038/nature02638. [DOI] [PubMed] [Google Scholar]
- Jormakka M, Yokoyama K, Yano T, Tamakoshi M, Akimoto S, Shimamura T, et al. Molecular mechanism of energy conservation in polysulfide respiration. Nat Struct Mol Biol. 2008;15:730–737. doi: 10.1038/nsmb.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap DR, Botero LM, Franck WL, Hassett DJ, McDermott TR. Complex regulation of arsenite oxidation in Agrobacterium tumefaciens. J Bacteriol. 2006;188:1081–1088. doi: 10.1128/JB.188.3.1081-1088.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempe S, Kazmierczak J. A terrestrial model for an alkaline martian hydrosphere. Planet Space Sci. 1997;45:1493–1499. [Google Scholar]
- Kocar BD, Fendorf S. Thermodynamic constraints on reductive reactions influencing the biogeochemistry of arsenic in soils and sediments. Environ Sci Technol. 2009;43:4871–4877. doi: 10.1021/es8035384. [DOI] [PubMed] [Google Scholar]
- Kocar BD, Polizzotto ML, Benner SG, Ying SC, Ung M, Ouch K, et al. Integrated biogeochemical and hydrologic processes driving arsenic release from shallow sediments to groundwaters of the Mekong Delta. Appl Geochem. 2008;23:3059–3071. [Google Scholar]
- Kulp TR, Hoeft SE, Asao M, Madigan MT, Hollibaugh JT, Fisher JC, et al. Arsenic(III) fuels anoxygenic, photosynthesis in hot spring biofilms from Mono Lake, California. Science. 2008;321:967–970. doi: 10.1126/science.1160799. [DOI] [PubMed] [Google Scholar]
- Lavergne J, Vermeglio A, Joliot P. Functional coupling between reaction centers and cytochrome bc1 complexes. In: Hunter CN, Daldal F, Thurnauer MC, Thomas Beatty J, editors. The Purple Phototrophic Bacteria. New York: Springer; 2009. pp. 509–536. [Google Scholar]
- Lett MC, Muller D, Lievremont D, Silver S, Santini J. Unified nomenclature for genes involved in Prokaryotic aerobic arsenite oxidation. J Bacteriol. 2011;194:207–208. doi: 10.1128/JB.06391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Masscheleyn PH, Delaune RD. Effect of redox potential and pH on arsenic speciation and solubility in a contaminated soil. Environ Sci Technol. 1991;25:1414–1419. [Google Scholar]
- Nickson R, McArthur J, Burgess W, Ahmed KM, Ravenscroft P, Rahman M. Arsenic poisoning of Bangladesh groundwater. Nature. 1998;395:338. doi: 10.1038/26387. [DOI] [PubMed] [Google Scholar]
- O'Day PA, Vlassopoulos D, Root R, Rivera N. The influence of sulfur and iron on dissolved arsenic concentrations in the shallow subsurface under changing redox conditions. Proc Natl Acad Sci USA. 2004;101:13703–13708. doi: 10.1073/pnas.0402775101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland RS. The ecology of arsenic. Science. 2003;300:939–944. doi: 10.1126/science.1081903. [DOI] [PubMed] [Google Scholar]
- Oremland RS, Stolz JF. Arsenic, microbes and contaminated aquifers. Trends Microbiol. 2005;13:45–49. doi: 10.1016/j.tim.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Oremland RS, Saltikov CW, Wolfe-Simon F, Stolz JF. Arsenic in the evolution of earth and extraterrestrial ecosystems. Geomicrobiol J. 2009;26:522–536. [Google Scholar]
- Overmann J. Ecology of phototrophic sulfur bacteria. In: Hell R, Dahl C, Knaff DB, Leustek T, editors. Sulfur Metabolism in Phototrophic Organisms. New York: Springer; 2008. pp. 375–391. [Google Scholar]
- Robert B. Spectroscopic properties of antenna complexes from purple bacteria. In: Hunter CN, Daldal F, Thurnauer MC, Thomas Beatty J, editors. The Purple Phototrophic Bacteria. New York: Springer; 2009. pp. 199–212. [Google Scholar]
- Saltikov CW. Regulation of arsenic metabolic pathways in prokaryotes. In: Stolz J, Oremland R, editors. Microbial Metal and Metalloid Metabolism. Washington, DC: ASM Press; 2011. pp. 195–210. [Google Scholar]
- Saltikov CW, Newman DK. Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci USA. 2003;100:10983–10988. doi: 10.1073/pnas.1834303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltikov CW, Wildman RA, Newman DK. Expression dynamics of arsenic respiration and detoxification in Shewanella sp. strain ANA-3. J Bacteriol. 2005;187:7390–7396. doi: 10.1128/JB.187.21.7390-7396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiwal S, Santini JM, Osborne TH, Djordjevic S. Characterization of a two-component signal transduction system that controls arsenite oxidation in the chemolithoautotroph NT-26. FEMS Microbiol Lett. 2010;313:20–28. doi: 10.1111/j.1574-6968.2010.02121.x. [DOI] [PubMed] [Google Scholar]
- Sessions AL, Doughty DM, Welander PV. The continuing puzzle of the Great Oxidation Event. Curr Biol. 2009;19:567–574. doi: 10.1016/j.cub.2009.05.054. [DOI] [PubMed] [Google Scholar]
- Sforna MC, Philippot P, Somogyi A, van Zuilen MA, Medjoubi K, Schoepp-Cothenet B, et al. Evidence for arsenic metabolism and cycling by microorganisms 2.7 billion years ago. Nat Geosci. 2014;7:811–815. [Google Scholar]
- Smith AH, Lingas EO, Rahman M. Contamination of drinking-water by arsenic in Bangladesh: A public health emergency. Bull World Health Organ. 2000;78:1093–1103. [PMC free article] [PubMed] [Google Scholar]
- Stuckey JW, Schaefer MV, Kocar BD, Benner SG, Fendorf S. Arsenic release metabolically limited to permanently water-saturated soil in Mekong Delta. Nat Geosci. 2015;9:70–76. [Google Scholar]
- Wallis J, Wickramasinghe NC. Proceedings of SPIE Optical, Instruments, Methods, and Missions for Astrobiology XVII, 96060V. 2015. Evidence of ancient microbial activity on Mars. [Google Scholar]
- Widdel F, Schnell S, Heising S, Ehrenreich A, Assmus B, Schink B. Ferrous iron oxidation by anoxygenic phototrophic bacteria. Nature. 1993;362:834–836. [Google Scholar]
- Zargar K, Hoeft S, Oremland R, Saltikov CW. Identification of a novel arsenite oxidase gene, arxA, in the haloalkaliphilic, arsenite-oxidizing bacterium Alkalilimnicola ehrlichii strain MLHE-1. J Bacteriol. 2010;192:3755–3762. doi: 10.1128/JB.00244-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zargar K, Conrad A, Bernick DL, Lowe TM, Stolc V, Hoeft S, et al. ArxA, a new clade of arsenite oxidase within the DMSO reductase family of molybdenum oxidoreductases. Environ Microbiol. 2012;14:1635–1645. doi: 10.1111/j.1462-2920.2012.02722.x. [DOI] [PubMed] [Google Scholar]
- Zehr JP, Harvey RW, Oremland RS. Big Soda Lake (Nevada). 1. Pelagic bacterial heterotrophy and biomass. Limnol Oceanogr. 1987;32:781–793. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Phototrophic growth curve analysis of BSL-9 wildtype and ARXA1 (arxA mutant) with acetate and thiosulfate. Strains were grown under photoautotrophic conditions in the presence of thiosulfate (diamonds) and photoheterotrophic conditions in the presence of acetate (circles) and compared with samples lacking electron donors as a negative control (triangles). No general growth defects were observed in BSL-9 wild-type and ARAX1 when providing thiosulfate (diamonds) or acetate (circles) as electron donors. No growth was observed in the absence of an electron donor (triangles) relative to acetate or thiosulfate. Data points and error bars in corresponding figures represent the averages and standard deviation of replicate samples.
Fig. S2. Transcription analysis of arx genes in BSL-9 wildtype and BSL-9 arxA mutant strain (ARXA1). To investigate the polar effects and impact of arxA gene insertion on downstream genes such as arxB1CD, we analyzed activity of the arxB2AB1CD gene cluster. BSL-9 ARXA1 and wildtype cells were grown (in triplicates) photosynthetically on 20 mM acetate to early exponential phase (OD600nm 0.2) and induced with 300 μM arsenite for 2 hours. Panels (A) and (B) are agarose gel electrophoresis (1%) images of the RT-PCR analyses for a single representative culture: BSL-9 wild-type (lane 2, 4, 6, 8, 10, 12, 14) and ARXA1 mutants (lane 3, 5, 7, 9, 11, 13, 15). The other two replicates showed identical results. cDNAs were generated with (A) and without (B) reverse transcriptase and analyzed for the presence of arxB2 (lane 2-3), a region flanking the arxA before (lane 4-5) and after (lane 6-7) the plasmid insertion mutation, arxB1 (8-9), arxC (10-11), arxD genes (lanes 12-13), 16S rRNA gene as a positive control for bacterial mRNA expression (lanes 14-15), and No DNA samples as a control for contamination. Genes were down regulated in BSL-9 ARXA1 mutant (lanes 7, 9, 11, 13) relative to BSL-9 wild-type cells (lanes 6, 8, 10, 12). Panel (C) shows PCR results using genomic DNA for BSL-9 wild-type and BSL-9 ARXA1 mutant as a positive control for gene amplification Detection was observed for all genes in the arxB2AB1CD gene cluster (same primer sets were used in RT-PCR).
Table S1. Bacterial strains and plasmids used in this study.
Table S2. Primers and probes PCR parameters.
Table S3. Metagenomic arxA detection in non-extreme environments. The BSL-9 arxA gene was used in a BLAST search of the JGI metagenome data sets. The sequences that were similar where then BLAST searched in NCBI to verify their similarties to BSL-9 wild-type strain and other arxA-like sequences. The data reported here are the identities/similarties to the BSL-9 arxA gene (BLASTn) and predict ArxA protein sequence (BLASTx).
See Excel sheet: Table_3S_arxA_metagenomes.xlsx
See Fasta File: Table_3S_arxA_metagenomes.fna


