Abstract
Glioblastoma (GBM) is comprised of distinct subtypes characterized by their molecular profile. Mesenchymal identity in GBM has been associated with a comparatively unfavorable prognosis, primarily due to inherent resistance of these tumors to current therapies. The identification of molecular determinants of mesenchymal transformation could potentially allow for the discovery of new therapeutic targets. Zinc Finger and BTB Domain Containing 18 (ZBTB18/ZNF238/RP58) is a zinc finger transcriptional repressor with a crucial role in brain development and neuronal differentiation. Here, ZBTB18 is primarily silenced in the mesenchymal subtype of GBM through aberrant promoter methylation. Loss of ZBTB18 contributes to the aggressive phenotype of glioblastoma through regulation of poor prognosis-associated signatures. Restitution of ZBTB18 expression reverses the phenotype and impairs tumor-forming ability. These results indicate that ZBTB18 functions as a tumor suppressor in GBM through the regulation of genes associated with phenotypically aggressive properties.
Implications
This study characterizes the role of the putative tumor suppressor ZBTB18 and its regulation by promoter hypermethylation, which appears to be a common mechanism to silence ZBTB18 in the mesenchymal subtype of GBM and provides a new mechanistic opportunity to specifically target this tumor subclass.
Keywords: Glioblastoma, ZBTB18, DNA methylation
Introduction
Glioblastoma (GBM) is the most malignant primary brain tumor, characterized by a highly invasive nature, poor prognosis, and resistance to aggressive therapies [1]. Over the past decade, gene expression profiling has contributed to the identification of multiple GBM subclasses with distinct molecular and clinical characteristics [2, 3]. In particular, the mesenchymal (MES) and the proneural (PN) groups appear as the most consistent subclasses reported in both studies [2, 3]. The MES subtype is characterized by resistance to radiotherapy [4].
Using bioinformatics tools [5, 6], several transcription factors have been identified as master regulators of a “mesenchymal gene expression signature” (MGES) in GBM, including STAT3, CEBPB and WWTR1 (a.k.a. TAZ) [7, 8]. More recently, a role of NF-κB in controlling the expression of the three master regulators and consequent mesenchymal differentiation was reported [4]. Notably, the transcriptional repressor Zinc Finger And BTB Domain Containing 18 (ZBTB18; formerly ZNF238) was identified as a potential negative regulator of the MGES in GBM [8]. ZBTB18 is a transcription factor that belongs to the Broad complex, Tramtrack, Bric à brac [BTB] or poxvirus and zing finger [POZ]-zinc finger (BTB/POZ-ZF) protein family and plays a crucial role in brain development and neuronal differentiation [9-12]. Previous findings revealed that ZBTB18 is downregulated or lost in mouse gliomas and human GBM cell lines and have implicated ZBTB18 as a putative tumor suppressor in the brain [11]. However, the mechanism of ZBTB18 downregulation in GBM remains to be defined.
Mounting evidence suggests that alteration of methylation pathways, which can induce silencing of tumor suppressor genes, is one of the earliest events in carcinogenesis that grant a predisposition to mutational changes [13, 14]. The DNA repair gene O-6-methylguanine-DNA methyltransferase (MGMT) is one such gene for which promoter methylation has been shown to result in gene silencing in many cancers [15-17].
Here, we have characterized the role of ZBTB18 as a transcriptional repressor of gene signatures associated with aggressive properties and poor prognosis in GBM. Our results indicate that ZBTB18 serves as a tumor suppressor in the brain and is silenced by DNA methylation in mesenchymal GBM.
Materials and Methods
Tumor samples and culture of GBM derived cells
Glioblastoma and cortical samples from epilepsy surgery were collected at the Department of Neurosurgery of the University Medical Center Freiburg in accordance with an Institutional Review Board (IRG)-approved protocol. Informed consent was obtained from all patients, in accordance with the declaration of Helsinki. Patient-derived glioblastoma stem cells (BTSCs) were prepared from tumor specimens as previously described [18]. For passaging, neurospheres were incubated in non-enzymatic cell dissociation solution (Sigma) and mechanically dissociated by pipetting. Proneural cells 3047, 3082 and 3111 were generated at the University of Uppsala [19]. Glioblastoma cell lines (SNB19 and LN229) and HEK 293T cells were routinely grown in DMEM with 10% FBS. SNB19 and LN229 cells have been authenticated on 2.3.2017 by PCR-single-locus-technology (Eurofins Medigenomix). All cells were mycoplasma-free.
Classification of brain tumor stem cells (BTSC)
The classification of BTSCs was performed using 510 genes out of the 840 classifier genes used by Verhaak et al. to classify 260 glioblastoma samples (Verhaak et al., 2010), and 529 glioblastoma tissue samples from TCGA with assigned subtypes for reference (Cancer Genome Atlas Research Network, 2008). The 510 genes were selected such that they reliably classify the extended set of 529 TCGA samples and were represented on the Illumina HumanHT-12v3 expression BeadChip arrays. The expression levels for these genes on the Illumina arrays and in the TCGA data set were converted into z-scores and the combined matrix was used to classify each BTSC sample based on a k-nearest neighbors (k=10) and voting procedure, in which a subtype was assigned based on the majority subtype among the 10 TCGA samples with highest correlation coefficients for these genes with respect to the BTSC sample. All data manipulations were performed in R (R Core Team, 2012) and MATLAB (The MathWorks, Inc., Natick, MA, United States).
The classification of BTSCs was performed using a method of hierarchical clustering with a Euclidean distance metric to cluster the samples alongside the TCGA samples. Two mRNA data sets with subtype assignment were used as references: an mRNA data set of 529 patient samples from TCGA assignment and a subset of the 810 signature genes published by Verhaak and colleagues [3]. Gliomas (BT) and BTSC classification is reported in Supplementary Table 1.
ZBTB18 vector construction, lentiviral production and infection
ZBTB18 coding sequence was PCR amplified from normal brain RNA using the following primers: BstXI-flag-hZBTB18v1-sense (TGGCCACAACCATGGACTAC AAGGACGACGATGACAAGTGTCCTAAAGGTTATGAAGACAG) and PmeI-hZBTB18-antisense (GCCTTGGTTTAAACTTATTTCCAAAGTTCTTGAGAG). The PCR product was first cloned into pDrive cloning vector (QIAGEN), sequence validated and subsequently transferred in the pCHMWS-EGFP lentiviral vector vector (kind gift from Veerle Baekelandt, University of Leuven). Lentiviral infections were performed as previously described [18].
Proteomic analysis
For protein analysis of the 30kDa band detected by the ZBTB18 antibody, SNB19 cells transduced with either control or FLAG-ZBTB18 lentiviral vector were subjected to immunoprecipitation using the M2 FLAG antibody (SIGMA). The immunoprecipitated proteins were analyzed by gel electrophoresis and stained by comassie blue staining. The 30kDa band was cut from the gel and analyzed by MS (Agilent 6520 Q-TOF) at the Core Facility Proteomics of the Center for Biological System Analysis (ZBSA) at the University of Freiburg. The identified peptides were aligned using the mascot software (Matrix science, Boston, USA).
DNA/RNA extraction and quantitative real time-PCR
DNA and RNA were extracted from human cortex, tumor tissue or cell culture using All Prep DNA/RNA Mini Kit or miRNeasy Mini Kit (Qiagen). First strand cDNA synthesis was generated using the Superscript cDNA synthesis kit (Invitrogen). Quantitative RT-PCR was performed using SYBR Green (Applied Biosystem) and analyzed relative to 18sRNA (housekeeping) using the ΔCt method. Primer sequences are listed in Supplementary Table 2.
Immunoblotting
Total protein extracts were prepared in RIPA buffer supplemented with protease inhibitor cocktail (Thermo Scientific), phosphatase inhibitor cocktail (Sigma) and PMSF (Sigma). The following antibodies were used: mouse anti-FLAG M2 (Sigma), rabbit anti- ZBTB18 (Abcam, ab118471), and mouse-anti β-actin (Abcam, ab7291).
Microarray expression
For microarray expression profiling, total RNA was prepared using the RNeasy kit or the all Prep DNA/RNA/Protein mini kit (Qiagen) and quantified using 2100 Bioanalyzer (Agilent). 1.5 μg of total RNA was processed and analyzed at the Deutsches Krebsforschungszentrum (DKFZ) in Heidelberg (Germany). Hybridization was carried out on Illumina HumanHT-12v4 expression BeadChip. Microarray data were analyzed using the GSEA software (http://www.broadinstitute.org/gsea/index.jsp). Microarray gene assession numer: GSE97350 (subseries GSE97347 and GSE97349).
Methylation analysis and pyrosequencing
Genomic DNA from tissues and cell lines was bisulfite modified using the EZ DNA methylation–gold kit (The Epigenetics Company) according to the manufacturer's instructions. A pool of normal brain tissues or glioma samples were subjected to PCR to amplify ZBTB18 promoter CpG islands using PyroMark PCR kit (Qiagen) and primers summarized in Supplementary Table 3. Non-CpG containing primers, methylation-specific primers (MSP) and un-methylation-specific primers (USP) were designed to cover the entire region. PCR products were cloned into pDrive cloning vector (Qiagen) and submitted for sequencing with SP6 primer and T7 primer (GATC).
Pyrosequencing analysis was performed using a PyroMark Q96 instrument (Qiagen), following the manufacturers protocol. Primers are summarized in Supplementary Table 3. The results were analyzed by PyroMark CpG software (Qiagen). The methylation index for each sample was calculated as the average value of CpG methylation in the CpG examined.
In vitro methylation and luciferase reporter assay
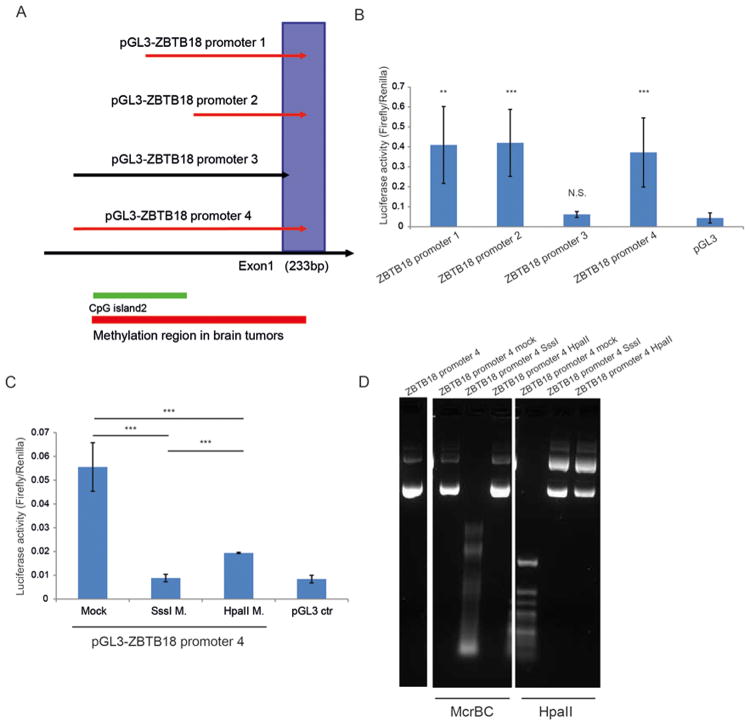
Different regions of the ZBTB18 promoter were amplified by PCR from brain (cortex)-derived RNA and cloned in the pGL3 vector (Promega). The activity of each promoter was measured by dual luciferase reporter assay system (Promega) according to the manufacturer's instructions. Cells were seeded at 50% confluence in 6-well plates 24 hours prior to transfection. Cells at 80-90% confluence were transiently transfected with 2.5μg of each pGL3-ZBTB18 promoter vector or pGL3 as a negative control, together with 0.5 μg of pRL-TK (Renilla luciferase reporter, Promega). Transfections were done using Lipofectamine 2000 as directed by the manufacturer (Life Technologies) in serum-free medium. All transfections were done in triplicate. After 48 hours cells were lysed with passive lysis buffer (Promega). Luciferase activity of each sample was determined in technical triplicate using a Thermo Scientific Appliskan luminometer. All data were reported relative to luciferase activity (firefly/renilla). 20 μg of pGL3-ZBTB18 promoter 4 vector was methylated with SssI methylase or with HpaII methylase (all from New England Biolabs, 2.5 U/μg DNA) in the presence of 160 μM S-Adenosylmethionine (SAM; New England Biolabs), in a manufacturer-supplied buffer at 37 °C for 2 h. The unmethylated DNA was treated as above but without methylases or SAM. The plasmid DNA was extracted by phenol/chlroroform, ethanol precipitated and quantified using a Nanodrop Spectrophotometer (Peqlab). The completion of methylation reaction was controlled by digesting both methylated and unmethylated DNA using the methylation sensitive restriction enzymes Hpa II and the methylation insensitive restriction enzymes McrBC.
Chromatin Immunoprecipitation (ChIP)
Promoter analysis was performed with the MatInspector software (www.Genomatix.de). Primers were designed using the Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and are listed in Supplementary Table 4. ChIP was performed as previously described [20] with some modifications. SNB19 cells expressing ZBTB18 were first crosslinked with 1% formaldehyde (Polysciences) for 30 minutes at room temperature and quenched by addition of 125mM Glycine for 15 minutes. Lysates were sonicated for 15 min (30 sec on / 30 sec off) using Branson digital sonifier and centrifuged at 14.000 rpm for 15 minutes at 4C. 50ug of sonicated chromatin per IP was pre-cleared and incubated with primary antibody (4 ug) anti-ZBTB18 (Abcam, ab118471). Chromatin-Antibody complexes were eluted by two 15 minute incubations at 30°C with 250 μl Elution Buffer (1% SDS, 100 mM NaHCO3). Chromatin was reverse-crosslinked by adding 20 μl of NaCl 5M and incubated at 65°C for 12 hours. DNA was extracted by phenol-chloroform after RNase and proteinase K digestion. Quantitative RT-PCR was performed using SYBR Green (Applied Biosystems).
Methylation array
For methylation, DNA was prepared using All Prep DNA/RNA/Protein mini kit (Qiagen) and quantified using NanoDrop2000c (Thermo Scientific). 1.5 μg of DNA for each sample was processed at DKFZ (Heidelberg, Germany) by Illumina HumanMethylation450 (Illumina). Data analysis was performed using Integrative Genomics Viewer (IGV) software (Broad Institute) [21, 22].
Migration, invasion and proliferation assay
Migration and invasion assays were performed as described before [18]. Images of migrating cells were taken every 24 hours. For BTSC233 and JX6 cells, laminin coated (Invitrogen, 4μg/ml) 60mm dishes containing a culture insert (Ibidi) were used. Cell migration was calculated using the following formula: (Pre-Migration Area – Migration Area)/Pre-Migration Area X100).
For invasion assay, 1×105 BTSC233, JX6 or SNB19 (both 2.5×104) cells were seeded in triplicates in the upper compartment. PDGF-BB (20ng/ml, R&D) was used as a chemoattractant. Pictures were acquired using an Axioimager 2 Microscope (Zeiss). The assays were validated in two independent experiments.
Cell proliferation was assessed using a commercially available kit for EdU detection (EdU Cell proliferation assay, base click). Cells were plated at a density of 2.0×104 per well in a 24 well microplates containing laminin-coated coverslips. After 24 hours of seeding, cells were incubated with EdU solution overnight following the manufacturer's instructions. Upon EdU detection, images were acquired using a fluorescent microscope (Axiovert, Zeiss).
5-AZA-2′-deoxycytidine (5-aza-dC) treatment
100 mM stock solutions of 5-AZA-dC (Sigma-Aldrich) were prepared by dissolving the substances in DMSO (GIBCO) and stored at −80°C. Immediately before treatment, stock solutions were diluted in cold PBS and added to the cell culture medium. LN229 cells not expressing ZBTB18 were treated with 10 μM 5-aza-dC for 2, 3, 4, 5 and 8 days. JX6 cells and BTSC161s cells were treated with equal amounts of 5-aza-dC for 3 and 6 days. Medium containing fresh drug was changed every 24 hours.
Intracranial injection and Immunohistochemistry
Intracranial injections were performed in NOD/SCID mice (Charles River Laboratory) in accordance with the directive 86/609/EEC of the European Parliament, following approval by regional authorities. Experiments were performed as described before [8]. Animals were monitored daily until the development of neurological symptoms by a blinded operator. Histology was performed as previously described [23]. For MIB1 staining, primary antibody MIB1 (1:50) (Dako, M7240) was used.
Statistical analysis
Linear regression analyses and graphical model validation were executed using R software. Scatterplots and locally weighted least squares (LOWESS) smooths were used to confirm the suitability of linear regression analyses, and statistical significance of these relationships was assessed according to the p value for the estimated slope of the regression line. A multiple linear regression model was computed based on the ordinary least squares method. Expression analysis of the TCGA data was performed by R software. Publicly available Level 3 TCGA (https://tcga-data.nci.nih.gov/tcga/) data were used for analysis. Data were downloaded at the UCSC Cancer Genome Browser. Only patients with full datasets (expression, methylome, and clinical information) were included. Expression analysis was performed based on Agilent array data (TCGA GBM G4502A) for high-grade glioma and RNA-seq data (TCGA LGG HiSeqV2 PANCAN) for low-grade glioma. Both datasets were normalized and log2 transformed. Methylation data from Infinium HumanMethylation450 BeadChip for lower-grade and high-grade glioma was used for further analysis. For TCGA Expression/Methylation Analysis, normalized expression values were analysed in tumour subtypes (Mesenchymal, Proneural, Classical and Neural) by a one-way ANOVA model. Survival analysis was performed by Cox proportional hazards model and plotted by Kaplan-Meier survival statistics. Patients without survival data were censored. Robustness was ensured by 10-fold cross validation. Methylation of the cg23829949 was extracted and analysed by Wilcox regression model and one-way ANOVA. Significant level was define as p<0.001. Analysis was performed by survival package included in R-Software. Different tumor numbers used in the analyses reflects sample availability in different genomic datasets and their overlap.
Results
ZBTB18 is downregulated in high-grade gliomas
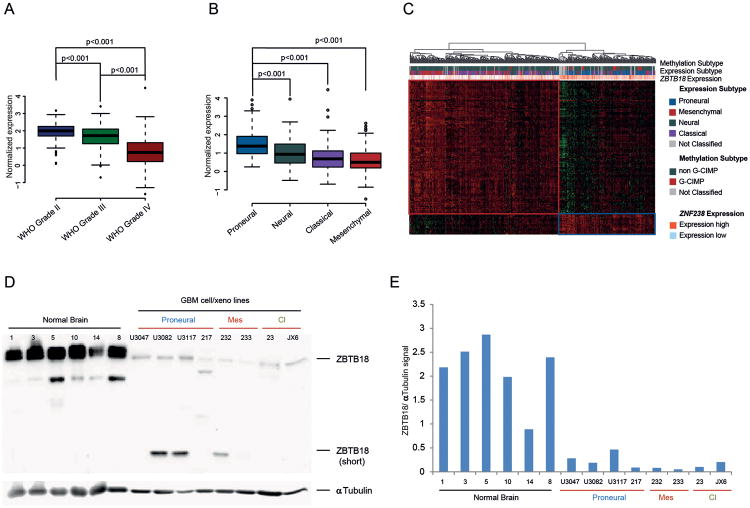
To study the role of ZBTB18 in gliomas, we looked at its expression in 1161 low and high grade gliomas from TCGA. ZBTB18 expression was lower in Glioblastoma (WHO grade IV) compared to grade II and grade III gliomas (p<0.001) (Figure 1A). Interestingly, ZBTB18 expression appeared strongly associated with the proneural subclass when we focused our analysis on high-grade gliomas only (n=561), (Figure 1B). This finding was confirmed by cluster analysis of ZBTB18 correlating genes in high grade gliomas (Figure 1C). ZBTB18 was mostly associated with the proneural subtype and expressed at lower levels in the mesenchymal and classical subtype (Figure 1C), consistent with our previous identification of ZBTB18 as a putative transcriptional repressor of the MGES described by Phillips and colleagues [2, 8]. ZBTB18 protein expression analysis showed low expression of ZBTB18 in GBM-derived cells compared to normal brain samples (cortex) (Figure 1D-E), further reinforcing the notion that ZBTB18 is downregulated in high grade gliomas. Interestingly, a lower band around 30 kDa was also detected.
Figure 1.
ZBTB18 is highly expressed in low grade gliomas and proneural GBMs. (A) Analysis of ZBTB18 expression in low and high-grade glioma samples from TCGA. (B) Analysis of ZBTB18 expression in glioblastoma subtypes from TCGA samples. (C) Cluster analysis showing association between expression of ZBTB18-correlating genes with methylation and, tumor subtype, in glioblastoma samples from TCGA. ZBTB18 appears to be mostly expressed in proneural GBMs. (D) Western blot showing ZBTB18 expression in normal brain tissues, and GBM-derived cells. The low band represents a shorter ZBTB18 isoform. α-Tubulin was used as loading control. (E) Densitometric analysis of the western blot displayed in (D). The ZBTB18 signal was normalized to the α-Tubulin signal.
ZBTB18 directly regulates MGES genes
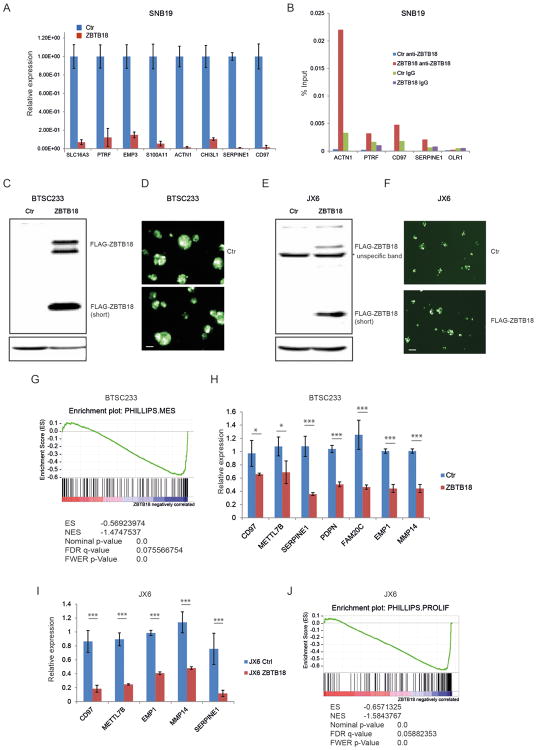
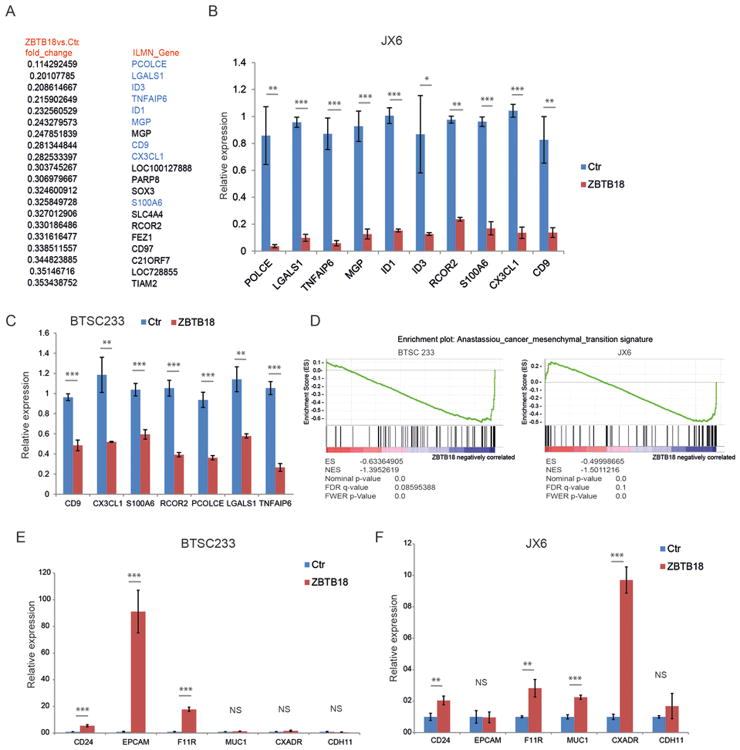
Our previous study identified ZBTB18 as a putative negative regulator of the MGES of glioblastoma [8]. Moreover, cluster analysis shown in Figure 1B indicates that ZBTB18 is downregulated in both mesenchymal and classical GBMs, suggesting that low ZBTB18 expression is associated with the more aggressive GBM subtypes. To validate ZBTB18 repressive function, we first analyzed the expression of mesenchymal genes previously predicted to be negatively connected to ZBTB18 in the ARACNE network [8], using SNB19 glioblastoma cell lines transduced with FLAG-ZBTB18 or control vector (Figure 2A and Supplementary Figure 3A-B). Among 13 putative targets analyzed, 8 genes were clearly downregulated upon ZBTB18 overexpression (Figure 2A). Chromatin immunoprecipitation revealed ZBTB18 binding at ACTN1, PTRF, SERPINE1 and CD97 promoters (Figure 2B), indicating that, at least for a subset of targets, ZBTB18 is directly involved in gene repression. To better understand the role of ZBTB18 in high grade gliomas, we performed overexpression studies and subsequent gene set enrichment analysis (GSEA http://software.broadinstitute.org/gsea/index.jsp) on mesenchymal patient-derived brain tumor stem cell like cell line (BTSC233, Figure 1D-E and Figure 2C-D) and a patient-derived GBM xenoline (JX6) [24] which was classified as classical according to the Verhaak study (Figure 1D-E and Figure 2E-F). Interestingly, we also detected a lower band similar to those observed in GBM-derived cells at around 30 kDa (Figure 2C, E and Figure 1D). Sequence analysis by Mass Spectrometry (MS) confirmed that the peptides correspond to the N-terminal portion of ZBTB18 up to around amino acid 270 (Supplemenentary Figure S1A-B). Western blot analysis of the transduced cells with a polyclonal anti-ZBTB18 antibody showed almost no expression of ZBTB18 in the control cells; the shorter band was weakly recognized since the antibody is directed to central region of ZBTB18 sequence (amino acids 228-498) (Supplemenentary Figure S1C-D). GSEA analysis for the gene signatures described by Phillips and colleagues [2], revealed a strong enrichment for mesenchymal genes in BTSC233 cells expressing a control GFP vector compared to BTSC233 cells expressing ZBTB18 (Figure 2G and Supplementary Figure S2A). Surprisingly, the same analysis using gene signatures from the Verhaak classification [3] did not show any specific signature enrichment (Supplementary Figure S2B). Validation by qRT-PCR in BTSC233 expressing ectopic ZBTB18 confirmed the downregulation of several mesenchymal genes (Figure 2H). The repressive function of ZBTB18 on a subset of genes was further validated in JX6 cells (Figure 2I). GSEA showed a strong downregulation of the Phillips proliferative signature in JX6 transduced with a ZBTB18 expressing lentiviral vector compared to the control vector, but again no significant change was induced in the Verhaak signatures (Figure 2J and Supplementary Figure S3A). Since the classification by Phillips is based on gene expression data of GBMs and grade III gliomas, with the goal of identifying survival-associated genes, we reasoned that the different results might indicate a role of ZBTB18 in the negative regulation of genes associated with poor survival (i.e. proliferative and mesenchymal genes). Interestingly, some of the validated ZBTB18 targets have been previously associated with unfavorable prognosis in glioma [25-27]. Further examination of the top downregulated genes by ZBTB18 in JX6 cells by gene expression array highlighted many genes previously reported to play a role in epithelial-to-mesenchymal transition (EMT) (Figure 3A) [28-34]. ID1 and ID3 were also downregulated, as consistent with previous findings (Figure 3A) [35]. The repressive role of ZBTB18 was validated by q-RT PCR in JX6 (Figure 3B) and BTSC233 (Figure 3C) cells. Many of the validated genes have been reported as part of a multi-cancer gene expression signature associated with prolonged time-to-recurrence in glioblastoma [36, 37]. As such, GSEA showed a strong loss of this multi-cancer signature (Anastassiou_cancer_mesenchymal_transition signature) upon ZBTB18 overexpression (Figure 3D). These data further suggest that ZBTB18 downregulation in high-grade gliomas leads to re-expression of genes associated with malignant features and poor outcome. Interestingly, ZBTB18 re-expression in BTSC233 and JX6 cells led to upregulation of epithelial markers which are often repressed during EMT [38-43] (Figure 3 E-F), further reinforcing the idea that ZBTB18 could play a role in suppressing an EMT-like phenotype in GBM. Consistent with our data and with its previously reported tumor suppressive role [11], ZBTB18 re-expression in SNB19 affected cell proliferation, migration and invasion (Supplementary Figure S4). The same effect on cell proliferation, migration and invasion was confirmed in BTSC233 cells (Supplementary Figure S5A, C-D). In JX6 cells, ZBTB18 overexpression also reduced cell proliferation and migration (Supplementary Figure S5B, E), although no clear effect on invasion was observed, probably due to the higher invasive properties of those cells (data not shown).
Figure 2.
Ectopic expression of ZBTB18 in GBM cells induces repression of poor prognosis signature genes. (A) Representative qRT-PCR analysis showing gene expression changes of ZBTB18 mesenchymal targets previously predicted by ARACNE in SNB19 cells expressing ectopic ZBTB18 (n=3 PCR replicates; error bars ± s.d). Gene expression was normalized to 18sRNA. (B) Representative chromatin immunoprecipitation experiment in SNB19 cells expressing ectopic ZBTB18. The panel shows ZBTB18 binding to the promoter of a subset of mesenchymal targets (n=3 PCR replicates) expressed as percentage of the initial DNA amount in the immune-precipitated fraction. The OLR1 gene which does not contain putative ZBTB18 binding sites was used as negative control. (C) Western blot showing ectopic expression of ZBTB18 in BTSC233. The low band represents a shorter ZBTB18 isoform. (D) Representative images of GFP positive BTSC233 cells expressing control vector or FLAG-ZBTB18 construct after lentiviral infection. The scale bar represents 100 μm. (E) Western blot showing ectopic expression of ZBTB18 in JX6. The shorter ZBTB18 isoform is indicated. (F) Representative images of GFP positive JX6 cells transduced with control vector or FLAG-ZBTB18 lentiviral vector. The scale bar represents 100 μm. (G) GSEA enrichment plot for mesenchymal genes in the comparison of 233 cells expressing control vector vs. FLAG-ZBTB18. (H-I) Validation by qPCR of selected mesenchymal genes in BTSC233 (H) or JX6 (I) expressing either control vector or FLAG-ZBTB18 construct (n=3; error bars ± s.d). *p < 0.05, **p < 0.01, ***p < 0.001. Gene expression was normalized to 18sRNA. J, GSEA enrichment plot for proliferative genes in the comparison of JX6 cells expressing control vector vs. FLAG-ZBTB18.
Figure 3.
Ectopic expression of ZBTB18 in GBM cells affects a cancer-mesenchymal transition signature. (A) List of the top 20 genes downregulated by ZBTB18 overexpression, in JX6 cells, analyzed by gene expression array. Genes selected for validation are in blue. (B-C) Validation by qRT-PCR of selected genes listed in (A) in BTSC233 (B) or JX6 (C) expressing either control vector or FLAG-ZBTB18 construct (n=3; error bars ± s.d). *p < 0.05, **p < 0.01, ***p < 0.001. Gene expression was normalized to 18sRNA. (D) GSEA enrichment plot for the Anastassiou_cancer_mesenchymal_transition signature described in [37]. (E) qRT-PCR analysis of epithelial genes in BTSC233 (E) and JX6 (F) transduced with either control vector or FLAG-ZBTB18.
ZBTB18 inhibits brain tumor growth in vivo
We next addressed the role of ZBTB18 in tumor formation in vivo. Immunocompromised (NOD/SCID) mice were intracranially injected with JX6 or BTSC233 cells stably expressing either ZBTB18 or control-GFP vector. Histology analysis of the mouse brains revealed that mice injected with JX6 cells expressing control vector developed bulky tumors. Conversely, only one mouse injected with JX6 cells expressing ZBTB18 formed a very small tumor (Figure 4A and data not shown). In accord, overall survival was significantly increased in the ZBTB18 group (Figure 4B). The same experiment in BTSC233 confirmed the effect of ZBTB18 on survival even though the mice still developed tumors (Figure 4C-D). Ki67/MIB1 staining showed a high level of proliferation in BTSC233 transduced with the control-GFP vector whereas, cells expressing ZBTB18 appeared to be less proliferative or confined in small satellite areas, suggesting that ZBTB18 might somehow impair proliferation or restrict it to specific tumor regions (Figure 4E). These results suggest that expression of ZBTB18 prolongs animal survival by delaying or inhibiting tumor formation and that the extent of tumor inhibition might depend on the cell background.
Figure 4.
Expression of ZBTB18 affects tumor formation and survival in vivo. (A) H&E staining of representative tumors resulting from intracranial injection of JX6 cells infected with either control (left) or FLAG-ZBTB18 overexpressing vector (right) in immunocompromised mice. The scale bar represents 1mm. (B) Kaplan-Meier survival curves of mice intracranially injected with JX6 cells expressing full length ZBTB18 or control vector (n=8). (C) H&E staining of representative mice tumors upon intracranial injection of BTSC233 cells infected with either control (left) or FLAG-ZBTB18 overexpressing vector (right) in immunocompromised mice. The scale bar represents 1mm. (D) Kaplan-Meier survival curves of mice intracranially injected in C (n=9). (E) Representative images of mice tumors resulting from intracranial injection of BTSC233 transduced with either control or FLAG-ZBTB18 stained with anti-MIB1 antibody. The scale bar represents 200 μm. A reduced proliferation or pattern of proliferating cells is observed upon ZBTB18 overexpression.
Promoter methylation silences ZBTB18 in GBM
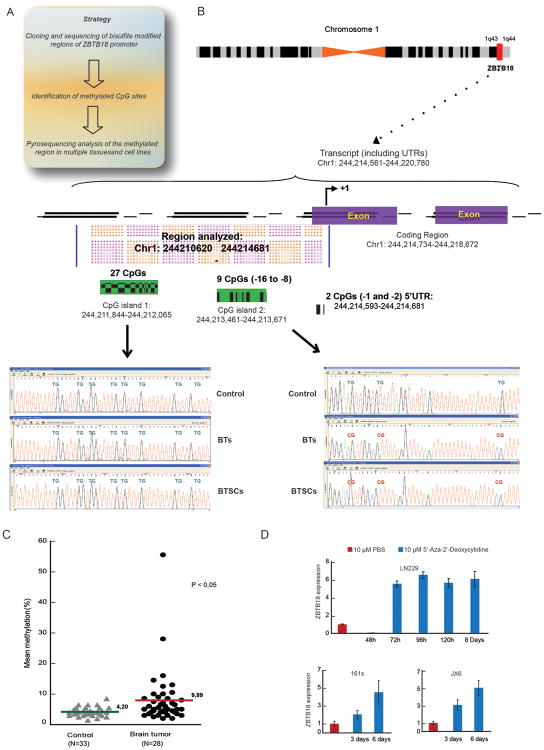
In order to elucidate the mechanism by which ZBTB18 is downregulated in GBM, we examined the ZBTB18 promoter in silico (http://genome.ucsc.edu). The analysis revealed the presence of two CpG islands (Figure 5A-B), suggesting that DNA hypermethylation could play a role in ZBTB18 transcriptional repression. To verify this hypothesis, we first cloned several promoter regions covering the two CpG islands from a pool of normal brains, GBM samples (BTs), and GBM cell lines after bisulfite modification (Figure 5A-B). Sequence analysis of the cloned DNA fragments revealed no change in DNA methylation in the more upstream CpG island (CpG1, containing 27 CpGs) (Figure 5B). Conversely, higher methylation in CpG island 2 (CpG2, containing 9 CpGs) was detected in the pool of tumor samples (Figure 5B). Pyrosequencing of promoter regions containing 6 CpGs located in CpG2 and 2 more downstream CpGs located in the 5′UTR revealed that, although not statistically significant, methylation of ZBTB18 CpG2 tended to be higher in the glioma samples compared to normal brain (Supplementary Figure S6A). Furthermore, methylation of the 2 CpGs in the 5′UTR (5′UTR CpG -1 and 5′UTR CpG -2) was higher in gliomas compared to normal brain samples (p = 0.0184, unpaired t-test) (Supplementary Figure S6B-C). The significance was even higher when only GBM samples where included in the analysis (p = 0.0017, unpaired t-test) (Figure 5C).
Figure 5.
ZBTB18 promoter is methylated in a group of GBM. (A) Schematic representation of strategy used for methylation analysis of ZBTB18 promoter. (B) Detailed analysis of ZBTB18 promoter showing the region analyzed, the CpGs islands identified and their relative methylation status in normal tumor samples (Control), brain tumors (BTs) and brain tumor derived cells (called MBs). (C) Comparison of the 2 CpG islands (-7 -6) methylation status between controls (N=33) and grade 4 brain tumors (n=28). (D) Expression of ZBTB18 in LN229, JX6 and BTSC161s cells after treatment with 5-Aza-2′-deoxycytidine and relative control. Representative image of an experiment performed in triplicate (n=3; error bars ± s.d).
Treatment with the hypomethylating agent 5′-Aza-2′-Deoxycytidine (5-Aza-2-dC; Decitabine) in GBM cell line LN229 and two patient-derived mesenchymal GBM cell lines (BT161s, JX6), all showing downregulation of ZBTB18 and hypermethylation of the ZBTB18 promoter, resulted in re-expression of ZBTB18 after 72 hours (Figure 5D). Pyrosequencing analysis confirmed concomitant reduction of 5′UTR CpG -1 and 5′UTR CpG -2 in LN229 cells (Supplementary Figure S6C). Together, these data support a role of promoter hypermethylation in the silencing of ZBTB18 in GBM.
To further prove that the ZBTB18 region including 5′UTR CpGs -1 and -2 is important for ZBTB18 promoter activity, we cloned several ZBTB18 promoter regions and analyzed their activity by luciferase reporter assay. As shown in Fig. 6A-B, the ZBTB18 promoter region 3 which does not contain the core promoter and the 5′UTR CpGs -1 and -2, had no promoter activity compared to other cloned promoter regions in which the core promoter and 5′UTR CpGs -1 and -2 were included (Figure 6 A-B). Next, we investigated the effect of DNA methylation on the luciferase reporter activity controlled by the ZBTB18 promoter region with the highest activity. As displayed in Figure 6 C-D, the promoter activity was completely inhibited by the SssI methylase, an enzyme that methylates all CGs, and to a less extent by HpaII methylase, which methylates only CGs in the CCGG context (Figure 6 C-D). These results further indicate that the ZBTB18 promoter region which includes the core promoter and 5′UTR CpGs -1 and -2 is responsible for promoter activity and sensitive to DNA methylation. This is consistent with the expected role of methylated CpGs close to the TSS in gene expression regulation [44].
Figure 6.
The core promoter region of ZBTB18 is essential for promoter activity and is sensitive to DNA methylation. (A) Schematic representation of the ZBTB18 promoter regions cloned in the pGL3 luciferase reporter vector. (B) Analysis of ZBTB18 promoter constructs activity by dual luciferase assay. (C) In vitro methylation assay of ZBTB18 promoter 4. The plasmid DNA was methylated with SssI or HpaII methylase. (D) Control restriction digestion of the methylase reaction using the methylation sensitive (HpaII) and the methylation insensitive (McrBC) restriction enzymes.
ZBTB18 promoter methylation is a hallmark of mesenchymal GBMs
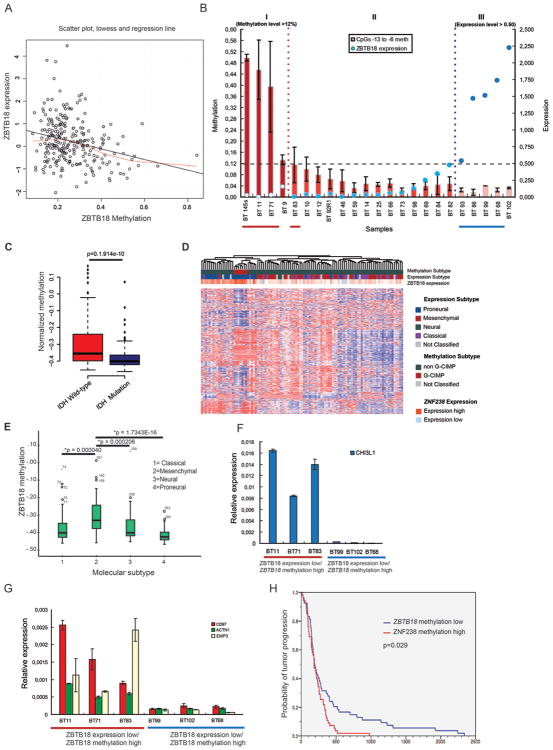
Correlation analysis between ZBTB18 expression and DNA methylation in 251 GBM samples from The Cancer Genome Atlas (TCGA, HumanMethylation27 platform, http://cancergenome.nih.gov) revealed a significant inverse correlation between ZBTB18 expression and promoter methylation (p = 1.96×10-05, linear regression) (Figure 7A). Interestingly, the examined probe (cg23829949) mapped into the same region analyzed by pyrosequencing (CpG island 2). Consistently, in our set of patient-derived glioma samples, ZBTB18 promoter methylation of 6 CpGs located at the 3′ end of CpG island 2 (Figure 7B) and of 5′UTR CpGs -1 and -2 (Supplementary Figure S6B) measured by pyrosequencing was higher in a subset of samples showing low ZBTB18 expression (high grade gliomas, red line), while low promoter methylation was detected in samples with higher ZBTB18 expression (low grade gliomas, blue line (Figure 7B). Thus, methylation of CpG island 2 and 5′UTR CpGs -1 and -2 correlates with ZBTB18 expression, at least in a subset of glioblastoma. However, a fraction of glioma samples that did not show promoter hypermethylation still had low ZBTB18 expression, suggesting that DNA hypermethylation is not the only mechanism regulating ZBTB18 expression, or alternatively, that additional methylated regions might be involved.
Figure 7.
Methylation at the ZBTB18 promoter causes ZBTB18 repression in GBM. (A) Linear regression of ZBTB18 expression on methylation status in n=251 samples (p=0.0000196). (B) Methylation level of ZBTB18 in a series of glioma samples assessed by pyrosequencing (n=3; error bars ± s.d). ZBTB18 expression level measured by qRT-PCR is indicated by blue circles (high expression and low expression is indicated by dark blue and blue circles respectively). Red line highlights the high methylation/low ZBTB18 expression samples and blue line highlights the low methylation/high ZBTB18 expression samples. BTSC 145 (GBM-derived cell line) was analyzed instead of the corresponding BT 145 GBM sample which was no longer available. (C) Association between ZBTB18 methylation and IDH1 mutation status. (D) Cluster analysis of methylation profiles of the G-CIMP and non-G-CIMP GBMs using the 450/27 K BeadChip from TCGA. The association between expression of ZBTB18 and tumor subtypes (methylation or expression) is shown. ZBTB18 appears to be mostly expressed in G-CIMP GBMs. (E) Association analysis between ZBTB18 promoter methylation and GBM subtypes. (F-G) Representative experiment showing expression of mesenchymal targets in samples of brain tumors highlighted in (A) assessed by qRT-PCR (n=3 PCR replicates; error bars ± s.d). Red line= high methylation and blue line= low methylation. (H) Kaplan-Meier estimates of time to tumor progression in 109 glioblastoma patients from TCGA, with patients stratified according to low versus high (relative to the median) ZBTB18 methylation (log-rank p = 0.029).
Promoter methylation analysis of the subset of gliomas with low ZBTB18 expression/high promoter methylation (indicated in red) vs. high ZBTB18 expression/low promoter methylation (indicated in blue) previously analyzed using the Infinium HumanMethylation450 BeadChip [45] (Supplementary Figure S7D) confirmed differential methylation of CpG island 2 between the two tumor groups covering CpGs located in the same region analyzed by pyrosequencing (probes cg19698993 and cg12869659). Intriguingly, tumor samples with high ZBTB18 expression and low ZBTB18 promoter methylation analyzed by DNA methylation array showed high levels of global methylation [45], suggesting that silencing of ZBTB18 by promoter methylation could be a hallmark of non G-CIMP gliomas. Consistent with this hypothesis, ZBTB18 methylation correlated with G-CIMP and IDH1 mutation status so that ZBTB18 was more methylated in IDH1 wild type compared to IDH1 mutant gliomas (Figure 7C p= 1.914e-10, Welch Two Sample t-test). Consistent with this, ZBTB18 expression was higher in G-CIMP GBMs (Figure 7D). We then investigated the relationship between ZBTB18 methylation (HumanMethylation27 platform) and GBM subclasses in 283 TCGA GBM samples. This analysis revealed a highly significant association between ZBTB18 methylation in a region confirmed by pyrosequencing (probe cg23829949) and mesenchymal GBM subtype (Figure 7E). Accordingly, regression analysis of ZBTB18 expression on ZBTB18 methylation revealed a strong correlation only in mesenchymal tumors but not in non-mesenchymal tumors (all GBMs: p = 0.000026; mesenchymal tumors: p = 0.0136; non-mesenchymal tumors: p = 0.13, linear regression), further highlighting that methylation-induced silencing of ZBTB18 might be particularly important in the mesenchymal subclass (Supplementary Figure S7A-C). This is consistent with previous analysis indicating that the mesenchymal tumors are usually non-G-CIMP [46] [47]. Expression analysis of selected mesenchymal genes (CD97, ACTN1, EMP3 and CHI3L1) revealed an inverse association with ZBTB18 promoter methylation (Figure 7E-F), further indicating that silencing of ZBTB18 through promoter hypermethylation could play a role in mesenchymal differentiation in GBM.
Survival modeling did not show a statistically significant association with patient survival (data not shown). Instead, we observed a significant link between time to tumor progression and ZBTB18 methylation in a two-class model of 109 GBM patients stratified according to lower-than-median versus higher-than-median ZBTB18 methylation (log-rank p = 0.029), such that patients with high methylation demonstrated a comparatively unfavorable outcome (Figure 7G). This association between ZBTB18 methylation and time to tumor progression was also evident in a continuous univariate Cox model (univariate Cox model p = 0.037, HR for tumor progression with methylated ZBTB18: 8.30, 95%-CI: 1.13-60.80) and prevailed in a multivariate Cox model that included GBM subclass (classical, mesenchymal, neural, proneural) as a co-variate (multivariate Cox model p = 0.048, HR: 7.49, 95%-CI: 1.01-65.32), suggesting that ZBTB18 methylation portends a more aggressive tumor phenotype.
Discussion
Here we describe a new role for the transcriptional repressor ZBTB18 as a negative regulator of signatures associated with poor survival in GBM and we propose DNA methylation as a mechanism to silence ZBTB18 in the mesenchymal tumor subtype. Our finding is in accordance with a previous study reporting that ZBTB18 is lost in established human GBM cells and identifying ZBTB18 as a brain tumor suppressor gene [11]. We demonstrate that ZBTB18 is mostly expressed in low-grade gliomas and proneural GBMs but less expressed in mesenchymal GBMs. This is consistent with our previous identification of ZBTB18 as a transcription factor negatively associated with mesenchymal GBMs [8]. ZBTB18 re-expression in primary BTSCs dampens the adherence poor-prognosis proliferative and mesenchymal signatures, which were identified as mutually exclusive to proneural in the subtype classification by Phillips and colleagues. Although the association between patient survival and mesenchymal subtype has not been confirmed in the previous TCGA study involving GBMs only [18], mouse models for glioma show that sequential mutations causing a shift from proneural to mesenchymal GBM are also associated with reduced survival [19, 20]. Moreover, since tumors often show mixed subtype profiles [20, 21], it is possible that this might mask an association with survival. This was clearly shown by Patel and colleagues who demonstrated that pure IDH1 wild-type proneural tumors were associated with a better survival compared to highly heterogeneous proneural tumors containing cells with other subtypes [21]. Similarly, tumor purity was recently shown to be an important parameter to determine a positive association of MGMT methylation with patient survival [17]. We show that ZBTB18 attenuates the expression of genes associated with EMT and with time-to-tumor progression in glioblastoma [36, 37]. This is in line with our analysis that presented a strong link between ZBTB18 methylation and time to tumor progression of GBM but only a trend of association with patient overall survival (data not shown). This finding is in accordance with the prevailing argument that time to tumor progression might relate more closely to tumor repopulation, aggressiveness, and therapy resistance, which are biological properties also associated with mesenchymal differentiation of GBM. Given this link to tumor progression and the fact that the mesenchymal phenotype is more prevalent in recurrent GBM [2], ZBTB18 hypermethylation might play a role in both the mesenchymal differentiation characteristics of the de novo tumor and the progression towards a more mesenchymal phenotype. However, as reported in our previous study {Carro, 2010 #5105} and also confirmed by others {Bhat, 2011 #3333}, several transcription factors usually cooperate to regulate specific glioblastoma subclasses. So, it would be interesting to study how deregulation of ZBTB18 fits in the previously described regulation of mesenchymal genes by other master regulators (i.e. STAT3, CEBPB and TAZ). The recent report that, at the single cell level, a GBM often consist of mosaic of subtype makes the picture even more complicated [48]. Still, identifying regulators of different glioblastoma subtype could be important since regulators of different subclasses co-existing in the tumor could be targeted.
The mechanism leading to ZBTB18 downregulation in GBM remained to be defined. Our data reveal a link between ZBTB18 promoter methylation and loss of ZBTB18 expression. However, we observed that, in some cases, low expression of ZBTB18 also occurs in the absence of promoter hypermethylation, suggesting that additional molecular mechanisms downregulating ZBTB18 are potentially operative in GBM. We further demonstrate a strong association between ZBTB18 promoter hypermethylation and the mesenchymal subtype of GBM, implying that silencing of ZBTB18 by promoter methylation is a particular hallmark of this unfavorable GBM subtype. We also report a strong correlation between ZBTB18 methylation and IDH1 wild type, which is consistent with previous studies indicating that the majority of mesenchymal GBMs are non G-CIMP [46, 47]. Consistently, we show that key mesenchymal genes [2, 8], which are also differentially methylated in G-CIMP versus non G-CIMP gliomas [46], are highly expressed in GBM exhibiting promoter hypermethylation/low expression of ZBTB18.
Collectively, our data identify ZBTB18 as a candidate tumor suppressor and transcriptional regulator of poor prognosis-associated signatures in GBM. We have identified promoter hypermethylation as a common mechanism to silence ZBTB18 in the mesenchymal subtype of GBM, which provides a new mechanistic opportunity to specifically target this tumor subclass.
Supplementary Material
Acknowledgments
We thank M. Oberle for mice brain histology; M. Lübbert for providing access to pyrosequencing facility; T. Feuerstein for normal brain tissues, C. Stein for technical assistance, R. Claus for help with methylation data analysis and D. Ó hAilín for editing the manuscript, (all University of Freiburg), V. D.Marinescu for BTSCs classification (University of Uppsala), S. Nozell for input on the manuscript (UAB, Birmingham), Y. Gillespie (UAB, Birmingham) for providing JX6 cells and V. Baekelandt (Katholieke Universiteit Leuven) for pLV-eGFP lentiviral vector.
This study was supported by Marie Curie International Reintegration Grant (MC IRG 268303), Deutsche Forschungsgemeinschaft Grant (DFG, CA 1246/2-1 (both to MSC) and German Cancer Aid Grant Award (107714, M. Bredel). MP was supported by the DFG (SFB 992).
Footnotes
Conflicts of Interest: The authors declare no conflict of interest.
References
- 1.Dunn GP, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26(8):756–84. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phillips HS, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 3.Verhaak RG, et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhat KP, et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–46. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basso K, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37(4):382–90. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 6.Margolin AA, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7 Suppl 1:S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhat KP, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25(24):2594–609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carro MS, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–25. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohtaka-Maruyama C, et al. The 5'-flanking region of the RP58 coding sequence shows prominent promoter activity in multipolar cells in the subventricular zone during corticogenesis. Neuroscience. 2012;201:67–84. doi: 10.1016/j.neuroscience.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Okado H, et al. The transcriptional repressor RP58 is crucial for cell-division patterning and neuronal survival in the developing cortex. Dev Biol. 2009;331(2):140–51. doi: 10.1016/j.ydbio.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 11.Tatard VM, et al. ZNF238 is expressed in postmitotic brain cells and inhibits brain tumor growth. Cancer Res. 2010;70(3):1236–46. doi: 10.1158/0008-5472.CAN-09-2249. [DOI] [PubMed] [Google Scholar]
- 12.Xiang C, et al. RP58/ZNF238 directly modulates proneurogenic gene levels and is required for neuronal differentiation and brain expansion. Cell Death Differ. 2012;19(4):692–702. doi: 10.1038/cdd.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cadieux B, et al. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66(17):8469–76. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- 14.Esteller M, et al. hMLH1 promoter hypermethylation is an early event in human endometrial tumorigenesis. Am J Pathol. 1999;155(5):1767–72. doi: 10.1016/S0002-9440(10)65492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esteller M, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59(4):793–7. [PubMed] [Google Scholar]
- 16.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22(1):9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulze Heuling E, et al. Prognostic Relevance of Tumor Purity and TERT Promoter Mutations on MGMT Promoter Methylation in Glioblastoma. Mol Cancer Res. 2017 doi: 10.1158/1541-7786.MCR-16-0322. [DOI] [PubMed] [Google Scholar]
- 18.Ferrarese R, et al. Lineage-specific splicing of a brain-enriched alternative exon promotes glioblastoma progression. J Clin Invest. 2014;124(7):2861–76. doi: 10.1172/JCI68836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie Y, et al. The Human Glioblastoma Cell Culture Resource: Validated Cell Models Representing All Molecular Subtypes. EBioMedicine. 2015;2(10):1351–63. doi: 10.1016/j.ebiom.2015.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izzo A, et al. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep. 2013;3(6):2142–54. doi: 10.1016/j.celrep.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldmann T, et al. USP18 lack in microglia causes destructive interferonopathy of the mouse brain. EMBO J. 2015;34(12):1612–29. doi: 10.15252/embj.201490791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han X, et al. The role of Src family kinases in growth and migration of glioma stem cells. Int J Oncol. 2014;45(1):302–10. doi: 10.3892/ijo.2014.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy A, et al. Glioma-derived plasminogen activator inhibitor-1 (PAI-1) regulates the recruitment of LRP1 positive mast cells. Oncotarget. 2015;6(27):23647–61. doi: 10.18632/oncotarget.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ernst A, et al. Genomic and expression profiling of glioblastoma stem cell-like spheroid cultures identifies novel tumor-relevant genes associated with survival. Clin Cancer Res. 2009;15(21):6541–50. doi: 10.1158/1078-0432.CCR-09-0695. [DOI] [PubMed] [Google Scholar]
- 27.Safaee M, et al. Overexpression of CD97 confers an invasive phenotype in glioblastoma cells and is associated with decreased survival of glioblastoma patients. PLoS One. 2013;8(4):e62765. doi: 10.1371/journal.pone.0062765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bacigalupo ML, et al. Galectin-1 triggers epithelial-mesenchymal transition in human hepatocellular carcinoma cells. J Cell Physiol. 2015;230(6):1298–309. doi: 10.1002/jcp.24865. [DOI] [PubMed] [Google Scholar]
- 29.Tang J, et al. CX3CL1 increases invasiveness and metastasis by promoting epithelial-to-mesenchymal transition through the TACE/TGF-alpha/EGFR pathway in hypoxic androgen-independent prostate cancer cells. Oncol Rep. 2016;35(2):1153–62. doi: 10.3892/or.2015.4470. [DOI] [PubMed] [Google Scholar]
- 30.Sakamaki M, GB M, Abe T. Legs and trunk muscle hypertrophy following walk training with restricted leg muscle blood flow. J Sports Sci Med. 2011;10(2):338–40. [PMC free article] [PubMed] [Google Scholar]
- 31.Anand V, et al. Functional characterization of Mammary Gland Protein-40, a chitinase-like glycoprotein expressed during mammary gland apoptosis. Apoptosis. 2016;21(2):209–24. doi: 10.1007/s10495-015-1196-z. [DOI] [PubMed] [Google Scholar]
- 32.Wang HX, et al. EWI-2 negatively regulates TGF-beta signaling leading to altered melanoma growth and metastasis. Cell Res. 2015;25(3):370–85. doi: 10.1038/cr.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X, et al. S100 calcium-binding protein A6 promotes epithelial-mesenchymal transition through beta-catenin in pancreatic cancer cell line. PLoS One. 2015;10(3):e0121319. doi: 10.1371/journal.pone.0121319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taube JH, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107(35):15449–54. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diotel N, et al. Differential expression of id genes and their potential regulator znf238 in zebrafish adult neural progenitor cells and neurons suggests distinct functions in adult neurogenesis. Gene Expr Patterns. 2015;19(1-2):1–13. doi: 10.1016/j.gep.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Anastassiou D, et al. Human cancer cells express Slug-based epithelial-mesenchymal transition gene expression signature obtained in vivo. BMC Cancer. 2011;11:529. doi: 10.1186/1471-2407-11-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng WY, et al. A multi-cancer mesenchymal transition gene expression signature is associated with prolonged time to recurrence in glioblastoma. PLoS One. 2012;7(4):e34705. doi: 10.1371/journal.pone.0034705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lubeseder-Martellato C, et al. Membranous CD24 drives the epithelial phenotype of pancreatic cancer. Oncotarget. 2016;7(31):49156–49168. doi: 10.18632/oncotarget.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vannier C, et al. Zeb1 regulates E-cadherin and Epcam (epithelial cell adhesion molecule) expression to control cell behavior in early zebrafish development. J Biol Chem. 2013;288(26):18643–59. doi: 10.1074/jbc.M113.467787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raschperger E, et al. The coxsackie and adenovirus receptor (CAR) is required for renal epithelial differentiation within the zebrafish pronephros. Dev Biol. 2008;313(1):455–64. doi: 10.1016/j.ydbio.2007.10.049. [DOI] [PubMed] [Google Scholar]
- 41.Li L, et al. The human cadherin 11 is a pro-apoptotic tumor suppressor modulating cell stemness through Wnt/beta-catenin signaling and silenced in common carcinomas. Oncogene. 2012;31(34):3901–12. doi: 10.1038/onc.2011.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guaita S, et al. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J Biol Chem. 2002;277(42):39209–16. doi: 10.1074/jbc.M206400200. [DOI] [PubMed] [Google Scholar]
- 43.Gracz AD, et al. Brief report: CD24 and CD44 mark human intestinal epithelial cell populations with characteristics of active and facultative stem cells. Stem Cells. 2013;31(9):2024–30. doi: 10.1002/stem.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin JC, et al. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12(5):432–44. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kling T, et al. Integrative Modeling Reveals Annexin A2-mediated Epigenetic Control of Mesenchymal Glioblastoma. EBioMedicine. 2016;12:72–85. doi: 10.1016/j.ebiom.2016.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ozawa T, et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26(2):288–300. doi: 10.1016/j.ccr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.