Abstract
Background Renal water excretion is controlled by vasopressin, in part through regulation of the transcription of the aquaporin-2 gene (Aqp2).
Methods To identify enhancer regions likely to be involved in the regulation of Aqp2 and other principal cell–specific genes, we used several next generation DNA-sequencing techniques in a well characterized cultured cell model of collecting duct principal cells (mpkCCD). To locate enhancers, we performed the assay for transposase-accessible chromatin using sequencing (ATAC-Seq) to identify accessible regions of DNA and integrated the data with data generated by chromatin immunoprecipitation followed by next generation DNA-sequencing (ChIP-Seq) for CCCTC binding factor (CTCF) binding, histone H3 lysine-27 acetylation, and RNA polymerase II.
Results We identified two high-probability enhancers centered 81 kb upstream and 5.8 kb downstream from the Aqp2 transcriptional start site. Motif analysis of these regions and the Aqp2 promoter identified several potential transcription factor binding sites, including sites for two b-ZIP transcription factors: CCAAT/enhancer binding protein-β (C/EBPβ) and cAMP-responsive element binding protein (CREB). To identify genomic binding sites for both, we conducted ChIP-Seq using well characterized antibodies. In the presence of vasopressin, C/EBPβ, a pioneer transcription factor critical to cell-specific gene expression, bound strongly at the identified enhancer downstream from Aqp2. However, over multiple replicates, we found no detectable CREB binding sites within 390 kb of Aqp2. Thus, any role for CREB in the regulation of Aqp2 gene transcription is likely to be indirect.
Conclusions The analysis identified two enhancer regions pertinent to transcriptional regulation of the Aqp2 gene and showed C/EBPβ (but not CREB) binding.
Keywords: ChIP-Seq, transcription factors, water transport, water channels, aquaporin-2, ATAC-Seq
Vasopressin regulates water excretion largely through control of water permeability in the renal collecting duct. At a molecular level, water transport is controlled through regulation of the water channel protein, aquaporin-2 (AQP2). AQP2 is regulated in at least two ways: (1) control of trafficking of AQP2 to the apical plasma membrane of collecting duct principal cells1,2 and (2) control of the absolute abundance of the AQP2 protein.3 The latter process results largely from the regulation of Aqp2 gene transcription,4–7 although regulation of AQP2 protein half life also plays a role.8–10 Defective regulation of Aqp2 gene transcription has been found to be responsible for many acquired water balance disorders,11 including polyuric disorders (e.g., lithium-induced nephrogenic diabetes insipidus, hypercalcemic polyuria, hypokalemic polyuria) and dilutional hyponatremia as seen with the syndrome of inappropriate antidiuresis and congestive heart failure. To understand the pathophysiologic basis of these disorders, it is important to discover the normal physiologic mechanisms by which vasopressin, working by binding and activation of the vasopressin V2 receptor, stimulates Aqp2 gene transcription.
The V2 receptor couples to the heterotrimeric G-protein α-subunit Gαs, which in collecting duct principal cells, activates adenylyl cyclase 6, thereby increasing intracellular cAMP levels. Actions of cAMP are mediated chiefly through activation of one or both protein kinase A catalytic proteins (coded by Prkaca and Prkacb genes). When these two genes were deleted from mpkCCD cells using CRISPR-Cas9, AQP2 mRNA and protein were nearly undetectable even in the presence of vasopressin, suggesting that Aqp2 gene transcription depends on protein kinase A.12
The mouse mpkCCD cell line has been used extensively in the investigation of regulation of collecting duct function.6,13–15 It manifests both major elements of vasopressin-mediated regulation of AQP2, namely regulated AQP2 trafficking and regulated Aqp2 gene transcription.15 On the basis of RNA-Seq and RNA polymerase II chromatin immunoprecipitation followed by DNA sequencing (ChIP-Seq) in mpkCCD cells, it seems that vasopressin mediates highly selective regulation of Aqp2 gene transcription, requiring the joint action of at least four DNA binding transcription factors.4 An intermediate goal on the way to an understanding of the mechanism of vasopressin-mediated Aqp2 gene transcription is to identify these transcription factors. A number of hypothesis-driven studies have proposed roles for a variety of transcription factors, including cAMP-responsive element binding (CREB) protein,16,17 AP-1,16 NF-κB,18 GATA family,19–21 ETS family,15,22 NFAT5, and NFATc proteins.23–25 However, a general solution to the problem is challenging, because the mammalian genome codes for approximately 1500 transcription factors and a trial and error approach are unlikely to be definitive. In this paper, we present a systems-level analysis using both prior data (histone H3 acetylation at lysine 27 [H3K27Ac] ChIP-Seq and RNA polymerase II ChIP-Seq) and new data from assay for transposase-accessible chromatin using sequencing (ATAC-Seq) experiments to identify enhancers near the Aqp2 gene. Enhancers are segments of DNA that are typically a few hundred base pairs in length, and they are occupied by multiple transcription factors that can recruit RNA polymerase II and transcriptional coactivators to target genes.26 ATAC-Seq is a new methodology designed to precisely identify regions of DNA that are open, allowing access to transcription factors and other DNA binding proteins.27 After enhancer identification, transcription factor ChIP-Seq experiments focused on two b-ZIP transcription factors, CCAAT/enhancer binding protein-β (C/EBPβ) and CREB, that are predicted to bind to either the identified enhancers or the Aqp2 promoter.
Methods
Cell Culture
AQP2-expressing mpkCCD clone 11 (mpkCCD) cells were grown on permeable membrane supports as described previously.15 Cells were exposed to 0.1 nM [deamino-Cys1, D-Arg8]-Vasopressin (dDAVP) or vehicle for 30 minutes.
Preparation of Nuclear and Cytosolic Fractions
Nuclear and cytosolic fractions were prepared from mpkCCD cells using the NE-PER Nuclear and Cytoplasmic Extraction Reagent Kit (Pierce) as described previously.28
Immunoblotting
Immunoblotting was performed as described.29 Primary antibodies were CREB (17–600; Millipore), C/EBPβ (sc-150; Santa Cruz), p-CREB (S133, 17–10131; Millipore), ATF1 (sc-243; Santa Cruz), β-catenin (9562; Cell Signaling), and p-β-catenin (S552, 9566; Cell Signaling).
ATAC-Seq
ATAC-Seq was performed following a method published by Buenrostro et al.27 Nuclear pellets (from 50,000 cells) were incubated in “transposition reaction mixture” using Tagment DNA Buffer (FC-121–1030; Illumina) and Tn5 transposase (FC-121–1030) for 30 minutes at 37°C. The transposed DNA fragments were purified and amplified using PCR. Purified DNA libraries were sequenced on an Illumina HiSeq 3000 platform.
ChIP-Seq
Chromatin immunoprecipitation (ChIP) was performed using the truChIP Chromatin Shearing Reagent Kit (Covaris) and the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling) following the manufacturer’s protocols. Protein/DNA was crosslinked using 1.1% formaldehyde. Nuclei were isolated and sheared using an ultrasonicator. The sheared chromatin was incubated with 4 μg of antibody (CREB, 17–600 or C/EBPβ, sc-150) and additionally incubated with Protein G magnetic beads. After elution of ChIP products, DNA was purified and subjected to library preparation using the Ovation SP Ultralow Library System (NuGEN) following the manufacturer’s protocol. The DNA libraries were gel purified and sequenced on an Illumina HiSeq 2000 platform.
Data Processing
The sequence reads were mapped on the mouse reference genome (mm10) using Burrows–Wheeler Aligner30 and Samtools.31 All uniquely mapped reads were used in downstream peak calling (MACS2; Bedtools), motif analysis (MEME-ChIP),32 and bioinformatic analysis (R package ChIPpeakAnno).33
For ATAC-Seq data processing, adapter sequences were trimmed for both forward and reverse reads using cutadapt and aligned on mouse reference genome mm10 using Bowtie2. Duplicates were removed from mapped reads using Picard. Peaks were identified using MACS2. Read distribution was visualized on the UCSC genome browser using UCSC bedClip and bedGraphToBigWig.
Data Mining and Bioinformatic Analyses
A dataset of CCCTC binding factor (CTCF) ChIP-Seq was downloaded from the mouse ENCODE database (GSE36027).34 mpkCCD ChIP-Seq datasets were downloaded from GSE95009 for histone H3K27 acetylation ChIP-Seq12 and GSE79584 for RNA polymerase II ChIP-Seq.4 Data from ATAC-Seq, H3K27Ac ChIP-Seq, and RNA polymerase II ChIP-Seq were transformed to obtain minimum Bayes factors (range, 0–1) as described35 on the basis of signal-to-noise ratios as a function of genomic position. The noise level for a given insulated region was estimated as twice the median signal across the region. The data from ATAC-Seq, H3K27Ac ChIP-Seq, and RNA polymerase II ChIP-Seq were integrated by multiplying the complements of their minimum Bayes factors.
ATAC-Seq and ChIP-Seq data have been deposited at Gene Expression Omnibus (GEO; GSE98076 and GSE108786).
Results
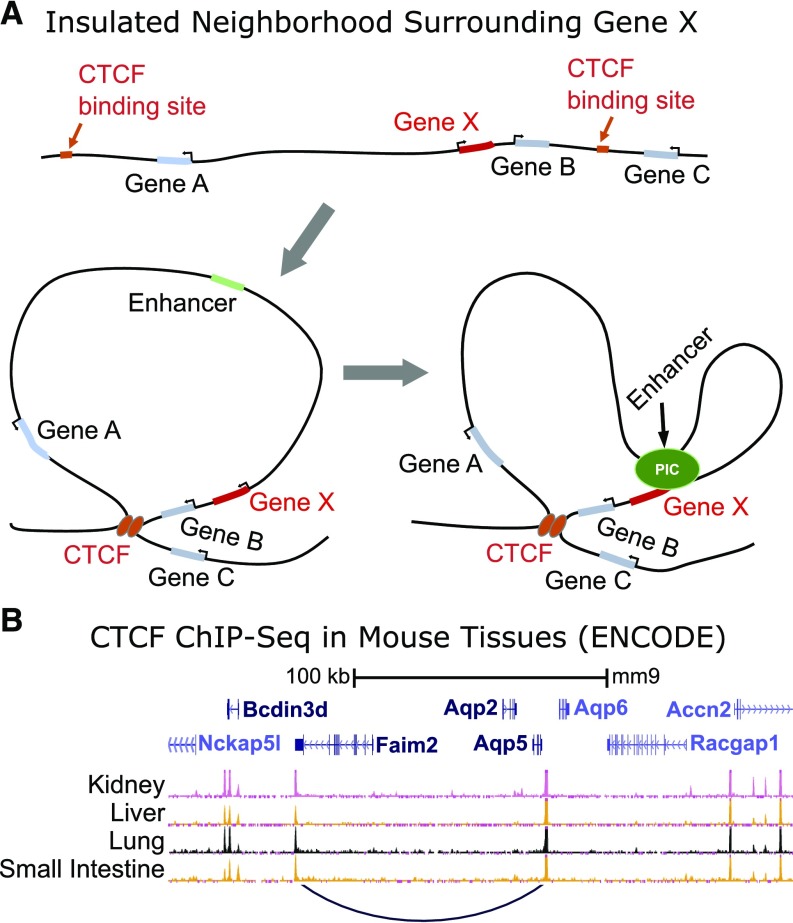
Transcription factor proteins that regulate transcription of Aqp2 and other genes in the renal collecting duct may bind directly to gene promoters or enhancers that are not immediately adjacent to the regulated gene. Recent studies summarized by Hnisz et al.36 provide information that narrows the possible locations of transcription factor binding sites that regulate a particular gene, such as Aqp2. This constraint is on the basis of the concept of CTCF-bounded loops that define insulated neighborhoods. CTCF is a DNA binding protein that binds to DNA as a monomer during interphase but homodimerizes with upstream or downstream DNA-bound CTCF molecules to create CTCF loops (Figure 1A). The locations of these loops are highly conserved among mammalian species and relatively invariant among cell types.36 There are approximately 13,000 such insulated loops across the genome, with an average of two to three genes per insulated loop. The enhancers that regulate a given gene are found within the same CTCF loop. Prior studies done as part of the mouse ENCODE project have used ChIP-Seq to identify CTCF binding sites.34 These data provide the information needed to identify the insulated loop that contains Aqp2 (Figure 1B). As expected, CTCF binding sites were the same among kidney, liver, lung, and small intestine. The loop that contains Aqp2 is 99 kb in length and also contains two other genes: Faim2 and Aqp5. Thus, enhancers that control Aqp2 gene transcription are highly likely to reside within this 99-kb insulated loop.
Figure 1.
CCCTC binding factor (CTCF) insulated loops. (A) Hypothetical insulated loop. Two adjacent DNA-bound CTCF molecules dimerize to create a loop. Genes within an insulated loop are regulated by their own promoters plus enhancers (and repressors) within the same insulated loop. (B) CTCF binding sites flanking the aquaporin-2 (Aqp2) gene from mouse ENCODE data. Two other genes, Faim2 (neuron specific) and Aqp5 (specific to secretory epithelia), are within the defined insulated loop. ChIP-Seq, chromatin immunoprecipitation followed by DNA sequencing.
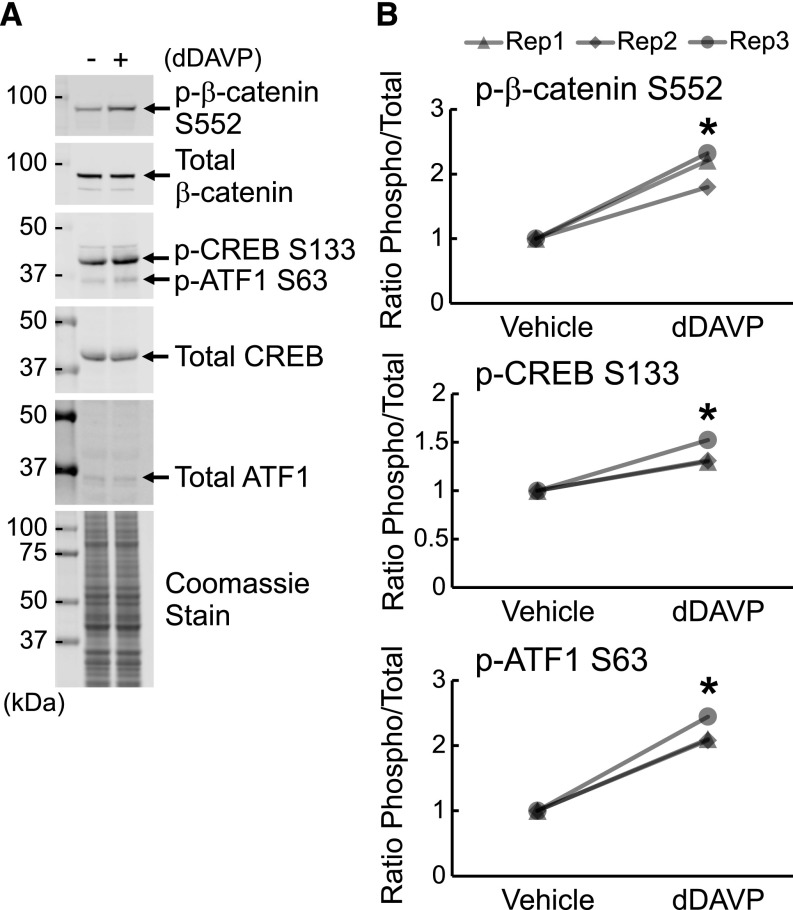
To assure that the mpkCCD cells used in this study respond appropriately to vasopressin, we carried out Western blotting for known vasopressin-regulated phosphorylation sites. We tested the ability of dDAVP (0.1 nM for 30 minutes) to increase phosphorylation of the transcriptional coregulator β-catenin at Ser552, a vasopressin-responsive site12,28,37 (Figure 2). Band density corresponding to this target was more than doubled. In the same samples, we probed for protein kinase A target sites on CREB and ATF1, which are recognized by the same antibody but can be discriminated on the basis of molecular weight. Both sites showed increases in phosphorylation in response to vasopressin (Figure 2), confirming the viability and vasopressin responsiveness of the cells.
Figure 2.
The mpkCCD cells are responsive to vasopressin. (A) Example immunoblots for β-catenin phosphorylated at S552, total β-catenin, cAMP-responsive element binding protein (CREB) phosphorylated at S133, ATF1 phosphorylated at S63, total CREB, and total ATF1 as well as a Coomassie blue–stained gel showing equality of loading. (B) Quantification of immunoblots (n=3 for each). *Statistically significant difference was found between cells treated with vehicle or [deamino-Cys1, D-Arg8]-Vasopressin (dDAVP) for 30 minutes (P<0.05 by t test).
We previously found evidence that 714 distinct transcription factors are expressed in mpkCCD cells12 (Supplemental Dataset 1). All could be considered candidates to play roles in regulation of Aqp2 gene transcription. However, 714 transcription factors are too many to allow efficient one by one evaluation by ChIP-Seq. To narrow the possible candidates, it is desirable to identify subregions of the Faim2-Aqp2-Aqp5 insulated loop that are likely to contain enhancers. Doing so will allow us to identify potential transcription factor binding sites in these regions. We present three types of data that are informative in this regard, viz. ATAC-Seq, histone H3K27Ac ChIP-Seq, and RNA polymerase II ChIP-Seq (explained below). Relevant profiles for these measures in mpkCCD cells are shown in Figure 3.
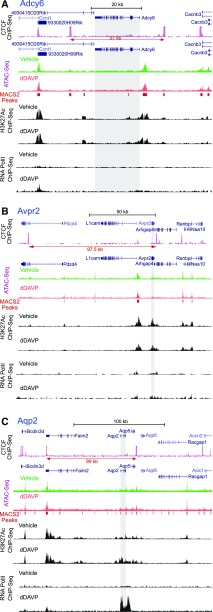
Figure 3.
Mapping ChIP-Seq data to three collecting duct genes: (A) adenylyl cyclase 6 (Adcy6), (B) V2 vasopressin receptor (Avpr2), and (C) aquaporin-2 (Aqp2). The ATAC-Seq sites indicating accessible DNA are indicated by the track MACS2 Peaks. (General feature of ATAC-Seq analysis are in Supplemental Figure 1.) Read distribution normalized by signal per million reads was visualized on the UCSC genome browser using two-reference genome information: mouse mm9 for CTCF ChIP-Seq in kidney downloaded from mouse ENCODE database and mouse mm10 for ATAc-Seq, H3K27Ac ChIP-Seq, and RNA polymerase II ChIP-Seq in mpkCCD cells. Gray shading indicates the region corresponding to gene body. dDAVP, [deamino-Cys1, D-Arg8]-Vasopressin. CTCF, CCCTC binding factor; ATAC-Seq, assay for transposase-accessible chromatin using sequencing; H3K27Ac, histone H3 acetylation at lysine 27; ChIP-Seq, chromatin immunoprecipitation followed by DNA sequencing; RNA PolII, RNA polymerase II.
ATAC-Seq is a recently devised method27 for assessing DNA accessibility, providing better spatial resolution than DNase I hypersensitivity profiling. Figure 3 shows ATAC-Seq data for mpkCCD cells mapped to the regions around three collecting duct genes: Adcy6, Avpr2, and Aqp2. Adcy6 and Avpr2 are strongly expressed in collecting duct principal cells, but their transcription is not regulated by vasopressin.4 Mouse ENCODE data for CTCF ChIP-Seq are included to provide a reference. The 37-kb putative CTCF insulated region surrounding Adcy6 encloses a major ATAC-Seq peak centered 1200 bases upstream from Adcy6 plus several less prominent peaks that are indicated by the “MACS2 Peaks” track. The 97.5-kb putative CTCF insulated region surrounding Avpr2 encloses three discernable ATAC-Seq peaks centered at 11,300, 43,800, and 67,200 bases upstream from Avpr2. The Faim2-Aqp2-Aqp5 insulated loop contains a major peak overlapping the unexpressed Faim2 gene 81 kb upstream from the Aqp2 transcriptional start site. It also contains several less prominent peaks between Faim2 and Aqp2 as well as between Aqp2 and Aqp5. Full ATAC-Seq data can be downloaded from GEO (GSE108786).
Figure 3 also shows data from H3K27Ac ChIP-Seq experiments for the same three regions. (A small subset of these data was previously reported in the work by Isobe et al.12) H3K27Ac mapping is widely used to identify open regions of DNA overlapping both promoters and enhancers.38 To a large extent, the H3K27Ac peaks overlap the ATAC-Seq peaks, providing redundancy for identification of potential enhancers. In response to dDAVP, there was a demonstrable increase in H3K27 acetylation in the Aqp2 promoter, gene body, and downstream from the gene body.
Figure 3 also shows binding sites for RNA polymerase II (Polr2a subunit) mined from Sandoval et al.4 RNA polymerase II has been shown to localize not only to gene bodies of actively transcribed genes but also, to active enhancers. dDAVP increases RNA polymerase II occupancy over the Aqp2 gene body as previously shown4 and also in two other regions of the Faim2-Aqp2-Aqp5 insulated loop, namely a region just downstream from Aqp2 and a region overlapping the last exon of Faim2. (Of note, the Faim2 mRNA is not detectable in mpkCCD cells.4)
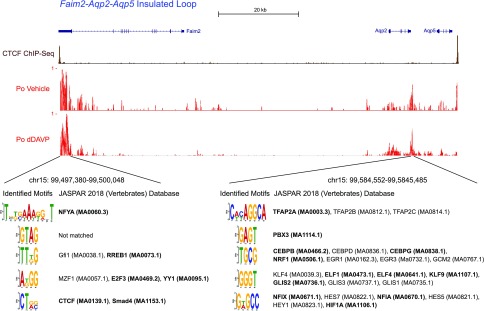
In Figure 4, we integrate the three measures (ATAC-Seq, histone H3K27Ac ChIP-Seq, and RNA polymerase II ChIP-Seq) to obtain an aggregate “open probability” (Po) indicating the regions in the Faim2-Aqp2-Aqp5 insulated loop that are most likely to be enhancers. Among the open regions, two stand out as being the densest and longest as highlighted in Figure 4. We analyzed these two enhancers using the JASPAR database of transcription factor binding motifs to identify DNA sequence motifs associated with specific transcription factors. In the upstream enhancer, potential sites for several transcription factors were identified, including some known to be expressed in mpkCCD cells viz. NFYA, RREB1, E2F3, YY1, CTCF, and SMAD4 (Supplemental Dataset 1). In the identified enhancer between the Aqp2 and Aqp5 genes, potential sites for the transcription factors TFAP2A, PBX3, CEBPB, CEBPG, NRF1, ELF1, ELF4, KLF9, GLIS2, NFIX, NFIA, and HIF1A were identified. Some of these have previously been implicated in regulation of Aqp2 gene transcription: AP2 (TFAP2A),15 ETS family (ELF1 and ELF4),15 and KLF (KLF9).39 Among the sites found, C/EBPβ is a so-called “pioneer” transcription factor that can initiate regulated changes in chromatin structure before binding of other transcription factors.40,41
Figure 4.
Identification of enhancer elements for aquaporin-2 (Aqp2) gene. High open probability regions in the Faim2-Aqp2-Aqp5 insulated loop were identified using minimum Bayes factors to integrate ATAC-Seq, histone H3K27Ac ChIP-Seq, and RNA polymerase II ChIP-Seq data into single “open probability” (Po) values as described in Supplemental Methods. The calculation is on the basis of the signal-to-noise ratio for each measure at each point in the loop. Noise values for this calculation were estimated as two times the median read density value across the loop. (Supplemental Figures 2 and 3 show sensitivity analysis.) The Po calculation identified two regions of high probability as indicated. These regions were analyzed using MEME-ChIP to identify JASPAR transcription factor motifs present. CTCF, CCCTC binding factor; dDAVP, [deamino-Cys1, D-Arg8]-Vasopressin. ATAC-Seq, assay for transposase-accessible chromatin using sequencing; H3K27Ac, histone H3 acetylation at lysine 27; ChIP-Seq, chromatin immunoprecipitation followed by DNA sequencing; RNA PolII, RNA polymerase II.
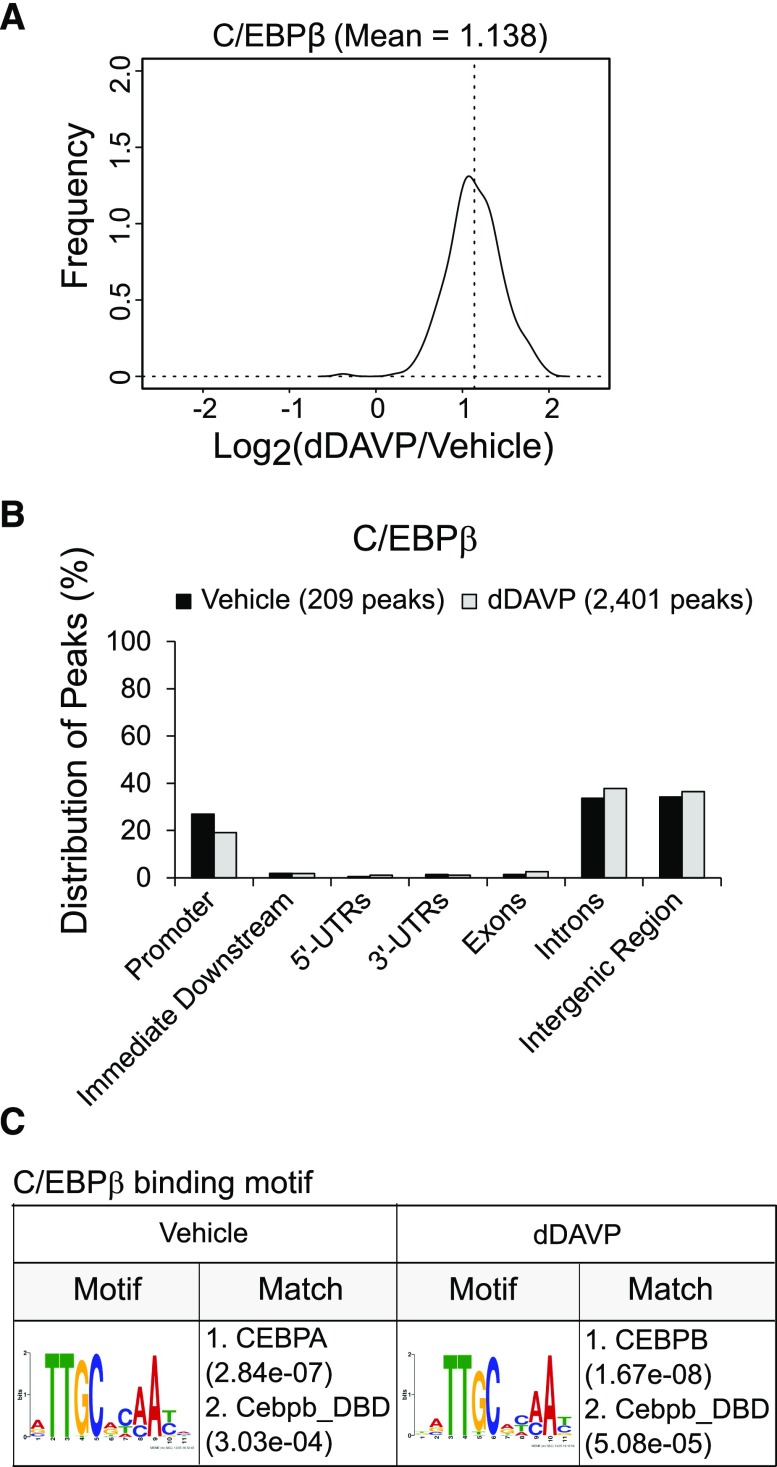
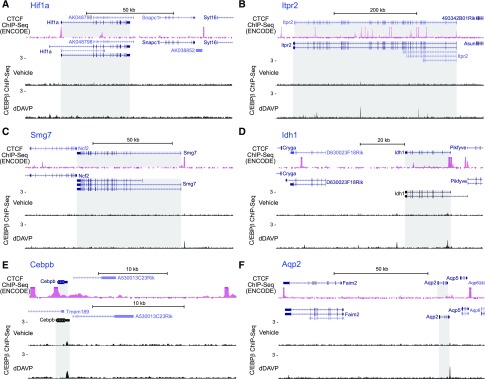
To identify whether C/EBPβ binds anywhere in the Faim2-Aqp2-Aqp5 insulated loop or elsewhere in the genome (Supplemental Figure 4), we carried out ChIP-Seq experiments (Figure 5) using a well tested C/EBPβ antibody.42 The full datasets are provided permanently at https://hpcwebapps.cit.nih.gov/ESBL/Database/ChIP-Seq_CREB_CEBPB/ and the GEO (GSE98076). As shown in Figure 5, 2401 C/EBPβ binding sites were identified in dDAVP-treated cells (0.1 nM, 30 minutes), whereas 209 reproducible sites were found in vehicle-treated cells. Read densities for many of the 209 sites common to the dDAVP and vehicle samples were increased in the dDAVP-treated versus vehicle-treated cells: mean log2(dDAVP/vehicle) =1.14 (Figure 5A) as expected on the basis of the observation that dDAVP causes translocation of C/EBPβ into the nucleus of mpkCCD cells (Supplemental Figure 4) (https://hpcwebapps.cit.nih.gov/ESBL/Database/QuantNucProteomics/). In mpkCCD cells, C/EBPβ binding sites are not limited to gene promoters, but they are also frequently found in intergenic regions and introns (Figure 5B), contrasting with CREB (see below). The consensus motif identified from C/EBPβ binding sites matched canonical binding motifs for the C/EBP family. These data support the fidelity of the ChIP-Seq methodology applied to C/EBPβ binding site localization. Vasopressin-sensitive C/EBPβ binding sites were identified in the vicinity of several known C/EBPβ-regulated genes, namely Hif1a,43 Itpr2,44 Smg7,42 and Idh145 (Figure 6, A–E), further supporting the fidelity of the method.
Figure 5.
C/EBPβ ChIP-Seq: general findings. (A) Quantification of peak read densities in dDAVP–treated samples and vehicle controls (n=2). (These observations were made with 30 minutes of exposure to dDAVP or vehicle; additional observations at 24 hours are reported in Supplemental Figure 5.) (B) Locations of peaks relative to the nearest gene. The majority of C/EBPβ binding sites were found within the promoters and introns of genes and intergenic regions. UTR, untranslated region. (C) C/EBPβ binding motif derived from ChIP-Seq data. Motifs matched previously reported motifs for C/EBPα and C/EBPβ found in the JASPAR motif database; e values in parentheses are the motif P values (computed by a Fisher exact test for enrichment of the motif in the sequences) times the number of candidate motifs tested. dDAVP, [deamino-Cys1, D-Arg8]-Vasopressin; C/EBPβ, CCAAT/enhancer binding protein-β; ChIP-Seq, chromatin immunoprecipitation followed by DNA sequencing.
Figure 6.
C/EBPβ ChIP-Seq: examples of binding sites found. Read distribution normalized by signal per million reads was visualized on the UCSC genome browser using two-reference genomes: mouse mm9 for CTCF ChIP-Seq in kidney downloaded from the mouse ENCODE database and mouse mm10 for C/EBPβ ChIP-Seq in mpkCCD cells. Gray shading indicates the region corresponding to gene body. (A–E) Results confirmed previous findings for Hif1a, Itpr2, Smg7, Idh1, and Cebpb. (F) A C/EBPβ binding site was identified just 400 bp downstream from the aquaporin-2 (Aqp2) gene in the dDAVP–treated sample. C/EBPβ, CCAAT/enhancer binding protein-β; CTCF, CCCTC binding factor; dDAVP, [deamino-Cys1, D-Arg8]-Vasopressin; ChIP-Seq, chromatin immunoprecipitation followed by DNA sequencing.
A C/EBPβ binding site was identified just 400 bp downstream from Aqp2, and the read density appeared to be increased by dDAVP (Figure 6F). This corresponds to a predicted C/EBPβ binding site in one of the high open probability DNA regions identified in Figure 4. The same site was identified in two additional samples treated for 24 hours with dDAVP (Supplemental Figure 5). Supplemental Tables 1 and 2 list all transcription factors with C/EBPβ binding sites at their promoters as well as protein kinase and transporter genes with C/EBPβ binding sites (Supplemental Figure 6).
The fraction of C/EBPβ binding sites associated with collecting duct–specific genes was greater than in the whole transcriptome, suggesting a role in tissue-specific gene expression (Supplemental Figure 7). Tissue-specific genes tend to have TATA boxes rather than CpG islands in their promoter proximal regions.46,47 Consistent with this, genes with TATA boxes are more frequent among genes with C/EBPβ binding sites than would be expected from a random selection of expressed genes, whereas genes with promoter proximal CpG islands are less frequent (Supplemental Figure 7). These observations lend support to the possible role of C/EBPβ in cell-specific gene expression in collecting duct principal cells.
Because vasopressin signals chiefly through increases in cAMP in collecting duct principal cells, it has been widely assumed that the transcription factor CREB is responsible for vasopressin-mediated regulation of Aqp2 gene transcription.11,48–51 A cAMP response element has been reported to be present within the promoter of Aqp2,16,17 although CREB binding to this site has not been directly shown. To identify CREB binding sites throughout the genome, we have carried out ChIP-Seq experiments in mpkCCD cells (four control-dDAVP pairs done on different days).
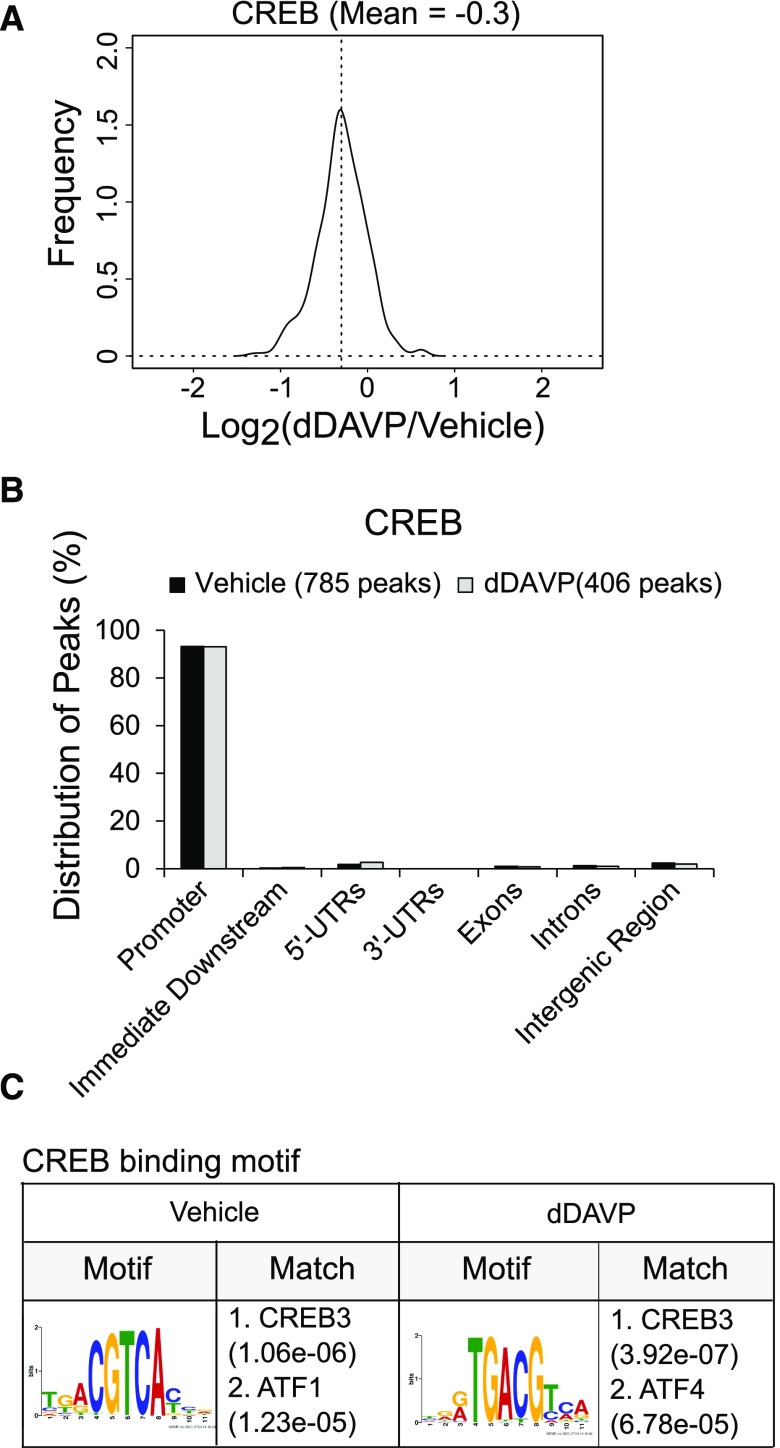
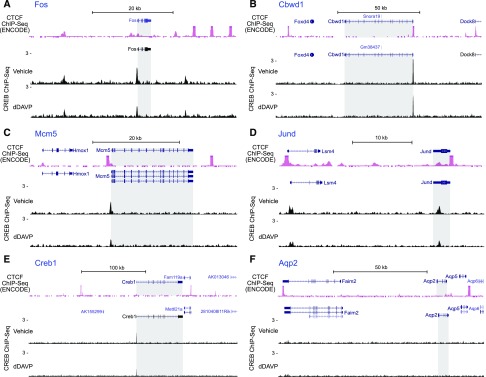
For ChIP-Seq, we used a CREB antibody previously used for ChIP-Seq in other tissues52 (Supplemental Figure 4). Binding sites were identified as genomic positions in which mapped reads for ChIP samples significantly exceeded the input control using a stringent cutoff (q<0.01 using MACS2). The full datasets are provided at https://hpcwebapps.cit.nih.gov/ESBL/Database/ChIP-Seq_CREB_CEBPB/ and at GEO (GSE98076). A total of 793 CREB binding sites were identified in mpkCCD, and most of them were seen under both conditions. In general, normalized read densities were virtually unchanged or slightly reduced in the dDAVP-treated cells (0.1 nM for 30 minutes) versus vehicle-treated cells (Figure 7A). This finding is consistent with prior studies showing that CREB is regulated chiefly by changes in phosphorylation in its “pKID domain” rather than by differential binding or nuclear translocation.53,54 Over 90% of total CREB binding sites were located at promoter regions (1 kb upstream to +100 bp downstream from the transcription start site) (Figure 7B). Sequences extracted from binding site peak centers were used to identify binding motifs (Figure 7C). A consensus half-cAMP response element sequence was identified that matches prior data for b-ZIP transcription factors CREB3, ATF1, and ATF4, confirming the fidelity of the method. The motif is the same for vehicle- and dDAVP-treated cells, although shifted by three bases. CREB binding sites identified in our ChIP-Seq analysis matched known target genes (Figure 8). For example, a canonical CREB binding site in the promoter of Fos55 matches the binding site found in this study (Figure 8A). CREB binding was also found at previously identified sites in the promoters of Mcm5,56 Cbwd1,57 and Jund57 (Figure 8, B–D). Interestingly, CREB also bound the promoter of the Creb1 gene itself (Figure 8E), suggesting a feedback relationship.
Figure 7.
CREB ChIP-Seq: general findings. (A) Quantification of peak read densities in dDAVP–treated samples and vehicle controls (n=4). There was no significant difference in peak read densities between dDAVP- and vehicle-treated samples (30 minutes of exposure). (B) Locations of peaks relative to nearest gene. The majority of CREB binding sites were found within the promoters of genes. UTR, untranslated region. (C) CREB binding motif derived from ChIP-Seq data. Motifs were the same for dDAVP- and vehicle-treated samples, although shifted by three bases. Motifs matched previously reported motifs for CREB3, ATF1, and ATF4 found in the JASPAR motif database; e values in parentheses are the motif P values (computed by a Fisher exact test for enrichment of the motif in the sequences) times the number of candidate motifs tested. CREB, cAMP-responsive element binding protein; dDAVP, [deamino-Cys1, D-Arg8]-Vasopressin; ChIP-Seq, chromatin immunoprecipitation followed by DNA sequencing.
Figure 8.
CREB ChIP-Seq: examples of binding sites found. Read distribution normalized by signal per million reads was visualized on the UCSC genome browser using two-reference genomes: mouse mm9 for CTCF ChIP-Seq in kidney downloaded from the mouse ENCODE database and mouse mm10 for CREB ChIP-Seq in mpkCCD cells. Gray shading indicates the region corresponding to gene body. (A–E) Results confirmed previous findings for Fos, Mcm5, Cbwd1, Jund, and Creb1. (F) No CREB binding sites were found within the Faim2-aquaporin-2 (Aqp2)-Aqp5 insulated loop. (The closest CREB binding site from the Aqp2 gene was 390 kb upstream.) CREB, cAMP-responsive element binding protein; CTCF, CCCTC binding factor; dDAVP, [deamino-Cys1, D-Arg8]-Vasopressin; ChIP-Seq, chromatin immunoprecipitation followed by DNA sequencing.
Interestingly, no CREB binding sites were found within 390 kb of the Aqp2 gene, including the 99-kb Faim2-Aqp2-Aqp5 insulated loop (Figure 8F), indicating that any role of CREB in transcriptional regulation of Aqp2 is likely to be indirect. For example, its regulatory role may be through regulation of expression of other transcription factors. Supplemental Tables 3 and 4 list transcription factors with CREB binding sites at their promoters as well as protein kinase and transporter genes with CREB binding sites (Supplemental Figure 8).
Discussion
Studies in model organisms have shown a high level of complexity in transcriptional networks with multiple crossregulated transcription factors affecting transcription.58 Sandoval et al.4 have estimated that a minimum of four transcription factors are required to explain selective regulation of Aqp2 gene transcription in collecting duct cells. mpkCCD cells have been found to express >700 transcription factors, making a protein by protein identification of relevant transcription factors using ChIP-Seq and CRISPR-Cas9 deletions impractical. Here, we have used an unbiased, systems-level approach using several next generation sequencing modalities to identify likely enhancers within the same insulated neighborhood as Aqp2. These Aqp2-associated enhancers are likely to bind the transcription factors involved in cell type–specific expression and regulation by vasopressin. Motif analysis of the enhancers identified relatively few transcription factors, thus reducing the number of candidates that will require investigation in future studies (Figure 4). We have carried out transcription factor ChIP-Seq for two candidates, viz. CREB and C/EBPβ, identifying a strong binding site for the latter in one of the identified enhancer regions. We provide comprehensive genome-wide binding data as an online resource for investigators interested in regulation of genes other than Aqp2. Future studies can use next generation sequencing techniques, advanced reporter systems, and CRISPR-Cas9 to examine roles of all transcription factor candidates in the Aqp2 gene regulatory network.
Disclosures
None.
Supplementary Material
Acknowledgments
Next generation sequencing was done in the National Heart, Lung and Blood Institute DNA Sequencing Core Facility (Yuesheng Li, Director).
The work was primarily funded by Division of Intramural Research, National Heart, Lung, and Blood Institute projects ZIA-HL001285 (to M.A.K.) and ZIA-HL006129 (to M.A.K.).
All authors approved the final version of the manuscript.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2017050545/-/DCSupplemental.
References
- 1.Nielsen S, Chou CL, Marples D, Christensen EI, Kishore BK, Knepper MA: Vasopressin increases water permeability of kidney collecting duct by inducing translocation of aquaporin-CD water channels to plasma membrane. Proc Natl Acad Sci U S A 92: 1013–1017, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamamoto T, Sasaki S, Fushimi K, Ishibashi K, Yaoita E, Kawasaki K, et al.: Vasopressin increases AQP-CD water channel in apical membrane of collecting duct cells in Brattleboro rats. Am J Physiol 268: C1546–C1551, 1995 [DOI] [PubMed] [Google Scholar]
- 3.DiGiovanni SR, Nielsen S, Christensen EI, Knepper MA: Regulation of collecting duct water channel expression by vasopressin in Brattleboro rat. Proc Natl Acad Sci U S A 91: 8984–8988, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandoval PC, Claxton JS, Lee JW, Saeed F, Hoffert JD, Knepper MA: Systems-level analysis reveals selective regulation of Aqp2 gene expression by vasopressin. Sci Rep 6: 34863, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsumura Y, Uchida S, Rai T, Sasaki S, Marumo F: Transcriptional regulation of aquaporin-2 water channel gene by cAMP. J Am Soc Nephrol 8: 861–867, 1997 [DOI] [PubMed] [Google Scholar]
- 6.Hasler U, Vinciguerra M, Vandewalle A, Martin PY, Féraille E: Dual effects of hypertonicity on aquaporin-2 expression in cultured renal collecting duct principal cells. J Am Soc Nephrol 16: 1571–1582, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Ecelbarger CA, Nielsen S, Olson BR, Murase T, Baker EA, Knepper MA, et al.: Role of renal aquaporins in escape from vasopressin-induced antidiuresis in rat. J Clin Invest 99: 1852–1863, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandoval PC, Slentz DH, Pisitkun T, Saeed F, Hoffert JD, Knepper MA: Proteome-wide measurement of protein half-lives and translation rates in vasopressin-sensitive collecting duct cells. J Am Soc Nephrol 24: 1793–1805, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nedvetsky PI, Tabor V, Tamma G, Beulshausen S, Skroblin P, Kirschner A, et al.: Reciprocal regulation of aquaporin-2 abundance and degradation by protein kinase A and p38-MAP kinase. J Am Soc Nephrol 21: 1645–1656, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moeller HB, Praetorius J, Rützler MR, Fenton RA: Phosphorylation of aquaporin-2 regulates its endocytosis and protein-protein interactions. Proc Natl Acad Sci U S A 107: 424–429, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielsen S, Frøkiaer J, Marples D, Kwon TH, Agre P, Knepper MA: Aquaporins in the kidney: From molecules to medicine. Physiol Rev 82: 205–244, 2002 [DOI] [PubMed] [Google Scholar]
- 12.Isobe K, Jung HJ, Yang CR, Claxton J, Sandoval P, Burg MB, et al.: Systems-level identification of PKA-dependent signaling in epithelial cells. Proc Natl Acad Sci U S A 114: E8875–E8884, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vuagniaux G, Vallet V, Jaeger NF, Pfister C, Bens M, Farman N, et al.: Activation of the amiloride-sensitive epithelial sodium channel by the serine protease mCAP1 expressed in a mouse cortical collecting duct cell line. J Am Soc Nephrol 11: 828–834, 2000 [DOI] [PubMed] [Google Scholar]
- 14.Hallows KR, Wang H, Edinger RS, Butterworth MB, Oyster NM, Li H, et al.: Regulation of epithelial Na+ transport by soluble adenylyl cyclase in kidney collecting duct cells. J Biol Chem 284: 5774–5783, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu MJ, Miller RL, Uawithya P, Rinschen MM, Khositseth S, Braucht DW, et al.: Systems-level analysis of cell-specific AQP2 gene expression in renal collecting duct. Proc Natl Acad Sci U S A 106: 2441–2446, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasui M, Zelenin SM, Celsi G, Aperia A: Adenylate cyclase-coupled vasopressin receptor activates AQP2 promoter via a dual effect on CRE and AP1 elements. Am J Physiol 272: F443–F450, 1997 [DOI] [PubMed] [Google Scholar]
- 17.Hozawa S, Holtzman EJ, Ausiello DA: cAMP motifs regulating transcription in the aquaporin 2 gene. Am J Physiol 270: C1695–C1702, 1996 [DOI] [PubMed] [Google Scholar]
- 18.Hasler U, Leroy V, Jeon US, Bouley R, Dimitrov M, Kim JA, et al.: NF-kappaB modulates aquaporin-2 transcription in renal collecting duct principal cells. J Biol Chem 283: 28095–28105, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rai T, Uchida S, Marumo F, Sasaki S: Cloning of rat and mouse aquaporin-2 gene promoters and identification of a negative cis-regulatory element. Am J Physiol 273: F264–F273, 1997 [DOI] [PubMed] [Google Scholar]
- 20.Uchida S, Matsumura Y, Rai T, Sasaki S, Marumo F: Regulation of aquaporin-2 gene transcription by GATA-3. off. Biochem Biophys Res Commun 232: 65–68, 1997 [DOI] [PubMed] [Google Scholar]
- 21.Yu L, Moriguchi T, Souma T, Takai J, Satoh H, Morito N, et al.: GATA2 regulates body water homeostasis through maintaining aquaporin 2 expression in renal collecting ducts. Mol Cell Biol 34: 1929–1941, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grassmeyer J, Mukherjee M, deRiso J, Hettinger C, Bailey M, Sinha S, et al.: Elf5 is a principal cell lineage specific transcription factor in the kidney that contributes to Aqp2 and Avpr2 gene expression. Dev Biol 424: 77–89, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasler U, Jeon US, Kim JA, Mordasini D, Kwon HM, Féraille E, et al.: Tonicity-responsive enhancer binding protein is an essential regulator of aquaporin-2 expression in renal collecting duct principal cells. J Am Soc Nephrol 17: 1521–1531, 2006 [DOI] [PubMed] [Google Scholar]
- 24.Hatem-Vaquero M, Griera M, Giermakowska W, Luengo A, Calleros L, Gonzalez Bosc LV, et al.: Integrin linked kinase regulates the transcription of AQP2 by NFATC3. Biochim Biophys Acta 1860: 922–935, 2017 [DOI] [PubMed] [Google Scholar]
- 25.Li SZ, McDill BW, Kovach PA, Ding L, Go WY, Ho SN, et al.: Calcineurin-NFATc signaling pathway regulates AQP2 expression in response to calcium signals and osmotic stress. Am J Physiol Cell Physiol 292: C1606–C1616, 2007 [DOI] [PubMed] [Google Scholar]
- 26.Atchison ML: Enhancers: Mechanisms of action and cell specificity. Annu Rev Cell Biol 4: 127–153, 1988 [DOI] [PubMed] [Google Scholar]
- 27.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ: Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolger SJ, Hurtado PA, Hoffert JD, Saeed F, Pisitkun T, Knepper MA: Quantitative phosphoproteomics in nuclei of vasopressin-sensitive renal collecting duct cells. Am J Physiol Cell Physiol 303: C1006–C1020, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pisitkun T, Jacob V, Schleicher SM, Chou CL, Yu MJ, Knepper MA: Akt and ERK1/2 pathways are components of the vasopressin signaling network in rat native IMCD. Am J Physiol Renal Physiol 295: F1030–F1043, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Durbin R: Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al.; 1000 Genome Project Data Processing Subgroup : The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machanick P, Bailey TL: MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, et al.: ChIPpeakAnno: A Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11: 237, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, et al.; Mouse ENCODE Consortium : A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue Z, Chen JX, Zhao Y, Medvar B, Knepper MA: Data integration in physiology using Bayes’ rule and minimum Bayes’ factors: Deubiquitylating enzymes in the renal collecting duct. Physiol Genomics 49: 151–159, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hnisz D, Day DS, Young RA: Insulated neighborhoods: Structural and functional units of mammalian gene control. Cell 167: 1188–1200, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schenk LK, Bolger SJ, Luginbuhl K, Gonzales PA, Rinschen MM, Yu MJ, et al.: Quantitative proteomics identifies vasopressin-responsive nuclear proteins in collecting duct cells. J Am Soc Nephrol 23: 1008–1018, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith E, Shilatifard A: Enhancer biology and enhanceropathies. Nat Struct Mol Biol 21: 210–219, 2014 [DOI] [PubMed] [Google Scholar]
- 39.Tchapyjnikov D, Li Y, Pisitkun T, Hoffert JD, Yu MJ, Knepper MA: Proteomic profiling of nuclei from native renal inner medullary collecting duct cells using LC-MS/MS. Physiol Genomics 40: 167–183, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaret KS, Carroll JS: Pioneer transcription factors: Establishing competence for gene expression. Genes Dev 25: 2227–2241, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, et al.: A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell 47: 810–822, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jakobsen JS, Waage J, Rapin N, Bisgaard HC, Larsen FS, Porse BT: Temporal mapping of CEBPA and CEBPB binding during liver regeneration reveals dynamic occupancy and specific regulatory codes for homeostatic and cell cycle gene batteries. Genome Res 23: 592–603, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schoenen H, Huber A, Sonda N, Zimmermann S, Jantsch J, Lepenies B, et al.: Differential control of Mincle-dependent cord factor recognition and macrophage responses by the transcription factors C/EBPβ and HIF1α. J Immunol 193: 3664–3675, 2014 [DOI] [PubMed] [Google Scholar]
- 44.Yu L, Zhang YD, Zhou J, Yao DM, Li X: Identification of target genes of transcription factor CEBPB in acute promyelocytic leukemia cells induced by all-trans retinoic acid. Asian Pac J Trop Med 6: 473–480, 2013 [DOI] [PubMed] [Google Scholar]
- 45.Yang X, Du T, Wang X, Zhang Y, Hu W, Du X, et al.: IDH1, a CHOP and C/EBPβ-responsive gene under ER stress, sensitizes human melanoma cells to hypoxia-induced apoptosis. Cancer Lett 365: 201–210, 2015 [DOI] [PubMed] [Google Scholar]
- 46.Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ Jr: Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol 6: R33, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu J, He F, Hu S, Yu J: On the nature of human housekeeping genes. Trends Genet 24: 481–484, 2008 [DOI] [PubMed] [Google Scholar]
- 48.Kortenoeven ML, Fenton RA: Renal aquaporins and water balance disorders. Biochim Biophys Acta 1840: 1533–1549, 2014 [DOI] [PubMed] [Google Scholar]
- 49.Bouley R, Hasler U, Lu HA, Nunes P, Brown D: Bypassing vasopressin receptor signaling pathways in nephrogenic diabetes insipidus. Semin Nephrol 28: 266–278, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pearce D, Soundararajan R, Trimpert C, Kashlan OB, Deen PM, Kohan DE: Collecting duct principal cell transport processes and their regulation. Clin J Am Soc Nephrol 10: 135–146, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bockenhauer D, Bichet DG: Pathophysiology, diagnosis and management of nephrogenic diabetes insipidus. Nat Rev Nephrol 11: 576–588, 2015 [DOI] [PubMed] [Google Scholar]
- 52.Telese F, Ma Q, Perez PM, Notani D, Oh S, Li W, et al.: LRP8-reelin-regulated neuronal enhancer signature underlying learning and memory formation. Neuron 86: 696–710, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayr B, Montminy M: Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol 2: 599–609, 2001 [DOI] [PubMed] [Google Scholar]
- 54.Everett LJ, Le Lay J, Lukovac S, Bernstein D, Steger DJ, Lazar MA, et al.: Integrative genomic analysis of CREB defines a critical role for transcription factor networks in mediating the fed/fasted switch in liver. BMC Genomics 14: 337, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, et al.: Defining the CREB regulon: A genome-wide analysis of transcription factor regulatory regions. Cell 119: 1041–1054, 2004 [DOI] [PubMed] [Google Scholar]
- 56.Laresgoiti U, Apraiz A, Olea M, Mitxelena J, Osinalde N, Rodriguez JA, et al.: E2F2 and CREB cooperatively regulate transcriptional activity of cell cycle genes. Nucleic Acids Res 41: 10185–10198, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lesiak A, Pelz C, Ando H, Zhu M, Davare M, Lambert TJ, et al.: A genome-wide screen of CREB occupancy identifies the RhoA inhibitors Par6C and Rnd3 as regulators of BDNF-induced synaptogenesis. PLoS One 8: e64658, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davidson EH: Emerging properties of animal gene regulatory networks. Nature 468: 911–920, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.