Abstract
Background Ischemia during kidney transplant causes chronic allograft injury and adversely affects outcome, but the underlying mechanisms are incompletely understood. In tumors, oxygen shortage reduces the DNA demethylating activity of the ten-11 translocation (TET) enzymes, yielding hypermethylated genomes that promote tumor progression. We investigated whether ischemia similarly induces DNA hypermethylation in kidney transplants and contributes to chronic injury.
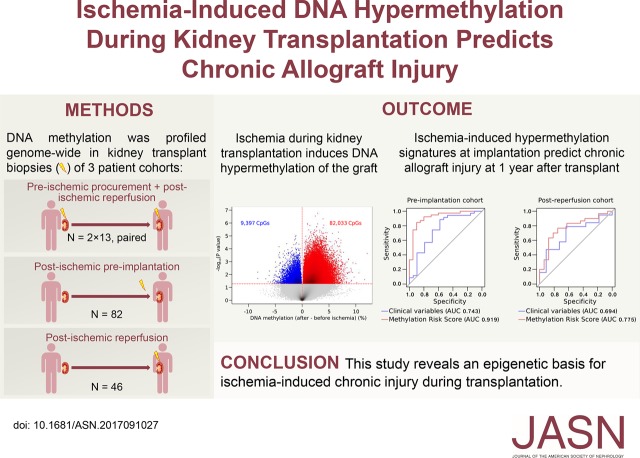
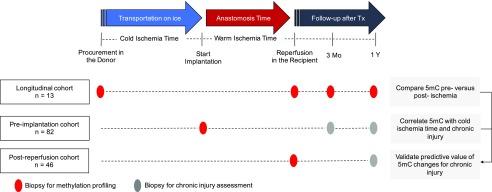
Methods We profiled genome-wide DNA methylation in three cohorts of brain-dead donor kidney allograft biopsy specimens: a longitudinal cohort with paired biopsy specimens obtained at allograft procurement (preischemia; n=13), after implantation and reperfusion (postischemia; n=13), and at 3 or 12 months after transplant (n=5 each); a cross-sectional cohort with preimplantation biopsy specimens (n=82); and a cross-sectional cohort with postreperfusion biopsy specimens (n=46).
Results Analysis of the paired preischemia and postischemia specimens revealed that methylation increased drastically in all allografts on ischemia. Hypermethylation was caused by loss of 5-hydroxymethylcytosine, the product of TET activity, and it was stable 1 year after transplant. In the preimplantation cohort, CpG hypermethylation directly correlated with ischemia time and for some CpGs, increased 2.6% per additional hour of ischemia. Hypermethylation preferentially affected and reduced the expression of genes involved in suppressing kidney injury and fibrosis. Moreover, CpG hypermethylation in preimplantation specimens predicted chronic injury, particularly fibrosis and glomerulosclerosis, 1 year after transplant. This finding was validated in the independent postreperfusion cohort, in which hypermethylation also predicted reduced allograft function 1 year after transplant, outperforming established clinical variables.
Conclusions We highlight a novel epigenetic basis for ischemia-induced chronic allograft injury with biomarker potential.
Keywords: renal transplantation, ischemia, fibrosis, epigenetics, chronic allograft nephropathy, DNA methylation
In kidney transplantation, cold ischemia time is directly proportional to delayed functioning of grafted kidneys,1 overall reduced allograft function,2 and chronic allograft injury.3 Despite intensive research, the pathophysiologic mechanisms underlying ischemia-induced chronic allograft injury are still insufficiently characterized. Experimental studies have highlighted that cold ischemia can trigger a complex set of events that delay graft function and sustain renal injury. For instance, acute ischemia can lead to chronic activation of the host immune response to the allograft.4 Nevertheless, there are currently no biomarkers to predict or effective treatment options to avoid ischemia-associated allograft injury.
Recently, DNA methylation changes affecting the Ras oncoprotein inhibitor RASAL1 have been proposed to underlie kidney fibrosis, which is a key pathologic feature contributing to chronic allograft injury after kidney transplantation.5 DNA methylation is the attachment of a methyl group to cytosines located in a CpG dinucleotide context, creating a 5-methylcytosine (5mC). CpG dinucleotides (CpGs) tend to cluster in so-called CpG islands, mostly within enhancers, the promoter, or first exon of genes, and when they are methylated, this correlates with transcriptional silencing of the affected gene. DNA methylation represents a relatively stable but reversible epigenetic mark.6 Its removal can be initiated by ten-11 translocation (TET) enzymes, which convert 5mC to 5-hydroxymethylcytosine (5hmC) in an oxygen-dependent manner.7 We recently showed that hypoxia reduces TET activity, leading to the accumulation of 5mC and the loss of 5hmC. In cancer cells, this caused hypermethylation at promoters of tumor suppressor genes.8 Specifically, we observed that, because cancer cells are highly proliferative and subject to strong genetic selection, these hypermethylation events are strongly selected for and progressively accumulate in cancer cells. Other medical conditions are, however, also characterized by long-lasting oxygen shortage, but in these affected tissues, they are far less proliferative, raising the questions of whether DNA demethylation activity is impaired and whether this similarly results in the hypermethylation driving disease progression.9
However, other than the report on DNA methylation changes affecting RASAL1 and one other report showing a relationship between epigenetic modifications and graft fibrosis,10 DNA methylation is only poorly characterized in the context of kidney transplantation. Specifically, epigenome-wide studies assessing the effects of ischemia on kidneys have never been performed, and also, the potential link between renal ischemia, epigenetic changes, and kidney allograft injury has never been addressed. Therefore, because ischemia during kidney transplantation is a major cause of chronic allograft injury and because kidneys have the unique advantage that they are amenable to repeated biopsies, allowing pre- versus postischemic DNA methylation changes to be accurately assessed within a single kidney, we here explore whether DNA hypermethylation underlies ischemia-induced chronic kidney allograft injury.
Methods
Study Design and Patients
We subjected three different cohorts of kidney transplants to genome-wide DNA methylation profiling: a longitudinal cohort of 13×2 paired procurement (preischemia) and postreperfusion (postischemia) kidney transplant biopsies, with an additional biopsy 3 or 12 months after transplantation in a subgroup (n=2×5); a second preimplantation cohort of biopsies obtained immediately before implantation (n=82); and a third cohort of postreperfusion biopsies (n=46; postreperfusion cohort). We additionally collected ten postreperfusion biopsies (five from living donor kidney transplantations versus five from deceased donor transplantations with long cold ischemia times) to validate DNA hydroxymethylation changes through liquid chromatography/mass spectrometry (LC/MS). Machine-perfused kidneys were excluded from all cohorts. All transplant recipients gave written informed consent, and the study was approved by the Ethical Review Board of the University Hospitals Leuven (S53364). Renal biopsies were performed within the University Hospital and centrally scored by pathologists dedicated to renal transplant pathology following the same standard procedures.
Epigenome-Wide Methylation Profiling
Genomic DNA was extracted from all biopsies using the Allprep DNA/RNA/miRNA Universal Kit (Qiagen, Hilden, Germany). For genome-wide methylation analysis, DNA was bisulphite converted using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA) and subsequently probed for DNA methylation levels using the Illumina EPIC array (for the longitudinal and preimplantation cohort) or the 450K array (for the postreperfusion cohort). TET-assisted bisulphite conversion was used for hydroxymethylation analysis as described.8 Quality control consisted of removal of probes for which any sample did not pass a 0.01 detection P value threshold, bead cutoff of 0.05, and removal of probes on sex chromosomes. Probe annotation was performed using Minfi.11
Gene Expression Profiling
RT-PCR was performed using OpenArray technology, a real-time PCR-based solution for high-throughput gene expression analysis (Quantstudio 12K Flex Real-Time PCR system; Thermofisher Scientific, Ghent, Belgium), for 70 transcripts that corresponded to the protein-coding genes associated with the 66 CpG islands that were hypermethylated on ischemia at false discovery rate (FDR)<0.05 in both cohorts and the DNA methylation modifiers TET1, TET2, TET3, DNMT1, DNMT3A, DNMT3B, and DNMT3L. Five housekeeping genes (B2M, 18S, TBP, RPL13A, and YWHAZ) were selected according to the literature, of which 18S, TBP, and YWHAZ were considered adequate on the basis of the gene expression changes pre- versus postischemia. Five of 70 transcripts failed.
Statistical Analyses
Statistical analyses were performed using RStudio (version 0.99). Raw methylation data were normalized using BMIQ and batch corrected using Combat with the ChAMP pipeline.12 Methylation levels (β-values) were logarithmically transformed to M values for all statistical tests unless stated otherwise. Results are presented as P values and FDR values using the Benjamini and Hochberg method. LC/MS to determine unmethylated cytosine (C), 5mC, and 5hmC concentrations in the transplant genome was performed as described.8 In the longitudinal cohort, we compared DNA methylation and hydroxymethylation levels pre- versus postischemia overall using Wilcoxon signed rank and paired t tests, respectively, and subsequently at CpG site level. In the preimplantation cohort, we examined the effect of cold ischemia time expressed as a continuous variable (in hours) on DNA methylation for all CpGs using linear regression adjusted for donor age and sex, because age and sex are major determinants of the DNA methylome. In addition, individual CpGs were grouped according to their associated CpG island (including shores and shelves), and similar analyses were performed for CpG islands: in the longitudinal cohort by paired t tests per island and in the preimplantation cohort using a linear mixed model adjusted for donor age and sex and with transplant identifier as a random effect. To evaluate locus specifically whether changes in 5mC are mirrored by inverse changes in 5hmC in the longitudinal cohort, 5mC levels for this particular analysis were estimated by subtracting 5hmC from 5mC as described previously,8 because 5mC and 5hmC are both measured as 5mC after bisulphite conversion.
Hyper- versus hypomethylation events were compared using binomial tests. Overlap between cohorts was investigated by chi-squared analysis. We annotated ischemia-hypermethylated probes in both cohorts to their chromatin state using chromHMM data annotated for human fetal kidney.13 Pathway analysis was performed using DAVID, and gene ontology enrichment was performed using topGO in R.
Gene expression in each postischemia sample was calculated relative to the expression of the reference preischemia sample using the ∆∆Ct method with log2 transformation.
Ischemia-induced hypermethylation was correlated with the Chronic Allograft Damage Index (CADI) score in protocol-specified allograft biopsies obtained at 3 months and 1 year after transplantation. Analyses were done unadjusted and adjusted for donor age (the major determinant of chronic injury)14 and donor sex (which influences DNA methylation), and a separate analysis was also done for cold and warm ischemia times. A methylation risk score (MRS) was developed to predict chronic injury (CADI score more than two) at 1 year after transplantation, and it was calculated as the sum of methylation values at each CpG in 66 ischemia-hypermethylated CpG islands weighted by marker-specific effect size. The effect size was calculated using ridge regression (Supplemental Material). Only CpGs that were also measured by the 450K arrays were considered to enable validation in the postreperfusion cohort. The DNA MRS was correlated to allograft function at 1 year after transplantation using the eGFR calculated by the Modification of Diet in Renal Disease formula.15
Results
DNA Hypermethylation of Kidney Allografts after Ischemia
To evaluate DNA methylation changes arising during cold ischemia, we set up a prospective clinical study to collect paired preischemic procurement and postischemic reperfusion biopsies of 13 brain-dead donor kidney transplants (Figure 1). This paired design minimized interindividual differences, such as genetic differences, age, and sex, which are known to profoundly influence DNA methylation levels. The average cold ischemia time was 10.1±4.1 hours. Table 1 summarizes other donor, transplant, and recipient characteristics.
Figure 1.
Schematic overview of the study cohorts to identify ischemia-induced DNA hypermethylation during kidney transplantation and evaluate its functional implications. 5mC, 5-methylcytosine; Tx, transplant.
Table 1.
Donor, transplant, and recipient characteristics of the transplants included in the different cohorts
| Characteristic | Longitudinal Cohort, n=13 | Preimplantation Cohort, n=82 | Postreperfusion Cohort, n=46 |
|---|---|---|---|
| Donor | |||
| Men/women | 8/5 | 43/39 | 18/28 |
| Age, yr | 43±13 | 47±15 | 50±15 |
| Last serum creatinine, mg/dl | 0.81±0.25 | 0.74±0.25 | 0.71±0.26a |
| Expanded versus standard criteria donation | 1/12 | 25/52b | 17/26c |
| Diabetes mellitus, N | 0 (0%) | 2 (2%) | 2 (6%)d |
| Transplant | |||
| Cold ischemia time, h | 10.08±4.15 | 13.09±3.96 | 14.59±4.68 |
| Anastomosis time, min | 32±5 | 36±9 | 33±6 |
| Recipient | |||
| Men/women | 8/5 | 54/28 | 32/14 |
| Age, yr | 55±11 | 55±12 | 57±11 |
| No. of HLA mismatches | 3±1 | 3±1c | 2±1e |
| Diabetes mellitus, N | 5 (38%) | 17 (21%) | 6 (13%) |
| Post-transplant | |||
| CADI score at baseline | 2±2b | 2±2f | 2±2g |
| Delayed graft function, N | 3 (23%) | 9 (11%) | 11 (24%) |
| Acute rejection, N | 4 (31%) | 24 (29%) | 12 (26%) |
| CADI score at 3 mo | 3±2h | 3±2i | 2±2e |
| eGFR at 3 mo, ml/min per 1.73 m2 | 42±13 | 43±16c | 44±17e |
| Serum creatinine at 3 mo, mg/ml | 1.77±0.56 | 1.82±0.71c | 1.82±0.78e |
| CADI score at 1 yr | 4±2b | 4±2j | 3±2 |
| eGFR at 1 yr, ml/min per 1.73 m2 | 45±13b | 52±14k | 50±20 |
| Serum creatinine at 1 yr, mg/ml | 1.70±0.45b | 1.44±0.40k | 1.62±0.60 |
Results are presented as mean±SD, N (%), or ratio. CADI, Chronic Allograft Damage Index.
Thirteen missing values.
Five missing values.
Three missing values.
Fifteen missing values.
One missing value.
Fourteen missing values.
Six missing values.
Two missing values.
Ten missing values.
Twenty-three missing values.
Eleven missing values.
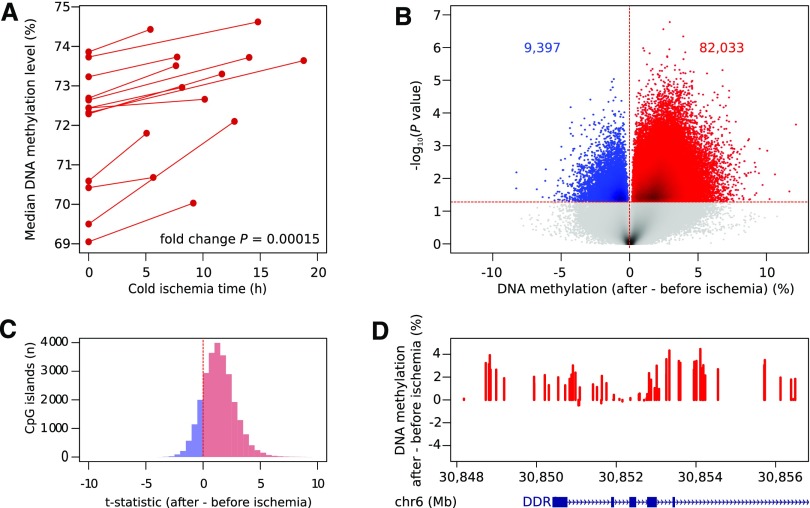
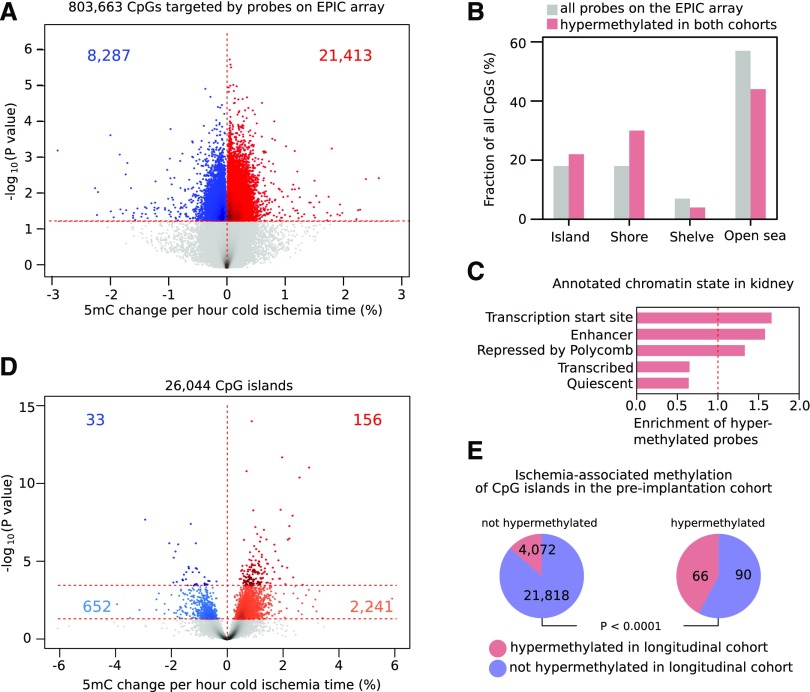
DNA methylation levels were analyzed for >850,000 CpGs using Illumina EPIC beadchips microarrays,16 and after normalization, pre- versus postischemia levels were compared in a pairwise fashion. First, we evaluated global DNA methylation levels averaged across all probes. We observed an increase in each transplant pair after ischemia (median increase: 1.3%±0.9%; P<0.001) (Figure 2A). Second, we assessed which individual CpGs were affected by ischemia. We identified 91,430 differentially methylated sites (P<0.05), most of which showed hypermethylation in the postreperfusion biopsy (82,033 CpG sites; 90%; P<0.001) (Figure 2B). Methylation levels of these CpGs increased up to 12.1% after ischemia. Significantly hypermethylated CpGs were frequently found near CpG islands, particularly within CpG island shores (20.2% versus 17.8% by random chance; P<0.001). We, therefore, grouped methylation of individual CpGs per CpG island: the vast majority of CpG islands (22,001 of 26,046; 84.5%) were hypermethylated after ischemia (Figure 2C), of which 8018 were hypermethylated after ischemia at P<0.05. When correcting for multiple testing (FDR<0.05), 4156 of 26,046 islands analyzed (16.0%) were differentially methylated, 4138 (99.6%) of which showed hypermethylation after ischemia. These islands corresponded to 2388 unique genes. Interestingly, the CpG island with the highest increase in methylation was located in the DDR1 promoter, a gene known to be involved in apoptosis and kidney fibrosis (Figure 2D).17
Figure 2.
DNA of kidney transplants becomes hypermethylated after ischemia. (A) Median overall DNA methylation levels of kidney transplants before and after ischemia. The increase in methylation is significant for each individual transplant pair (P<0.001; paired Mann–Whitney U test) and also when comparing the fold increase between median preischemic and postischemic biopsies (P<0.001). (B) Logarithmic P values of changes in methylation at individual CpG dinucleotides (CpGs) in paired kidney transplants comparing post- with preischemia conditions. Peaks with a gain (red) or loss (blue) in 5-methylcytosine are highlighted at P<0.05. (C) Distribution of the T statistics of paired tests on CpGs combined per island for all islands, showing the skewing toward hypermethylation of kidney transplants after ischemia. (D) Difference in DNA methylation after ischemia in and around the CpG island chr6:30852102–30852676 located in the promoter of DDR1, showing diffuse hypermethylation of this region.
Loss of DNA Hydroxymethylation on Ischemia
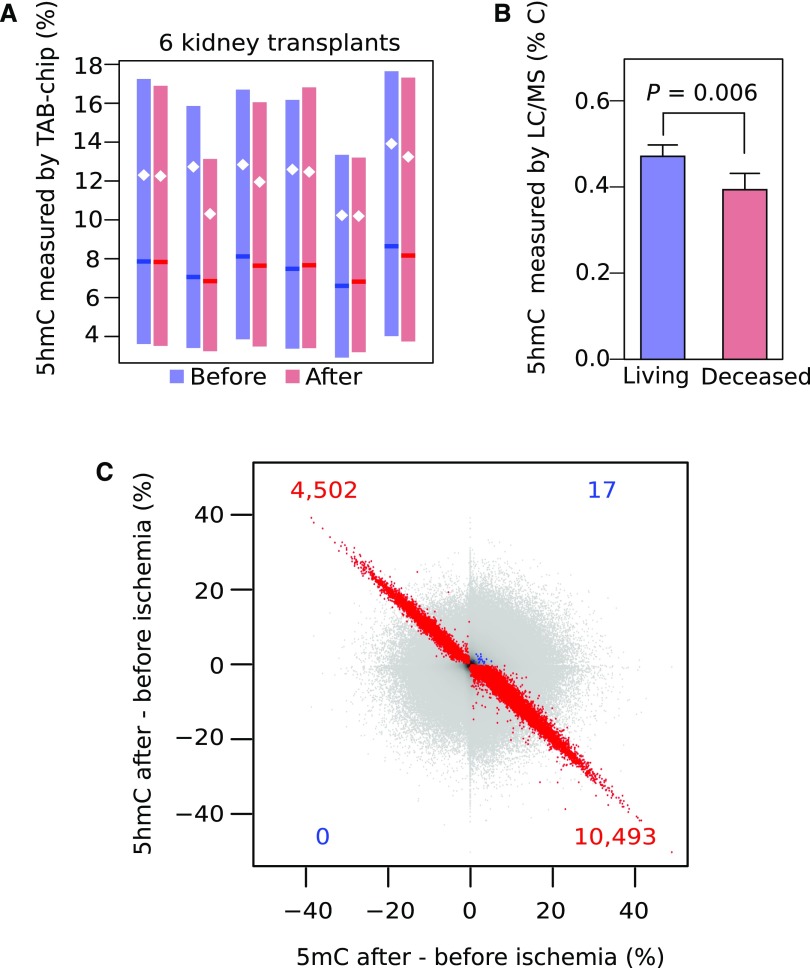
Because we recently showed that low oxygen levels in tumors inhibit DNA demethylation by reducing TET activity8 and because in postischemic biopsies, hypermethylation was enriched near CpG islands, which are preferential targets of TET enzymes,7 we measured the product of TET activity (i.e., 5hmC). Specifically, we determined 5hmC levels genome wide at >850,000 CpGs in six paired biopsies from our longitudinal cohort. Mean 5hmC levels were lower in post- versus preischemia transplants (P<0.001 for all transplants) (Figure 3A), indicating that ischemia reduces 5hmC levels in the kidney. We then evaluated locus specifically whether changes in 5hmC are mirrored by inverse changes in 5mC. 5hmC was indeed decreased in 351,966 of 427,724 (82.3%) CpGs with 5mC levels that increased after ischemia. When considering CpGs at P<0.05 for both the 5hmC and the 5mC comparisons, this relationship was even more striking: 10,493 of 10,510 (99.8%) of these CpGs with a 5mC increase showed 5hmC loss (Figure 3C). Reductions in 5hmC were not due to changes in TET expression, because expressions of TET1, TET2, and TET3 were unaltered in paired pre- versus postischemic biopsies (P>0.05). Likewise, expression of DNA methyltransferases (i.e., DNMT1, DNMT3A, DNMT3B, and DNMT3L) was unchanged.
Figure 3.
DNA hydroxymethylation levels decrease in kidney transplants on ischemia. (A) Overall DNA hydroxymethylation levels of transplants before (blue) and after (red) ischemia. The decrease in hydroxymethylation is significant for all transplants (P<0.001; paired t test). Boxes are interquartile ranges, means are white dots, and medians are darker lines. (B) 5-Hydroxymethylcytosine (5hmC) levels measured by liquid chromatography/mass spectrometry (LC/MS) show a significant loss of 5hmC in kidney transplant biopsies from deceased donation (mean of 17 hours cold ischemia time; n=5) compared with living donation (<1 hour; n=5). (C) Changes in 5-methylcytosine (5mC) levels against changes in 5hmC after ischemia. Colored points depict CpG dinucleotides for which the changes in 5hmC and 5mC are significant at P<0.05, with red used for the inverse relationship between 5mC and 5hmC and blue used for the direct relationship.
Finally, we confirmed the loss of 5hmC on ischemia using LC/MS by comparing five postreperfusion biopsies obtained from brain-dead donors characterized by long ischemia time (17.9±4.4 hours) with five biopsies obtained from living donors undergoing minimal ischemia (32±6 minutes). Warm ischemia (anastomosis) times were comparable between both groups. 5hmC levels in kidney transplants from deceased donors were, on average, 16.4%±4.4% lower compared with kidney transplants from living donors (P<0.01) (Figure 3B). Together, these findings suggest that, on ischemia, kidney allografts become hypermethylated due to reduced TET activity.
Dose Dependency of Ischemia-Induced DNA Methylation Changes
Each additional hour of cold ischemia time increases the risk of chronic allograft failure.18 Therefore, we assessed whether a similar correlation exists between cold ischemia time and the extent to which ischemia-induced methylation changes occur. We assembled a second independent cross-sectional cohort of 82 postischemic preimplantation biopsies (Figure 1, Table 1). In preimplantation biopsies, DNA methylation levels cannot be affected by warm ischemia or reperfusion, and therefore, cell composition changes cannot occur, excluding the possibility that changes in cell type composition underlie the methylation changes.
Cold ischemia time ranged from 4.7 to 26.7 hours. Genome-wide DNA methylation levels analyzed using Illumina EPIC beadchips were correlated with cold ischemia time using a linear regression adjusted for donor sex and age. Methylation levels correlated with cold ischemia time for 29,700 CpG sites (P<0.05), the bulk of these (21,413 CpGs; 72.1%) showing ischemia time–dependent hypermethylation (P<0.001) (Figure 4A). In some CpGs, methylation increased up to 2.6% with each 1-hour increase in cold ischemia time. These CpGs were also more likely to be hypermethylated in the postischemic biopsies analyzed in the longitudinal cohort (P<0.001). Particularly, up to 2932 CpGs were hypermethylated in both cohorts (P<0.05), and this mainly affected CpG islands and shores and less frequently affected shelves and open sea regions (Figure 4B). When classifying these 2932 CpGs on the basis of kidney chromatin state, these CpGs were predominantly found at enhancers and gene promoters (Figure 4C), which is in line with known TET binding sites.7
Figure 4.
Cold ischemia time correlates with the extent of ischemia-induced methylation changes. (A) Logarithmic P values obtained for individual CpG dinucleotides (CpGs) that were correlated with the duration of cold ischemia time while adjusting for donor age and sex. Peaks with a gain (red) or loss (blue) in 5-methylcytosine (5mC) are highlighted at P<0.05. (B) Distribution of the CpGs hypermethylated on ischemia in both cohorts (red) versus all probes (gray) according to their relationship with CpG islands. (C) Observed/expected fraction of ischemia-hypermethylated CpGs overlapping different kidney chromatin states. (D) Logarithmic P values obtained for CpG islands, which were correlated with the duration of cold ischemia time while adjusting for donor age and sex. Peaks gaining (red) and losing (blue) are highlighted at false discovery rate (FDR)<0.05 and P<0.05 (light colors). (E) CpG islands hypermethylated in the preimplantation cohort were also more likely to be hypermethylated in the longitudinal cohort.
At the CpG island level, cold ischemia time significantly correlated with methylation levels of 189 CpG islands (FDR<0.05 adjusted for age and sex). The vast majority of these were hypermethylated (156 islands; 82.5%) (Figure 4D). Of these 156 CpG islands, 66 (42.3%) were also hypermethylated at an FDR<0.05 threshold in the longitudinal cohort (versus 15.9% expected by random chance; P<0.001) (Figure 4E). We thus identified 66 CpG islands that were consistently hypermethylated at a stringent multiple correction threshold in both cohorts.
Expression Changes Due to Ischemia-Induced Hypermethylation
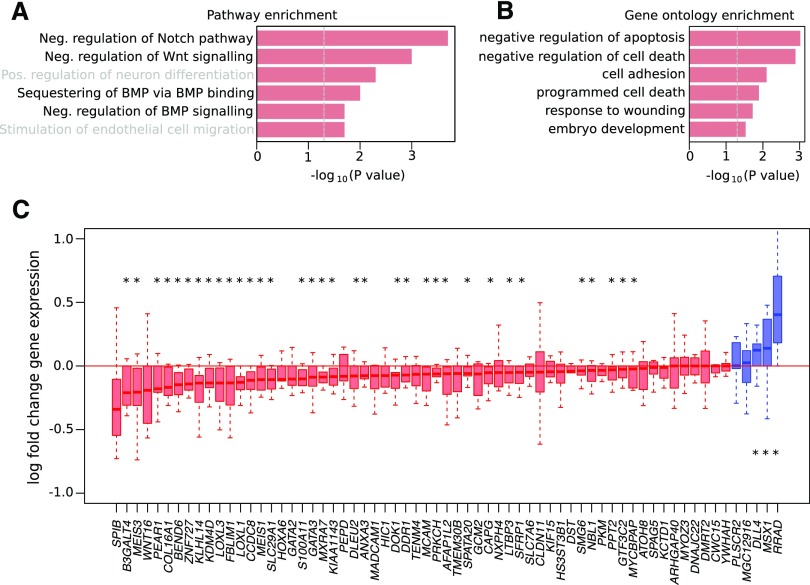
Interestingly, pathway analysis on the 81 genes associated with these 66 CpG islands revealed that genes involved in the negative regulation of the Notch and Wnt pathways, which are strongly implicated in kidney fibrosis and allograft injury,19 were enriched (Figure 5A). Other genes also played a role in the negative regulation of apoptosis and cell death (Figure 5B).
Figure 5.
DNA hypermethylation preferentially affects genes involved in suppression of kidney fibrosis and injury. (A) Pathway enrichment and (B) gene ontology enrichment of the genes associated with the 66 CpG islands that were hypermethylated after ischemia in both the longitudinal and preimplantation cohorts. (C) Log fold change in the expression of hypermethylated genes after versus before ischemia in the longitudinal cohort (n=2×13). Each boxplot represents one transcript: red indicates that expression is reduced after ischemia (median log fold change below one), and blue indicates that expression in increased after ischemia (median log fold change above one). BMP, bone morphogenetic protein. *P<0.05 by Wilcoxon test.
To evaluate whether hypermethylation of these 66 CpG islands also translates into gene expression changes within the allograft, we assessed expression of corresponding genes in the paired pre- versus postischemia biopsies of the longitudinal cohort. Of the 65 genes for which we could reliably assess expression changes, 60 (92.3%) were characterized by decreased expression in kidney transplants on ischemia and reperfusion (34 at P<0.05) (Figure 5C). These CpG islands were mainly located in gene promoters, consistent with hypermethylation suppressing gene expression. Three genes (MSX1, RRAD, and DLL4) were characterized by increased expression (P<0.05), but the corresponding hypermethylated CpG islands overlapped either completely (MSX1) or partly (RRAD and DLL4) with gene bodies. Overall, these findings indicate that methylation occurring on ischemia affects genes in biologically relevant pathways and mostly decreases expression of the associated gene.
Ischemia-Induced Hypermethylation and Chronic Allograft Injury
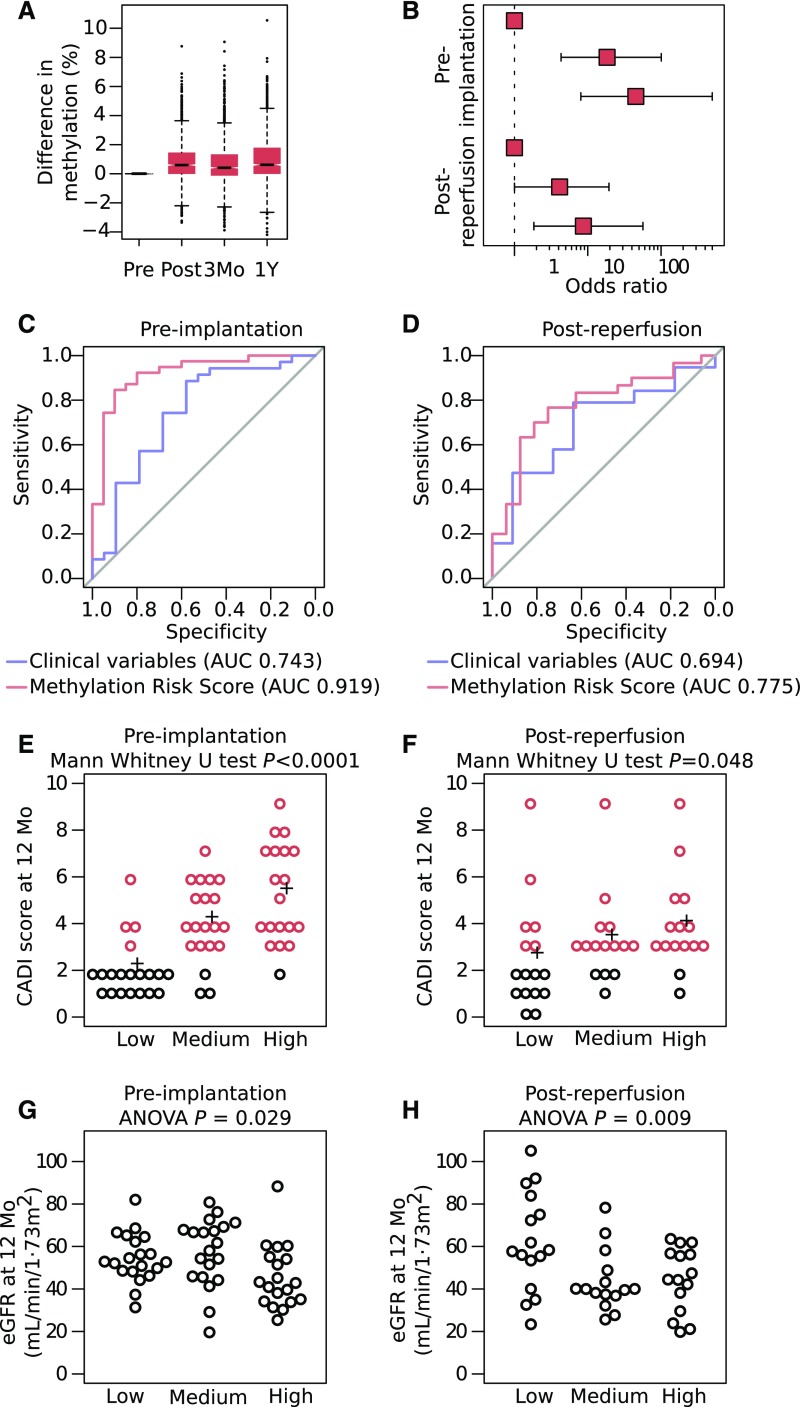
Next, we assessed whether these methylation changes are stably embedded in the kidney methylome. We, therefore, measured DNA methylation in biopsies obtained several months after transplantation (longitudinal cohort) and assessed hypermethylation in the 66 CpG islands. Interestingly, we observed that CpGs located in these islands were still hypermethylated at 3 months and 1 year after transplantation (Figure 6A).
Figure 6.
DNA hypermethylation predicts chronic allograft injury. (A) Average DNA methylation changes of CpG dinucleotides in the 66 CpG islands of kidney transplants postischemia and postreperfusion at 3 months and 1 year after transplantation in the longitudinal cohort compared with their preischemia procurement samples, showing the stability of the hypermethylation. (B) Relative risk of chronic allograft injury at 1 year after transplantation after stratifying patients into tertiles on the basis of the methylation risk score. Odds ratios are shown for the preimplantation cohort and replicated in the postreperfusion cohort. (C and D) Receiver-operating characteristic curves for the methylation risk score (red) to predict chronic injury at 1 year after transplantation compared with baseline clinical variables (donor age, donor last serum creatinine, expanded versus standard criteria donation, cold and warm ischemia times, and number of HLA mismatches; blue). Curves are shown for (C) the preimplantation cohort and replicated in (D) the postreperfusion cohort (P<0.01). (E and F) Chronic Allograft Damage Index (CADI) score for each tertile on the basis of the methylation risk score in the preimplantation and postreperfusion cohort. (G and H) Allograft function by tertile of methylation risk score in the preimplantation and postreperfusion cohort. Although eGFR levels at 12 months were available for 71 patients, CADI scores were available for only 59 patients. AUC, area under the curve.
We then investigated whether ischemia-induced hypermethylation observed at the time of transplantation correlates with chronic allograft injury (calculated by the CADI score20) (Table 1). Of the 1634 CpGs in the 66 islands, 487 (30%) CpGs were positively correlated with CADI score at 3 months (P<0.05; 332 CpGs survive multiple testing correction), and 402 (25%) CpGs were associated with CADI at 1 year (P<0.05; 135 CpGs survive multiple testing correction) (Supplemental Figure 1). This was significantly more than the 48 and 14 CpGs negatively correlating (P<0.05) with CADI at 3 months and 1 year, respectively. When adjusting for donor age and sex, similar effects were observed. The bias toward a direct correlation between hypermethylation and future injury was also not detected at baseline injury, because only 43 of 75 (57%; P>0.05) CpGs correlated positively with CADI at baseline. Also, when adjusting for cold and warm ischemia time (Supplemental Figure 2), DNA methylation correlated better with future injury than with injury already evident at the time of transplantation. Similarly, adjusting for acute rejection when correlating methylation to future injury did not influence these results (Supplemental Figure 3).
DNA Hypermethylation Predicts Chronic Allograft Injury
Having shown that ischemia-induced hypermethylation of kidney transplants correlates with chronic allograft injury, we tested whether a methylation-based risk score at the time of transplantation could predict chronic injury 1 year after transplantation. First, we developed an MRS reflecting DNA methylation in the 66 CpG islands weighted for their correlation with chronic injury at 1 year after transplant in the preimplantation cohort. We defined chronic injury as a CADI>2, representing a threshold that predicts graft survival at 1 year after transplantation.20 Patients with an MRS in the highest tertile had an increased risk (odds ratio, 45; 95% confidence interval, 8 to 499; P<0.001) to develop chronic injury relative to patients in the lowest tertile (Figure 6, B and E). The score had an area under the curve value of 0.919 to predict chronic injury, thereby outperforming baseline clinical risk factors, including donor age, donor criteria, donor last serum creatinine, cold ischemia time, anastomosis time, and the number of HLA mismatches (combined area under the curve of 0.743) (Figure 6C). The sensitivity, specificity, and positive and negative predictive value of MRS-based ROC curves were 90%, 90%, 95%, and 82%, respectively. Because CADI combines six different histopathologic lesions, we additionally evaluated MRS for each lesion individually. MRS was higher in recipients with interstitial fibrosis (P<0.001), vascular intima thickening (P=0.003) and glomerulosclerosis (P<0.001) on the 1-year protocol-specified biopsies. In contrast, MRS did not differ in recipients with or without inflammation (P=0.82), tubular atrophy (P=0.13), or mesangial matrix increase (P=0.77).
Second, because chronic injury was used as an outcome variable to assign coefficients to individual CpGs in MRS and at the same time, considered an outcome variable when assessing the predictions of the MRS, we validated our MRS score (on the basis of the same coefficients) in an independent cross-sectional cohort of 46 postreperfusion brain-dead donor kidney biopsies (Table 1). We deliberately selected biopsies taken at the postreperfusion time point to ensure robustness and clinical validity of our observations. The highest versus lowest tertile of patients had a ninefold increased risk to develop chronic injury (CADI>2; 95% confidence interval, 2 to 57; P<0.01) (Figure 6, B and F). Likewise, MRS yielded a better area under the curve than baseline clinical risk factors combined (area under the curve of 0.775 versus 0.694, respectively) (Figure 6D). The sensitivity, specificity, and positive and negative predictive value were 82%, 75%, 94%, and 46%, respectively.
Interestingly, patients in the highest MRS tertile also correlated with lower eGFR levels at 1 year (ANOVA; P=0.05 in the preimplantation cohort; P=0.01 in the postreperfusion cohort) (Figure 6, G and H). We additionally evaluated the correlation of MRS with earlier post-transplant events. We observed no correlation with delayed graft function or acute rejection (Supplemental Material). Furthermore, although MRS correlated with CADI at 3 months in both cohorts, MRS failed to predict CADI>2 at 3 months in the postreperfusion cohort. Likewise, there was no significant correlation with eGFR at 3 months (Supplemental Material). Overall, this suggests that MRS predicts changes occurring at 12 months post-transplant but does not predict those at 3 months post-transplant or baseline. Interestingly, when considering CADI at the time of implantation, MRS was higher in patients who experienced an increase in CADI at 1 year compared with those who remained stable (P=0.02), and the highest tertile of MRS had a significantly increased risk of CADI progression (P=0.03) (Supplemental Material). Patients with higher MRS also experienced a steeper decrease in eGFR between 3 and 12 months after transplant (Supplemental Material).
Discussion
In this multicohort, epigenome-wide study, we showed that cold ischemia occurring during kidney transplantation induced DNA hypermethylation of allografts through reduced TET DNA demethylation activity. The observed hypermethylation changes remained stable for months after transplantation, downregulated expression of associated genes, and preferentially affected genes involved in suppression of kidney fibrosis and injury. Importantly, the resulting methylation signature predicted future chronic allograft injury, and it did so with a predictive power that is superior compared with a combination of clinical variables routinely monitored in clinical practice.
In some CpGs, the observed DNA hypermethylation was quite substantial, with changes mounting up to 2.6% for each additional hour of cold ischemia time. With cold ischemia for some transplants lasting over 24 hours, the cumulative effect on the DNA methylome thus could become quite impactful. DNA hypermethylation was moreover observed in different cohorts involving biopsies obtained at different time points (e.g., preimplantation versus postreperfusion), thereby underscoring the robustness of our findings. Although our data are purely correlative by nature and therefore, fail to formally prove a causal relationship between ischemia-induced hypermethylation and chronic allograft injury, several observations suggest that DNA hypermethylation does causally contribute to chronic allograft injury. For instance, we observed ischemia-induced hypermethylation predominantly near genes involved in the “negative” regulation of fibrosis and cell death. Hypermethylation further silenced expression of affected genes, suggesting that it triggers allograft injury. The ischemia-induced hypermethylation was also evident up to 1 year after transplantation, which is a prerequisite for DNA methylation to induce long-term histologic changes in kidney transplants. Moreover, when correlating methylation levels of CpG islands measured at baseline to chronic injury, there was a strong correlation to future injury at 12 months but not to injury already present at baseline.
Notably, the concept of DNA hypermethylation being involved in chronic allograft injury also changes our thinking on the pathophysiology underlying ischemia-induced chronic allograft injury. Hitherto, chronic allograft injury has mainly been considered to be driven indirectly by a host immune response to acute injury.4 Our data support a more direct and lasting effect of ischemia on graft fibrosis and suggest that inhibitors of DNA methylation represent promising therapeutic agents to prevent chronic allograft injury. Indeed, DNA methylation changes are generally considered to be reversible, and DNA methylation inhibitors are already approved for the treatment of hematologic malignancies.21 Another possibility is to use compounds that induce TET activity, thereby preventing TET suppression on ischemia, prohibiting DNA hypermethylation of the graft, and protecting it from future chronic injury. Although all of these therapies might entail important side effects, there is the possibility to specifically treat grafts during their preservation, thereby preventing systemic side effects in the recipient. Importantly, however, epigenome-wide methylation profiling of whole-kidney biopsies did not allow us to assess in which cells methylation changes occurred. Recently developed single-cell methylation analyses22 are needed to overcome this limitation. Furthermore, the relevance of the methylation changes could be assessed by determining the accessibility of the affected genes to transcription (for instance, by chipping with an RNA pol2 antibody).23
Our findings also bear important biomarker potential. Indeed, we reliably predicted chronic allograft injury 1 year after transplantation by assessing methylation at the time of transplantation in those CpG islands becoming consistently hypermethylated on ischemia. In an independent replication cohort, the tertile of patients with the highest MRS exhibited a ninefold increased risk of developing allograft injury relative to patients with the lowest risk. Currently, the risk of developing chronic allograft injury is estimated on the basis of clinical risk factors, such as donor age and ischemia time, but in a head to head comparison, our MRS outperformed the combined predictive effect of these baseline clinical variables.
Mechanistically, our findings build on previous observations that we had in solid tumors, in which reduced TET DNA demethylation activity led to DNA hypermethylation of gene promoters and enhancers.8 TET enzymes are Fe2+ and α-ketoglutarate–dependent dioxygenases that oxidize 5mC to 5hmC,24 which is then further oxidized to other demethylation intermediates and subsequently, replaced by an unmodified cytosine, leading to DNA demethylation.25 In line with these findings, we observe in kidney allografts subjected to cold ischemia that DNA hypermethylation was also enriched in regions known to be TET binding sites (i.e., gene promoter and enhancer regions).7 Furthermore, each hypermethylation event was mirrored by an inverse change in 5hmC, indicating that DNA hypermethylation occurs through reduced TET activity. Although the underlying mechanisms in transplanted kidneys thus seem to be akin to those operating in tumors, our observations are quite surprising. Indeed, in transplanted kidneys, oxygen levels are lower than in tumors (0.1% versus 0.3%–0.5%), but ischemia time is much shorter (on average, 24 hours during transplantation versus months to even years in tumors). Furthermore, cancer cells are highly proliferative and can select for epigenetic changes conferring a survival benefit. In contrast, kidneys are characterized by low levels of cell proliferation, which reduce the potential for stabilization of epigenetic changes through cellular selection. Interestingly, the functional implications of our findings may be translated to other fields of medicine. Indeed, other than obvious implications in other transplant settings, they may be of relevance for other ischemic diseases, for which it would be less straightforward to show similar mechanisms. Performing paired biopsies in patients is indeed nearly impossible in other ischemic diseases, such as stroke or myocardial infarction, and also, the correlation of epigenetic changes with ischemia time would be challenging, because the exact onset of ischemia is almost impossible to determine in these pathologies.
In conclusion, we here describe a novel epigenetic mechanism that links ischemia at the time of kidney transplantation with progressive chronic allograft injury after transplantation. Because DNA methylation is generally considered to be reversible, our results can have further therapeutic applications for the prevention of chronic allograft injury, a disease that is currently lacking therapeutic options.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank Thomas Van Brussel, Jetty De Loor, Tamara Coopmans, Marc Dekens, and Jana Paulissen for technical assistance and the transplant coordinators of University Hospitals Leuven for coordinating the prospective collection of longitudinal biopsies at procurement in the donor in the donor center and after implantation and reperfusion in the recipient in our transplant center.
This work was supported by funding from the Research Foundation Flanders (11M9314N to L.H.), from the European Research Council (617595 to D.L.), and from the Klinische Onderzoeks-en Opleidingsraad (to B.S.).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2017091027/-/DCSupplemental.
References
- 1.Ojo AO, Wolfe RA, Held PJ, Port FK, Schmouder RL: Delayed graft function: Risk factors and implications for renal allograft survival. Transplantation 63: 968–974, 1997 [DOI] [PubMed] [Google Scholar]
- 2.Salahudeen AK, Haider N, May W: Cold ischemia and the reduced long-term survival of cadaveric renal allografts. Kidney Int 65: 713–718, 2004 [DOI] [PubMed] [Google Scholar]
- 3.Yilmaz S, McLaughlin K, Paavonen T, Taskinen E, Monroy M, Aavik E, et al.: Clinical predictors of renal allograft histopathology: A comparative study of single-lesion histology versus a composite, quantitative scoring system. Transplantation 83: 671–676, 2007 [DOI] [PubMed] [Google Scholar]
- 4.Perico N, Cattaneo D, Sayegh MH, Remuzzi G: Delayed graft function in kidney transplantation. Lancet 364: 1814–1827, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Bechtel W, McGoohan S, Zeisberg EM, Müller GA, Kalbacher H, Salant DJ, et al.: Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat Med 16: 544–550, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachman M, Uribe-Lewis S, Yang X, Williams M, Murrell A, Balasubramanian S: 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat Chem 6: 1049–1055, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PAC, Rappsilber J, et al.: TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473: 343–348, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu M, Hon GC, Szulwach KE, Song CX, Jin P, Ren B, He C: Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat Protoc 7: 2159–2170, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye D, Xiong Y: Cancer: Suffocation of gene expression. Nature 537: 42–43, 2016 [DOI] [PubMed] [Google Scholar]
- 10.Bontha SV, Maluf DG, Archer KJ, Dumur CI, Dozmorov MG, King AL, et al.: Effects of DNA methylation on progression to interstitial fibrosis and tubular atrophy in renal allograft biopsies: A multi-omics approach. Am J Transplant 17: 3060–3075, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al.: Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30: 1363–1369, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, et al.: ChAMP: 450k Chip analysis methylation pipeline. Bioinformatics 30: 428–430, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al.; Roadmap Epigenomics Consortium : Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stegall MD, Park WD, Larson TS, Gloor JM, Cornell LD, Sethi S, et al.: The histology of solitary renal allografts at 1 and 5 years after transplantation. Am J Transplant 11: 698–707, 2011 [DOI] [PubMed] [Google Scholar]
- 15.Poggio ED, Wang X, Weinstein DM, Issa N, Dennis VW, Braun WE, et al.: Assessing glomerular filtration rate by estimation equations in kidney transplant recipients. Am J Transplant 6: 100–108, 2006 [DOI] [PubMed] [Google Scholar]
- 16.Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, et al.: Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol 17: 208, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borza CM, Pozzi A: Discoidin domain receptors in disease. Matrix Biol 34: 185–192, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Debout A, Foucher Y, Trébern-Launay K, Legendre C, Kreis H, Mourad G, et al.: Each additional hour of cold ischemia time significantly increases the risk of graft failure and mortality following renal transplantation. Kidney Int 87: 343–349, 2015 [DOI] [PubMed] [Google Scholar]
- 19.Edeling M, Ragi G, Huang S, Pavenstädt H, Susztak K: Developmental signalling pathways in renal fibrosis: The roles of Notch, Wnt and Hedgehog. Nat Rev Nephrol 12: 426–439, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yilmaz S, Tomlanovich S, Mathew T, Taskinen E, Paavonen T, Navarro M, et al.: Protocol core needle biopsy and histologic Chronic Allograft Damage Index (CADI) as surrogate end point for long-term graft survival in multicenter studies. J Am Soc Nephrol 14: 773–779, 2003 [DOI] [PubMed] [Google Scholar]
- 21.Baylin SB, Jones PA: A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 11: 726–734, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angermueller C, Clark SJ, Lee HJ, Macaulay IC, Teng MJ, Hu TX, et al.: Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods 13: 229–232, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, et al.: A high-resolution map of active promoters in the human genome. Nature 436: 876–880, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al.: Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, Fung HL, et al.: Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153: 692–706, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.