Abstract
O-linked β-N-acetylglucosamine (O-GlcNAc) is emerging as an essential protein post-translational modification in a range of organisms. It is involved in various cellular processes such as nutrient sensing, protein degradation, gene expression, and is associated with many human diseases. Despite its importance, identifying O-GlcNAcylated proteins is a major challenge in proteomics. Here, using peracetylated N-azidoacetylglucosamine (Ac4GlcNAz) as a bioorthogonal chemical handle, we described a gel-based mass spectrometry method for the identification of proteins with O-GlcNAc modification in A549 cells. In addition, we made a labeling efficiency comparison between two modes of azide-alkyne bioorthogonal reactions in click chemistry: copper-catalyzed azide-alkyne cycloaddition (CuAAC) with Biotin-Diazo-Alkyne and stain-promoted azide-alkyne cycloaddition (SPAAC) with Biotin-DIBO-Alkyne. After conjugation with click chemistry in vitro and enrichment via streptavidin resin, proteins with O-GlcNAc modification were separated by SDS-PAGE and identified with mass spectrometry. Proteomics data analysis revealed that 229 putative O-GlcNAc modified proteins were identified with Biotin-Diazo-Alkyne conjugated sample and 188 proteins with Biotin-DIBO-Alkyne conjugated sample, among which 114 proteins were overlapping. Interestingly, 74 proteins identified from Biotin-Diazo-Alkyne conjugates and 46 verified proteins from Biotin-DIBO-Alkyne conjugates could be found in the O-GlcNAc modified proteins database dbOGAP (http://cbsb.lombardi.georgetown.edu/hulab/OGAP.html). These results suggested that CuAAC with Biotin-Diazo-Alkyne represented a more powerful method in proteomics with higher protein identification and better accuracy compared to SPAAC. The proteomics credibility was also confirmed by the molecular function and cell component gene ontology (GO). Together, the method we reported here combining metabolic labeling, click chemistry, affinity-based enrichment, SDS-PAGE separation, and mass spectrometry, would be adaptable for other post-translationally modified proteins in proteomics.
Keywords: Bioorthogonal chemistry, Biotin-Diazo-Alkyne, Biotin-DIBO-Alkyne, O-GlcNAc, Proteomics
1 Introduction
O-GlcNAc, a monosaccharide modification of nucleic and cytoplasmic proteins on serine and threonine residues, plays an important role in modulation of cellular transcription, translation and signaling processes, as well as in diseases such as cancer, diabetes and Alzheimer’s disease [1–4]. Contrast to its important role, detection and enrichment of O-GlcNAc modified proteins remain difficulty with traditional tools [5], leading to the consequence of missing remarkable O-GlcNAc modified proteins via proteomics analysis. Bioorthogonal click chemistry was first proposed by Carolyn R. Bertozzi in 2003 and offered an elite chemical handle for biological manipulation and glycomics studies [6–11].
N-azidoacetylmannosamine was the first metabolic precursor used in the corresponding sialic acid research [10]. In brief, cells were incubated with N-azidoacetylmannosamine, which will be incorporated into sialic acid biosynthetic pathway, followed by Staudinger reaction with a probe bearing complementary bioorthogonal functionality. However, the slow kinetics of the Staudinger reaction has hindered the development of labeling. Based on 1,3-dipole [3+2] cycloaddition with alkynes, two kinds of azide-based biorthogonal click chemistry were then developed in glycobiology researches [10, 11]. One type was copper-catalyzed azide-alkyne cycoaddition (CuAAC) and the other one was strain-promoted alkyne-azide cycoaddition (SPAAC). CuAAC was coined by K. Barry Sharpless and Morten Meldal in separate efforts [12, 13] and widely utilized in bio-conjugation. However, the free Cu (I) ion in the reaction is toxic to organisms. So, Carolyn R. Bertozzi developed SPAAC which can directly react with azide-labeled cells without negative additives and extended click chemistry in live cells imaging, biomolecules tracking and target protein identification [14–18]. Although these two methods were frequently used in the target molecule labeling and enrichment, less was reported about the comparison in aspects of target protein labeling efficiency and identification in O-GlcNAc proteomics.
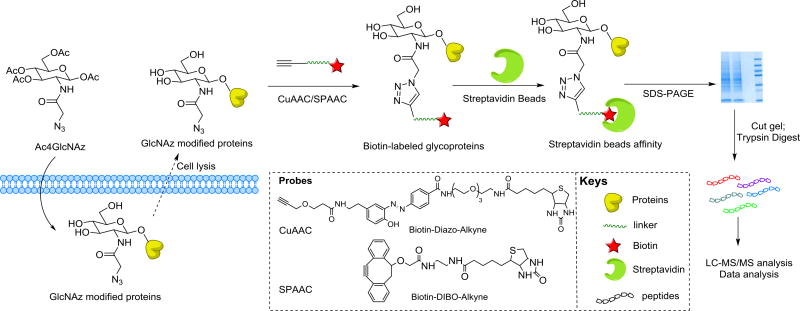
Herein, we report a comparative study for labeling O-GlcNAcylated proteins with different biotinylated alkyne probes (Fig. 1). The strategy involves feeding cells an azido GlcNAc analog that is mostly metabolically incorporated into O-GlcNAcylated proteins. The modified glycoproteins bearing azides can then be selectively labeled using Biotin-Diazo-Alkyne via CuAAC or Biotin-DIBO-Alkyne via SPAAC, permitting their enrichment from proteomes with immobilized streptavidin. The study provides a comparison between CuAAC and SPAAC methods in glycoprotein labeling and O-GlcNAc proteomics.
Figure 1.
Strategy for metabolic labeling of O-GlcNAcylated proteins with an azido GlcNAc analog (Ac4GlcNAz) for proteomics analysis by means of Biotin-Diazo-Alkyne or Biotin-DIBO-Alkyne probes.
2 Materials and methods
2.1 Chemicals and reagents
Biotin-DIBO-Alkyne was purchased from Molecular Probes. Biotin-Diazo-Alkyne was purchased from Click Chemistry Tools. Ac4GlcNAz, NuPAGE LDS sample buffer (4×), PageRuler prestained protein ladder, iBlot gel transfer stacks PVDF membrane, fetal bovine serum, bovine serum albumin, DMEM, streptavidin agarose resin, BCA protein assay kit, enhanced chemiluminescence, dithiothreitol, iodoacetamide, RIPA lysis buffer, protease inhibitor cocktail and other analytical-grade solvents were purchased from Thermo Fisher Scientific. Coomassie brilliant blue R-250 staining solution was purchased from Bio-Rad. Streptavidin HRP (ab7403) was purchased from Abcam. Antibiotics antimycotic solution (100×), DMSO were purchased from Sigma-Aldrich. Sequencing grade modified trypsin was purchased from Promega Corporation.
2.2 Culturing of A549 cells and metabolic labeling conditions
A549 cells were maintained in 5% CO2 at 37°C, and grown in DMEM with 4.5 mg/mL glucose supplemented with 10% fetal bovine serum, 0.1% antibiotics antimycotic solution (100×). One hundred micromolar Ac4GlcNAz in DMSO was delivered into 40 mL DMEM with 1.0 mg/mL glucose when the cell densities maintained at 70–80% in 175 cm2 flask. Control cultures without Ac4GlcNAz were treated in the same manner.
2.3 Protein extraction
Cells were harvested and pooled by centrifugation, washed with PBS, and lysized with cold lysis buffer supplemented with 1% protease inhibitor. Centrifugation of the cell lysates yielded a supernatant that was used as a sample for click chemistry. BCA assay was performed to dilute a final protein concentration at 1 mg/mL using lysis buffer.
2.4 Click chemistry and biotin enrichment
As previously described [19], supernatant was incubated separately with 1 mM Biotin-DIBO-Alkyne in TBS buffer (PH 8.0) at 25°C water bath for 1 h, or Biotin-Diazo-Alkyne cocktail in dark at room temperature for 1 h. Captured proteins were precipitated with complex of methanol, water, and chloroform, followed by air dry. The pallet of proteins was resuspended in buffer (6 M urea, 2 M thiourea, 10 mM HEPES, pH 8.0), reduced with 10 mM dithiothreitol at 56°C for 1 h, alkylated with 90 µM iodoacetamide in dark for 45 min, and then incubated with prewashed streptavidin beads. Bound proteins were eluted from the beads by treating with NuPAGE LDS sample buffer and boiling for 5 min.
2.5 SDS-PAGE and Western blot
Eluted proteins were electrophoresed on 0.75 mm SDS-12% polyacrylamide gels in Mini-Protean Tetra cell (Bio-Rad) according to the manufacturer’s instructions. After electrophoresis, separated proteins were either stained with Coomassie blue R-250 (Bio-Rad), or transferred to PVDF membranes (0.2 µm pore size) in an iBlot dry blotting system (Invitrogen) as described by the manufacturer. Blots were blocked for 2 h at room temperature with TBST containing 3% BSA and then incubated at 4°C overnight with anti-biotin antibody diluted 1:10 000 in TBST-3% BSA. After three times washing in TBST, blots were developed with enhanced chemi-luminescence. Target bands shown with Coomassie blue R-250 were cut into pieces for trypsin digestion as previous reported [20].
2.6 Nano RP HPLC-MS analysis and data analysis
Nano RP HPLC-MS experiments were performed on an LTQ-Orbitrap Elite mass spectrometer equipped with EASY-spray source and nano-LC UltiMate 3000 high performance liquid chromatography system as described previously [21]. Briefly, EASY-Spray PepMap C18 Column (15 cm; particle size, 2 µm; pore size, 100 Å) was used for separation. Separation was achieved with a linear gradient from 3 to 40% buffer B for 80 min at a flow rate of 300 nL/min (mobile phase A: 1.95% ACN, 97.95% H2O, 0.1% FA; mobile phase B: 79.95% ACN, 19.95% H2O, and 0.1% FA). LTQ-Orbitrap Elite MS was operated in the data-dependent mode. A full-scan survey MS experiment (m/z range from 400 to 1600; automatic gain control target, 1 000 000 ions; resolution at 200 m/z, 60 000; maximum ion accumulation time, 50 ms) was acquired by the Orbitrap mass spectrometer, and ten most intense ions were fragmented by CID. The other conditions used were: capillary temperature of 200°C, collision energy of 35 ev.
The raw file was identified using pFind 2.1 software [22] to perform the database searching against Uniprot-Swiss HUMAN.fasta (2015_09 release, 20 196 reviewed entries). Searching parameters were used as follows: fixed modification, carbamidomethyl (Cys); variable modifications, deamination (Asn), Gln→pyro-Glu (N-terminus), and oxidation (Met). Trypsin was selected as the enzyme, and two missed cleavages were allowed. The mass tolerance for the precursor ions and the fragment ions was set to 20 ppm and 0.5 Da, respectively. A false discovery rate (FDR) of 1% was applied to all data sets at the peptide level. Furthermore, pBuild 2.0 was used to remove redundant protein entries and integrate the related proteins into a single group entry. The PANTHER (http://www.pantherdb.org) [23] functional analysis and gene ontology (GO) classification tool was applied to classify the molecular function of the identified proteins.
3 Results and discussion
3.1 Comparative analysis of protein labeling efficiency with different biotinylated alkyne probes
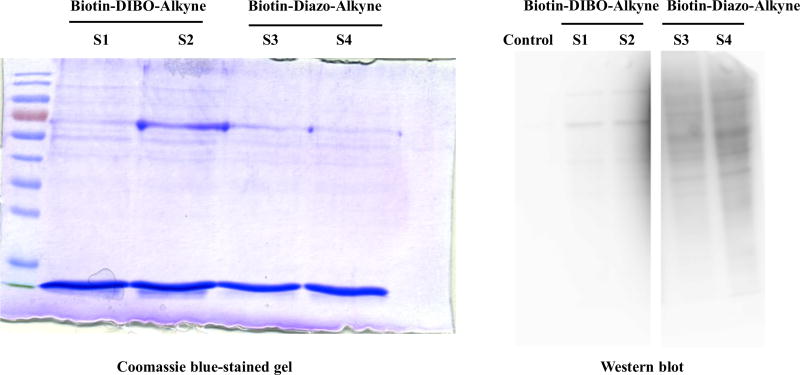
Lysates of cells treated with Ac4GlcNAz were reacted with Biotin-Diazo-Alkyne via CuAAC and Biotin-DIBO-Alkyne via SPAAC, and then precipitated to remove excess click reagents. Air-dried protein pellets were dissolved, reduced, alkylated, and incubated with prewashed streptavidin resin. After extensive washing, streptavidin bead-bound proteins were boiled from beads in the presence of LDS sample buffer and loaded onto two SDS-PAGEs. The larger one was stained with Coomassie blue R-250 and performed ingel digestion and proteomics. The other one was transferred to PVDF membrane and reacted with streptavidin-HRP for biotin detection. From SDS-PAGE (Fig. 2), more proteins were displayed in the sample with Biotin-DIBO-Alkyne conjugates. However, an opposite result was obtained from Western blot with streptavidin-HRP. The accuracy of Western blot was undoubted since streptavidin-HRP specifically bound to biotinylated alkyne probes which were covalently linked to the GlcNAz modified proteins. Nonspecific proteins leftover in the loading sample were the reasons of inconsistencies between SDS-PAGE and Western blot. Similar observations have been reported [24–26]. Strained alkyne in SPAAC is highly reactive toward cysteine-containing proteins in biological systems and results in high amount of background in click chemistry. Therefore, Cu (I)-catalyzed click reaction outperformed the stain-promoted one in the proteomics identification of labeled targets from cell lysates. The LC-MS/MS proteomics data analysis further confirmed these results.
Figure 2.
SDS-PAGE and Western blot of Biotin-DIBO-Alkyne and Biotin-Diazo-Alkyne labeled A549 cells proteomes. The SDS-PAGE and Western blot images showed the elution profiles of metabolically labeled proteins that were reacted with either probe Biotin-DIBO-Alkyne or Biotin-Diazo-Alkyne. S1, the elution of Biotin-DIBO-Alkyne conjugated proteins; S2, the repeat of S1; S3, the elution of Biotin- Diazo-Alkyne conjugated proteins; S4, the repeat of S3. Sequentially, Western blot was carried out with streptavidin-HRP. The amount of proteins in each line shown above was equally loaded.
3.2 Comparison analysis of MS-identified O-GlcNAz modified proteins
Using biotinylated alkyne probes for protein enrichment combined with in gel digestion, we characterized the O-GlcNAz modified proteomics in A549 cells. After processing with click chemistry and separating with SDS-PAGE, bands of proteins were cut out and performed LC-MS/MS analysis.
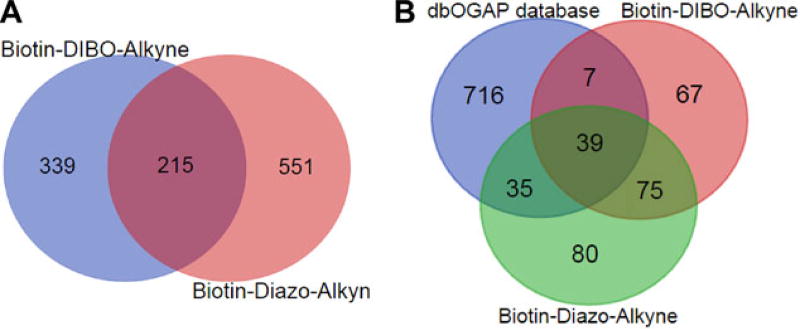
Using these method, 229 proteins were identified from Biotin-Diazo-Alkyne conjugated sample (Supporting Information Table 1), including many known O-GlcNAcylated proteins, such as pyruvate kinase, O-GlcNAcase, and glucose-6-phosphate 1-isomerase. 188 proteins were identified from Biotin-DIBO-Alkyne conjugated sample, among which 114 proteins were overlapping with each other (Supporting Information Table 2). Compared with the previous report [27], 359 proteins were identified with GlcNAz metabolic labeling in HEK293 cells, which seemed higher than our results. This should be reasonable given that different cells used and thus representing different O-GlcNAcylation profiles. We next examined the proteomics accuracy by comparing our proteins lists with the O-GlcNAc proteins database dbOGAP (http://cbsb.lombardi.georgetown.edu/hulab/OGAP.html). As described in Fig. 3B, Biotin-Diazo-Alkyne probe had higher proteomics accuracy with 74 proteins found in the database than Biotin-DIBO-Alkyne’s results of 46.
Figure 3.
Proteomics analysis of O-GlcNAz modified proteins using Biotin-Diazo-Alkyne and Biotin-DIBO-Alkyne methods. (A) Venn diagram of O-GlcNAz modified peptides identified by two probes. (B) Venn diagram of the O-GlcNAz modified proteins by two methods and O-GlcNAcylated proteins database (http://cbsb.lombardi.georgetown.edu/hulab/OGAP.html).
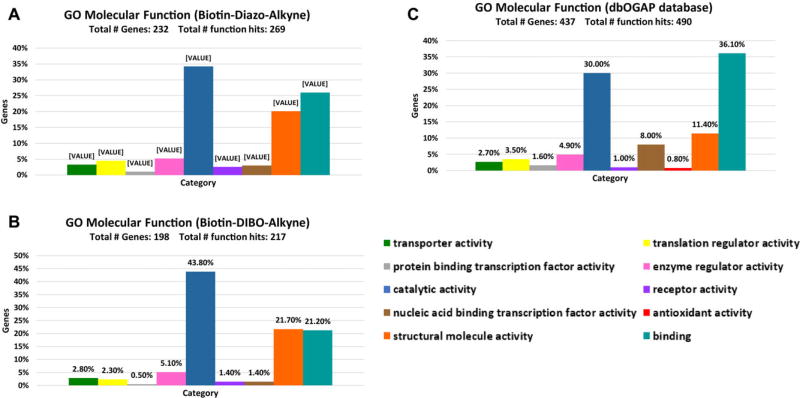
The LC-MS/MS proteomics results further confirmed the conclusion that CuAAC had higher labeling efficiency and specificity than SPAAC. Cell component gene ontology (GO) analysis [28] was consisted of the common notion that O-GlcNAc was primarily a nucleic and cytoplasmic modification, and 0.7% extracellular proteins were identified from Biotin-DIBO-Alkyne conjugates (Supporting Information Fig. 1), while none from Biotin-Diazo-Alkyne conjugates. As previously reported [27], 5–8% extracellular and membrane proteins were detected with Ac4GlcNAz labeling in HEK293 cells, which were presumably not O-GlcNAc proteins but O- or N-linked glycoproteins as GlcNAz could be incorporated into O- or N-linked glycoprotein biosynthesis pathway. Further molecular function GO analysis revealed that proteins identified from CuAAC and SPAAC had a good match with O-GlcNAc proteins database, with similar percentage in each category (Fig. 4). The identified proteins mainly distributed in the category of catalytic activity, structure molecule activity and binding, in accordance with the GO result of O-GlcNAc proteins database.
Figure 4.
O-GlcNAz modified proteins’ distribution in molecular function GO categories. Each bar represents the number of proteins (percentage indicated) assigned to a given GO category. (A–C) Molecular function GO of O-GlcNAz modified proteins using Biotin-Diazo-Alkyne, Biotin-DIBO-Alkyne, and O-GlcNAcylated proteins database in dbOGAP. We performed the molecular function GO on the website: http://www.pantherdb.org/ and analysis in software Excel.
4 Concluding remarks
O-GlcNAc is one of the post-translational modifications thought to modulate the function and activity of proteins in eukaryotes [1]. In spite of its biological function, the systematic identification of O-GlcNAc proteins in proteomics is still challenging [5]. Azide–alkyne cycloaddition is the widely recognized example of click chemistry and has been rapidly applied in glycobiology [11, 14]. Several different click reagents have been developed and used in O-GlcNAc proteomics researches [27, 29], but the efficiency and activity were rarely discussed. Here, using Ac4GlcNAz as a metabolic probe, we compared the efficiency of two biorthogonal click methods, namely SPAAC and CuAAC. In brief, Ac4GlcNAz was fed into cells, and converted into UDP-N-azidoacetylglucosamine through hexosamine salvage pathway, and then attached to serine or threonine sites of proteins by O-GlcNAc transferase. GlcNAz–labeled proteins were extracted, conjugated with biotin by CuAAC or SPAAC separately, enriched with streptavidin beads, followed by separating with SDS-PAGE, ingel digestion and MS analysis. We were able to make several observations: (i) Both CuAAC and SPAAC can efficiently tag azido-modified glycoproteins for proteomics application. However, proteomics accuracy analysis verified more potential of CuAAC as a highly effective ligation tool for biological applications in vitro, which was in concordance with the results of Western blot. High background of SPAAC in click reaction may be due to a thiol-yne reaction between cysteine containing proteins and the strained alkyne [26]. (ii) In addition, an overlapping but not identical list of identified glycoproteins was obtained from CuAAC or SPAAC prepared samples. These results suggested that to some extent both biotinylated alkyne probes should be used as complementary methods to maximize the identification of O-GlcNAcylated proteins.
Due to the limited resolution of SDS-PAGE, this gel-based MS analysis is more time-intensive, and is most applicable to the depth analysis of dozens, rather than hundreds, of samples [30]. In summary, the methodology we described here provides a generally applicable guide for alkyne-azide based click chemistry, and therefore should be of used in many other situations.
Supplementary Material
Acknowledgments
This work was supported by grants from National Institute of General Medical Sciences (R01GM085267), Natural Science Foundation of China (NO. 31000371, NO. 21372130) and Natural Science Foundation of Tianjin (15JCYBJC29000).
Abbreviations
- Ac4GlcNAz
peracetylated N-azidoacetylglucosamine
- Biotin-DIBO-Alkyne
biotindibenzocyclooctyne-alkyne
- CuAAC
copper-catalyzed alkyne-azide 1, 3-dipolar cycloaddition
- GO
gene ontology
- O-GlcNAc
O-linked β-N-acetylglucosamine
- SPAAC
strain-promoted alkyne-azide cycloaddition
Footnotes
Additional supporting information may be found in the online version of this article at the publisher’s web-site
The authors have declared no conflict of interest.
References
- 1.Varki A, Lowe JB. In: Essentials of Glycobiology. Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Cold Spring Harbor, NY, USA: 2009. [PubMed] [Google Scholar]
- 2.Wells L, Vosseller K, Hart GW. Science. 2001;291:2376–2378. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- 3.Hart GW. Annu. Rev. Biochem. 1997;66:315–335. doi: 10.1146/annurev.biochem.66.1.315. [DOI] [PubMed] [Google Scholar]
- 4.Hart GW, Kreppel LK, Comer FI, Arnold CS, Snow DM, Ye Z, Cheng X, DellaManna D, Caine DS, Earles BJ, Akimoto Y, Cole RN, Hayes BK. Glycobiology. 1996;6:711–716. doi: 10.1093/glycob/6.7.711. [DOI] [PubMed] [Google Scholar]
- 5.Cecioni S, Vocadlo DJ. Curr. Opin. Chem. Biol. 2013;17:719–728. doi: 10.1016/j.cbpa.2013.06.030. [DOI] [PubMed] [Google Scholar]
- 6.Sletten EM, Bertozzi CR. Angew. Chem. Int.Ed. Engl. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. Proc. Natl. Acad. Sci. USA. 2003;100:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertozzi CR. Acc. Chem. Res. 2011;44:651–653. doi: 10.1021/ar200193f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hang HC, Yu C, Kato DL, Bertozzi CR. Proc. Natl. Acad. Sci. USA. 2003;100:14846–14851. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxon E, Bertozzi CR. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 11.Agard NJ, Prescher JA, Bertozzi CR. J. Am. Chem. Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 12.Tornoe CW, Christensen C, Meldal M. J. Org. Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 13.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew. Chem. Int. Ed. Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 14.Chang PV, Chen X, Smyrniotis C, Xenakis A, Hu T, Bertozzi CR, Wu P. Angew. Chem. Int. Ed. Engl. 2009;48:4030–4033. doi: 10.1002/anie.200806319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jewett JC, Bertozzi CR. Chem. Soc. Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Besanceney-Webler C, Jiang H, Zheng T, Feng L, Soriano del Amo D, Wang W, Klivansky LM, Marlow FL, Liu Y, Wu P. Angew. Chem. Int. Ed. Engl. 2011;50:8051–8056. doi: 10.1002/anie.201101817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. ACS Chem. Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 18.Wu P, Feldman AK, Nugent AK, Hawker CJ, Scheel A, Voit B, Pyun J, Frechet JM, Sharpless KB, Fokin VV. Angew. Chem. Int. Ed. Engl. 2004;43:3928–3932. doi: 10.1002/anie.200454078. [DOI] [PubMed] [Google Scholar]
- 19.Zaro BW, Hang HC, Pratt MR. Methods Mol. Biol. 2013;951:57–67. doi: 10.1007/978-1-62703-146-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 21.Ma C, Qu J, Meisner J, Zhao X, Li X, Wu Z, Zhu H, Yu Z, Li L, Guo Y, Song J, Wang PG. Anal. Chem. 2015;87:7833–7839. doi: 10.1021/acs.analchem.5b02177. [DOI] [PubMed] [Google Scholar]
- 22.Li D, Fu Y, Sun R, Ling CX, Wei Y, Zhou H, Zeng R, Yang Q, He S, Gao W. Bioinformatics. 2005;21:3049–3050. doi: 10.1093/bioinformatics/bti439. [DOI] [PubMed] [Google Scholar]
- 23.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Nat. Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Speers AE, Cravatt BF. Chem. Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 25.van der Linden WA, Li N, Hoogendoorn S, Ruben M, Verdoes M, Guo J, Boons GJ, van der Marel GA, Florea BI, Overkleeft HS. Bioorg. Med. Chem. 2012;20:662–666. doi: 10.1016/j.bmc.2011.06.037. [DOI] [PubMed] [Google Scholar]
- 26.Yang Y, Yang X, Verhelst SH. Molecules. 2013;18:12599–12608. doi: 10.3390/molecules181012599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hahne H, Sobotzki N, Nyberg T, Helm D, Borodkin VS, van Aalten DM, Agnew B, Kuster B. J. Proteome Res. 2013;12:927–936. doi: 10.1021/pr300967y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gurcel C, Vercoutter-Edouart AS, Fonbonne C, Mortuaire M, Salvador A, Michalski JC, Lemoine J. Anal. Bioanal. Chem. 2008;390:2089–2097. doi: 10.1007/s00216-008-1950-y. [DOI] [PubMed] [Google Scholar]
- 30.Speers AE, Cravatt BF. Curr. Protoc. Chem. Biol. 2009;1:29–41. doi: 10.1002/9780470559277.ch090138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.