Abstract
Exploring neuroanatomical sex differences using a multivariate statistical learning approach can yield insights that cannot be derived with univariate analysis. While gross differences in total brain volume are well-established, uncovering the more subtle, regional sex-related differences in neuroanatomy requires a multivariate approach that can accurately model spatial complexity as well as the interactions between neuroanatomical features. Here, we developed a multivariate statistical learning model using a support vector machine (SVM) classifier to predict sex from MRI-derived regional neuroanatomical features from a single-site study of 967 healthy youth from the Philadelphia Neurodevelopmental Cohort (PNC). Then, we validated the multivariate model on an independent dataset of 682 healthy youth from the multi-site Pediatric Imaging, Neurocognition and Genetics (PING) cohort study. The trained model exhibited an 83% cross-validated prediction accuracy, and correctly predicted the sex of 77% of the subjects from the independent multi-site dataset. Results showed that cortical thickness of the middle occipital lobes and the angular gyri are major predictors of sex. Results also demonstrated the inferential benefits of going beyond classical regression approaches to capture the interactions among brain features in order to better characterize sex differences in male and female youths. We also identified specific cortical morphological measures and parcellation techniques, such as cortical thickness as derived from the Destrieux atlas, that are better able to discriminate between males and females in comparison to other brain atlases (Desikan-Killiany, Brodmann and subcortical atlases).
Keywords: Sex difference, Cortical morphology, Multivariate modeling, SVM, GLM, FreeSurfer
Introduction
The study of sex differences is of considerable scientific interest. Previous work has discovered links between sex differences and many phenotypic traits, such as behavior and susceptibility to disease (Gobinath et al., 2017; Rutter et al., 2003). In fact, several neuropsychiatric and developmental disorders manifest differently in males and females. For example, autism spectrum disorders (ASD), attention deficit and hyperactivity disorder (ADHD) and oppositional defiant disorder are more common in males (Baron-Cohen et al., 2011; Munkvold et al., 2011; Nøvik et al., 2006); while depression and anxiety are more prevalent in females (Schuch et al., 2014; Altemus et al., 2014). Moreover, because cognitive processes are rooted in neuronal architecture, the evaluation of sex differences in brain structure may provide a neuroanatomical basis for the sex differences in behavior and susceptibility to certain psychiatric disorders (Baron-Cohen et al., 2005; Gur et al., 1999; Gur and Gur, 2016). Specifically, identification of neurological structures underlying sexually dimorphic relationships may provide important insight into disease etiology and potential targets for treatment.
Previous studies of brain structure in vivo using magnetic resonance imaging (MRI) have revealed consistent differences in whole brain tissue volume between the sexes, with total brain volume significantly larger in males compared to females across all ages (Giedd et al., 1997; Goldstein, 2001; Gur and Gur, 2016; Ingalhalikar et al., 2014; Nopoulos et al., 2000; Ritchie et al., 2017). While these gross neuroanatomical differences are well-documented, more subtle regional differences in brain architecture are unclear. Previous studies using univariate parametric approaches, such as voxel-based morphometry (VBM), have yielded mixed results in cortical and subcortical structures, such as the amygdala (Andreano and Cahill, 2009; Hines, 2010; Marwha et al., 2017; Ruigrok et al., 2014), hippocampus (Cahill, 2006; Neufang et al., 2009; Ruigrok et al., 2014; Tan et al., 2016), and thalamus (Koolschijn and Crone, 2013; Ruigrok et al., 2014; Sowell et al., 2002). Discrepancies between studies could be due to differences in methodology, such as using different age ranges or different sample sizes. However, perhaps more fundamentally, the differences between studies could be due to the limitations inherent in using a univariate approach to studying sex differences. Because univariate methods neglect interactions between neuroanatomical features, they fail to account for differences with high spatially complexity (Davatzikos, 2004). This limitation could be overcome by employing a multivariate model; however, incorporating too many covariates into a generalized linear model (GLM) is not recommended, because high-dimensional modeling requires prohibitively large number of observations (Bellman, 1957; Hastie et al., 2009).
Compared to GLM approaches, multivariate statistical learning may have several advantages in establishing neuroanatomical differences between various groups, including the sexes. Specifically, multivariate statistical learning is theoretically a better approach since the problem of dimensionality can be overcome by considering the high-dimensional morphological profile as a single entity and optimizing parameters in order to reduce dimensionality (Davatzikos, 2004; Rosenblatt, 2016). Specifically, linear support vector machine (SVM) classifiers have been used to identify group differences in neuroimaging features for several neurological disorders (Bendfeldt et al., 2012; Ecker et al., 2010; Wendler, 2013). Also, studies using SVM classifiers have shown a correlation between age-related and sex-related differences in brain connectivity and cognition (Satterthwaite et al., 2015; Tunc et al., 2016). Thus, a multivariate approach using SVM may be especially useful for identifying neuroimaging features that reflect distinct neuroantomic differences between the sexes (Chekroud et al., 2016; Del et al., 2016; Rosenblatt, 2016), not previously detected using explanatory analysis (Joel et al., 2015).
To date, only a few studies have used multivariate classification approach to look at neuroanatomical differences between males and females. Wang et al. established discriminative neuroanatomical maps between sexes from anatomical and functional neuroimaging datasets of 140 healthy subjects (70 females, age range: 18–26) utilizing an SVM voxel-wise approach (i.e. each voxel was treated as a feature) (Wang et al., 2012). Similarly, Feis et al. created sex discriminative maps from anatomical and diffusion imaging datasets of 121 healthy subjects (67 females, age range: 20–30) using a voxel-based SVM approach (Feis et al., 2013). Both studies evaluated accuracy of the model in predicting an individual's sex using cross-validation (CV) on the same cohort (CV accuracies of 96% and 89%, respectively). However, limitations common to both of these studies were relatively small sample size and failure to test the model on an independent dataset. Furthermore, while both studies examined features on a voxel-wise level, it may also be valuable to identify regional neuroanatomical differences that can be used to discriminate between sexes.
The current study aims to expand upon previous findings in order to identify differences in regional neuroanatomical features between the sexes derived from structural MRI datasets of 967 youth (age range: 8–22) using a multivariate model tested on an independent multi-site cohort of 682 children and youth (age range: 3–21). Specifically, we built a linear SVM classifier comprised of cortical features, including curvature, thickness, volume and surface area, extracted from standard atlases. The SVM model for sex classification was first developed using the large single-site Philadelphia Neurodevelopmental Cohort (PNC) study (Satterthwaite et al., 2016, 2014), and then validated by applying it to the independent, multi-site Pediatric Imaging, Neurocognition and Genetics (PING) dataset (http://ping.chd.ucsd.edu/). The statistical parameters derived from applying our model to this dataset were compared against those derived from GLM. In summary, the methodology outlined in this study aims to do the following: quantify neuroanatomical differences between sexes using a multivariate SVM classifier model based on cortical morphology, determine to what extent these sex-related differences derived from this multivariate approach coincide and/or differ with those obtained from a GLM-based approach.
Materials and methods
We utilized the Big Data for Discovery Science (BDDS: http://bd2k.ini.usc.edu) (Toga et al., 2015) toolset to pre-process datasets from two independent cross-sectional youth cohorts (one single-site and one multi-site). Support vector machine (SVM) classification with a linear kernel was applied to the single-site cohort dataset to build a model for sex classification based solely on neuroimaging features. The generalizability of this model was tested on the independent, multi-site dataset (n = 682). We then compared the parameters derived from this SVM model to the statistical measures obtained from conventional generalized linear models (GLM). The source code for all the statistical analyses, including multivariate statistical learning and independent validation on the PING dataset, as well as GLM analysis is available on GitHub (https://github.com/sepehrband/Mining_NeuroAnat).
Datasets
Inferential and exploratory (i.e. training) analyses were performed on the Philadelphia Neurodevelopmental Cohort (PNC) dataset. The Pediatric Imaging, Neurocognition and Genetic (PING) dataset was used only as an independent dataset for testing the generalizability of the multivariate statistical learning model.
PNC dataset
Cross-sectional neuroimaging data from 997 healthy subjects from the PNC, ages 8–21 years (mean age ± SD = 14.64 ± 3.44 y), including 512 females, were acquired through the database of Genotypes and Phenotypes (dbGaP) (Satterthwaite et al., 2016, 2014). Detailed acquisition parameters are described elsewhere (Satterthwaite et al., 2014). For this study, we used the three-dimensional (3D) T1-weighted structural MRI scans, acquired using a T1-weighted magnetization prepared, rapid-acquisition gradient-echo (MPRAGE) sequence with the following parameters: TR = 1810 ms, TE = 3.5 ms, FOV = 180 × 240 mm2, matrix = 256 × 192, 160 slices, TI = 1100 ms, flip angle = 9°, effective voxel resolution = 0.9 × 0.9 × 1 mm3. For the PNC subjects, all data were collected using the same protocol on the same scanner (3T Siemens Tim Trio whole-body MRI, Erlangen, Germany; with 32-channel head coil). Of the 997 subjects, 30 subjects (14 females) were excluded because of missing demographic data, poor raw image quality, or failure in pre-processing, leaving 967 subjects for the present analysis. Demographics of the subjects included is presented in Table 1.
Table 1.
Demographics of the subjects included from the Philadelphia Neurodevelopmental Cohort (PNC) and the Pediatric Imaging, Neurocognition and Genetic (PING) datasets.
| Dataset | Number | Mean age (SD) | Age range |
|---|---|---|---|
| PNC | 967 | 14.7 (3.4) | 8.3–22.6 |
| Female | 498 | 15.0 (3.4) | 8.6–22.6 |
| Male | 469 | 14.4 (3.5) | 8.3–21.7 |
|
| |||
| PING | 682 | 12.0 (5.0) | 3.2–21.0 |
| Female | 322 | 12.1 (5.1) | 3.2–21.0 |
| Male | 360 | 12.0 (4.9) | 3.2–21.0 |
PING dataset
Cross-sectional structural T1-weighted MRI images were acquired from the PING study (http://pingstudy.ucsd.edu/). Data from 777 healthy subjects, ages 3–21 years (mean age ± SD = 12.29 ± 5.03 y), including 368 females were acquired at twelve sites using a standardized high-resolution 3D T1-weighted protocol (http://pingstudy.ucsd.edu/resources/neuroimaging-cores.html). MRI machines from multiple different vendors were used, however, the acquisition protocols were nearly identical. After applying the same pre-processing steps as the PNC dataset, the final N was 682 subjects (Table 1).
Data preparation using BBDS tools
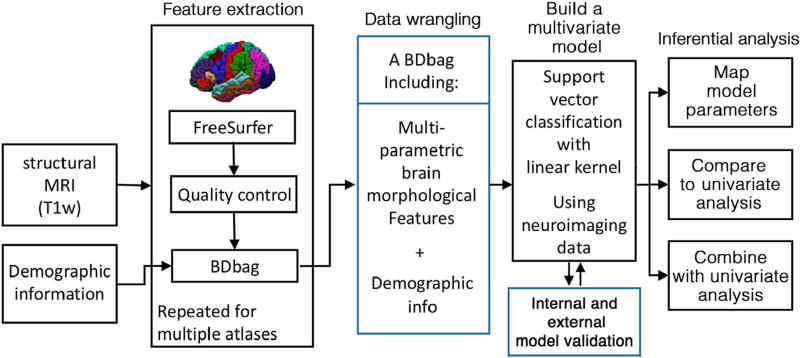
The BDDS based approach utilized is summarized in Fig. 1. Brain morphological features were extracted using the FreeSurfer (v5.3.0) software package (http://surfer.nmr.mgh.harvard.edu/) (Fischl, 2012). Data processing was performed using the Laboratory of Neuro Imaging (LONI) pipeline system (http://pipeline.loni.usc.edu) (Dinov et al., 2010, 2009; Moon et al., 2015; Torri et al., 2012). For each subject, the segmented cortex was visually inspected, and any inaccuracies were manually corrected. Data management was carried out using the Deriva Scientific Data Asset management system (http://bd2k.ini.usc.edu/tools/deriva/), and neuroimaging data and demographic information was then retrieved using the BDbag tool (http://bd2k.ini.usc.edu/tools/bdbag/).
Fig. 1.
Flowchart of the BDDS based approach implemented.
Features
For each region created by the included FreeSurfer atlases (Destrieux, Desikan-Killiany, Brodmann and subcortical atlases), the following steps were taken: 1) if the region was a cortical region, the following morphometries were computed: volume (mm3), surface area (mm2), average cortical thickness (mm), cortical thickness standard deviation (mm), integrated rectified mean curvature (1/mm), integrated rectified Gaussian curvature (1/mm2), folding index (unitless) and intrinsic curvature index (unitless), 2) if the region was a non-cortical region, e.g. gray matter nucleus or a ventricle, only the volume of the structure was computed. Neuroanatomical features, obtained from the Destrieux, Desikan-Killiany, Brodmann and subcortical atlases (Brodmann, 1909; Desikan et al., 2006; Destrieux et al., 2010; Fischl et al., 2002) were combined into single matrix for each dataset. The matrix for the PNC dataset had 967 × 2087 dimensions (967 subjects and 2087 features) and the matrix for the PING dataset had 682 × 2087 dimensions (682 subjects and 2087 features). Sex of the subject was assigned based on the prediction “outcome” (female = 1 and male = −1).
Multivariate statistical learning
A support vector machine (SVM) classifier was used as the basis of our multivariate model of neuroanatomical differences between the sexes. We used SVM with a linear kernel and a regularized solver because (Hearst et al., 1998; Schölkopf et al., 2001; Schölkopf and Smola, 2002): 1) using SVM with a linear kernel preserves the correspondence between neuroimaging data and underlying neuroanatomy, which allows for the choice of sex classification metrics directly derived from neuroanatomical differences (i.e. interpreting results directly based on their discriminative power), 2) using SVM with a regularized solver routine is able to handle wide datasets (features > instances). A linear SVM kernel was selected because it preserves, to a large degree, the one-to-one correspondence between the differences seen in neuroimaging datasets and the underlying neuroanatomical differences they represent. An SVM classifies groups (i.e. ‘male’ and ‘female’) by finding a hyperplane with an optimum (largest) margin between the two classes (Cristianini and Shawe-Taylor, 2000). The data for modeling sex-difference is a set of neuroanatomical measures of features (xi) along with sex as the label (yi = ±1, where ‘female’ = 1). To model sex-differences that are not mediated by development, age was included in the model as one of the features. Self-identified race was also included in the model, because differences were seen in the mean eTIV between races in the PNC study (524 African American, 443 European American). However, no sex-related differences were seen in race. The constraint optimization model.
aims to find the optimum β (beta coefficients) and b (intercept) that minimize β, such that:
In the current study, all derived neuroanatomical features (see above) were used for modeling sex differences. Age and estimated total intracranial volume (eTIV) were also concatenated to the feature space. All features were standardized to have zero mean and unit standard deviation. To maintain correspondence between neuroimaging features and underlying neuroanatomy, and to obtain a map of the beta coefficients for the whole brain, no feature-engineering algorithm (dimensionality reduction or feature transformation) was applied. Soft margins (overlapping margins) were chosen for finding the optimum hyperplane in order to account for the high degree of overlap between male and female neuroanatomical measurements (Supplementary Fig. 1, highlights the overlap in eTIV measures between males and females). A soft margin optimization routine finds a hyperplane that separates many, but not all, data points (Cristianini and Shawe-Taylor, 2000), which can be formulated as follows (where λ is the slack variable and C is the box constraint):
such that
Sequential minimal optimization (Platt, 1998) with 1000 iterations was utilized to minimize the cost function, approximating a solution to the above equation. Equal weights were assigned to all features (C = 1). Internal (5-fold) cross-validation (CV) was used to evaluate classification performance and to avoid overfitting. The CV routine was permuted 200 times. Performance of the classifier was evaluated based on the prediction accuracy, sensitivity (true positive rate), specificity (1- false positive rate), and the area under the receiver operation characteristic curve (AUC). Calculations involving the SVM model and the assessment of its performance were performed using MATLAB (Release, 2016a; The MathWorks, Inc.,) statistics and machine learning toolbox.
Feature extraction, through brute-force searching
The parcellation atlas and the neuroanatomical measures (e.g. cortical thickness) were chosen after evaluating their performance on modeling differences in sexes of PNC dataset. Multiple SVM classifiers were trained on the FreeSurfer outputs from different cortical atlases. Then, classification performances were evaluated and compared across neuroanatomical measures and atlases. The same parameters as above were used to train and evaluate these classifiers. In addition, different combinations of neuroanatomical features (e.g. cortical thickness and cortical surface area) were used to create sub-datasets from the entire feature space. Classification performances of these sub-datasets were compared against the full dataset. One-way analysis of variance (ANOVA) with Tukey–Kramer post hoc correction (Toothaker, 1993; Williams and Abdi, 2010) was used to compare the performances of the classifiers.
Testing the classifier on an independent dataset
The classifier with the highest score was selected as the multivariate model used to determine neuroanatomical sex differences. In order to examine the validity and generalizability of the chosen predictive, it was tested on the independent PING dataset. The modeling and testing steps were repeated 100 times. For each repetition, prediction accuracy, true and false positive rates and true and false negative rates were calculated. Two-sided t-tests were then used to validate the prediction accuracy of the independent test data against 1) the CV accuracy of a baseline model built, which was built on the eTIV of the PING subjects, and 2) the CV accuracy of the gold standard model that was trained on the neuroanatomical features of the PING subjects (same neuroanatomical features that were used to build the model on the PNC dataset). The baseline model represents a univariate model of the sex difference (using eTIV). The gold standard model represents a multivariate model, which uses the same neuroanatomical features as our model of the sex differences. Both the baseline and gold standard models used PING dataset, but our sex difference model was built on the PNC dataset. Therefore, a comparison of the model performance on the PING dataset against the baseline and gold standard models could determine the generalizability of the model.
Generalized linear model (GLM)
We used a GLM with robust linear regression as the conventional technique for identifying sex differences in the PNC dataset. We used robust linear regression to minimize the influence of outliers by implementing the statsmodels. RLM module in Python 3.5.3 (StatsModels version 0.8.0–other Python packages that were used are Pandas version 0.20.3 and SciPy version 1.13.1). Robust linear regressions with an error term (ε) was formulated as follows:
Model parameters (β) were derived by minimizing the Huber loss function (ρ(.)) (Huber, 1964), using a least trimmed squared maximum likelihood estimator (M) (Huber, 2011; Venables and Ripley, 2013). Multiple robust regressions were fitted to all 2087 FreeSurfer outputs, one neuroanatomical feature (yi) at a time. For every instance, sex, eTIV, age and self-identified race were included as predictors (xi). Regression statistics with respect to sex were extracted from each model, including beta coefficients, t-statistics and p-values. Based on the Bonferroni criterion, regression coefficients were considered significant for p-values smaller than 5 × 10−6.
Correlation with brain size
For each neuroanatomical feature, correlation between the feature and the brain size was also calculated across the PNC cohort, in order to find neuroanatomical features that exhibit sex differences that are independent of brain size. To obtain the correlation with brain size, we calculated the Spearman rank-order correlation coefficient of each feature with eTIV. The Spearman correlation is a nonparametric measure of monotonicity of the relationship between two features, and, unlike the Pearson correlation, it does not make parametric assumptions about normally distributed data. The correlation coefficient varies between −1 and +1, where correlation coefficients of 0, −1 and +1 imply no, exact negative, and exact positive correlations, respectively.
GLM and SVM
Comparing GLM and SVM
Summary statistics of neuroanatomical sex differences derived from GLM and SVM were visualized using 2D plots and on 3D surfaces in order to depict the distribution and relationship of neuroanatomical features with significant sex-related differences. Specifically, the maps represent significant neuroanatomical differences between sexes, as derived from both univariate and multivariate modeling techniques. Given that the sign of both the t-statistics of the GLM and the beta coefficients of the SVM model indicate the particular sex that correlates with the observed neuroanatomical feature (negative for male and positive for female), we mapped these onto the corresponding region on the population averaged cortical surface obtained from FreeSurfer.
Combining GLM and SVM
GLM and SVM model reveal associative and discriminative sex differences, respectively. Yet both models identify neuroanatomical features that have a dependence on sex. Given the metrics derived from these two models complement each other, it might be beneficial to combine the statistical measures derived from these techniques, to enhance the model specificity. To this end, we plotted the following three statistical properties of the neuroanatomical sex-differences together: 1) discriminative power of the feature in the multivariate model (i.e., SVM beta coefficient), 2) significance of the feature in the univariate association model (i.e., GLM p-value), 3) correlation of the feature with brain size. Visualization was performed using Python visualization libraries (matplotlib 2.0.0 and Potly 2.0.7). An online version of the plot is also provided at the Plotly website (https://plot.ly/~sepehrband/50/neuroanatomy-of-sex-difference/), enabling interactive data mining of neuroimaging measures using the mixed-method approach.
Results
SVM model of neuroanatomical sex differences
Building the SVM base model of neuroanatomical sex differences
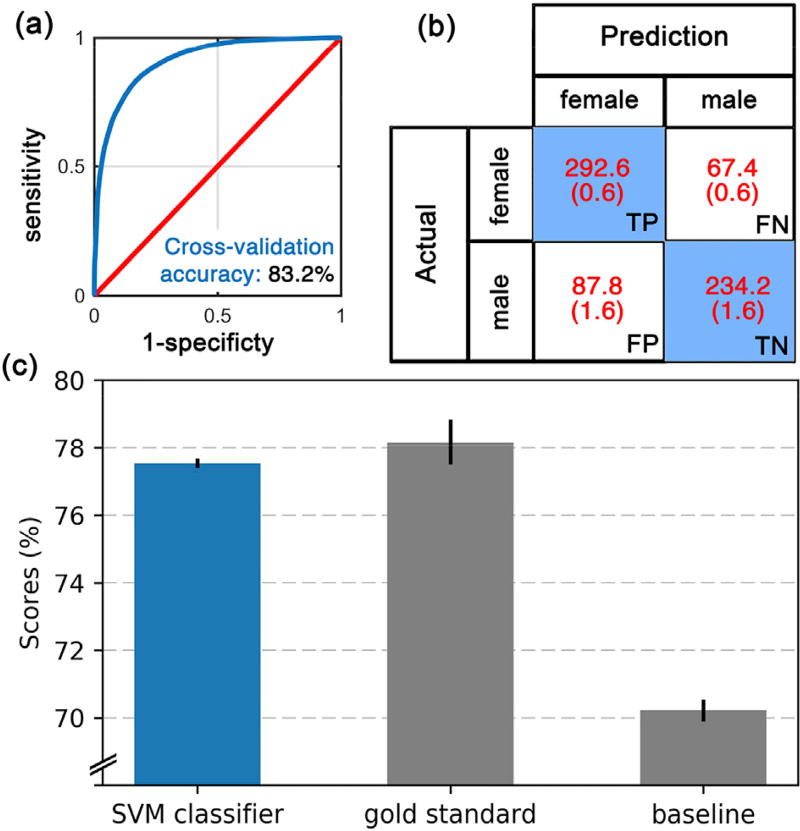
Of the twenty-four combinations (three atlases and eight neuroanatomical features) that are plotted in Fig. 2, seven features had cross-validation (CV) accuracy higher than the SVM baseline model (i.e. eTIV-only model): cortical thickness, volume, mean curvature and surface area from the Destrieux atlas, cortical surface area and volume from the Desikan-Killiany atlas, and cortical surface area from the Brodmann atlas. When a brute force comparison of different possible combinations of neuroanatomical features was conducted, the best performing model contained cortical thickness, volume, mean curvature and surface area from the Destrieux atlas. This combination resulted in a CV accuracy of 83.2 ± 0.6% (AUC: 0.89 ± 0.004, false positive rate: 0.17 ± 0.03, true positive rate: 0.82 ± 0.02), which was significantly higher (p < 0.0001) than any other combination. Our final SVM model of neuroanatomical sex-differences was therefore built using the above combination from the Destrieux atlas and was used throughout this work. The Receiver operating characteristic (ROC) curve of this classifier is shown in Fig. 3 a. The CV accuracy from subcortical (74.8 ± 0.6%) or white matter (75.7 ± 0.6%) regions were significantly lower than that compared to the Destrieux atlas (p's < 0.0001). Other metrics used to evaluate the model (i.e. AUC, sensitivity and specificity) showed the same trend (which are presented in Supplementary Table 1).
Fig. 2.
Violin plots of cross validation (CV) accuracy of sex-difference models over 200 permutations (y-axis), for different neuroanatomical features derived from Destrieux, Desikan-Killiany and Brodmann atlases (x-axis). White dots represent the median, gray bar and line represent interquartile range and 95% confident interval, respectively. Wider sections of the violin plot represent a higher density. The highest prediction accuracy was achieved when cortical volumes from the Destrieux atlas were used. Corresponding area under the receiver operating characteristic curve values of this figure is presented in Supplementary Fig. 2.
Fig. 3.
Evaluation of the SVM model of neuroanatomical sex differences on an independent dataset (PING). (a) Receiver operating characteristic (ROC) curves from cross-validation accuracy test of the model building stage on the PNC dataset. Blue line is the ROC curve of the model of neuroanatomical sex differences and red line presents chance. (b) Confusion matrix based on the performance of the model in predicting the sex of subjects of the PING dataset. TP: true positive, FP: false positive, FN: false negative, TN: true negative. (c) prediction accuracies (y-axis) of the following (x-axis): the model of sex differences (blue bar), the gold standard and the baseline models (gray bars). For all 3 bars, the mean and standard deviation of 100 repetitions are plotted.
Testing the SVM model of neuroanatomical sex differences on an independent dataset
The classification of the sex of subjects in the PING dataset was predicted with 77.2 ± 0.2% accuracy using the neuroanatomical sex-difference model (Fig. 3 b and 3. c). The model's prediction accuracy was notably higher than chance. In addition, the model had significantly higher prediction power than the baseline model that was trained on the eTIV of PING subjects (70.3 ± 0.4%), at t(99) = 167.81, p < 0.0001, with Cohen'd of 23.73. Interestingly, the prediction accuracy of the model was close to the CV accuracy of the gold standard model (77.9 ± 0.8%), but significantly lower, at t(99) = −8.9, p < 0.0001, with Cohen'd of −1.26. Note that, the gold standard was defined as the model that was trained on the neuroanatomical features of the PING data.
GLM analyses of sex differences
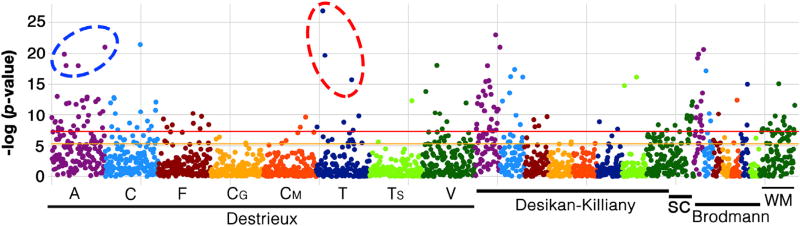
GLM analysis (multiple univariate robust linear regressions) across all neuroanatomical features, identified 162 covariates that were statistically significant at the Bonferroni adjusted p < 0.0001 (the negative log of p-value > 7.3). A plot of the negative log of the p-values (Fig. 4) shows that the majority of these covariates were related to cortical surface area (violet dots) and volume (dark green dots), followed by cortical curvature index (blue dots) and thickness (navy dots). The entire list of covariates with summary statistics are included in Supplementary file 1 (a separate list of statistical summaries from regions of only the Destrieux atlas is included in Supplementary file 2).
Fig. 4.
Important neuroanatomical features identified by GLM when modeling sex differences in the PNC dataset. Plot shows the negative log of p-values (y-axis) of neuroanatomical features (x-axis) from the GLM analysis. The orange and red lines are the Bonferroni corrected statistically significant line for p = 0.01 and p = 0.0001. Note: the red and blue dashed ovals show the neuroanatomical features that were used in an additional logistic regression (see text). A: surface area, C: curvature index, F: folding, CG: Gaussian curvature, CM: mean curvature, T: thickness, TS: thickness standard deviation, V: volume. SC: subcortical, WM: white matter.
Within the cortical thickness results, the following three regions stood out, with their structure being strongly influenced by the subject's sex (dashed red line in Fig. 4): the left occipital middle gyrus, the left and right angular gyri (with the negative log of p-value was 27.7, 20 and 17.1, respectively). When these three regions were combined into a single model to determine unique explained variance, the left angular gyrus did not exhibit any independent variance (p = 0.15). However, the right angular gyrus and left occipital middle gyrus showed a significant sex-related difference with p = 0.045 and p < 0.0001, respectively. Similar second level tests were performed on the top four regions from surface area estimates: The planum temporale of the left superior temporal gyrus, the posterior ramus of the lateral sulcus, and the total left and right hemisphere white matter (dashed blue line in Fig. 4). The GLM fit resulted in p-values of 0.1, 0.04, 0.14 and 0.002, respectively. This highlights that the univariate GLM could result in type I error, if significant p-values from independent tests are interpreted as discoveries.
GLM and SVM
Comparing GLM and SVM estimates
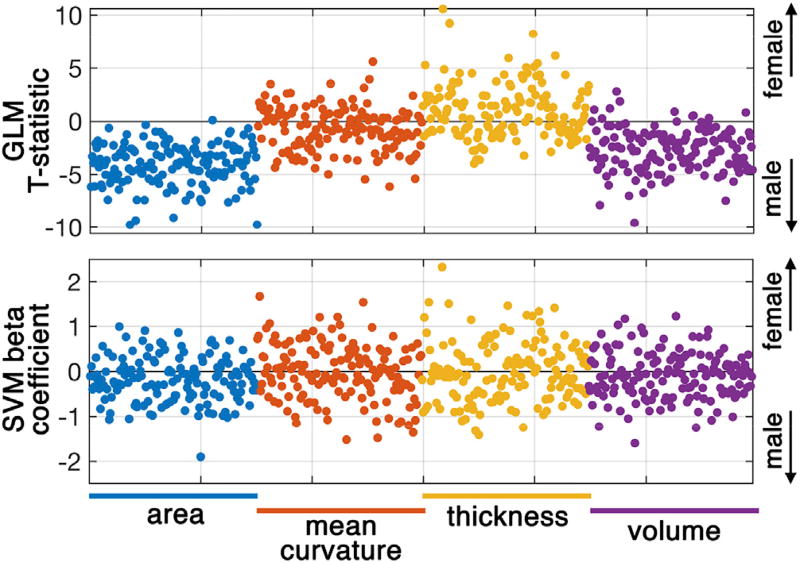
Comparing the t-statistics of the GLM analysis with the beta coefficients of the SVM demonstrates that they each carry distinct statistical information about neuroanatomical features and each have their contributions in detecting sex differences (Fig. 5). T-statistic values from the GLM, which reflect mean group differences in neuroanatomical feature between sex, were shifted toward the sex that has the higher mean value. For example, as expected, the t-statistics of cortical surface areas and volumes were shifted towards the male class (negative t-statistics), whereas cortical thickness values were shifted toward the female class (positive t-statistics). Beta coefficients from the SVM, which reflect the discriminative power of the model variable, were scattered across groups.
Fig. 5.
Statistical inferences as derived from the generalized linear model (GLM) and support vector machine (SVM) model. T-statistics of GLM (top row) and beta coefficients of SVM (bottom row) are derived from the cortical surface area, mean curvature, cortical thickness and cortical volume using the Destrieux atlas. Supplementary Files 2 and 3 contain full lists of the GLM and the SVM statistics, respectively.
A significant positive correlation between beta coefficients of the SVM and the t-statistics of the GLM were observed: cortical surface area: Pearson r = 0.65, p < 0.0001, cortical mean curvature: Pearson r = 0.64, p < 0.0001, cortical thickness: Pearson r = 0.74, p < 0.0001, cortical volume: Pearson r = 0.62, p < 0.0001) (see Supplementary Fig. 3, joint plots across all four morphological features of Fig. 5). For instance, a significant positive correlation between the GLM and the SVM on cortical surface area results, in which t-statistics are smaller than zero, means that regions with high positive SVM beta value (highly discriminative) have t-statistics near zero (non-significant association). This highlights that the GLM could lead to type II error, if covariates with non-significant p-values are interpreted as non-predictive discoveries.
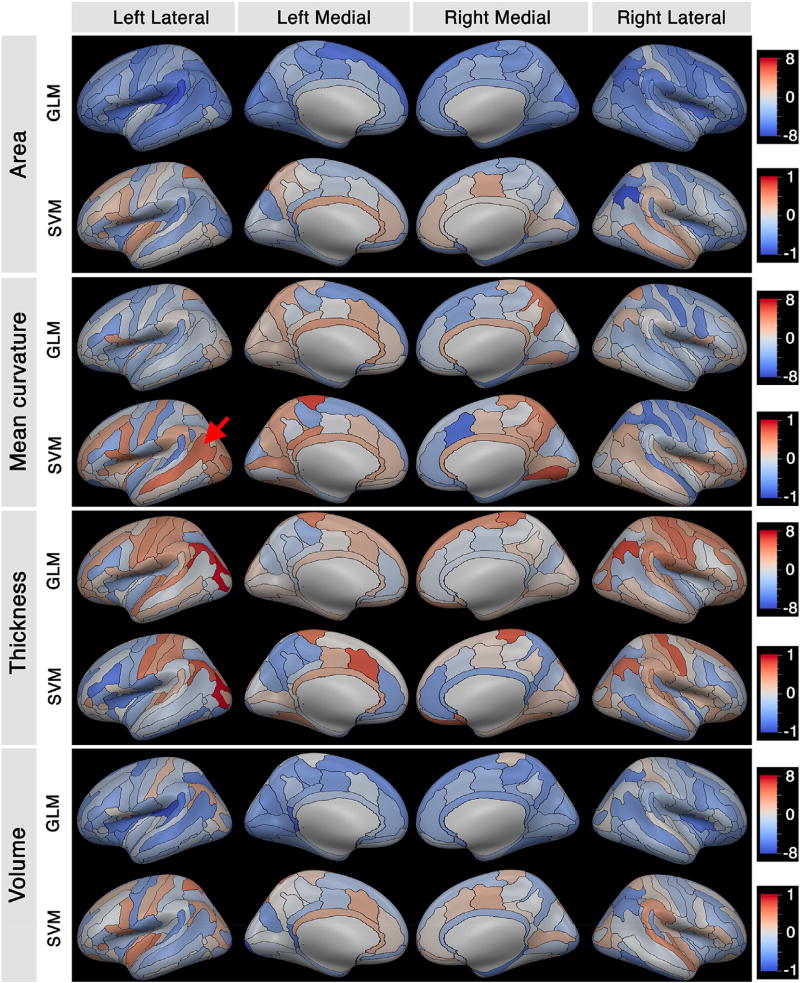
Maps of the above GLM t-statistics and SVM beta coefficients (Fig. 6) illustrate the same distribution pattern as seen in Fig. 5. For cortical surface area and cortical volume measures, the GLM results were shifted toward the male group (blue), but the SVM results were distributed between both groups. Maps show that the overall pattern of the SVM beta coefficients and the t-statistics of the GLM analysis are similar. However, there are regions that were identified as having high discriminatory power when using the SVM model, but were identified as non-significant using GLM (i.e. regions with near-zero GLM t-statistics, but with high absolute SVM beta value). For example, the left superior temporal sulcus (red arrow in Fig. 6) had a SVM beta value of 1.34 (top 2% of all prediction features) and t-statistic of −0.7 (with p > 0.05). These are regions that are highly discriminative of sex, but would not be statistically significant between sexes using GLM. Regions with high absolute SVM beta coefficient values (top 10%) with t-statistics in the range of [−1,1] are presented in Table 2 (further statistical details can be found in Supplementary Files 2 and 3).
Fig. 6.
Maps of brain regions showing sex-related differences in measures of cortical surface area, mean curvature, thickness and volume on the Destrieux atlas obtained (generated from FreeSurfer (Fischl, 2012)). For each morphological feature, the GLM t-statistic and SVM beta coefficient are mapped in the red-blue color scale shown on the right. The red arrow points to left superior temporal sulcus with high SVM discriminatory index, but near-zero t-statistic (p > 0.05). Destrieux label keys are provide in Table 1 of (Destrieux et al., 2010).
Table 2.
Neuroanatomical features with high predictive power to determine sex that exhibited nonsignificant associations in the GLM analysis.
| Neuroanatomical regions | Beta (SVM) |
t-stat (GLM) |
|---|---|---|
| cortical mean curvature of the inferior part of the left precentral sulcus | 0.94 | −0.91 |
| cortical mean curvature of the left superior temporal sulcus | 1.34 | −0.71 |
| cortical mean curvature of the right fronto-marginal gyrus (of Wernicke) and sulcus | 1.03 | 0.82 |
| cortical thickness of the left Precuneus (medial par of P1) | −0.94 | −0.82 |
| cortical thickness of the left inferior temporal gyrus | −0.96 | 0.37 |
| cortical thickness of the vertical ramus of the anterior segment of left lateral sulcus | −1.07 | −0.58 |
| cortical volume of left superior parietal lobule (lateral part of P1) | 1.02 | −0.46 |
| cortical volume of left anterior transverse temporal gyrus | 1.16 | −0.21 |
| cortical volume of the right transverse frontopolar gyri and sulci | 1.18 | −0.01 |
| cortical volume of the right angular gyrus | −1.00 | −0.70 |
| cortical volume of the lateral aspect of the right superior temporal gyrus | 0.97 | 0.30 |
Combining predictive and explanatory analysis
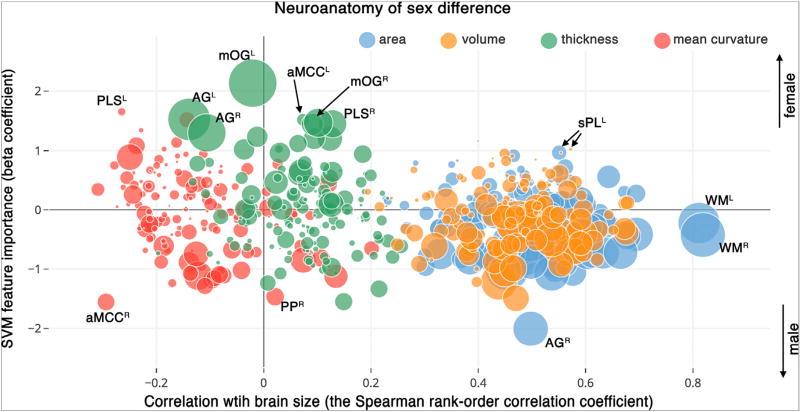
Visualization of the statistical summaries, using the mixed-method approach (Fig. 7), shows a distinction between measures of cortical volume and cortical surface area (high correlation with eTIV) and measures of cortical thickness and cortical curvature (low/no correlation with eTIV). Measures of cortical volume and cortical surface area were all significantly and positively correlated with the estimated brain size, eTIV (cortical volume measures: r ranging in 0.19–0.68 and Bonferroni adjusted p < 0.0001, cortical surface area measures: r ranging in 0.23–0.82 and Bonferroni adjusted p < 0.0001). Among these neuroanatomical measures, those with positive values of SVM beta coefficients (i.e. features that are predictors of the ‘female’ sex) had low univariate association with sex (depicted by smaller bubble size in Fig. 7). However, those with negative values of the SVM beta coefficients (i.e. predictors of the 'male' group) almost all had high univariate association with sex (depicted by large bubble size in Fig. 7). Two brain features with the highest correlation with eTIV were the left and the right hemisphere white matter surface areas (note that they have relatively low SVM-derived discriminative indices). Cortical thickness and mean curvature measures showed diverse correlation with eTIV. The majority of the regions were uncorrelated, and the remainder showed both negative and positive correlations. Interestingly, cortical mean curvature measures on average were not positively correlated with eTIV (r = −0.13 ± 0.09). Interactive exploration of the neuroanatomical features is available in the online version of the graph. Below are few instances of neuroanatomical features highly associated with sex:
Fig. 7.
Visualization of neuroanatomical differences of sex by combining the following three statistical values: Correlation of the neuroanatomical features with brain size as assessed by estimating Spearman's correlation with estimated total intracranial volume (x-axis), sex-related discriminatory indices derived from the SVM model (y-axis), and the univariate sex-related differences obtained from the GLM analysis (radius of spheres = negative log of the p-value). Pls: Paracentral lobule and sulcus, aMCC: Middle-anterior part of the cingulate cortex, mOG: Medial occipital gyrus, AG: angular gyrus, PP: Planum polare of the superior temporal gyrus, sPL: Superior parietal lobe, WM: white matter hemisphere. Superscripts refers to left (L), right (R) hemispheres. Interactive version of the plot is presented online on the Plotly website (https://plot.ly/~sepehrband/50/neuroanatomy-of-sex-difference/).
Cortical thickness of the left middle occipital gyrus (mOG): this region had the highest discriminative power of sex from multivariate modeling, was the most significant covariate from multiple univariate modeling, and had no correlation with eTIV (r = 0.02, p = 0.51). The right middle occipital gyrus was also an important predictor and a significant covariate of GLM, but to a lesser degree (SVM beta = 0.47, GLM p < 0.0001, Spearman's r = 0.02, p = 0.45).
Thickness of middle-anterior part of the cingulate cortex (aMCC): this region was not correlated with eTIV (r = 0.07, p = 0.02) and was not a significant covariate in univariate modeling (GLM p = 0.014). Yet, the SVM beta coefficient for this region was within top 1% values for predicting sex (SVM beta = 1.52).
Cortical mean curvature of the left paracentral lobule and sulcus (PLS): this region was negatively correlated with eTIV (r = −0.28, p < 0.0001) and within the top 1% of the SVM beta coefficients (SVM beta = 1.65), and was a non-significant covariate of the GLM analysis.
Discussion
Using both the SVM and the GLM, the current study provides a new understanding of sex differences in the brains of youth. The model was built on a cohort of 967 subjects with 83% cross-validation accuracy and correctly predicted the sex of 77% of subjects of an independent multisite cohort (n = 682). The testing accuracy of our SVM based multivariate model was close to the validation accuracy of the gold standard model and was significantly more accurate than the model that was built on brain volume (Fig. 3 c), highlighting the importance of multivariate inclusion of the neuroanatomical features.
Neuroanatomy of sex differences using multivariate modeling
Similar to previous studies (Gur et al., 1999; Gur and Gur, 2016; Koolschijn and Crone, 2013; Ruigrok et al., 2014), we observed that volume and surface areas of cortical regions were, in general, larger in males as compared to female youths (Figs. 4–7). However, these regions were strongly correlated with intracranial size (p < 0.0001). Therefore, sex differences in these regions may not be useful in helping to understand neuroanatomical differences between males and females beyond the well-established intracranial size difference. However, by using multivariate analysis, we were able to identify novel features with high discriminative power, regardless of the group-level mean differences. In other words, features that may show no significant difference between the sexes using GLM, could be identified as a discriminative sex feature with multivariate modeling. The left superior parietal lobule is a prime example of such a feature (p-value of GLM = 0.57, but SVM beta of 1.02). In addition to those identified by SVM alone, there were also regions that appeared informative, regardless of the employed statistical technique.
The cortical thickness of the middle occipital lobes and the angular gyri were identified as important predictors of sex (the left medial occipital lobe was the most discriminative feature). Sowell et al. (Sowell et al., 2006), similarly reported cortical thickness differences in the medial occipital lobes and the posterior temporal inferior parietal regions. Sex differences in the cortical thickness of the occipital lobes have been also reported in mice based on sex-linked genes (Markham et al., 2003). Animal studies also suggest sex differences in dendritic branching, dendritic spine density, and myelination (Kozorovitskiy et al., 2005), although these findings cannot be assessed using MRI.
In the current study, we also found that cortical thickness of the left middle-anterior part of the cingulate cortex (aMCC) and mean curvature of right aMCC were among major predictors of sex. These findings are inline with previously reported sex difference in the activation of the anterior cingulate cortex, which was correlated with personality rating in a sex-specific manner (Liu et al., 2012). However, the sex differences in the anterior cingulate cortex described in this study was only identified by multivariate analysis, showing that in general, multivariate modeling may allow for a more comprehensive description of the neuroanatomical sex differences compared to conventional univariate analysis. Moving forward, multivariate modeling may be considered in trying to understand how the identified sex differences in neuroanatomy relate to behavior and mental health by incorporating the interaction between brain regions, and perhaps providing a more accurate model of the true nature of an altered mental trait.
Methodological considerations when examining sex differences in neuroanatomy
Our findings highlight the importance of statistical approaches and neuroanatomical atlases in identifying sex differences in youth. The differences and similarities of the results from the SVM model and the GLM highlighted above and in Fig. 7, suggest that these techniques yield both distinct and overlapping information. However, the current study also highlights that the SVM-based approach has some additional technical and practical advantages over the traditional GLM technique for modeling sex differences. That is, multivariate statistical learning approaches, such as those using a SVM classifier, are able to model the complexity of the brain more accurately than classical GLM analysis because the multivariate approach can model all the neuroanatomical features at once. For a given preprocessing stream, (e.g. FreeSurfer, to derive the cortical surface), there are multiple choices of brain atlases to parcellate the surface and related, yet unique, neuroanatomical features can be extracted to quantify a given region, e.g. surface area, volume, and thickness. An advantage of statistical learning is the capability to explore many of these unique features simultaneously, without the need for manual pre-selection of covariates. In addition, incorporating multiple neuroanatomical features provides additional discriminative power compared to classifiers that were built on only one neuroanatomical feature. The SVM model incorporated the interaction between neuroanatomical features and optimizes the model based on their discriminative power. In contrast, the GLM ignores the interaction between neuroanatomical features (unless directly modeled as an interaction term). For this reason, GLM may be susceptible to inferential fallacies that could arise from detecting a “significant” p-value, because it incorrectly treats the neuroanatomical features as independent measures. Thus, by ignoring this interdependence, GLM testing can lead to type I errors (false discovery) or type II errors (missed discovery), where a metric can be erroneously interpreted as significant or not simply because of an unincorporated covariate or collinearity with another neuroanatomical feature. The GLM also relies heavily on the predefined model and selected covariates, while the statistical learning approach derives the model from the data, without a priori assumptions. As such, a multivariate model (e.g. an SVM model) that can accurate identify complex structural differences and considers their interaction was found to identify a number of sex-related differences in neuroanatomy in the brains of youths that would have otherwise been missed by GLM.
Of note, the predictive ability of neuroanatomical features differed by atlas. Features from the Destrieux atlas were found to be more predictive of sex differences than other atlases used, as shown by the highest cross-validation (CV) score using regional values of cortical thickness, volume, mean curvature and surface area from the Destrieux atlas as compared to the other atlases. Arslan et al. (Arslan et al., 2017). recently showed that the Destrieux atlas has more homogeneity and reliability compared to the Desikan-Killiany atlas and has higher agreement with myeloarchitecture. This may explain the higher prediction power that was achieved using the Destrieux atlas. Moreover, conclusions regarding differences in the discriminative power of morphologic features (e.g. cortical surface area has higher prediction accuracy compared to folding index) may also reflect a difference in the reliability of these morphological estimates via the processing pipeline rather than true sex differences. Thus, future studies may also choose to examine alternative processing pipelines, such as the one included in Advanced Normalization Tools software package (ANTs; http://stnava.github.io/ANTs/) (Avants et al., 2009), in order to determine how this may influence the performance of the classifier and SVM performance (Das et al., 2009; Mikhael et al., 2017; Tustison et al., 2014). Also, future studies that compare even finer parcellation of cerebral cortex (Glasser et al., 2016) could provide additional insight about the effect of morpho-functional segmentation of the brain on the discriminative power of sex neuroanatomy in youth.
It should be noted that we only utilized SVM for multivariate statistical learning. However, there are several alternative techniques that can potentially handle neuroimaging datasets, which are usually underpowered and contain large numbers of overlapping features (Button et al., 2013; Krahe et al., 2016; Nichols et al., 2017).
Lastly, to aid others in exploring these ideas and in hopes of improving reproducibility, the source code was made available on (https://github.com/sepehrband/Mining_NeuroAnat), alongside with descriptive demo code and interactive plots. Thus, the statistical learning routine can be applied to other MRI-derived quantitative features with only minor modification.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Biomedical Imaging and Bioengineering (P41EB015922 and U54EB020406), the Eunice Kennedy Shriver National Institute of Child Health and Human Development (R00HD065832), the National Institute of Mental Health (R01MH094343; K01MH1087610), National Institute of Diabetes and Digestive and Kidney Diseases (P30DK089503), National Institute of Neurological Disorders and Stroke (P30DK089503), National Institute of Nursing Research (P20 NR015331). This work was partially supported by NSF grants 1734853, 1636840, 1416953, 0716055 and 1023115. Many colleagues, who are part of the Big Data Discovery Science (BDDS) community, contributed indirectly to this research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIBIB, NICHD, NIMH, NIDDK, NINDS, NINR or NIH.
Data collection and sharing for this project was funded by the Philadelphia Neurodevelopmental Cohort (PNC) (Supported by RC2 grants from the National Institute of Mental Health MH089983 and MH089924, as well as T32 MH019112) and the Pediatric Imaging, Neurocognition and Genetics Study (PING) (National Institutes of Health Grant RC2DA029475). PING is funded by the National Institute on Drug Abuse and the Eunice Kennedy Shriver National Institute of Child Health & Human Development. PING data are disseminated by the PING Coordinating Center at the Center for Human Development, University of California, San Diego. The investigators within PING contributed to the design and implementation of PING and/or provided data but did not participate in analysis or writing of this report. A complete listing of PING investigators can be found at http://pingstudy.ucsd.edu/investigators.html.
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at https://doi.org/10.1016/j.neuroimage.2018.01.065.
References
- Altemus M, Sarvaiya N, Epperson CN. Sex differences in anxiety and depression clinical perspectives. Front. Neuroendocrinol. 2014;35(3):320–330. doi: 10.1016/j.yfrne.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano JM, Cahill L. Sex influences on the neurobiology of learning and memory. Learn. Mem. 2009;16:248–266. doi: 10.1101/lm.918309. https://doi.org/10.1101/lm.918309. [DOI] [PubMed] [Google Scholar]
- Arslan S, Ktena SI, Makropoulos A, Robinson EC, Rueckert D, Parisot S. Human brain mapping: a systematic comparison of parcellation methods for the human cerebral cortex. Neuroimage. 2017 doi: 10.1016/j.neuroimage.2017.04.014. https://doi.org/10.1016/j.neuroimage.2017.04.014. [DOI] [PubMed]
- Avants BB, Tustison N, Song G. Advanced normalization tools (ANTS) Insight J. 2009:1–35. [Google Scholar]
- Baron-Cohen S, Knickmeyer RC, Belmonte MK. Sex differences in the brain: implications for explaining autism. Science. 2005;310:819–823. doi: 10.1126/science.1115455. (80) [DOI] [PubMed] [Google Scholar]
- Baron-Cohen S, Lombardo MV, Auyeung B, Ashwin E, Chakrabarti B, Knickmeyer R. Why are Autism Spectrum conditions more prevalent in Males? PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001081. https://doi.org/10.1371/journal.pbio.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellman RE. Dynamic Programming. Princeton University Press; 1957. [Google Scholar]
- Bendfeldt K, Klöppel S, Nichols TE, Smieskova R, Kuster P, Traud S, Mueller-Lenke N, Naegelin Y, Kappos L, Radue EW, Borgwardt SJ. Multivariate pattern classification of gray matter pathology in multiple sclerosis. Neuroimage. 2012;60:400–408. doi: 10.1016/j.neuroimage.2011.12.070. https://doi.org/10.1016/j.neuroimage.2011.12.070. [DOI] [PubMed] [Google Scholar]
- Brodmann K. Vergleichende Lokalizationslehre der Grosshirnrinde in ihren Prinzipien Dargestellt auf Grund der Zellenbaues. Barth; 1909. [Google Scholar]
- Button KS, Ioannidis JPA, Mokrysz C, Nosek BA, Flint J, Robinson ESJ, Munafò MR. Power failure: why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 2013;14:365–376. doi: 10.1038/nrn3475. https://doi.org/10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
- Cahill L. Why sex matters for neuroscience. Nat. Rev. Neurosci. 2006;7:477–484. doi: 10.1038/nrn1909. https://doi.org/10.1038/nrn1909. [DOI] [PubMed] [Google Scholar]
- Chekroud AM, Ward EJ, Rosenberg MD, Holmes AJ. Patterns in the human brain mosaic discriminate males from females 113, 2016. 2016 doi: 10.1073/pnas.1523888113. https://doi.org/10.1073/pnas.1523888113. [DOI] [PMC free article] [PubMed]
- Cristianini N, Shawe-Taylor J. An Introduction to Support Vector Machines and Other Kernel-based Learning Methods. Cambridge university press; 2000. [Google Scholar]
- Das SR, Avants BB, Grossman M, Gee JC. Registration based cortical thickness measurement. Neuroimage. 2009;45:867–879. doi: 10.1016/j.neuroimage.2008.12.016. https://doi.org/10.1016/j.neuroimage.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C. Why voxel-based morphometric analysis should be used with great caution when characterizing group differences. Neuroimage. 2004;23:17–20. doi: 10.1016/j.neuroimage.2004.05.010. https://doi.org/10.1016/j.neuroimage.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Del M, Lippa RA, Puts DA, Bailey DH, Bailey JM. Joel et al.’s method systematically fails to detect large, consistent sex differences 113, 2016. 2016 doi: 10.1073/pnas.1525534113. https://doi.org/10.1073/pnas.1525534113. [DOI] [PMC free article] [PubMed]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. https://doi.org/10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Destrieux C, Fischl B, Dale A, Halgren E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage. 2010;53:1–15. doi: 10.1016/j.neuroimage.2010.06.010. https://doi.org/10.1016/j.neuroimage.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinov I, Lozev K, Petrosyan P, Liu Z, Eggert P. Neuroimaging study designs, computational analyses and data provenance using the LONI pipeline. PLoS One. 2010;5:e13070. doi: 10.1371/journal.pone.0013070. https://doi.org/10.1371/journal.pone.0013070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinov ID, Van Horn JD, Lozev KM, Magsipoc R, Petrosyan P, Liu Z, MacKenzie-Graham A, Eggert P, Parker DS, Toga AW. Efficient, distributed and interactive neuroimaging data analysis using the LONI pipeline. Front. Neuroinf. 2009;3 doi: 10.3389/neuro.11.022.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C, Rocha-Rego V, Johnston P, Mourao-Miranda J, Marquand A, Daly EM, Brammer MJ, Murphy C, Murphy DG. Investigating the predictive value of whole-brain structural MR scans in autism: a pattern classification approach. Neuroimage. 2010;49:44–56. doi: 10.1016/j.neuroimage.2009.08.024. https://doi.org/10.1016/j.neuroimage.2009.08.024. [DOI] [PubMed] [Google Scholar]
- Feis DL, Brodersen KH, von Cramon DY, Luders E, Tittgemeyer M. Decoding gender dimorphism of the human brain using multimodal anatomical and diffusion MRI data. Neuroimage. 2013;70:250–257. doi: 10.1016/j.neuroimage.2012.12.068. https://doi.org/10.1016/j.neuroimage.2012.12.068. [DOI] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, Van Der Kouwe A, Killiany R, Kennedy D, Klaveness S. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Castellanos FX, Rajapakse JC, Vaituzis AC, Rapoport JL. Sexual dimorphism of the developing human brain. Prog. Neuro Psychopharmacol. Biol. Psychiatr. 1997;21:1185–1201. doi: 10.1016/s0278-5846(97)00158-9. https://doi.org/10.1016/S0278-5846(97)00158-9. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, Smith SM, Van Essen DC. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536:171–178. doi: 10.1038/nature18933. https://doi.org/10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobinath AR, Choleris E, Galea LAM. Sex, hormones, and genotype interact to influence psychiatric disease, treatment, and behavioral research. J. Neurosci. Res. 2017;95:50–64. doi: 10.1002/jnr.23872. [DOI] [PubMed] [Google Scholar]
- Goldstein JM. Normal sexual dimorphism of the adult human brain assessed by in vivo magnetic resonance imaging. Cerebr. Cortex. 2001;11:490–497. doi: 10.1093/cercor/11.6.490. https://doi.org/10.1093/cercor/11.6.490. [DOI] [PubMed] [Google Scholar]
- Gur RC, Turetsky BI, Matsui M, Yan M, Bilker W, Hughett P, Gur RE. Sex differences in brain gray and white matter in healthy young adults: correlations with cognitive performance. J. Neurosci. 1999;19:4065. doi: 10.1523/JNEUROSCI.19-10-04065.1999. LP-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur RE, Gur RC. Sex differences in brain and behavior in adolescence: findings from the Philadelphia neurodevelopmental cohort. Neurosci. Biobehav. Rev. 2016;70:159–170. doi: 10.1016/j.neubiorev.2016.07.035. https://doi.org/10.1016/j.neubiorev.2016.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. Springer Series in Statistics. 2009 https://doi.org/10.1007/b94608.
- Hearst MA, Dumais P, Susan T, Osman E, Platt J, Scholkopf B. Support vector machines. Intell Syst Appl IEEE. 1998;13 https://doi.org/10.1109/5254.708428. [Google Scholar]
- Hines M. Sex-related variation in human behavior and the brain. Trends Cognit. Sci. 2010;14:448–456. doi: 10.1016/j.tics.2010.07.005. https://doi.org/10.1016/j.tics.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber PJ. International Encyclopedia of Statistical Science. Springer; 2011. Robust statistics; pp. 1248–1251. [Google Scholar]
- Huber PJ. Robust estimation of a location parameter. Ann. Math. Stat. 1964;35:73–101. [Google Scholar]
- Ingalhalikar M, Smith A, Parker D, Satterthwaite TD, Elliott MA, Ruparel K, Hakonarson H, Gur RE, Gur RC, Verma R. Sex differences in the structural connectome of the human brain. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:823–828. doi: 10.1073/pnas.1316909110. https://doi.org/10.1073/pnas.1316909110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel D, Berman Z, Tavor I, Wexler N, Gaber O, Stein Y, Shefi N, Pool J, Urchs S, Margulies DS, Liem F, Hänggi J, Jäncke L, Assaf Y. Sex beyond the genitalia: the human brain mosaic. Proc. Natl. Acad. Sci. Unit. States Am. 2015;112:15468–15473. doi: 10.1073/pnas.1509654112. https://doi.org/10.1073/pnas.1509654112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolschijn PCMP, Crone EA. Sex differences and structural brain maturation from childhood to early adulthood. Dev. Cogn. Neurosci. 2013;5:106–118. doi: 10.1016/j.dcn.2013.02.003. https://doi.org/10.1016/j.dcn.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozorovitskiy Y, Gross CG, Kopil C, Battaglia L, McBreen M, Stranahan AM, Gould E. Experience induces structural and biochemical changes in the adult primate brain. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17478–17482. doi: 10.1073/pnas.0508817102. https://doi.org/10.1073/pnas.0508817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahe C, Drabek M, Paloyelis Y, Fotopoulou A. Affective touch and attachment style modulate pain: a laser-evoked potentials study. Philos. Trans. R. Soc. B. 2016:1–22. doi: 10.1098/rstb.2016.0009. https://doi.org/10.1098/not. [DOI] [PMC free article] [PubMed]
- Liu J, Zubieta J-K, Heitzeg M. Sex differences in anterior cingulate cortex activation during impulse inhibition and behavioral correlates. Psychiatr. Res. 2012;201:54–62. doi: 10.1016/j.pscychresns.2011.05.008. https://doi.org/10.1016/j.pscychresns.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham JA, Jurgens HA, Auger CJ, De Vries GJ, Arnold AP, Juraska JM. Sex differences in mouse cortical thickness are independent of the complement of sex chromosomes. Neuroscience. 2003;116:71–75. doi: 10.1016/s0306-4522(02)00554-7. https://doi.org/10.1016/S0306-4522(02)00554-7. [DOI] [PubMed] [Google Scholar]
- Marwha D, Halari M, Eliot L. Meta-analysis reveals a lack of sexual dimorphism in human amygdala volume. Neuroimage. 2017;147:282–294. doi: 10.1016/j.neuroimage.2016.12.021. https://doi.org/10.1016/j.neuroimage.2016.12.021. [DOI] [PubMed] [Google Scholar]
- Mikhael S, Hoogendoorn C, Valdes-Hernandez M, Pernet C. A critical analysis of neuroanatomical software protocols reveals clinically relevant differences in parcellation schemes. Neuroimage. 2017:1–17. doi: 10.1016/j.neuroimage.2017.02.082. https://doi.org/10.1016/j.neuroimage.2017.02.082. [DOI] [PubMed]
- Moon SW, Dinov ID, Kim J, Zamanyan A, Hobel S, Thompson PM, Toga AW. Structural neuroimaging genetics interactions in Alzheimer's disease. J. Alzheim. Dis. 2015;48:1051–1063. doi: 10.3233/JAD-150335. https://doi.org/10.3233/JAD-150335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold LH, Lundervold AJ, Manger T. Oppositional defiant disorder-gender differences in co-occurring symptoms of mental health problems in a general population of children. J. Abnorm. Child Psychol. 2011;39:577–587. doi: 10.1007/s10802-011-9486-6. https://doi.org/10.1007/s10802-011-9486-6. [DOI] [PubMed] [Google Scholar]
- Neufang S, Specht K, Hausmann M, Güntürkün O, Herpertz-Dahlmann B, Fink GR, Konrad K. Sex differences and the impact of steroid hormones on the developing human brain. Cerebr. Cortex. 2009;19:464–473. doi: 10.1093/cercor/bhn100. https://doi.org/10.1093/cercor/bhn100. [DOI] [PubMed] [Google Scholar]
- Nichols TE, Das S, Eickhoff SB, Evans AC, Glatard T, Hanke M, Kriegeskorte N, Milham MP, Poldrack RA, Poline J-B, Proal E, Thirion B, Van Essen DC, White T, Yeo BTT. Best practices in data analysis and sharing in neuroimaging using MRI. Nat. Neurosci. 2017;20:299–303. doi: 10.1038/nn.4500. https://doi.org/10.1038/nn.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nopoulos P, Flaum M, O'Leary D, Andreasen NC. Sexual dimorphism in the human brain: evaluation of tissue volume, tissue composition and surface anatomy using magnetic resonance imaging. Psychiatry Res. Neuroimage. 2000;98:1–13. doi: 10.1016/s0925-4927(99)00044-x. https://doi.org/10.1016/S0925-4927(99)00044-X. [DOI] [PubMed] [Google Scholar]
- Nøvik TS, Hervas A, Ralston SJ, Dalsgaard S, Rodrigues Pereira R, Lorenzo MJ, Baldursson G, Coghill D, Curatolo P, Döpfner M, Falissard B, Le Heuzey MF, Preuss U, Rasmussen P, Riley AW, Rothenberger A, Spiel G, Steinhausen HC, Vlasveld L. Influence of gender on attention-deficit/hyperactivity disorder in Europe - ADORE. Eur. Child Adolesc. Psychiatr. 2006;15 doi: 10.1007/s00787-006-1003-z. https://doi.org/10.1007/s00787-006-1003-z. [DOI] [PubMed] [Google Scholar]
- Platt JC. Sequential Minimal Optimization: a Fast Algorithm for Training Support Vector Machines. Advances in Kernel Methods. 1998 doi:10.1.1.43.4376. [Google Scholar]
- Ritchie SJ, Cox SR, Shen X, Lombardo MV, Reus LM, Alloza C, Harris MA, Alderson HL, Hunter S, Neilson E, Liewald DCM, Auyeung B, Whalley HC, Lawrie SM. Sex Differences in the Adult Human Brain: Evidence from 5,216 UK Biobank Participants. BioRxiv. 2017 doi: 10.1093/cercor/bhy109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt JD. Multivariate revisit to “sex beyond the genitalia. Proc. Natl. Acad. Sci. Unit. States Am. 2016;113:E1966–E1967. doi: 10.1073/pnas.1523961113. https://doi.org/10.1073/pnas.1523961113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruigrok ANV, Salimi-Khorshidi G, Lai M-C, Baron-Cohen S, Lombardo MV, Tait RJ, Suckling J. A meta-analysis of sex differences in human brain structure. Neurosci. Biobehav. Rev. 2014;39:34–50. doi: 10.1016/j.neubiorev.2013.12.004. https://doi.org/10.1016/j.neubiorev.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M, Caspi A, Moffitt TE. Using sex differences in psychopathology to study causal mechanisms: unifying issues and research strategies. J. Child Psychol. Psychiatry. 2003;44:1092–1115. doi: 10.1111/1469-7610.00194. [DOI] [PubMed] [Google Scholar]
- Satterthwaite TD, Connolly JJ, Ruparel K, Calkins ME, Jackson C, Elliott MA, Roalf DR, Hopsona R, Prabhakaran K, Behr M, Qiu H, Mentch FD, Chiavacci R, Sleiman PMA, Gur RC, Hakonarson H, Gur RE. The Philadelphia Neurodevelopmental Cohort: a publicly available resource for the study of normal and abnormal brain development in youth. Neuroimage. 2016;124:1115–1119. doi: 10.1016/j.neuroimage.2015.03.056. https://doi.org/10.1016/j.neuroimage.2015.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Elliott MA, Ruparel K, Loughead J, Prabhakaran K, Calkins ME, Hopson R, Jackson C, Keefe J, Riley M, Mentch FD, Sleiman P, Verma R, Davatzikos C, Hakonarson H, Gur RC, Gur RE. Neuroimaging of the Philadelphia neurodevelopmental cohort. Neuroimage. 2014;86:544–553. doi: 10.1016/j.neuroimage.2013.07.064. https://doi.org/10.1016/j.neuroimage.2013.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Wolf DH, Roalf DR, Ruparel K, Erus G, Vandekar S, Gennatas ED, Elliott MA, Smith A, Hakonarson H, Verma R, Davatzikos C, Gur RE, Gur RC. Linked sex differences in cognition and functional connectivity in youth. Cerebr. Cortex. 2015;25:2383–2394. doi: 10.1093/cercor/bhu036. https://doi.org/10.1093/cercor/bhu036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schölkopf B, Platt JC, Shawe-Taylor J, Smola AJ, Williamson RC. Estimating the support of a high-dimensional distribution. Neural Comput. 2001;13:1443–1471. doi: 10.1162/089976601750264965. https://doi.org/10.1162/089976601750264965. [DOI] [PubMed] [Google Scholar]
- Schölkopf B, Smola AJ. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and beyond. MIT press; 2002. [Google Scholar]
- Schuch JJ, Roest AM, Nolen WA, Penninx BW, de Jonge P. Gender differences in major depressive disorder: results from the Netherlands study of depression and anxiety. J. Affect. Disord. 2014;156:156–163. doi: 10.1016/j.jad.2013.12.011. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Peterson BS, Kan E, Woods RP, Yoshii J, Bansal R, Xu D, Zhu H, Thompson PM, Toga AW. Sex differences in cortical thickness mapped in 176 healthy individuals between 7 and 87 years of age. Cerebr. Cortex. 2006;17:1550–1560. doi: 10.1093/cercor/bhl066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell ER, Trauner Da, Gamst A, Jernigan TL. Development of cortical and subcortical brain structures in childhood and adolescence: a structural MRI study. Dev. Med. Child Neurol. 2002;44:4–16. doi: 10.1017/s0012162201001591. https://doi.org/10.1111/j.1469-8749.2002.tb00253.x. [DOI] [PubMed] [Google Scholar]
- Tan A, Ma W, Vira A, Marwha D, Eliot L. The human hippocampus is not sexually-dimorphic: meta-analysis of structural MRI volumes. Neuroimage. 2016;124:350–366. doi: 10.1016/j.neuroimage.2015.08.050. https://doi.org/10.1016/j.neuroimage.2015.08.050. [DOI] [PubMed] [Google Scholar]
- Toga AW, Foster I, Kesselman C, Madduri R, Chard K, Deutsch EW, Price ND, Glusman G, Heavner BD, Dinov ID, Ames J, Van Horn J, Kramer R, Hood L. Big biomedical data as the key resource for discovery science. J. Am. Med. Inf. Assoc. 2015;22:1126–1131. doi: 10.1093/jamia/ocv077. https://doi.org/10.1093/jamia/ocv077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toothaker L. Multiple Comparison Procedures. Sage; 1993. https://doi.org/10.4135/9781412985178. [Google Scholar]
- Torri F, Dinov ID, Zamanyan A, Hobel S, Genco A, Petrosyan P, Clark AP, Liu Z, Eggert P, Pierce J, Knowles JA, Ames J, Kesselman C, Toga AW, Potkin SG, Vawter MP, Macciardi F. Next generation sequence analysis and computational genomics using graphical pipeline workflows. Genes. 2012;3:545–575. doi: 10.3390/genes3030545. https://doi.org/10.3390/genes3030545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunc B, Solmaz B, Parker D, Satterthwaite TD, Elliott MA, Calkins ME, Ruparel K, Gur RE, Gur RC, Verma R. Establishing a link between sex-related differences in the structural connectome and behaviour. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2016;371:20150111. doi: 10.1098/rstb.2015.0111. https://doi.org/10.1098/rstb.2015.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison NJ, Cook PA, Klein A, Song G, Das SR, Duda JT, Kandel BM, van Strien N, Stone JR, Gee JC, Avants BB. Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements. Neuroimage. 2014;99:166–179. doi: 10.1016/j.neuroimage.2014.05.044. https://doi.org/10.1016/j.neuroimage.2014.05.044. [DOI] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. Modern Applied Statistics with S-PLUS. Springer Science & Business Media; 2013. [Google Scholar]
- Wang L, Shen H, Tang F, Zang Y, Hu D. Combined structural and resting-state functional MRI analysis of sexual dimorphism in the young adult human brain: an MVPA approach. Neuroimage. 2012;61:931–940. doi: 10.1016/j.neuroimage.2012.03.080. https://doi.org/10.1016/j.neuroimage.2012.03.080. [DOI] [PubMed] [Google Scholar]
- Wendler DS. Problems with the consensus definition of the therapeutic misconception. J. Clin. Ethics. 2013;24:387–394. https://doi.org/10.1038/jid.2014.371. [PMC free article] [PubMed] [Google Scholar]
- Williams LJ, Abdi H. Tukey's honestly significant difference (HSD) test. Encycl. Res. Des. 2010:1–5. https://doi.org/10.4135/9781412961288.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.