Abstract
Objective
This study proposes and evaluates a technique for in vivo deep-tissue super-resolution imaging in the light-scattering mouse brain at up to a 3.5 Hz two-dimensional imaging rate with a 21 × 21 µm2 field-of-view.
Methods
We combine the deep-tissue penetration and high imaging speed of resonant laser scanning two-photon microscopy with the super-resolution ability of patterned excitation microscopy. Using high frequency intensity modulation of the scanned two-photon excitation beam, we generate patterned illumination at the imaging plane. Using the principles of structured illumination, the high frequency components in the collected images are then used to reconstruct images with an approximate 2-fold increase in optical resolution.
Results
Using our technique, resonant two-photon (2P) Super-resolution Patterned Excitation Reconstruction (2P-SuPER) microscopy, we demonstrate our ability to investigate nanoscopic neuronal architecture in the cerebral cortex of the mouse brain at a depth of 120 µm in vivo and 210 µm ex vivo with a resolution of 119 nm. This technique optimizes the combination of speed and depth for improved in vivo imaging in the rodent neocortex.
Conclusion
This study demonstrates a potentially useful technique for super-resolution in vivo investigations in the rodent brain in deep tissue, creating a platform for investigating nanoscopic neuronal dynamics.
Significance
This technique optimizes the combination of speed and depth for improved super-resolution in vivo imaging in the rodent neocortex.
Index Terms: Two-photon microscopy, structured illumination, super-resolution imaging, neuron
I. Introduction
Neuronal components change in shape and size in response to electrical stimulation, development, and experience [1]. Morphological alterations in neuronal nanoarchitecture regulate synaptic function and influence cellular activity. They are implicated in memory formation and cognition [2]. Abnormal neuronal morphology and altered morphological dynamics impair cellular function, as is observed in many types of neurodegenerative disorders such as Parkinson’s and Alzheimer’s diseases [3], [4]. Therefore, visualizing nanoscopic neuronal morphology is important for future investigations aiming to further our understanding of mechanisms underlying neuronal plasticity and the associated disorders.
Optical microscopy is the method of choice for investigating neuronal morphology and dynamics due to the high spatial and temporal resolution granted by light. However, neuronal morphology and structural changes often occur on the nanoscopic scale, beyond the limited optical resolution of conventional microscopy. Recent advances in optical microscopy have led to the development of super-resolution light microscopy (SRLM) technologies, such as photon localization microscopy, stimulated emission depletion (STED) microscopy, high numerical aperture film-imbedded microspheres, and structured illumination microscopy (SIM) [5]-[8]. These SRLM imaging modalities increase optical resolution to visualize nanoscopic features beyond Abbe’s diffraction limit [9]. The increased resolution provided has proven valuable in biological studies, especially in evaluating neuronal dynamics and the morphology-function relationship of neurons [10]–[15]. Despite significant progress, SRLM technologies are constrained in scattering tissues by either low imaging speed or poor imaging depth, or both, consequently confining investigations primarily to cell cultures, as well as acute or organotypic brain slices [16]–[19].
One path to overcoming the limitation of optical scattering and improving SRLM imaging depth while maintaining sufficient imaging speed involves combining SIM with two photon laser scanning microscopy (2P-LSM) [20]–[25]. Canonical 2P-LSM increases imaging depth by using a near infrared (NIR) excitation laser beam. The NIR excitation laser beam has lower scattering in tissue and the 2P excitation process reduces background by confining excitation to the focal point of the objective. In the case of patterned illumination, 2P excitation can maintain illumination patterns deeper into scattering tissue when compared to single photon illumination. This combination of 2P and SIM has improved SRLM imaging depth up to 135 µm ex vivo in scattering tissues with imaging rates of ~1 Hz over a large field of view (< 500 µm2) [20], [21]. However, these imaging rates are still insufficient for many in vivo applications, especially those that involve imaging neuronal activity [26], [27]. Thus, in vivo studies in mouse models past the most superficial surface layers are still beyond the technological capabilities of most SRLM implementations.
Here, we apply our technique, resonant two-photon Super-resolution Patterned Excitation Reconstruction (2P-SuPER) microscopy, which combines the improved imaging depth, contrast and imaging speed of resonant scanning 2P-microscopy with the super-resolution ability of SIM (see Supplementary Material). Compared to other SRLM techniques, resonant 2P-SuPER improves imaging rate to 3.5 Hz and imaging depth past 100 µm in living scattering tissue while maintaining the two-fold resolution enhancement of SIM. These improvements allow for first-time investigation of nanoscopic neuronal architecture in vivo past the superficial layer of the brain. We demonstrate the capability of our technique by imaging eGFP in the cytosol of neurons in acute brain slices and in the neocortex of anesthetized mice.
II. Materials and Methods
A. Optical System
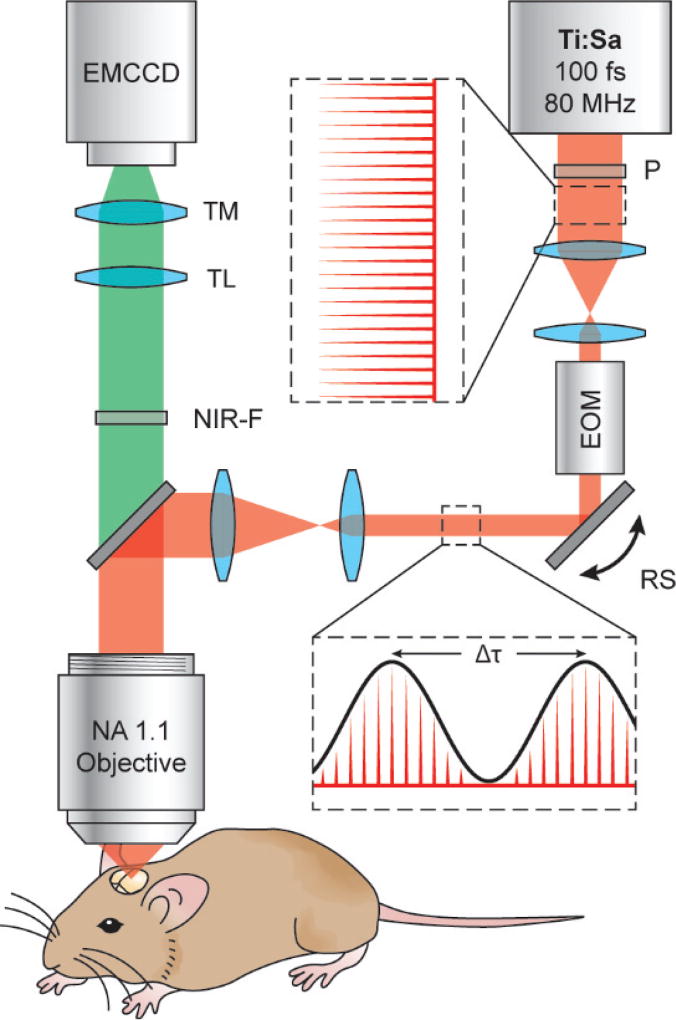
The optical schematic for resonant 2P-SuPER microscopy is shown in Fig. 1. The 850 nm output from an 80 MHz Ti:sapphire oscillator with a 100 fs pulse width (Mai Tai, Newport) was used as the source for 2P excitation. The beam first passed through a polarizer before being carefully aligned through the center of an electro-optic modulator (EOM) (350-160-02-BK KD*P, ConOptics) for beam intensity modulation. The EOM was modulated to generate a sinusoidal output beam with a controllable frequency 1/Δτ; where Δτ is the adjustable temporal distance between each consecutive crest of the sinusoidal output wave (Fig. 1). The modulated beam was directed through an achromatic beam compressor composed of 2 lenses (LA1251-B, LA1255-B, Thorlabs). After passing through the beam compressor the beam was directed into an X-Y resonant scanner assembly (SC-SYS-XYG, Electro-optical Products CORP). Next, the beam reflected from the X-Y resonant scanner assembly, passed through a beam expander comprised of 2 lenses (LA1027-B, LA1708-B, Thorlabs) and into the microscope (BX61WI, Olympus). Inside the microscope, the modulated beam was reflected off a dichroic mirror (69218, Edmund Optics) and onto the back aperture of a high NA water immersion objective (LUMFLN 60XW, Olympus). The beam was expanded to overfill the back aperture at all beam angles used for scanning, in order to maintain maximum resolution.
Fig. 1.
Schematic of the resonant 2P-SuPER microscopy experimental setup. The output of the laser is modulated with a frequency of 1/Δτ. The frequency is synchronized with the X-Y scan to generate a grid shaped illumination pattern at the imaging plane. Ti:Sa: titanium sapphire ultrafast laser; P: polarizer; EOM: electro-optical modulator; RS: closed loop X-Y resonant scanner assembly; NIR-F: near infrared filter; TL: tube lens; TM: tunable magnification optics; EMCCD: electron-multiplied CCD.
2P induced fluorescence was collected by the same high NA objective and passed through the NIR dichroic mirror for imaging. Collected emission light was filtered through a NIR dichroic shortpass filter (69–206, Edmund Optics) to remove stray laser light then passed through a tube lens and tunable magnification optics (UltraZoom 6000, Navitar) before finally being imaged on the electron-multiplied charge coupled device (EMCCD) (C9100-13, Hamamatsu).
To build the patterned illumination structure the beam modulation frequency (1/Δτ) was synchronized to the fixed scanning rate of the X-Y resonant scanner assembly. We phase shifted the illumination pattern 2-times by 2π/3 radians while holding the illumination angle constant and capturing images at each of the phases (0, 2π/3, and 4π/3). We repeated this process at angles of 120° and 240° from the original incident angle (50°, 170°, and 290°) to generate a total of 9 images to isotropically cover frequency space [7], [20]. The 9 images were then used to form a single super-resolution image using the principles of structured illumination reconstruction.
B. Data Acquisition and Imaging Parameters
We used an 850 nm wavelength for excitation and limited the average laser power to 27 mW at the back aperture of the objective lens, corresponding to laser fluence of 155 kW/mm2 (~1.94 mJ/mm2) at the imaging plane using the current objective (70% transmission efficiency at 850 nm). In tissue and three-dimensional phantom samples, the laser fluence at the imaging plane will be lower than 155 kW/mm2 due to optical scattering.
All resonant 2P-SuPER data were collected using custom control software written in LabVIEW (National Instruments). The software used the transistor-transistor logic (TTL) output signal from the resonant scanner to synchronize the modulation of the EOM to generate the desired patterns at the imaging plane with adjustable spatial frequency. The TTL signal is triggered each time the fast resonant scanner begins a new roundtrip cycle. A fast microcontroller (Teensy 3.2, Sparkfun) detects the TTL and generates a corresponding sample clock signal, which was then sent to a shielded connector block (BNC-2110, National Instruments) to synchronize the multichannel DAQ device (PCI-6115, National Instruments). The DAQ device generates analog signal corresponding to the designed sinusoid pattern to control the EOM. Meanwhile, a TTL pulse is sent to the camera at the beginning of each slow axis roundtrip (Y) to synchronize frame acquisition. The camera was cooled using a water chiller (Thermocube 200, Solid State Cooling Systems) to reach chip temperature of −90°C. Temperature of the camera and microscopy system was allowed to stabilize for 1 hour before all experiments.
C. Phantom Samples
We used fluorescein (F2456, Sigma-Aldrich) dissolved in water (0.1mg/dL) to visualize the illumination patterns at 850 nm excitation and determined their corresponding frequency domain distributions by performing Fast Fourier Transform (FFT).
We used 100-nm-diameter fluorescent nanospheres (Fluoresbrite plain YG 0.1 micron microspheres, Polyscience Inc.) to approximate the resolution of resonant 2P-SuPER and array 2P-LSM. Nanospheres were deposited onto a plasma-treated hydrophilic glass substrate via spin coating. The sample was allowed to dry before being rinsed 3 times with water to remove loose spheres. Before imaging, a droplet of water was placed over the top of the spheres for use with the water immersion objective. We imaged the spheres at 850 nm excitation wavelength with incident laser fluence of < 20 kW/mm2 (~0.25 mJ/mm2) (3 mW at the back aperture with 70% transmission efficiency).
For 3D resolution approximation in scattering media, we used the same 100-nm-diameter fluorescent nanospheres embedded in agarose (A6013-100-G, Sigma-Aldrich) formed gel. The agarose powder was dissolved in 50 mL of hot water (0.5µg/mL) using a magnetic stirrer to form a homogenous mixture. Before solidifying, we added the spheres (5µL/mL) and intralipid (I141-100ML, 20% emulsion, Sigma-Aldrich) (50µL/mL) to the solution. Before the solution solidified, 1 mL of the mixture was placed in a 29 mm glass bottom dish with a 20 mm bottom well (D29-20-0-N, In Vitro Scientific) and immediately placed in a 4°C environment to solidify. The final intralipid concentration composed ~1% of the phantom (by weight) and had a reduced scattering coefficient of ~10cm−1, which is similar to that of the mouse cortex [28], [29]. Imaging was conducted through the bottom of the glass bottom dish by placing a droplet of water on the glass bottom and directly placing the water objective lens in contact with the droplet. We imaged the sample from 0 to 110 µm into the agarose gel at axial steps of 400 nm. We used 850 nm excitation wavelength with incident laser fluence of less than 70 kW/mm2 (~0.875 mJ/mm2) to prevent saturation of the camera near the surface of the phantom while still allowing enough fluence for imaging spheres at all depths.
D. Image Reconstruction
We achieved super-resolution image reconstruction using custom routines written in Matlab software (R2015a, Mathworks, Inc.). To reconstruct a super-resolution image, we first recovered the unattenuated frequency spectrum by Weiner filtering each individual image. The parameters of the Weiner filter, including the system PSF and SNR, were predetermined based on image analysis of illumination patterns imaged from a fluorescein dye solution [20]. SNR parameters were readjusted based on the signal intensity. We then performed a two-dimensional Fourier transform to convert each image into k space. After a 4-fold up-sampling, we gained precise control of the frequency shift (up to 1/4 of the pixel resolution). The precise modulation angle and frequency were decoded by analyzing the location of the first-order harmonic peaks as they appeared in the two-dimensional spectrum image. Once we determined the exact modulation frequency and angle, the phase retardation was estimated by shifting and matching the theoretical cosine pattern against the original image in the spatial domain. The corresponding baseband spectrum and modulated high frequency components were recovered using matrix-based image algebra. The recovered frequency components were then reshaped into a 2D matrix and shifted back to their true positions for reassembly.
2P-LSM images were obtained by summing the patterned illumination images acquired at a single imaging plane.
E. Estimation of Imaging Resolution
FWHM measurements of intensity profiles across single spheres were used to estimate resolution. Standard error from the mean (SEM) was used to show variation in FWHM measurements. All intensity profile measurements were conducted using ImageJ [30]. Data were then plotted, interpolated, and FWHM measurements calculated using a Matlab function. Before analysis, background was subtracted from the images using a single image acquired without the 2P excitation laser beam exciting the sample. We were careful to avoid measuring structural artifacts that can occur in SIM reconstruction. These often appear as near background structures in reconstructed images.
F. Acute Slice Preparation
For cortical and hippocampal imaging, acute brain slices (300 µm-thick) were prepared from postnatal day 23–40 line M Tg(Thy1-EGFP)MJrs/J male and female mice (stock # 007788, Jackson Laboratory). Mice were deeply anesthetized by isoflurane inhalation and sacrificed. Brains were placed into ice-cold, continuously carbogenated (95% O2/ 5% CO2), artificial cerebrospinal fluid (ACSF) containing (in mM) 127 NaCl, 2.5 KCl, 25 NaHCO3, 1.25 NaH2PO4, 2.0 CaCl2, 1.0 MgCl2, and 25 Glucose (osmolality ~310 mOsm/L). Brains were sectioned on a Leica VT1000s (Leica Instruments, Nussloch, Germany) in a coronal or transverse orientation. Acquired slices were placed in a continuously carbogenated ACSF solution at 34°C for ~20–30 minutes. After recovery, slices were kept at room temperature until use. During imaging, individual brain slices were placed in a flow chamber with circulating carbogenated ACSF solution.
Imaging was targeted to eGFP-positive spiny pyramidal neurons in superficial layers of retrosplenial, and perirhinal/ectorhinal cortices region. We varied imaging depth from 170 µm to 250 µm and used 850-nm excitation wavelengths. Average beam power was kept at 27 mW at the objective back aperture, translating into maximum fluence of under 155 kW/mm2 (~1.94 mJ/mm2) at the imaging plane. During image acquisition, the beam was scanned over the entire field of view (21 × 21 µm2) of the 512 × 512 pixel detector at frame rates of 29 to 32 fps (imaging rates of 3.22 Hz to 3.55 Hz) with an average rate of approximately 31.5 fps (3.5 Hz). All animal procedures were approved by the Institutional Animal Care and Use Committee at Northwestern University and conform to the guidelines on the Use of Animals from the National Institutes of Health.
G. In Vivo Mouse Preparation
Male mice (line M Tg(Thy1-EGFP)MJrs/J, stock #007788, Jackson Laboratory), postnatal 25–32, were anesthetized with urethane and the pial arterioles exposed by craniotomy. The body temperature was maintained at 37°C during surgery. During surgery, the cerebral surface was continuously covered with phosphate buffered saline to prevent drying. To minimize brain motion, a silicone elastomer (Kwik-Sil, World Precision Instruments) was directly deposited on the exposed dura layer covering the cerebral surface. A glass coverslip was bonded to the silicone elastomer before elastomer solidification to form a support structure between the brain and the skull. After surgery and glass coverslip implantation, anesthetized mice were fitted with a metal headbar, which was attached to the exposed skull using glue and dental cement. Immediately after the cement solidified, the headbar was securely mounted in headbar constrains and placed under the microscope objective. During the experiment, we placed the mice in a custom holder with an open bottom to allow for free chest motion of the mouse, which helped reduce motion artifacts. We imaged cortical neurons sparsely expressing cytosolic eGFP. A total of 5 to 80 super-resolution images (45 to 720 acquisitions total) were acquired at each imaging plane.
III. Results & Discussion
To demonstrate the improved resolution of resonant 2P-SuPER, we imaged 100-nm green fluorescent spheres dispersed on a glass coverslip and immersed in water. We then compared the resonant 2P-SuPER image to the image captured by array 2P-LSM. Full-width at half-maximum (FWHM) measurements of intensity profiles across the center of imaged single fluorescent spheres gave a value of 121±4.5 nm for resonant 2P-SuPER microscopy (92% of the theoretical maximum resolution for patterned illumination). By comparison, array 2P-LSM resolution was approximately twice the value, 279±6.1 nm (91% of the theoretical maximum resolution for Abbe’s diffraction limit). Scan rate of the X-Y resonant scanner assembly fluctuated from 29 and 32 Hz, with an average scan rate of 31.5 Hz. Therefore, the 9-frame acquisition rate ranged from 3.22 to 3.55 Hz, with an average rate of 3.5 Hz. We compensated for the changing frequency by synchronizing the TTL output signal triggered by the resonant scanner at the beginning of each new roundtrip cycle with the EOM modulation frequency and EMCCD integration time.
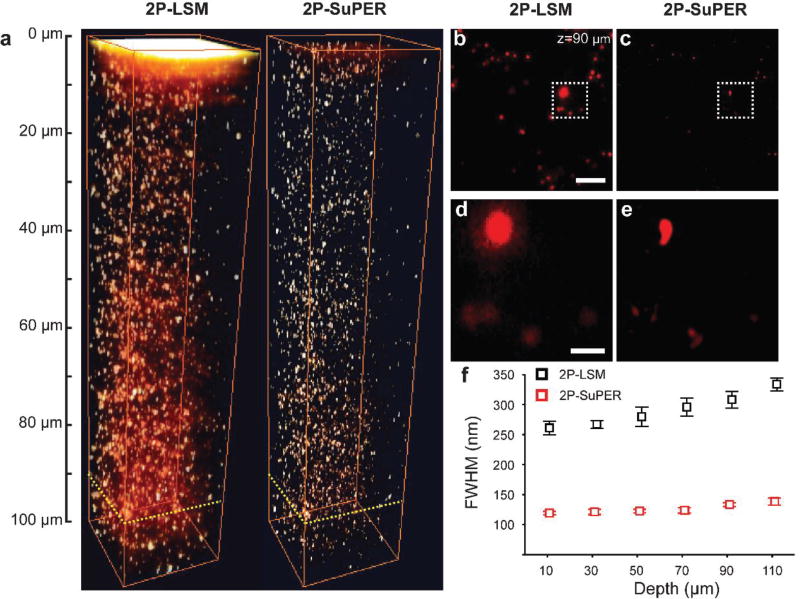
After verifying the approximate 2-fold improvement in resolution, we next evaluated the relationship between resolution and scattering, which is related to imaging depth. We imaged the same type of fluorescent spheres embedded in agarose gel containing a 2% concentration of intralipids. Figs. 2a–e shows a comparison of array 2P-LSM and resonant 2P-SuPER imaging in scattering samples. The FWHM measurements of intensity profiles across the center of single fluorescent spheres at different depths imaged by resonant 2P-SuPER microscopy and array 2P-LSM were used for resolution estimation. The FWHM measurements for resonant 2P-SuPER microscopy remained relatively depth-invariant (Z0=119±2.7; Z110=138±5.9 nm, +16%) at depths of up to 110 µm in the phantom sample. In contrast, array 2P-LSM the FWHM measurements of the spheres gradually increased (Z0=261±11.3; Z110=334±10 nm, +28%), Fig. 2f. The degradation in resolution for both techniques was likely caused by increased scattering since the measured signal from individual spheres was significantly above background for both techniques.
Fig. 2.
(a) 2P-LSM (left) and resonant 2P-SuPER (right) three-dimensional image of 100 nm green fluorescent nanospheres embedded in a scattering agarose gel. A comparison of images acquired by (b) 2P-LSM and (c) resonant 2P-SuPER at 90 µm from the surface of the agarose sample demonstrate the improved resolution of resonant 2P-SuPER (scale bar: 5 µm). A magnified comparison of (d) resonant 2P-LSM and (e) resonant 2P-SuPER resolution improvement (scale bar: 1 µm). (f) FWHM measurements from 10 spheres at each depth show relative independence of resolution from depth in resonant 2P-SuPER but not in 2P-LSM (error bars: sem).
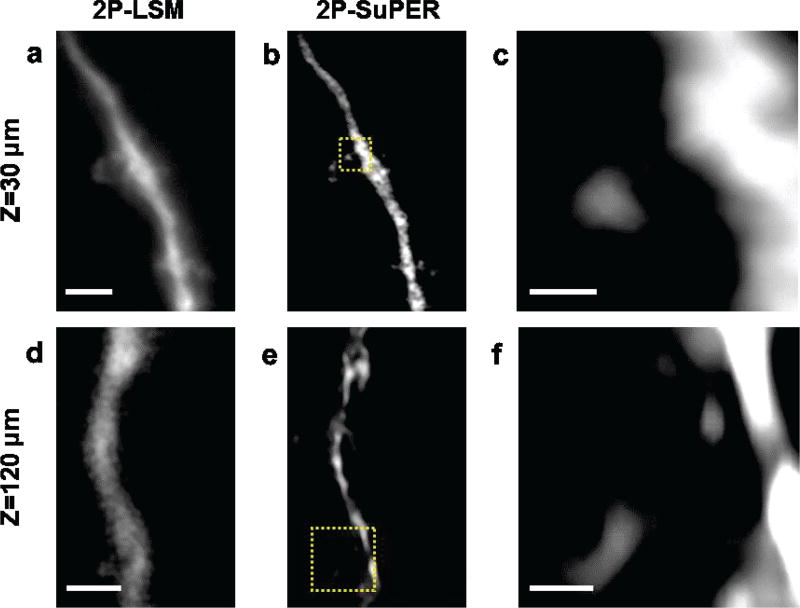
To test the imaging depth of resonant 2P-SuPER in living tissues, we imaged eGFP in the cytoplasm of neurons in 300 µm-thick acute coronal brain slices from line M mice Tg(Thy1-EGFP)MJrs/J (Fig. 3) [31]. To determine the maximum imaging depth, images were acquired between 170–250 µm below the surface of the slice at an average imaging rate of approximately 3.5 Hz. A total of 5 super-resolution images were acquired from each plane in 400 nm axial steps. The 5 super-resolution images (45 patterned illumination images) were then summed together (Fig. 3). Images of resonant 2P-SuPER confirm the super-resolution ability of resonant 2P-SuPER at depths up to 210 µm inside the acute brain slices (Figs. 3b and 3f, respectively).
Fig. 3.
Ex vivo comparison between (a) 2P-LSM and (b) resonant 2P-SuPER imaging a eGFP-positive neuron at 200 µm in an acute brain slice at 3.5 Hz. The corresponding magnified section of the (c) 2P-LSM (green box in panels a and d) resonant 2P-SuPER (blue box in b) images show that resonant 2P-SuPER maintains super-resolution in deep tissue and is more resilient to scattering than array based 2P-LSM. Comparisons between (e) 2P-LSM and (f) resonant 2P-SuPER imaging a eGFP neurons at 210 µm in the acute brain slice at 3.5 Hz showed an analogous resolution improvement. A magnified section of the (g) 2P-LSM (green box in panels e) and (h) resonant 2P-SuPER (blue box in panel f) images show that resonant 2P-SuPER maintained super-resolution at depths up to 210 µm from the cortical surface. Scale bars: 1 µm in (a) and (e), 500 nm in (c) and (g).
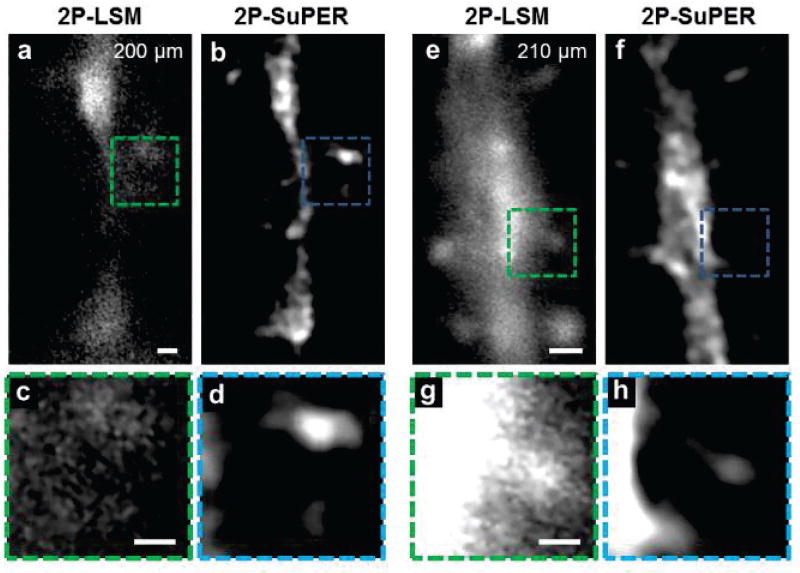
Next, we imaged sparsely expressed eGFP in neuronal architecture in the brains of anesthetized mice, as shown in Fig. 4. Imaging was performed at depths of up to 120 µm below the brain surface. We avoided imaging neurons near major blood vessels to minimize motion artifacts. We measured the heart rate of anesthetized mice by imaging the beating frequency of the brain, which ranged from < 2 Hz to 6 Hz, depending on amount of anesthesia. Between 5 and 80 super-resolution images (45 to 720 frames) were attempted for each plane. Due to fast micromotion (~2–6 Hz) of the brain caused by respiration and the heartbeat, neurons were rarely stationary for the necessary time (285 ms) to achieve 9-frame image acquisition using resonant 2P-SuPER microscopy [32]. Due to brain motion, image reconstruction was successful for approximately 15 percent of in vivo images (see Supplementary Material). Furthermore, imaged neurons rarely returned to the same nanoscopic position after each heartbeat, which prevented sum intensity Z-projections or volumetric rendering. Therefore, all in vivo 2P-SuPER images are from a single 9-frame reconstructions.
Fig. 4.
In vivo comparison between (a) 2P-LSM and (b) resonant 2P-SuPER microscopy image of a dendrite located 30 µm from the neocortical of the mouse brain and (b). (c) The magnified resonant 2P-SuPER image (indicated by the yellow box in panel b) shows the dendritic spine extending from the neuronal dendrite. Comparison between (d) 2P-LSM and resonant 2P-SuPER image of a dendrite located 120 µm from the neocortical surface. (f) The magnified resonant 2P-SuPER image (indicated by the yellow box in panel e) shows dendritic spines emerging from the neurite. Scale bars: 4 µm in (a), 1 µm in (c), 2 µm in (d), and 500 nm in (f).
Resonant 2P-SuPER attempts to fill the gap in methods for timely super-resolution microscopy for in vivo imaging in moving, light-scattering tissues. The lateral resolution was nearly depth-invariant in phantom samples created to mimic the scattering properties of tissues. The imaging rate was primarily limited by the scanning speed of the resonant scanner assembly and frame rate of the camera, suggesting potential for further improvements in imaging rate. In this work, we used a high quantum efficiency array detector to maximize the number of detected photons and to acquire the entire shape of the point-spread-function (PSF). In canonical 2P-LSM, point detection is generally applied to improve contrast that is lost in array detection from scattered emission photons. Since structured illumination requires array detection, the background from scattered photons increases with increasing imaging depth. However, we found that Fourier reconstruction can improve the contrast similar to point-detection in structured illumination techniques since the illumination pattern is predetermined. This allows us to reject scattered light outside of the known illumination field.
Spacing between the peak intensity of each line in the illumination patterns is limited by the nonlinear excitation point spread function (PSF). Unlike standard SIM, which uses optical interference to generate diffraction patterns on the imaging plane, resonant 2P-SuPER employs high frequency modulation and laser scanning to builds the desired pattern on the imaging plane. The limitation for spacing between each line in the grating and spatial modulation depth is limited by overlap between the excitation PSF ( ), which is theoretically closer than optical diffraction allows. However, overlap of nearby PSF degrades spatial modulation, which degrades image contrast and increases reconstruction artefacts [24]. In our experiments, we found that a spatial modulation depth of 30% was necessary for consistent image reconstruction, however reconstruction was still possible at modulation depths of 20% with reduced success.
Here we demonstrate the ability of resonant 2P-SuPER to image nanoscopic components of dendrites, including dendritic spines and spine necks. Dendritic spines are the postsynaptic sites of many excitatory synapses in the brain [33]. Dendritic spine neck width and length control synaptic potential, molecular diffusion, and regulate current flow into the parent dendrite [13], [33], [34]. Consequently, nanoscopic changes in dendritic spine structure alter the functions of dendritic spines and can affect synaptic integration and cellular activity. Imaging living dendritic spines in vivo can be accomplished by using canonical 2P-LSM. However, the smaller structures such as nanoscopic dendritic spine neck widths (~ 0.1–0.2 µm) cannot be resolved without SRLM imaging. Previous SRLM investigations were able to investigate dendritic spines at the surface of the rodent brain, but were not successful at imaging past 15 µm from the neocortical surface [35]. Thus, the lack of super-resolution morphological information compatible with in vivo investigations limits our understanding of nanoscopic morphological changes in neurons and their relationship to cellular function.
IV. Conclusion
Resonant 2P-SuPER overcomes previous limitations in SRLM techniques by improving imaging depth and imaging speed in scattering tissues, extending SRLM technology to in vivo mouse studies.
Supplementary Material
Acknowledgments
We thank Dr. Kevin Jia and Mrs. Julie Ives from Olympus for experimental assistance, Lindsey Butler for genotyping, Dr. Xian Zhang for conducting animal surgeries, and Dr. Jim Heys for expert advice on mouse craniotomy preparation.
We acknowledge support from the National Institutes of Health (R01EY026078, R24EY022883, and DP3DK108248), the National Science Foundation (DBI-1353952 and CBET-1055379), and the Chicago Biomedical Consortium Catalyst Award. YK acknowledges support from the William and Bernice E. Bumpus Foundation Innovation Award, the Beckman Young Investigator Award, and the NARSAD Young Investigator Grant and P&S Fund.
Contributor Information
Ben E. Urban, Department of Biomedical Engineering, Northwestern University, Evanston, IL 60208, USA
Lei Xiao, Department of Neurobiology, Northwestern University, Evanston, IL 60208, USA.
Siyu Chen, Department of Biomedical Engineering, Northwestern University, Evanston, IL 60208, USA.
Huili Yang, Department of Biomedical Engineering, Northwestern University, Evanston, IL 60208, USA.
Biqin Dong, Department of Biomedical Engineering, Northwestern University, Evanston, IL 60208, USA.
Yevgenia Kozorovitskiy, Department of Neurobiology, Northwestern University, Evanston, IL 60208, USA.
Hao F. Zhang, Department of Biomedical Engineering, Northwestern University, Evanston, IL 60208, USA
References
- 1.Turrigiano GG. Homeostatic plasticity in neuronal networks: the more things change, the more they stay the same. Trends Neurosci. 1999;22(5):221–227. doi: 10.1016/s0166-2236(98)01341-1. [DOI] [PubMed] [Google Scholar]
- 2.Deng W, et al. New neurons and new memories: how does adult hippocampal neurogenesis affect learning and memory. Nat. Rev. Neurosci. 2010 May;11(5):339–350. doi: 10.1038/nrn2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lian H, et al. NFκB-Activated Astroglial Release of Complement C3 Compromises Neuronal Morphology and Function Associated with Alzheimer’s Disease. Neuron. 2015;85(1):101–115. doi: 10.1016/j.neuron.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toy WA, et al. Treadmill exercise reverses dendritic spine loss in direct and indirect striatal medium spiny neurons in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) mouse model of Parkinson’s disease. Neurobiol. Dis. 2014;63:201–209. doi: 10.1016/j.nbd.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rust MJ, et al. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006 Oct.3(10):793–5. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klar TA, et al. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. U. S. A. 2000 Jul.97(15):8206–10. doi: 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gustafsson MGL. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 2000 May;198(2):82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 8.Darafsheh A, et al. Optical super-resolution imaging by high-index microspheres embedded in elastomers. Opt. Lett. 2015 Jan.40(1):5. doi: 10.1364/OL.40.000005. [DOI] [PubMed] [Google Scholar]
- 9.Abbe E. VII.-On the Estimation of Aperture in the Microscope. J. R. Microsc. Soc. 1881 Jun.1(3):388–423. [Google Scholar]
- 10.Ballesteros-Yáñez I, et al. Density and morphology of dendritic spines in mouse neocortex. Neuroscience. 2006 Jan.138(2):403–9. doi: 10.1016/j.neuroscience.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 11.Takasaki K, Sabatini BL. Super-resolution 2-photon microscopy reveals that the morphology of each dendritic spine correlates with diffusive but not synaptic properties. Front. Neuroanat. 2014 Jan.8:29. doi: 10.3389/fnana.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu. Rev. Neurosci. 2001 Jan.24:1071–89. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
- 13.Araya R, et al. Activity-dependent dendritic spine neck changes are correlated with synaptic strength. Proc. Natl. Acad. Sci. U. S. A. 2014 Jul.111(28):E2895–904. doi: 10.1073/pnas.1321869111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segal M. Dendritic spines and long-term plasticity. Nat. Rev. Neurosci. 2005 Apr.6(4):277–84. doi: 10.1038/nrn1649. [DOI] [PubMed] [Google Scholar]
- 15.Komendantov AO, Ascoli GA. Dendritic Excitability and Neuronal Morphology as Determinants of Synaptic Efficacy. J. Neurophysiol. 2009 Feb.101(4):1847–1866. doi: 10.1152/jn.01235.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izeddin I, Specht CG, Lelek M, Darzacq X, Triller A, Zimmer C, Dahan M. Super-resolution dynamic imaging of dendritic spines using a low-affinity photoconvertible actin probe. PLoS One. 2011 Jan.6(1):e15611. doi: 10.1371/journal.pone.0015611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bethge P, et al. Two-photon excitation STED microscopy in two colors in acute brain slices. Biophys. J. 2013 Feb.104(4):778–85. doi: 10.1016/j.bpj.2012.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tønnesen J, et al. Two-color STED microscopy of living synapses using a single laser-beam pair. Biophys. J. 2011 Nov.101(10):2545–52. doi: 10.1016/j.bpj.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Testa I, et al. Nanoscopy of Living Brain Slices with Low Light Levels. Neuron. 2012 Sep.75(6):992–1000. doi: 10.1016/j.neuron.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 20.Urban BE, et al. Super-resolution two-photon microscopy via scanning patterned illumination. Physical Review E. 2015 Apr.91(4):42703. doi: 10.1103/PhysRevE.91.042703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winter PW, et al. Two-photon instant structured illumination microscopy improves the depth penetration of super-resolution imaging in thick scattering samples. Optica. 2014 Sep.1(3):181–191. doi: 10.1364/OPTICA.1.000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ingaramo M, et al. Two-photon excitation improves multifocal structured illumination microscopy in thick scattering tissue. Proc. Natl. Acad. Sci. U. S. A. 2014 Apr.111(14):5254–9. doi: 10.1073/pnas.1314447111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeh CH, Chen SY. Two-photon-based structured illumination microscopy applied for superresolution optical biopsy. in SPIE BiOS. 2013:858826. [Google Scholar]
- 24.Yeh C-H, Chen S-Y. Resolution enhancement of two-photon microscopy via intensity-modulated laser scanning structured illumination. Appl. Opt. 2015 Mar.54(9):2309–17. doi: 10.1364/AO.54.002309. [DOI] [PubMed] [Google Scholar]
- 25.Orieux F, et al. Bayesian Estimation for Optimized Structured Illumination Microscopy. IEEE Trans. Image Process. 2012 Feb.21(2):601–614. doi: 10.1109/TIP.2011.2162741. [DOI] [PubMed] [Google Scholar]
- 26.Chen T-W, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013 Jul.499(7458):295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods. 2009 Dec.6(12):875. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacques SL. Optical properties of biological tissues: a review. Phys. Med. Biol. 2013 Jun.58(11):R37–R61. doi: 10.1088/0031-9155/58/11/R37. [DOI] [PubMed] [Google Scholar]
- 29.Lai P, et al. Dependence of optical scattering from Intralipid in gelatin-gel based tissue-mimicking phantoms on mixing temperature and time. J. of Biomed. Optics. 2014 Mar.19(3):035002–035002. doi: 10.1117/1.JBO.19.3.035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider CA, et al. NIH image to ImageJ: 25 years of image analysis. Nat. Methods. 2012 Jul.9(7):671–676. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng G, et al. Imaging Neuronal Subsets in Transgenic Mice Expressing Multiple Spectral Variants of GFP. Neuron. 2000 Oct.28(1):41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 32.Clark CG, et al. Be still my beating brain - reduction of brain micromotion during in vivo two-photon imaging. J. Physiol. 2013 May;591(10):2379–2380. doi: 10.1113/jphysiol.2013.253138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nimchinsky EA, et al. Structure and function of dendritic spines. Annu. Rev. Physiol. 2002 Jan.64:313–53. doi: 10.1146/annurev.physiol.64.081501.160008. [DOI] [PubMed] [Google Scholar]
- 34.Bloodgood BL, et al. Neuronal Activity Regulates Diffusion Across the Neck of Dendritic Spines. Science. 2005 Nov.310(5749):866–869. doi: 10.1126/science.1114816. [DOI] [PubMed] [Google Scholar]
- 35.Berning S, et al. Nanoscopy in a Living Mouse Brain. Science. 2012 Feb.335(6068):551–551. doi: 10.1126/science.1215369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.