Summary
O-linked β-N-acetyl-D-glucosamine (O-GlcNAc) transferase (OGT) regulates protein O-GlcNAcylation, an essential post-translational modification that is abundant in the brain. Recently, OGT mutations have been associated with intellectual disability, although it is not understood how they affect OGT structure and function. Using a multi-disciplinary approach we show that the L254F OGT mutation leads to conformational changes of the tetratricopeptide repeats and reduced activity, revealing the molecular mechanisms contributing to pathogenesis.
Keywords: O-GlcNAc transferase, intellectual disability, tandem repeat proteins, tetratricopeptide repeats, crystallography, molecular dynamics simulations
Graphical Abstract
Highlights
-
•
The intellectual disability L254F mutation in OGT affects activity
-
•
The L254F mutation leads to shifts up to 12 Å in the OGT structure
-
•
Thermal denaturing studies reveal reduction in TPR stability caused by L254F
-
•
Simulations suggest the presence of alternate TPRL254F conformations
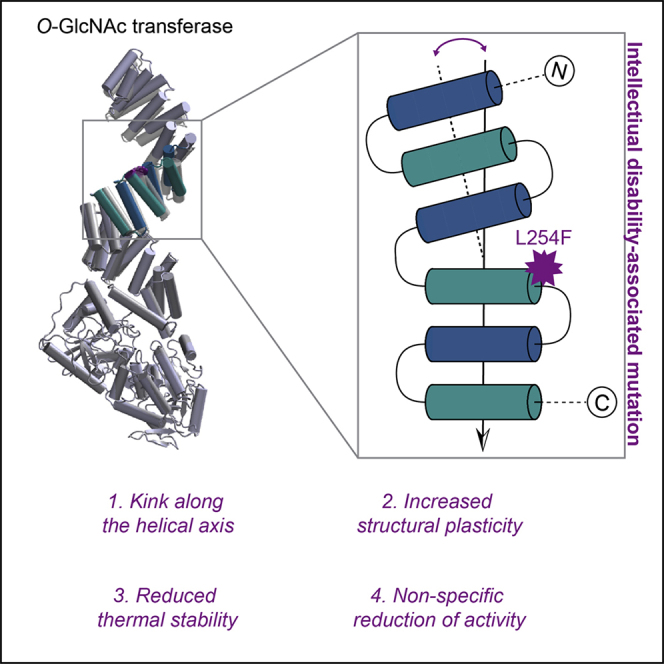
Gundogdu et al. demonstrate how the intellectual disability-associated mutation in O-GlcNAc transferase (OGT), L254F, leads not only to large shifts in OGT structure and altered dynamics in solution, but also a non-specific reduction in activity, providing the first molecular characterization of such a mutation.
Introduction
O-linked β-N-acetyl-D-glucosamine (O-GlcNAc) transferase (OGT) and hydrolase (OGA) control a dynamic, reversible, and tightly regulated post-translational modification termed O-GlcNAcylation (Yang and Qian, 2017). OGT is abundantly expressed in the brain (Okuyama and Marshall, 2003) and localizes to synaptosomes (Cole and Hart, 2001). Accordingly, OGT has been linked to regulation of axonal and dendritic morphology (Francisco et al., 2009), axonal transport of mitochondria (Pekkurnaz et al., 2014) and, at the behavioral level, to response and habituation to environmental stimuli (Timbers et al., 2017). These findings implicate O-GlcNAcylation in various brain functions, including learning and memory.
Intellectual disability (ID) is a disease defined by early-onset impairment of cognitive function and limitation of adaptive behavior (Ropers, 2010). The most common causes of ID are monogenic mutations in over 650 genes (Kochinke et al., 2016). Recently, mutations in the N terminus of human OGT have been associated with ID, namely, A319T, L254F, R284P, and Δ155-177 (Bouazzi et al., 2015, Niranjan et al., 2015, Willems et al., 2017) (Figure 1A). Two recent studies have reported that while some of these mutations affect OGT activity in vitro, O-GlcNAc homeostasis appears to be maintained in patient-derived cells by reduced OGA expression (Vaidyanathan et al., 2017, Willems et al., 2017). It is as yet unclear how these ID-associated mutations affect OGT structure and function and result in the ID phenotype.
Figure 1.
In Vitro Characterization of OGTL254F
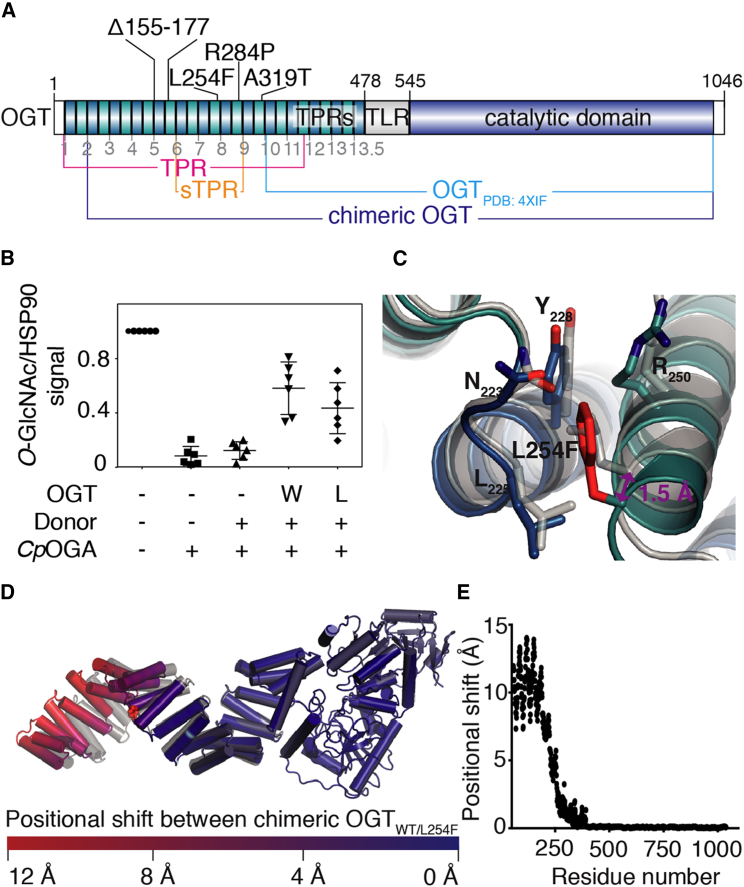
(A) Schematic representation of OGT highlighting the intellectual disability-associated mutations and all the constructs used in this study.
(B) Scatterplot showing OGT activity against deglycosylated HEK-293 cell lysate, with the data averaged from six replicates and the error bars showing SD. See also Figure S1B.
(C) Superposition of the TPRWT/L254F crystal structures at the site of mutation. The gray and colored cartoons are that of TPRWT (PDB: 1W3B; Jínek et al., 2004) and TPRL254F (PDB: 6EOU) structures, respectively.
(D) Overlay of the chimeric OGTWT/L254F structures. The wild-type structure is colored gray, while the mutant structure is colored to reflect the positional shift of each Cα atom between the two structures.
(E) Graph showing the positional shift between equivalent Cα atoms between chimeric OGTWT (PDB: 4XIF [Pathak et al., 2015] and PDB: 1W3B [Jínek et al., 2004]) and OGTL254F (PDB: 4XIF [Pathak et al., 2015] and PDB: 6EOU) as a function of residue number.
L, L254F; OGT, O-GlcNAc transferase; sTPR, simplified TPR; TPR, tetratricopeptide repeat; W, wild-type.
OGT possesses 13.5 tetratricopeptide repeats (TPRs) at the N terminus, which harbor all currently known ID-associated mutations (Lubas et al., 1997) (Figure 1A). TPR-containing proteins are ubiquitous and functionally versatile (Zeytuni and Zarivach, 2012). TPRs were first described in protein phosphatase 5 (PP5), where they negatively regulate catalytic activity in a ligand-dependent manner (Connarn et al., 2014). In OGT, the TPRs form a 120-Å superhelix, which serves as a potential interaction surface for substrates and binding partners (Jínek et al., 2004, Zeytuni and Zarivach, 2012). Two modes of interaction have been proposed. The central channel generated by the TPR superhelix has been shown to engage some substrates through an asparagine ladder, which interacts with the carbonyl and amide groups of the substrate backbone in a sequence-independent fashion (Lazarus et al., 2013, Rafie et al., 2017). In addition, the superhelical grooves on the TPR superhelix may interact with substrates, with different TPRs recruiting distinct target proteins in a sequence-specific fashion, in a manner similar to HSP70/90 organizing protein (Scheufler et al., 2000). These two models explain how OGT can modify thousands of intracellular proteins on specific sites while also serving as a scaffold in multi-protein complexes. Here, we demonstrate that the L254F mutation in OGT induces folding defects in the TPR superhelix, suggesting that changes in interactions with substrates and/or binding partners may underpin the ID disease phenotype.
Results and Discussion
The ID-associated mutation L254F (Vaidyanathan et al., 2017) is located on TPR helix 7, distant from the active site (Figure 1A). In vitro OGT activity was tested with a short acceptor peptide derived from the RB2 protein (Pathak et al., 2015). In this assay, the L254F mutation had no effect on steady-state kinetics of OGT (peptide KM = 0.6 mM, Vmax = 15 nM/s for both enzymes; Figure S1A). Next, we measured OGT activity against de-O-GlcNAcylated HEK-293 cell lysate containing a multitude of substrates for OGT, some of which may be recognized by the superhelical grooves found on OGT TPRs. We observed that addition of recombinant OGTL254F was not able to fully restore the amount of O-GlcNAc transfer to that of the OGTWT-treated control (Figures 1B and S1B). Thus, in our hands the ID OGT L254F mutation shows effects on in vitro activity.
A single TPR motif consists of an anti-parallel pair of α helices, named helix A and B, which are held together by interactions between conserved residues in the 34-amino-acid consensus sequence W4-L7-G8-Y11-A20-F24-A27-P32 (Jínek et al., 2004, Lamb et al., 1995). The W4-L7-G8-Y11 motif on helix A forms a hydrophobic pocket into which the bulky residue of the A20-F24-A27 motif on helix B is lodged. The OGT TPRs possess two additional features: helix A contains a ladder of conserved asparagines (N6) on TPRs 2–13.5; helix B contains a series of large aliphatic residues on TPRs 6–13.5 (Ψ30; where Ψ represents Leu, Ile, or Val). While the N6 ladder is involved in substrate recognition (Jínek et al., 2004, Lazarus et al., 2013, Rafie et al., 2017), the series of large aliphatic residues (Ψ30), of which Leu254 is part, interdigitate with aliphatic residues found at the first position within the TPR motifs (X1; where X represents Leu, Ala, or Pro). To investigate structural changes attributable to the L254F mutation, we determined the crystal structure of the mutant TPR domain (TPRL254F). Recombinant protein was obtained from Escherichia coli using the construct boundaries previously employed to crystallize the wild-type OGT TPR domain (TPRWT, Figure 1A) (Jínek et al., 2004). Diffraction data were collected to 1.75 Å (Table 1) and initial refinement starting from the TPRWT structure required substantial rebuilding of the terminal TPRs, an early indication of considerable conformational changes. Indeed, the overall root-mean-square deviation (RMSD) on 343 Cα atoms of the refined TPRL254F structure versus that of TPRWT was 1.6 Å.
Table 1.
Scaling and Model-Building Statistics of the TPRL254F Crystal Structure
| TPRL254F | |
|---|---|
| Data Collection | |
| Space group | C2221 |
| Cell dimensions | |
| α, β, γ (°) | 44.00, 203.16, 116.87 |
| a, b, c (Å) | 90.00, 90.00, 90.00 |
| Resolution (Å) | 46.58–1.75 (1.75–1.78) |
| Rsym or Rmerge | 0.05 (0.57) |
| I/σI | 13.00 (2.00) |
| Completeness (%) | 99.70 (99.80) |
| Redundancy | 4.1 (4.1) |
| Refinement | |
| Resolution (Å) | 46.58–1.75 (1.75–1.78) |
| No. of reflections | 54,404 (3,607) |
| Rwork/Rfree | 0.19/0.23 |
| No. of atoms | 3,002 |
| Protein | 2,722 |
| Ligand/ion | NA |
| Water | 280 |
| B factors | |
| Protein | 38.40 |
| Ligand/ion | NA |
| Water | 42.52 |
| RMSDs | |
| Bond lengths (Å) | 0.02 |
| Bond angles (°) | 2.00 |
Related to Figures 1C and 1D. Values in parentheses represent the highest-resolution shell.
In wild-type OGT, L254 occupies the interface between the helices of TPR7, with its side chain constricted in a pocket formed by the surrounding residues N223, L225, Y228, and R250 (Figure 1C). Mutation of L254 to the bulkier Phe appears to be accommodated in this pocket, however, by causing small changes in torsion angles of the residues lining the pocket and by displacing TPR helix 7B away from helix 7A by 1.5 Å (Figure 1C). We examined the effects of this change in the context of the full-length protein, modeled by merging the TPRWT/L254F structures and an OGT structure containing the catalytic domain (residues 325–1,038; PDB: 4XIF [Pathak et al., 2015]) using the overlapping TPRs (residues 325–381) as superposition anchor (Figures 1A and 1D). This reveals a deviation from the wild-type TPR geometry that is propagated toward the N terminus (Figures 2D and 2E). Increasing shifts of the TPRs starting from the mutation site lead to a maximum shift of 12 Å for the N-terminal TPR (Figures 2D and 2E). Thus, the L254F mutation causes a distortion of the TPR helix.
Figure 2.
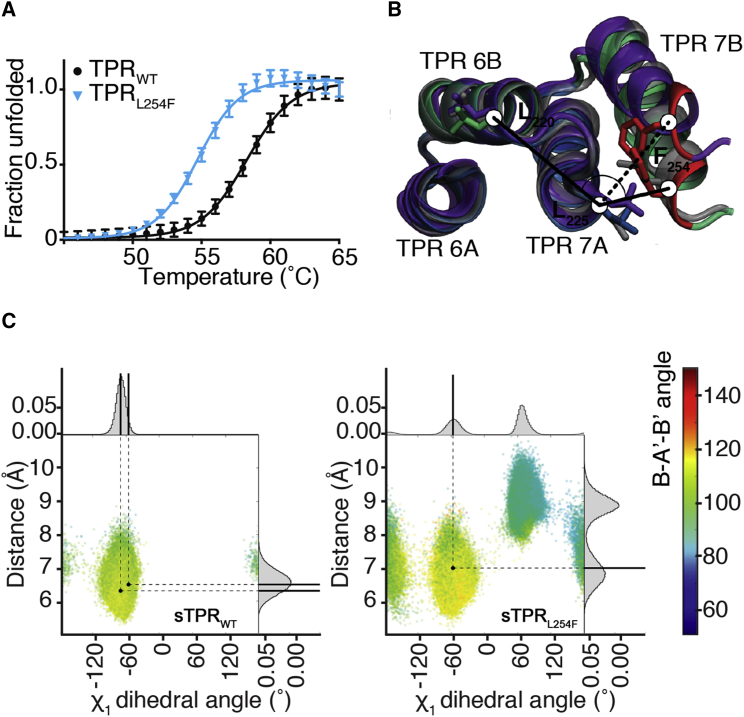
Characterization of the Effects of the ID-Associated Mutation on OGT TPR Stability and Dynamics
(A) Thermal denaturing curve showing fraction of unfolded TPRWT and TPRL254F constructs as a function of temperature. Data averaged from seven replicates were fitted to a Boltzmann sigmoidal curve equation, with error bars representing SD.
(B) Superposition of sTPRWT (gray), sTPRL254F-C1 (green), and sTPRL254F-C2 (purple), with the B-A′-B′ angle and intra-TPR distance demarcated with solid and dashed lines, respectively. See also Figures S2 and S3.
(C) Graphs of sTPRWT (left) and sTPRL254F (right) conformational populations in the molecular dynamics simulations, with the χ1 dihedral angle of residue 254 shown on the x axis, the intra-TPR repeat distance shown on the y axis, and the angle B-A′-B′ shown as a color scale. The B-A′-B′ values observed in the crystal structures are shown as black dots. Histograms attached to the graph show the distribution of χ1 dihedral angles. See also Figures S2 and S3.
sTPRL254F-C1 and sTPRL254F-C1, sTPRL254F conformations 1 and 2; sTPR, simplified TPR; TPR, tetratricopeptide repeats.
To investigate possible effects of the L254F mutation on stability of the TPRs, we determined the melting temperatures of TPRWT and TPRL254F using differential scanning fluorimetry. A monophasic, sigmoidal melting curve was obtained for TPRWT with an inflection point (Tm) of 58°C (Figure 2A). The melting curve for TPRL254F was shifted, yielding a Tm of 55°C, indicative of reduced thermal stability (Figure 2A). This is in agreement with previous characterization of OGTL254F in patient-derived cells, where the L254F mutation was found to reduce OGT half-life by 2-fold (Vaidyanathan et al., 2017). Thus it appears that the L254F mutation destabilizes OGT.
To explore the mechanisms underlying the reduction in thermal stability, we performed atomistic molecular dynamics simulations (2 μs) using a fragment of the TPRWT/L254F crystal structures, comprising TPRs 6, 7, and 8 (residues 189–294; hereafter sTPRWT and sTPRL254F, respectively; Figure 1A). In these simulations we monitored the intra-TPR distance between the Cα atoms of L225 (the first residue of helix 7A) and L254 (the last residue of helix 7B), the TPR6-7 B-A′-B′ angle formed between the Cα atoms of residues L220, L225, and L254, and the χ1 dihedral angle of residue 254 (Figure 2B). The intra-TPR distance and the angle B-A′-B′ were stable in the sTPRWT simulations (Figures 2C, S2, and S3), while in the sTPRL254F simulations higher conformational plasticity was observed, showing a partition between two major conformations (Figures 2B and 2C). In sTPRL254F conformation 1 (sTPRL254F-C1), the F254 side chain occupies the same position as observed in the TPRL254F crystal structure, while in sTPRL254F conformation 2 (sTPRL254F-C2), the F254 phenyl moiety interacts with the side chains of N223, L225, and Y228 and the backbone of the F224 and R250 (Figure 2B). In sTPRL254F-C2 the F254 side chain adopts a different conformation, with its aromatic ring positioned parallel to the N223 backbone amide in a manner that enables a phenyl-amide interaction. This is accompanied by a 140° ± 17° shift in the F254 χ1 dihedral angle, which in turn distorts the TPR geometry, increasing the intra-TPR distance by 2.1 ± 0.1 Å and shifting the B-A′-B′ angle by approximately 21° ± 7° (Figure 2B). Similar to the conformational rearrangement described for the TPRL254F crystal structure, these local changes propagate through subsequent TPR repeats and modify the overall geometry of the protein. Thus, the L254F mutation destabilizes the interface between TPRs 6 and 7.
The data presented here show that the L254F mutation causes a subtle structural distortion at the mutation site that propagates through the TPR superhelix, resulting in a substantial displacement of the N-terminal TPRs and a markedly increased structural plasticity compared with the TPRWT. Although in vitro assays show global effects on the O-GlcNAc proteome, they are modest. However, given that OGT is essential for life from stem cells to vertebrates and resides on the X chromosome, it is likely that only relatively subtle mutations are tolerated in males. Moreover, it is possible that the ID-associated mutations result in misrepresentation of a distinct subset of the O-GlcNAc proteome in different cell lineages, or under certain stimuli. In light of this, it is interesting to note that all OGT ID mutations reported to date are at some distance from the active site (Figure 1A). In addition to effects on the O-GlcNAc proteome, it is possible that the conformational changes we observe in the TPRs affect the OGT interactome. For example, mSin3A (Yang et al., 2002), TRAK1 (Iyer and Hart, 2003), TET2, and TET3 (Deplus et al., 2013) all rely on the six N-terminal TPRs for their recruitment onto OGT, while Atx-10 recruitment on OGT is mediated by TPRs 6–8 (März et al., 2006). The work described here forms a platform for the future dissection of these different roles of OGT.
Significance
O-GlcNAc was discovered more than three decades ago, and more than 1,000 proteins in the human proteome are known to be O-GlcNAc modified. However, there is still a substantial gap in our understanding of how O-GlcNAcylation regulates protein function and downstream cellular pathways. An inroad into this became possible with the recent discovery that patients with mutations in OGT suffer from ID, the clearest evidence yet linking O-GlcNAc to neuronal function, in addition to previous reports implying dysregulation of O-GlcNAc in neurodegeneration. Although several OGT mutations linked to ID have recently been reported, it is not understood how these mutations affect OGT at the molecular level. This article is the first to describe the substantial molecular consequences of such a mutation. The L254F mutation resides in the TPR helix, which facilitates OGT substrate recognition. Using X-ray crystallography we have uncovered that this mutation leads to shifts up to 12 Å in the TPR helix, which ties in with the observed reduction in activity. Furthermore, using differential scanning fluorimetry and molecular dynamics simulations we show that the TPR helix is significantly destabilized, leading to defects in substrate recognition. This is a major advance for the O-GlcNAc field as it provides a molecular understanding of this mutation and provides a platform for exploring effects on the O-GlcNAc proteome.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti O-GlcNAc (RL2) | Abcam | ab2739; RRID:AB_303264 |
| Rabbit polyclonal anti HSP90 | Cell Signalling | Cat#4874; RRID:AB_2121214 |
| Rabbit polyclonal anti HSP90 | Enzo Life Sciences | ADI-SPA-836-D; RRID:AB_991589 |
| Rabbit polyclonal anti OGT (H-300) | Santa Cruz Biotechnology | sc-32921 |
| Goat IRDye® 680RD anti-Mouse | LICOR | P/N 925-68070; RRID:AB_621840 |
| Donkey IRDye® 800CW anti-Rabbit | LICOR | P/N 925-32213; RRID:AB_621848 |
| Morpheus® HT-96 Crystal Screen | Molecular Dimensions | MD1-47 |
| Bacterial and Virus Strains | ||
| E. coli BL21-DE3 | New England Biotechnologies | C25271 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SYPRO® Orange dye | Sigma | S5692-50UL |
| GlcNAcstatin-G (OGA inhibitor) | GlycoBioChem | GBC10002 |
| RB2 (residues 410-422) | GlycoBioChem | N/A |
| CpOGA | GlycoBioChem | N/A |
| OGTWT/L254F | This paper | N/A |
| TPRWT/L254F | This paper | N/A |
| Deposited Data | ||
| TPRL254F structure | This paper | PDB: 6EOU |
| Experimental Models: Cell Lines | ||
| HEK-293 | ATCC | CRL-1573 |
| Oligonucleotides | ||
| Primer TPR: Forward CTGGGATCCGGCCCGATGGAACT GGGCTCATCGTGAAATATCAG |
This paper | N/A |
| Primer TPR: Reverse GATGCGGCCGCTTAGTCTTGCATT TCTTTCAGCGTATTAC |
This paper | N/A |
| Recombinant DNA | ||
| pGEX-6P-1 Vector | GE Healthcare | 28954648 |
| pHEX-6P-1 Vector | This paper | N/A |
| Software and Algorithms | ||
| Graphpad Prism v5.0 | GraphPad Software | https://www.graphpad.com/ |
| iMosflm | (Battye et al., 2011) | http://www.ccp4.ac.uk/ |
| CCP4 | (Winn et al., 2011) | http://www.ccp4.ac.uk/ |
| REFMAC | (Vagin et al., 2004), | http://www.ccp4.ac.uk/ |
| GROMACS | (Abraham et al., 2015). | http://www.gromacs.org/ |
| MDAnalysis | (Michaud-Agrawal et al., 2011) | https://www.mdanalysis.org/ |
| MDTraj | (McGibbon et al., 2015) | http://mdtraj.org/1.9.0/ |
| Other | ||
| Morpheus® HT-96 Crystal Screen | Molecular Dimensions | MD1-47 |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Daan M. F. van Aalten (dmfvanaalten@dundee.ac.uk).
Experimental Model Details
Cell Lines
Female HEK-293 cells were obtained from ATCC. Due to the use of HEK-293 lysates solely for biochemistry, the cell line was not further authenticated. The cells were tested negative for mycoplasma contamination (October 2017). HEK-293 cells were grown on 15 cm plates in DMEM (Life Technologies) supplemented with 2 mM L-glutamine (Sigma), 100 units/ml Penicillin and 100 μg/ml Streptomycin (Life Technologies), and 10% foetal calf serum (Labtech).
Method Details
Molecular Cloning
The full-length codon optimised OGT was obtained from GenScript and subcloned as a BamHI-NotI fragment into pHEX-6P-1 (modified version of pGEX-6P-1 which contains a 6His tag instead of GST). The L254F mutation was introduced using a method similar to the QuikChange site-directed mutagenesis kit by Agilent but using KOD polymerase and DpnI from Fermentas. All inserts were confirmed by DNA sequencing. The TPR region of OGT (residues 26-410) was amplified from both the OGTWT and OGTL254F expression constructs. These were cloned into pGEX-6P-1 as BamHI-NotI fragments and the inserts were confirmed by DNA sequencing. Forward and reverse primers used were CTGGGATCCGGCCCGATGGAACTGGGCTCATC-GTGAAATATCAG and GATGCGGCCGCTTAGTCTTGCATTTCTTTCAGCGTATTAC, respectively.
OGT Expression and Purification
Full length OGTWT and OGTL254F were expressed in E. coli BL21-DE3 as N-terminal His fusion proteins as described previously (Willems et al., 2017). Briefly, transformed E. coli cells were grown in autoinduction medium at 37°C with agitation until OD600 reached 0.8, at which point the temperature was lowered to 18°C for overnight incubation. Cells were harvested by centrifugation at 4°C (35 min 4,500 × g). Resulting cell pellet was resuspended in base buffer (0.1 M Tris-HCl, pH 7.5, 0.15 M NaCl, 0.5 mM TCEP (tris[2-carboxyethyl]phosphine) supplemented with 25 mM imidazole 0.1 mg/ml DNase I and protease inhibitor cocktail (1 mM benzamidine, 0.2 mM PMSF, 5 mM leupeptin), and lysed via continuous flow cell disruptor (three passes at 15,000 PSI). Lysate was clarified by centrifugation (30,000 g for 1 h at 4°C) followed by incubation with 1 ml per litre of culture of Ni2+-NTA agarose resin (GE Healthcare) for 2 h at 4°C. The resin was thoroughly washed and eluted with base buffer supplemented with 25 mM and 500 mM imidazole respectively. Eluted protein was dialyzed and cleaved from the His-tag overnight at 4°C in buffer A (0.1 M tris-HCl, pH 8.5, 25 mM NaCl) supplemented with PreScission protease (GE Healthcare), then passed through fresh Ni2+-NTA agarose resin. Dialyzed protein was loaded onto 5 ml HiTrap Q Sepharose FF anion exchange resin (GE Healthcare) and eluted with a linear gradient up to 60% of buffer B (0.1 M tris-HCl, pH 8.5, 500 mM NaCl). Peak fractions were pooled, concentrated and further purified via size exclusion chromatography using 300-ml prepacked Superdex™ 200 column (GE Healthcare) equilibrated with base buffer. The peak fractions were concentrated to 10 mg/ml, mixed 1:1 with 50% glycerol, snap-frozen and stored at -80°C until use.
TPR Expression and Purification
The TPR region of OGT (residues 26-410), either wild type (TPRWT) or bearing the L254F mutation (TPRL254F), was expressed and purified as N-terminally GST fusion proteins as described previously (Jínek et al., 2004). Briefly, E. coli BL21-DE3 cells were transformed, grown and harvested as described for the full length OGT constructs. Resulting cell pellet was resuspended in base buffer (20 mM Na-HEPES, pH 7.5, 200 mM NaCl, 2 mM DTT supplemented with 0.1 mg/ml DNase I and protease inhibitor cocktail (1 mM benzamidine, 0.2 mM PMSF, 5 mM leupeptin), prior to being lysed and clarified as described above. Clarified lysate was then incubation with 1 ml per litre of culture of Glutathione Sepharose 4B resin (GE Healthcare) for 2 h at 4°C. The resin was thoroughly washed with base and the recombinant proteins were cleaved on-resin by addition of PreScission protease (GE Healthcare) and overnight incubation at 4˚C. Cleaved protein was eluted, concentrated and further purified via size exclusion chromatography using 300-ml prepacked Superdex™ 200 column (GE Healthcare) equilibrated with base buffer. The peak fractions were concentrated to 30 mg/ml, snap-frozen in liquid nitrogen and stored at -80°C until use.
TPR Crystallisation and Structural Analysis
Crystallisation of TPRL254F was performed at 22°C using MRC 96-well sitting drop crystallization plates (Molecular Dimensions) by combining 0.2 μl TPRL254F (in 20 mM Na-HEPES-NaOH pH 7.5, 200 mM NaCl and 2 mM DTT) with 0.2 μl of reservoir solution (0.1 M Na-HEPES and 0.1 M MOPS-HCl pH 7,5, 0.04 M diethylene glycol, 0.04 M triethylene glycol, 0.04 M tetraethylene glycol, 0.04 M pentaethylene glycol, 20 % v/v ethylene glycol and 10 % w/v PEG 4000) (Morpheus®, Molecular Dimensions (Gorrec, 2015)). Orthorhombic rod and disc shaped crystals appeared within 1-2 days. Prior to diffraction experiments, individual crystals were flash-frozen in liquid nitrogen without prior cryoprotection. Diffraction data were collected at the European Synchrotron Radiation Facility beamline ID30A-1. Data were processed with iMosflm (Battye et al., 2011) and scaled to 1.75 Å using SCALA (Winn et al., 2011). The structure was solved by molecular replacement using the structure for TPRWT (PDB: 1W3B Jínek et al., 2004) as the search model. The resulting model was initially truncated at both N- and C-termini where the fit of the electron density and the model was poor, and manually rebuilt and refined using Coot (Winn et al., 2011) and REFMAC (Vagin et al., 2004), respectively. The editing and refinement of the model was iterated until it was in complete agreement with the data. Scaling and model building statistics can be seen in Table 1.
Thermal Denaturing Assay
Thermal denaturation experiments were performed in triplicate, using constructs encompassing the TPR domain (residues 26-410). 50 μl solutions contained 5 μM protein and 1.1x SYPRO® Orange dye (Sigma) in base buffer of 25 mM HEPES-NaOH pH 7.5, 150 mM NaCl and 0.5 mM TCEP. CFX Connect™ Real-Time System (BIO-RAD) was used to measure fluorescence (λex = 530 nm, λem = 560 nm) while temperature was increased from 25 to 95°C at 1 degree per minute increments. The data were transformed, normalised and fitted to a four-parameter Boltzmann sigmoidal curve using GraphPad Prism 5.0.
Molecular Dynamics Simulations
Truncated OGT TPR wild type and L254F constructs comprising TPRs 6-8 (sTPRWT and sTPRL254F; residues 189-294, Figure 1A) were used in molecular dynamics simulations, similar to an approach previously used to simulate sections of the alpha-solenoid HEAT repeat protein importin-β (Kappel et al., 2010). Appropriate capping groups were added to N- and C-terminal ends of both sTPRWT and sTPRL254F constructs. The major axes of the sTPR constructs were aligned to the z-axis of a triclinic simulation box a triclinic box of 62.5 x 62.5 x 82.5 Å and solvated using explicit water molecules. Na+ and Cl- ions were added in order to neutralise the system at the physiological NaCl concentration of 0.15 mM. The amber99SB-ildn force field (Lindorff-Larsen et al., 2010) and virtual sites for hydrogen atoms (Feenstra et al., 1999) were used. The TIP3P water model was used to model the solvent molecules and Joung and Cheatham III parameters (Joung and Cheatham, 2008) were used to model the counter ions. Simulations were carried out with the GROMACS molecular dynamics package, version 5.1.5 (Abraham et al., 2015). For each system, the geometry was minimized in four cycles that combined 3500 steps of steepest descent algorithm followed by 4500 of conjugate gradient. Thermalisation of the system was performed in 6 steps of 5 ns, where the temperature was gradually increased from 50 K to 298 K, while the protein was restrained with a force constant of 10 kJ mol-1 Å-2. Production runs consisted of four replicates of 500 ns simulations for each system (accounting for 2 μs of simulation time per system). Making use of virtual sites, the integration time-step was set to 4 fs. Temperature was kept constant by weakly coupling (t = 0.1 ps) protein and water and ions separately to a temperature bath of 298K with the velocity rescale thermostat of Bussi et al. (Bussi et al., 2007). The pressure was kept constant at 1 bar using semi-isotropic Berendsen coupling (Berendsen et al., 1984). Long-range electrostatic interactions were calculated using the smooth particle mesh Ewald method (Darden et al., 1993) beyond a short-range Coulomb cut-off of 10 Å. A 10-Å cut-off was also set for Lennard-Jones interactions. The LINCS algorithm (Hess et al., 1997) was used to restrain the bonds involving hydrogen and the SETTLE algorithm (Miyamoto and Kollman, 1992) was used to constrain bond lengths and angles of water molecules. Periodic boundary conditions were applied.
In Vitro O-GlcNAcylation Assays
Michaelis-Menten kinetics of OGT were measured using a fluorimetric assay as described previously (Borodkin et al., 2014), with the exception of reduced reaction volume of 25 μl and usage of 384-well plate. As acceptor substrate, a 13 amino acid long sequence from retinoblastoma-like protein 2 (RB2; 410KENSPAVTPVSTA422; GlycoBioChem) was used. Reactions for Michaelis-Menten kinetics contained 0-768 μM acceptor peptide substrate, 200 μM UDP-GlcNAc and 50 nM OGT in 50 mM HEPES-NaOH pH 7.5, 0.1 mg/ml BSA and 10 μM Na2S2O4. Reactions were stopped before 10% of the acceptor substrate was depleted by addition of 50 μl detection reagent (25 mM HEPES-NaOH pH 7.5, 10 mM NaCl, 15 μM xanthene based Zn(II) complex, 75 μM pyrocathecol violet and 50% methanol). The fluorescence was read using excitation and emission wavelengths of 485 nm and 530 nm, respectively. Data were background corrected and plotted using GraphPad Prism.
Additional O-GlcNAcylation assays were performed on de-O-GlcNAcylated HEK-293 lysate proteins. Cultured HEK-293 cells were washed twice with ice-cold PBS buffer (Life Technologies) prior to lysis. Cells were lysed by addition of lysis buffer (50 mM Tris-HCl, pH 7.4, 1 mM EGTA, 1 mM EDTA, 1% Triton-X100, 1 mM Na3VO4, 50 mM NaF, 5 mM Na4P2O7, 0.27 M sucrose) supplemented with 1 μM ß-mercaptoethanol, 1 mM benzamidine, 0.2 mM PMSF and 5 mM leupeptin. The lysate was transferred into microfuge tubes and clarified by centrifugation at 4°C (17,000 g for 15 min). The lysate was then treated with 120 μg CpOGA per mg of lysate protein and incubated for 90 minutes at 37°C. CpOGA and endogenous HEK-293 OGA were then neutralised by addition of 250 μM GlcNAcstatin-G, an OGA inhibitor. Reactions were then supplemented with OGTWT or OGTL254F (0.2 μM) in presence of 2 mM UDP-GlcNAc and incubated for an additional 2 h at 37°C. Proteins were resolved by SDS-PAGE (3-8% Tris-Acetate gels; Life Technologies), transferred onto nitrocellulose membrane (GE Healthcare), and probed using O-GlcNAc-RL2 (1:1,000 dilution; Abcam), HSP90 (1:5,000 dilution; Cell Signalling or Enzo Life Sciences) and OGT-H300 (1:1,000 dilution; Santa Cruz) primary antibodies and corresponding IRDye associated secondary antibodies (1:10,000 dilution; LI-COR). Resulting signal was quantified using a LI-COR Odyssey scanner and associated quantification software. Data were plotted using GraphPad Prism 5.0.
Quantification and Statistical Analysis
Michaelis-Menten kinetics of the OGT-catalysed reaction against the peptide substrate derived from RB2 was performed as three technical replicates and repeated two times (data presented in Figure S1A). The O-GlcNAc activity assay against deglycosylated HEK-293 cell lysate proteins was repeated six times (data presented in Figures 1B and S1B). The ThermoFluor assay was performed as seven technical replicates, and repeated two times (data presented in Figure 2A). For all activity and thermal stability assays, GraphPad Prism was used for calculation of statistics. Error bars represent the standard deviation of the mean in all presented data. The trajectories obtained by the molecular dynamics simulations were analysed with the MDAnalysis (RMSD and RMSF) (Gowers et al., 2016, Michaud-Agrawal et al., 2011) and MDtraj (distances and angles) (McGibbon et al., 2015) packages. The values reported in the results and discussion sections correspond to the mean values ± standard deviation of the mean.
Data and Software Availability
The crystallographic structure has been deposited in RCSB Protein Data Bank (http://www.rcsb.org/pdb) under ID code 6EOU.
Acknowledgments
This work funded by a Wellcome Trust Investigator award (110061) to D.M.F.v.A and a Wellcome Trust ISSF award (WT097818MF) to U.Z. M.G. was supported by a University of Dundee Translational Medical Research Fund Ph.D. fellowship. A.G. was supported by a Wellcome Trust PhD studentship. Diffraction experiments were performed on beamline ID30A-1 at the European Synchrotron Radiation Facility (ESRF), Grenoble, France. We are grateful to Matthew Bowler at the ESRF for providing assistance in using the beamline ID30A-1.
Author Contributions
D.M.F.v.A. and U.Z. conceived the study; M.G. performed all protein production, in vitro assays, and protein crystallography experiments; M.G. and A.G. performed the OGT activity assay against HEK-293 cell lysate; S.L. performed molecular dynamics simulations; A.T.F. performed molecular biology experiments; M.G., A.G., and S.L. analyzed the data; M.G. and D.M.F.v.A. interpreted the data and wrote the manuscript with input from all authors.
Declaration of Interests
The authors declare no competing interests.
Published: March 29, 2018
Footnotes
Supplemental Information includes three figures and can be found with this article online at https://doi.org/10.1016/j.chembiol.2018.03.004.
Supplemental Information
References
- Abraham M.J., Murtola T., Schulz R., Páall S., Smith J.C., Hess B., Lindah E. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- Battye T.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- Borodkin V.S., Schimpl M., Gundogdu M., Rafie K., Dorfmueller H.C., Robinson D.A., van Aalten D.M. Bisubstrate UDP-peptide conjugates as human O-GlcNAc transferase inhibitors. Biochem. J. 2014;457:497–502. doi: 10.1042/BJ20131272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouazzi H., Lesca G., Trujillo C., Alwasiyah M.K., Munnich A. Nonsyndromic X-linked intellectual deficiency in three brothers with a novel MED12 missense mutation [c.5922G>T (p.Glu1974His)] Clin. Case Rep. 2015;3:604–609. doi: 10.1002/ccr3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Cole R.N., Hart G.W. Cytosolic O-glycosylation is abundant in nerve terminals. J. Neurochem. 2001;79:1080–1089. doi: 10.1046/j.1471-4159.2001.00655.x. [DOI] [PubMed] [Google Scholar]
- Connarn J.N., Assimon V.A., Reed R.A., Tse E., Southworth D.R., Zuiderweg E.R., Gestwicki J.E., Sun D. The molecular chaperone Hsp70 activates protein phosphatase 5 (PP5) by binding the tetratricopeptide repeat (TPR) domain. J. Biol. Chem. 2014;289:2908–2917. doi: 10.1074/jbc.M113.519421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T., York D., Pedersen L. Particle mesh Ewald: an N log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089. [Google Scholar]
- Deplus R., Delatte B., Schwinn M.K., Defrance M., Méndez J., Murphy N., Dawson M.A., Volkmar M., Putmans P., Calonne E. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenstra K.A., Hess B., Berendsen H.J.C. Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. J. Comput. Chem. 1999;20:786–798. doi: 10.1002/(SICI)1096-987X(199906)20:8<786::AID-JCC5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Francisco H., Kollins K., Varghis N., Vocadlo D., Vosseller K., Gallo G. O-GlcNAc post-translational modifications regulate the entry of neurons into an axon branching program. Dev. Neurobiol. 2009;69:162–173. doi: 10.1002/dneu.20695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrec F. The MORPHEUS II protein crystallization screen. Acta Crystallogr. F Struct. Biol. Commun. 2015;71:831–837. doi: 10.1107/S2053230X1500967X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowers, R.J., Linke, M., Barnoud, J., Reddy, T.J.E., Melo, M.N., Seyler, S.L., Domański, J., Dotson, D.L., Buchoux, S., Kenney, I.M., et al. (2016). MDAnalysis: a Python package for the rapid analysis of molecular dynamics simulations. In: Proceedings of the 15th Python in Science Conference, pp. 98–105.
- Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- Iyer S.P., Hart G.W. Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J. Biol. Chem. 2003;278:24608–24616. doi: 10.1074/jbc.M300036200. [DOI] [PubMed] [Google Scholar]
- Jínek M., Rehwinkel J., Lazarus B.D., Izaurralde E., Hanover J.A., Conti E. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin α. Nat. Struct. Mol. Biol. 2004;11:1001–1007. doi: 10.1038/nsmb833. [DOI] [PubMed] [Google Scholar]
- Joung I.S., Cheatham T.E. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappel C., Zachariae U., Dölker N., Grubmüller H. An unusual hydrophobic core confers extreme flexibility to HEAT repeat proteins. Biophys. J. 2010;99:1596–1603. doi: 10.1016/j.bpj.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochinke K., Zweier C., Nijhof B., Fenckova M., Cizek P., Honti F., Keerthikumar S., Oortveld M.A., Kleefstra T., Kramer J.M. Systematic phenomics analysis deconvolutes genes mutated in intellectual disability into biologically coherent modules. Am. J. Hum. Genet. 2016;98:149–164. doi: 10.1016/j.ajhg.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J.R., Tugendreich S., Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem. Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- Lazarus M.B., Jiang J., Kapuria V., Bhuiyan T., Janetzko J., Zandberg W.F., Vocadlo D.J., Herr W., Walker S. HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science. 2013;342:1235–1239. doi: 10.1126/science.1243990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas W.A., Frank D.W., Krause M., Hanover J.A. O-linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J. Biol. Chem. 1997;272:9316–9324. doi: 10.1074/jbc.272.14.9316. [DOI] [PubMed] [Google Scholar]
- März P., Stetefeld J., Bendfeldt K., Nitsch C., Reinstein J., Shoeman R.L., Dimitriades-Schmutz B., Schwager M., Leiser D., Özcan S. Ataxin-10 interacts with O-linked β-N-acetylglucosamine transferase in the brain. J. Biol. Chem. 2006;281:20263–20270. doi: 10.1074/jbc.M601563200. [DOI] [PubMed] [Google Scholar]
- McGibbon R.T., Beauchamp K.A., Harrigan M.P., Klein C., Swails J.M., Hernandez C.X., Schwantes C.R., Wang L.P., Lane T.J., Pande V.S. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 2015;109:1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud-Agrawal N., Denning E.J., Woolf T.B., Beckstein O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S., Kollman P.A. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- Niranjan T.S., Skinner C., May M., Turner T., Rose R., Stevenson R., Schwartz C.E., Wang T. Affected kindred analysis of human X chromosome exomes to identify novel X-linked intellectual disability genes. PLoS One. 2015;10:e0116454. doi: 10.1371/journal.pone.0116454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama R., Marshall S. UDP-N-acetylglucosaminyl transferase (OGT) in brain tissue: temperature sensitivity and subcellular distribution of cytosolic and nuclear enzyme. J. Neurochem. 2003;86:1271–1280. doi: 10.1046/j.1471-4159.2003.01939.x. [DOI] [PubMed] [Google Scholar]
- Pathak S., Alonso J., Schimpl M., Rafie K., Blair D.E., Borodkin V.S., Schüttelkopf A.W., Albarbarawi O., van Aalten D.M.F. The active site of O-GlcNAc transferase imposes constraints on substrate sequence. Nat. Struct. Mol. Biol. 2015;22:744–750. doi: 10.1038/nsmb.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekkurnaz G., Trinidad J.C., Wang X., Kong D., Schwarz T.L. Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell. 2014;158:54–68. doi: 10.1016/j.cell.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafie K., Raimi O., Ferenbach A.T., Borodkin V.S., Kapuria V., van Aalten D.M.F. Recognition of a glycosylation substrate by the O-GlcNAc transferase TPR repeats. Open Biol. 2017;7 doi: 10.1098/rsob.170078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropers H.H. Genetics of early onset cognitive impairment. Annu. Rev. Genomics Hum. Genet. 2010;11:161–187. doi: 10.1146/annurev-genom-082509-141640. [DOI] [PubMed] [Google Scholar]
- Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F.U., Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Timbers T.A., Ardiel E.L., Mcdiarmid T.A., Lee K.C.Y., Safaei J., Pelech S.L., Rankin C.H. CaMK (CMK-1) and O-GlcNAc transferase (OGT-1) modulate mechanosensory responding and habituation in an interstimulus interval-dependent manner in Caenorhabditis elegans. BioRxiv. 2017 [Google Scholar]
- Vagin A.A., Steiner R.A., Lebedev A.A., Potterton L., McNicholas S., Long F., Murshudov G.N. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- Vaidyanathan K., Niranjan T., Selvan N., Teo C.F., May M., Patel S., Weatherly B., Skinner C., Opitz J., Carey J. Identification and characterization of a missense mutation in the O-linked β-N-acetylglucosamine (O-GlcNAc) transferase gene that segregates with X-linked intellectual disability. J. Biol. Chem. 2017;292:8948–8963. doi: 10.1074/jbc.M116.771030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems A.P., Gundogdu M., Kempers M.J.E., Giltay J.C., Pfundt R., Elferink M., Loza B.F., Fuijkschot J., Ferenbach A.T., van Gassen K.L.I. Mutations in N-acetylglucosamine (O-GlcNAc) transferase in patients with X-linked intellectual disability. J. Biol. Chem. 2017;292:12621–12631. doi: 10.1074/jbc.M117.790097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2017;18:452–465. doi: 10.1038/nrm.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Zhang F., Kudlow J.E. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;110:69–80. doi: 10.1016/s0092-8674(02)00810-3. [DOI] [PubMed] [Google Scholar]
- Zeytuni N., Zarivach R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure. 2012;20:397–405. doi: 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.