Abstract
Pathogenic bacteria cause various infections worldwide, especially in immunocompromised and other susceptible individuals, and are also associated with high infant mortality rates in developing countries. MicroRNAs (miRNAs), small non-coding RNAs with evolutionarily conserved sequences, are expressed in various tissues and cells that play key part in various physiological and pathologic processes. Increasing evidence implies roles for miRNAs in bacterial infectious diseases by modulating inflammatory responses, cell penetration, tissue remodeling, and innate and adaptive immunity. This review highlights some recent intriguing findings, ranging from the correlation between aberrant expression of miRNAs with bacterial infection progression to their profound impact on host immune responses. Harnessing of dysregulated miRNAs in bacterial infection may be an approach to improving the diagnosis, prevention and therapy of infectious diseases.
Microbiology: Tracking infection with tiny transcripts
Changes in production of tiny cellular RNAs in response to bacterial infection could guide the development of better diagnostics and therapies. MicroRNAs regulate other genes by binding to messenger RNA strands and controlling their translation into proteins. Xikun Zhou, Min Wu and colleagues of the University of North Dakota have now reviewed current knowledge about how microRNA levels shift during infection with various bacterial pathogens. These microRNAs can modulate the immune response as well as pathways that influence metabolic activity and cell survival. Increasing studies have indicated that shifts in microRNA levels in response to different infections could provide a potential bacterial ‘fingerprint’ for achieving accurate diagnosis. With deeper insight into how different microRNAs influence infection, it might one day day become possible to target these molecules with ‘antisense’ or ‘agonist’ drugs that modulate their activity.
Introduction
Pathogenic bacteria hold a wide range of strategies to invade, survive, and replicate in their hosts. These pathogens are the major causes of many deadly diseases and widespread epidemics in mammals, including humans. However, host immune systems have also developed extremely complex adaptations to counteract bacterial infection1. Host–pathogen interactions are one of the most complex themes involved in disease initiation, development and progression. An intact immune system is critical for host resistance to bacterial infections. There are many important regulators involved in a range of pathological processes during host defense against infection that modulate diverse biological processes. Host immune cells, such as lymphocytes, innate lymphoid cells, macrophages and neutrophils, are vital parts of innate immunity systems that find, process and clear invading microbes by phagocytosis, secreting cytokines and mounting inflammatory responses. Pathogen-associated molecular patterns (PAMPs) bind and talk with Toll-like receptors (TLRs), NOD-like receptors (NLRs) and other pattern-recognition receptors (PRRs) to activate a number of inflammatory signals and subsequently lead to proinflammatory cytokine production or inflammatory cell death. Each type of PAMP can be recognized by its specific receptor(s)2,3. Subsequently, the adaptive immune response is induced to promote and facilitate the removal of pathogenic bacteria4. Once these intruders are cleared, negative immunoregulatory cytokines and Th2 cells play a dominant role in balancing the extent of the immune response to avoid overreaction and tissue damage5. Recent studies provided some insight into the critical participation of microRNAs (miRNAs) in host immune defense against bacterial infection.
miRNAs are evolutionarily conserved small (~22 nucleotide) non-coding RNAs first discovered two decades ago6. The transcription of miRNAs is most commonly mediated by RNA polymerase II; they are then processed by two nucleases, Drosha and Dicer. After exporting to the cytoplasm, the functional mature miRNA is incorporated into and preferentially stabilized by the RNA-induced silencing complex (RISC). In most cases, the RISC converts the miRNA to a 6–8 nucleotide-long complementary region, named the ‘seed sequence’, on the 3′-Untranslated Region (3′-UTR) of its target mRNA and mediates its function. The partial or imperfect complementarity of a miRNA to a target mRNA may result in translational repression, while full or perfect complementarity binding sites cause target degradation at the posttranscriptional levels7. Certain miRNAs can also bind the 5′ untranslated region (5′-UTR) and amino-acid coding sequence (CDS) sites of their target mRNA, and several miRNAs may also induce gene expression8–10. Moreover, one mRNA might be modulated by numerous miRNAs, and a miRNA has the ability to modulate the expression of a number of target mRNAs. miRNAs have emerged as critical regulators in a great deal of biological processes, such as cell proliferation, differentiation, autophagy, metabolism and immune responses. The dysregulated expression of miRNAs has also been correlated with various diseases, including cancer, autoimmunity, and cardiovascular diseases, among others 11.
In this review, we first summarize the dysregulated miRNAs identified during different bacterial infections. Then, we describe the host signal transduction pathways utilized by bacterial effectors by which miRNA expression is dysregulated in mechanisms of modulation. Finally, we discuss the potential of miRNAs to serve as diagnosis biomarkers and treatment targets, and discuss the challenges facing miRNA studies.
MiRNAs affected by bacterial infections
Bacterial pathogens are thought to have complex interactions with relevant hosts, and the interactions between hosts and pathogens are becoming a forefront research area of infectious diseases. Latest studies have highlighted that the expression of miRNAs is profoundly impacted by a variety of bacterial pathogens and that likewise miRNAs impose strong pressure to the invading microorganisms.
Helicobacter pylori
H. pylori is particularly capable of colonization in human stomach and is thus responsible for various gastric diseases, such as chronic active gastritis, peptic ulcers, and gastric carcinoma worldwide12,13. Several studies have reported that infection of gastric epithelial cells with H. pylori could lead to altered expression of miRNAs, including let-714–16, miR-30b17, miR-21018, miR-128919, miR-152/miR-200b20, miR-15521–25, miR-16, and miR-146a24–26. Histological analysis has shown higher miR-155 levels in gastric mucosal tissue sections of patients infected with H. pylori. Potential binding sites for nuclear factor-κB (NF-κB) as well as activator protein-1 (AP-1) have been identified within the BIC/miR-155 promoter, and both NF-κB and AP-1 are the necessity for the induction of miR-155 upon H. pylori infection in gastric epithelial cells24. The expression of miR-155 might also be influenced by Foxp3 in H. pylori-infected T cells23. In addition to NF-κB signaling in bone marrow-derived macrophages (BMDMs), TLR2/4- and NOD1/2-independent upregulation of miR-155 was found to depend on the H. pylori type IV secretion system (T4SS)21. Several miR-155-targeted mRNAs, including tumor protein p53-inducible nuclear protein 1 (TP53INP1), tetraspanin 14 (Tspan14), lipin 1 (Lpin1), phorbol-12-myristate-13-acetate-induced protein 1 (Pmaip1), protein kinase (cAMP-dependent, catalytic) inhibitor alpha (PKIα), IκB kinase ε (IKK-ε), Sma- and Mad-related protein 2 (SMAD2), and Fas-associated death domain protein (FADD), have been linked to proapoptotic and immune responses21,23,24,27. miR-155 knockout mice failed to control H. pylori infection and had reduced protection from infection after H. pylori-specific vaccination than their wild-type counterparts as a result of impaired pathogen-specific T helper type 1 (Th1) and Th17 responses22. A multi-epitope vaccine, CTB-UE, could relieve the H. pylori-induced gastric inflammatory reaction by upregulating miR-155 to inhibit Th17 responses28. These results implied that the increase in miR-155 expression during H. pylori infection is involved in negative regulation of inflammation by attenuating NF-κB signaling, Th17/Th1 differentiation and cyclic adenosine monophosphate (cAMP) activity23,24. Furthermore, there was a strong link between miR-155 levels and immunohistochemical grades in H. pylori-positive patients, and miR-155 expression was downregulated in intestinal metaplasia individuals29.
Another miRNA, miR-146a, has been shown to be increased after H. pylori infection in gastric epithelial cells, as well as in gastric mucosal tissues, in an NF-κB-dependent manner. Subsequently, miR-146a can diminish the expression of target genes, e.g., TNF receptor-associated factor 6 (TRAF6) and IL1 receptor-associated kinase 1 (IRAK1). In addition, miR-146a may inhibit the expression of IL-8, growth-related oncogene (GRO)-α, and macrophage inflammatory protein (MIP)-3α, TNF-α, and IL1β by reducing NF-κB activity30,31. Additional studies have found that the overexpression of miR-146a results in significantly reduced Prostaglandin endoperoxide synthase 2 (PTGS2) production induced by H. pylori infection26.
H. pylori infection is thought to have a link with the early stages of gastric cancer pathogenesis via the induction of chronic gastritis16. This is also a hallmark of H. pylori, making it different from other bacteria. Cytotoxin-associated gene A (CagA), a key virulence factor of H. pylori, harms the gastric mucosa and is associated with an increased risk of atrophic gastritis, peptic ulcer and gastric cancer13,16. Hayashi et al. found that CagA drives epigenetic regulation to impede let-7 expression in H. pylori-related carcinogenesis. CagA augmented c-myc, DNA methyltransferase 3B (DNMT3B) and Enhancer of Zeste homolog 2 (EZH2) expression and reduced miR-26a and miR-101 levels, lowering let-7 expression by altering histone and DNA methylation16. CagA can also downregulate miR-370 levels, resulting in increased FoxM1, a positive modulator for cell growth32. miR-320a and miR-4496 attenuate the possibility of CagA-induced cancer-initiation and chemoresistance through influencing β-catenin and ATP-binding cassette subfamily G member 233. miR-210 was downregulated in the gastric epithelium in response to persistent H. pylori infection. Inflammation-induced epigenetic silencing of miR-210 augmented cell proliferation by activating two tumorigenesis-related proteins, STMN1 (stathmin1) and DIMT1 (demethyladenosine transferase 1)18. Several other miRNAs, such as miR-20334, miR-20435, miR-37536, and miR-27b37, were aberrantly expressed in H. pylori-positive tissues and cells and affecting neoplastic transformation and invasion. These studies suggest a role for miRNAs in regulating pathogenesis in various H. pylori-infected cell types.
Salmonella
Salmonella is a Gram-negative intracellular pathogen belonging to the family Enterobacteriaceae and can cause a number of diseases in humans and animals, such as gastroenteritis and typhoid fever38. Salmonella has three main serovars, typhi, typhimurium, and enteritidis, and can exert diverse effects to establish an intracellular niche for successful propagation39. A number of studies have reported that the dysregulation of miRNAs critically contributes to disease pathogenesis. Hoeke et al. showed that focal adhesion and organization of the actin cytoskeleton is regulated by miRNAs in an intestinal Salmonella typhimurium infection model. S. typhimurium infection upregulated the expression of miR-29a, which subsequently targeted Caveolin 2, a focal adhesion factor that is associated with uptake of bacterial pathogens, to modulate the activation state of the small Rho GTPase CDC4240. miR-146 decreased the induction of six members of the apolipoprotein gene family in S. typhimurium-infected zebrafish embryos. This suggested that miR-146 may play a role in regulating lipid metabolism during inflammation41. TLR4-sensing of bacterial LPS downregulated the expression of let-7 family miRNAs upon Salmonella infection. The downregulation of these miRNAs promoted the expression of the key cytokines IL6 and IL-1042. By a combination of high-throughput screening with a library of miRNA mimics and RNA-seq, Maudet et al. proposed that miRNAs are potential modulators in S. typhimurium infection and that distinct miRNAs impair various infection stages. They further found that downregulation of the miR-15 family upregulated cyclin D1 expression upon Salmonella infection. G2/M arrest of host cells dramatically increased Salmonella replication within hosts43. Moreover, miR-155 regulated the function of both lymphocytes and DCs, leading to an overall diminution of immune responses. Vaccination of miR-155-deficient mice with an attenuated vaccine against S. typhimurium failed to protect them against virulent S. Typhimurium 44.
Macrophage colony-stimulating factor (M-CSF, CSF1) is a cytokine for attracting macrophages to infection sites to defend against different pathogenic infections. Virulent Salmonella enteritidis modulates intestinal epithelial miR-128 levels, which inhibits epithelia-secreted M-CSF and impedes the subsequent recruitment of macrophage45. A combined study of differentially expressed miRNAs with mRNAs predicted miRNA targets, revealing miRNA-mRNA profiles. This analysis found that miR-214 and miR-331-3p could participate in host immunity against S. typhimurium. Salmonella-challenged pigs showed downregulated miR-214 expression and upregulated miR-331-3p expression in whole blood. While levels of the candidate targets (SLC11A1 and PIGE-108A11.3) of miR-214 were enhanced following challenge, the potential target (VAV2) of miR-331-3p was reduced46. Another binding site enrichment analysis of miRNAs responsible for deregulated mRNAs in peripheral blood suggested that miR-143 and miR-26 might be involved in the initiation and progression of Salmonella infection in pigs47. In addition, miRNAs may also modulate the innate immunity involving miRNAs to S. enteritidis infection in laying chicken ceca48. Thus, these deregulated miRNAs may be functionally important for manipulating Salmonella-induced inflammation.
Pseudomonas aeruginosa
Pseudomonas aeruginosa is an important opportunistic Gram-negative bacterium that infects a broad range of individuals, invading many different parts of the body, with corresponding symptoms and signs49. Multidrug-resistant Pseudomonas can be deadly for patients in intensive care units worldwide50. Up to date, few miRNAs have been reported in modulating inflammatory responses, and are likely TLR/NF-κB-responsive51. Our laboratory recently reported that miR-302b can be activated by TLR2 and TLR4 via ERK-p38-NF-κB pathways following P. aeruginosa infection. miR-302b, together with other members of the miR-302 family, is a crucial regulator of TLR-induced downstream NF-κB signaling, macrophage and epithelial cell activation, and respiratory inflammation via directly targeting of IRAK1, a member of the TLR/myeloid differentiation factor 88 (MyD88) complex that is critical for NF-κB activation52. We further identified another miRNA, miR-301b, can be induced via a TLR4/MyD88/NF-κB pathway against P. aeruginosa infection. miR-301b positively modulates the expression of the anti-inflammatory cytokines IL-4 and TGF-β1 and negatively regulates the expression of the proinflammatory cytokines MIP-1α and IL-17A. This function is exerted by repressing c-Myb expression, and the antimicrobial effect was potentiated by caffeine uptake. Moreover, repression of miR-301b resulted in elevated levels of neutrophil infiltration 53.
There are several other negative feedback miRNAs, such as miR-762 and miR-155, whose levels are enhanced upon P. aeruginosa infection to downregulate levels of immune response genes51,54. Using a P. aeruginosa-infected Caenorhabditis elegans model, Ren et al. reported that the let-7 family (let-7-Fam) acted in innate immune response pathways to timely induce strong immunity to reduce pathogen-induced stress. The developmental timing phenotypes of let-7-Fam miRNA mutants were modified by growth in pathogenic processes. The let-7-Fam miRNA activity was downmodulated during P. aeruginosa infection through the p38 MAPK signals. Furthermore, let-7-Fam miRNAs-reduced resistance to pathogens, also involving the p38 MAPK axis55. In C. elegans, let-7 may influence innate immunity against P. aeruginosa PA14 infection in both the intestine and the neurons56. Muraleedharan et al. found that the miR-183/96/182 cluster modulated the immune response in cornea to bacterial infection via influencing the neuroimmune axis. Expression of miR-183/96/182 in macrophages decreased, while reducing or blocking miR-183/96/182 in macrophages and polymorphonuclear neutrophils (PMNs) increased their ability to phagocytize and kill P. aeruginosa57.
One important feature of P. aeruginosa is its much higher frequency of infections in patients with cystic fibrosis (CF) than most other patient groups and healthy individuals58. miRNA profiles for CF bronchial epithelial IB3-1 cells after P. aeruginosa challenge demonstrated that miR-93, which is highly expressed in basal conditions, reduced along with increased IL-8 levels after infection. Specifically, in addition to increased IL-8 transcription upon NF-κB activation, IL-8 protein levels were modulated via IL-8 mRNA crosstalk with miR-93 at posttranscriptional levels59. The unfolded protein response (UPR) has been reported to play an important role in innate immunity and inflammation, involving the development, differentiation, and survival of immune cells60. P. aeruginosa infection upregulated the levels of miR-233 via p38 MAPK circuits. miR-233 reportedly also has an impact on innate immune response through activation of a UPR-associated protein, sarco/endoplasmic reticulum Ca2+-ATPase (SERCA)61. Future studies may elucidate whether miRNAs have therapeutic efficacies in the treatment of CF.
Mycobacterium
Tuberculosis (TB) is a common infectious disease, with morbidity and mortality rates exceeding tens of millions of people each year. Both the intracellular bacteria Mycobacterium tuberculosis (Mtb) and Mycobacterium bovis (M. bovis) can infect animals and humans and are the most well-investigated mycobacteria62. Concerning innate immune responses, Dorhoi et al. demonstrated that miR-223 controlled tuberculosis susceptibility via impacting recruitment of neutrophils through chemokine (C-C motif) ligand 3 (CCL3), chemoattractant chemokine (C-X-C motif) ligand 2 (CXCL2), and IL6 in myeloid cells. Deletion of miR-223 increased susceptibility to lung infection of Mtb-resistant mice63. Mtb could induce the expression of miR-99 in dendritic cells (DCs). Importantly, Inhibiting miR-99b in DCs dramatically augmented levels of proinflammatory cytokines including IL1β, IL-12, and IL6, and decreased the bacterial burden64. Regarding adaptive immune responses, infection of mice with Listeria monocytogenes or vaccine strain M. bovis bacillus Calmette-Guerin (BCG) largely reduced expression levels of miR-29 in CD8+ T cells, CD4+ T cells, and natural killer cells. miR-29 ihibited IFN-γ production via targeting of IFN-γ mRNA. Thus, miR-29 transgenic mice exhibited stronger Th1 responses and higher resistance to BCG or Mtb infection 65.
Hedgehog (HH) signaling is an important factor for cell fate decisions in various disease conditions. M. bovis BCG-specific TLR2 signaling influences the states of Sonic HH (SHH) signaling in macrophages through TNF-α secretion. Intriguingly, SHH signaling serves as a negative regulator to counteract TLR2 responses in mycobacterial invasion. SHH signaling drives miR-31 and miR-150 expression, which modulates TLR2 levels involving MyD88, a canonical adapter for TLR signaling 66.
As an important miRNA for immune reactions, miR-155 is also increased in macrophages after Mtb and M. bovis BCG infection. Enhancing miR-155 expression augments autophagic influx in macrophages, thereby facilitating mycobacterial phagosome maturation and ROS production and decreasing the survival rate of intracellular mycobacteria67,68. Mice with miR-155-deficiency died much earlier and showed drastically increased colony forming units (CFUs) in their lungs than wild-type mice after Mtb infection69. However, miR-155 could also facilitate the survival of Mtb in macrophages by directly attenuating the expression of BTB and CNC homology 1 (Bach1), a transcriptional repressor of haemoxygenase-1 (HO-1), and SH2-containing inositol 5′-phosphatase 1 (SHIP1), which is important for Mtb survival70. Rothchild et al. dissected the in vitro and in vivo function of miR-155 in impacting both innate and adaptive immunity systems. miR-155 helped improving survival of Mtb-infected macrophages but providing a niche supporting bacteria colonization. However, the miRNA also extended the survival and augmented the function of Mtb-specific T cells to upregulate adaptive immunity. Although miR-155 may render early defense, miR-155-deficient mice may succumb in the late stages of infection 71.
The dynamic expression and function of miRNAs and their isoforms in infection are an important focus in the field of host-Mtb interactions. Siddle et al. performed a genome-wide miRNA transcriptional analysis of human DCs exposed to mycobacteria and other bacteria with different virulence. They revealed some critical elements of miRNA variants in immune reaction against bacterial infection, particularly identifying miR-132/212 family as a vital responder to mycobacteria. Another insight is that infection might differentially impact the expression of each member of the same polycistronic family 72.
M1 and M2 macrophages are functionally polarized subsets of macrophages in various conditions including bacterial infection. Kruppel-like factor 4 (KLF4) is a key molecule for regulating this polarization73. During Mtb infection, downregulation of miR-26a upregulated KLF4, which in turn prevented trafficking of Mtb to lysosomes74. Additionally, infection of macrophages with Mtb and M. bovis BCG led to higher expression of miR-125a75, miR-13276, miR-26a76, miR-146a77,78, and miR-2179, which functioned as negative regulators, whereas miR-20680 as a positive regulator, decreased the expression of miR-let-7f81 in host defense. A nuclear body protein, Sp110 that has been linked to TB resistance, could modulate expression levels of miRNA in macrophages, hence modifying host immune response (miR-146a, miR-155, miR-27b and miR-29a) and apoptosis (miR-125a) in response to infection with Mtb82. Therefore, miRNAs may be emerging mediators of macrophage inflammatory responses to bacterial pathogens.
Listeria monocytogenes
Listeria monocytogenes is an intracellular bacterium that causes serious illness in immunocompromised individuals and pregnant women83. L. monocytogenes can evade miRNA-mediated host defense in various cells. miR-146b, miR-16, let-7a1, miR-145, and miR-155 were significantly dysregulated following Listeria infection in epithelial cells84. Schnitger et al. demonstrated that L. monocytogenes promoted significant changes in miRNA expression in macrophages. miR-146a, miR-155, miR-125a-3p/5p, and miR-149 were among the most altered miRNAs. miR-125a-3p/5p were found to be involved in the TLR2 axis, while transactivation of miR-155 upon infection was influenced by NF-κB p6583. Lind et al. further showed that CD8+ T cells with miR-155 deficiency exhibited unresponsiveness to the AKT signaling after T-cell receptor (TCR) cross-linking responses to L. monocytogenes infection. This suggested that miR-155 is necessary for agitating a proper CD8+ T-cell response85. Strikingly, miR-29 inhibited immune responses of natural killer cells, CD4+ T cells and CD8+ T cells to L. monocytogenes infection by targeting IFN-γ65. In contrast, in macrophages, miR-21 limited the uptake of listeria monocytogenes to control infection by impairing the intracellular niche.
The intestinal tract is thought to be the main reservoir of microbes in humans, but much remains to be determined about the role of the intestinal microbiota in modulating miRNA expression. Archambaud et al. showed that intestinal microbiota could alter the gut miR-143, miR-148a, miR-200b, miR-200c, and miR-378 responses after oral Listeria infection. Moreover, the expression levels of protein-coding target genes were inversely correlated with those of the above-mentioned miRNAs86. Thus, miRNAs may mediate the proinflammatory responses of host immune responses to L. monocytogenes infection.
Staphylococcus aureus
Staphylococcus aureus, a Gram-positive round-shaped bacterium, causes common skin infections, and occasionally causes pneumonia, endocarditis, and osteomyelitis, in humans87. S. aureus produces a spectrum of virulence factors and modifies the protein levels of TGF-β, which may limit the inflammation and tissue injury during infection88. TGF-β induces expression of miR-29b to promote murine alveolar macrophage dysfunction, and miR-29b compromises bacterial killing in macrophages through prostaglandin E2 (PGE2) signaling89. A network of miRNA-gene-pathway interactions could be induced in bovine mammary gland cells in response to invading S. aureus. The increase in bta-miR-223 and bta-miR-21-3p was found in the teat quarters following high dose S. aureus infection. Further analysis suggested critical roles of these two miRNAs in defending hosts against bacterial infection, probably through inhibiting CXCL14 and KIT proto-oncogene receptor tyrosine kinase (KIT)90. Skin wound healing after S. aureus infection might be impaired in miR-142-deficient mice compared to that in wild-type mice. These alterations may be associated with cytoskeletal function, and the levels of the small GTPases were thus drastically enhanced in miR-142-deficient neutrophils91. Bacterial pneumonia after influenza infection is dubbed with high mortality and morbidity92,93. In contrast to the results from other murine infection models44,85, mice with miR-155 deficiency were resistant to infection, with significantly reduced bacterial CFUs and no differences in viral load, along with augmented IL-23 and IL-17 compared to WT mice after sequentially challenged by virus and bacteria, respectively. A miR-155 antagomir application significantly reduced bacterial loads versus control antagomir treatment after sequentially infected by viruses and bacteria92. These studies indicated that the regulation of miR-155 may be modified under different infection conditions.
Other bacterial pathogens
In addition to the above-mentioned infection models, miRNAs have been reported to be involved in host immune responses against the other bacterial pathogens. Despite NF-κB activation being the necessity of miR-155 induction, much remains to be elucidated about the underlying mechanisms of miR-155 induction by various stimuli or pathogens. Cremer et al. showed that de novo synthesis of c-Jun and c-Fos upon NF-κB activation is needed for inducing miR-155 by Francisella novicida, Burkholderia cenocepacia, and Mycobacterium smegmatis stimuli in monocytes94. miR-155 is also critical for effective clearance of primary and secondary Streptococcus pneumoniae colonization via IL-17A and IFN-γ CD4+ T-cell responses95. Interestingly, miR-155 may not participate in the cytokine production induced following Francisella tularensis phagocytosis. Instead, this miRNA may be important for inhibiting endotoxin-stimulated TNF-α secretion96. In P. gingivalis-infected BMMs, mmu-miR-155-5p could markedly decrease the production of TNF-α97. Thus, these data demonstrate that miR-155 may act as a global negative regulator of inflammation during bacterial infection.
miR-15a and miR-16 have been reported to play an important role in bacterial infection-associated sepsis. Deletion of miR-15a/16 in myeloid cells significantly decreased E. coli-associated mortality in several mouse models of sepsis. Consistently, miR-15a/16 overexpression using miRNA mimics decreased both phagocytosis and production of mitochondrial reactive oxygen species. In addition, deficiency of miR-15a/16 boosted secretion of cytokine/chemokine of bone marrow-derived macrophages (BMDMs) at the initial phase of infections98. This is different from miR-15a/16, presumably a restriction factors for Salmonella infection via control of the G1/S phase transition43.
In Neisseria gonorrhoeae infection models, TLR4 instead of TLR3 is required for inducing miR-718 expression in macrophages. miR-718 can impact PI3K/AKT axis through direct downregulation of phosphatase and tensin homolog (PTEN), while increasing AKT phosphorylation and cytokine production. Decrease in miR-718 levels correlated to bacterial burdens during N. gonorrhoeae infection and thus altering the infection dynamics of N. gonorrhoeae in vitro99. Additionally, miR-214 could be significantly upregulated by Vibrio harveyi and LPS stimulation. Upregulating miR-214 subsequently inhibited the production of inflammatory cytokines by targeting MyD88 to avoid excessive inflammation100. However, Chlamydia muridarum infection in mouse genital tracts drastically reduced miR-214, while repressing the expression of intracellular adhesion molecule 1 (ICAM1). The alteration of ICAM1 by miR-214 in mice correlated with a reduction of neutrophil infiltration in genital tissues101. Filopodia are thin actin-rich cell protrusions, and forming host-cell filopodia is critical for phagocytosis and bacterial internalization. miR-29b-2-5p, by inhibiting its direct target UNC5C, rapidly increased filopodia in hosts upon Shigella flexneri infection102. These studies highlight the complicated transcriptional and posttranscriptional response mechanisms of host cells to bacterial infection.
Significance of altered miRNAs in bacterial infection
Host response to pathogens needs fine regulation of various cellular signals including immune signaling103. Given the abnormal miRNA expression in bacterial infection, it has been hypothesized that these dysregulated miRNAs could affect multiple cell physiological functions and pathological processes, depending on their target genes.
TLRs/NF-κB signaling
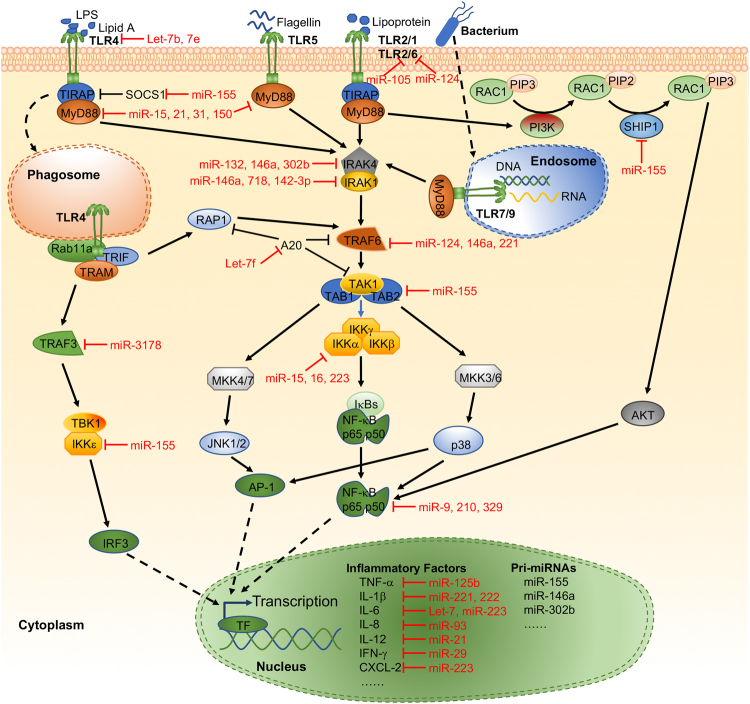
TLRs were the first identified and the most well-investigated PRRs. TLRs sit at the center of innate immunity against almost any pathogens through their PAMP recognition3. After recognizing the PAMPs of pathogens, TLRs can transduce downstream signaling through either MyD88 or TRIF. These TLR-mediated responses may induce secretion of inflammatory cytokines and miRNA expression (Fig. 1). TLR-4 signaling is necessary for enhancing miR-32-5p following Mtb infection. miR-32-5p significantly extended the survival of intracellular mycobacteria104. miR-124 was upregulated in the peripheral leukocytes of patients with pulmonary tuberculosis in both M. bovis- and BCG-infected macrophages in vitro and in vivo. Mechanistically, miR-124 can regulate TLR signaling pathways in macrophages in response to BCG infection105. Many subsets of TLRs and related signals, including TLR6, MyD88, TRAF6, and TNF-α, can be directly impacted by miR-124105,106. miR-3178 mitigated inflammatory response and gastric carcinogenesis that are facilitated by a H. pylori new toxin, Tip-α, via targeting TRAF3107. miR-223 may down-modulate NF-κB activation by inhibiting p65 phosphorylation and nuclear translocation108. miR-329 plays a major part in promoting trophoblast apoptosis induced by S. pneumoniae peptidoglycan (PDG) and inhibiting IL6 mRNA expression involving the NF-κB subunit p65109. miR-210 targets another subunit of NF-κΒ, p50, to inhibit LPS-induced expression of proinflammatory cytokines110. TLR4-activated NF-κB rapidly increases the expression of miR-9 to provide feedback to NF-κB-dependent responses by fine tuning the expression of the NF-κB subunit p50111. IKKα mRNA was targeted by miR-15a, miR-16, and miR-223, which then substantially decreased NF-κB p52 production112. miR-146a could also directly target TRAF6 after transcription and ameliorate the activation of NF-κB and p38 MAPK circuits during BCG challenge. miR-146a increase may block inducible nitric oxide (NO) synthase (iNOS) expression and NO generation, thereby enhancing mycobacterial survival in macrophages78. In E. coli, Mycobacterium and Helicobacter infection models, miR-146a downregulated the expression of the target genes IRAK1 and TRAF6, thus downregulating LPS levels and bacteria-induced production of cytokines and chemokines, endocytosis and lysosome trafficking30,31,113–115. Furthermore, enforced expression of miR-146a generates a tolerance to lipoprotein-, and leading to a significantly decreased expression of in IRAK-1 and phosphorylated IκBα in S. typhimurium-infected or control THP-1 cells 116.
Fig. 1.
Representative miRNAs in the regulation of TLR signaling. TLRs recognize different bacterial components and induce NF-κB signaling or activate other transcription factors through adapter molecules and downstream signaling molecules. Various inflammatory factors are transcribed that are initiated by different transcription factors. The transcription of miRNAs is most commonly mediated by RNA polymerase II, under the control of transcription factors, and transcripts are then processed by two nucleases, Drosha and Dicer. Then, the mature miRNAs will be incorporated into the RISC and guide the RISC to their target mRNA(s) in cytoplasm. Both early- and late-phase-activated TLRs induce different types and expression levels of inflammatory factors and miRNAs. TF, transcription factor
The involvement of miR-155 in TLR/NF-κB signaling has also been reported. miR-155 downregulated IKK-ε, and increased miR-155 levels downregulated the production of IL-8 and GRO-α in gastric epithelial cells after H. pylori infection24. SHIP1 is found to be a main target of miR-155, and knocking down endogenous SHIP1 led to AKT activation in response to LPS117. In addition, miR-142-3p may have a role in inhibiting proinflammatory mediators NF-κB p50, TNF-α, and IL6 in BCG-challenged macrophages, possibly via targeting IRAK-1118. Finally, the TLR adapter MyD88 may also serve as a target of miRNAs. miR-150 targets MyD88, thus leading to suppression of TLR responses in macrophages66. miR-214 inhibited the production of inflammatory cytokines by targeting MyD88 to avoid excessive inflammation in Vibrio harveyi-infected and LPS-treated fish 100.
Several miRNAs can directly target TLRs to modulate immunologic processes. Importantly, Let-7b is partially complementary to the TLR4 mRNA 3′-UTR and therefore influence TLR4 levels at the posttranscriptional levels in gastric epithelium. Overexpression of let-7b reduced TLR4 and subsequently mitigated H. pylori-induced activation of NF-κB, MyD88, NF-κB1/p50, and RelA/p6515. In macrophages, AKT1 regulates LPS-induced let-7e expression, and let-7e regulates endotoxin sensitivity and tolerance by interacting with the 3’-UTR of TLR4119. miR-124 could also play a negative regulatory role for inflammatory responses in macrophages upon mycobacterial infection by directly targeting TLR6105,106. In a Porphyromonas gingivalis infection model, miR-105 had complementarity for TLR-2 mRNA and inhibited TLR-2 protein translation in human gingival keratinocytes120. TLR2 could also be targeted by miR-23a-5p to modulate survival of mycobacteria and activation of autophagy pathways 121.
In addition to the well-investigated miRNAs targeting positive signaling cascade molecules, miRNAs could also target negative regulators during bacterial infection. For example, Mtb-triggered let-7f could target A20, which can inhibit NF-κB 81.
Autophagy
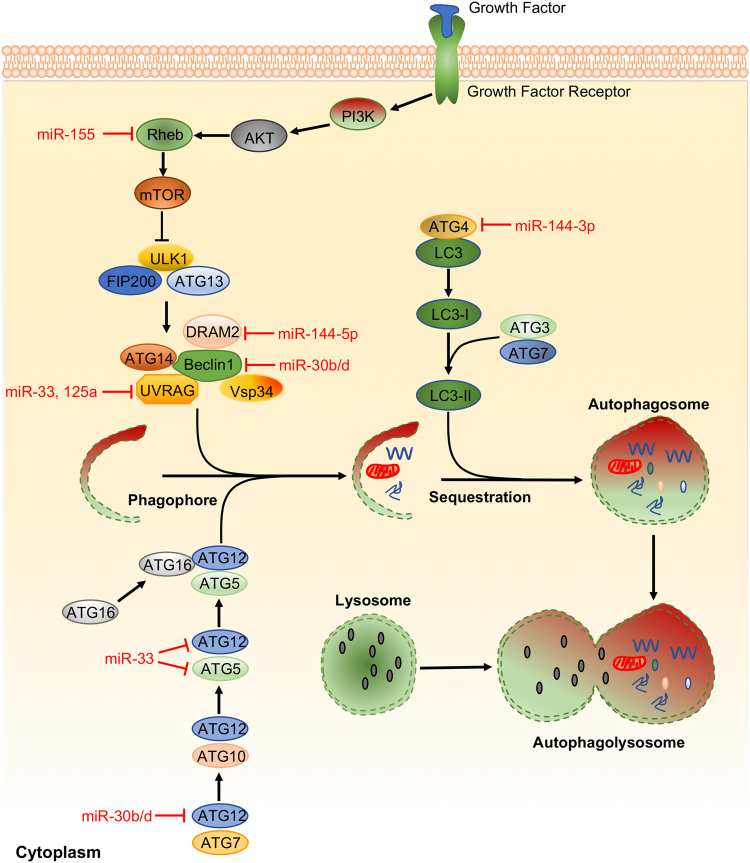
Autophagy is a vital measure of eukaryotic cells that maintains cellular homeostasis to recycle nutrients and degrade damaged or aged cytoplasmic constituents, which also impacts the survival of bacterial pathogens103. It has become increasingly recognized that abnormal autophagy may contribute to defense against bacterial infection (Fig. 2). BCL2-interacting coiled-coil protein (BECN1) and autophagy-related protein 12 (ATG12) are two important proteins that guide other autophagy proteins to the pre-autophagosomal membrane and subsequently facilitating phagophore elongation and autophagosomes maturation17. Two groups have reported that H. pylori infection increases miR-30b/d and that compromised autophagy by miR-30b/d promotes bacterial replication by targeting BECN1 and ATG1217,122. DNA damage-regulated autophagy modulator 2 (DRAM2) is shown to interact with BECN1 as a coordinator of autophagy activation. miR-144-5p inhibited antimicrobial responses against Mtb in human monocytes and macrophages by targeting DRAM2 123.
Fig. 2.
Representative miRNAs in the regulation of autophagy. Autophagy is an important immune response used to eliminate bacterial pathogens. In turn, bacterial pathogens have also developed the ability to subvert host autophagy by interfering with autophagy signaling. Deregulation of miRNAs can occur as the result of interplay between bacterial factors and autophagy components. miRNAs that target autophagy-related proteins function as a specialized immunologic effector and effectively regulate host innate immune responses for the elimination of pathogenic bacteria
These data demonstrated that mycobacterial challenge may augment miR-155 levels, and miR-155 increase induces autophagy through targeting Ras homolog enriched in brain (Rheb), a negative autophagy modulator. miR-155 activates autophagy and facilitating phagosome maturation after mycobacterial internalization into macrophages, thus killing intracellular mycobacteria67. However, another group reported no difference in formation of autophagosomes in WT control and miR-155−/− macrophages after virulent Mtb infection by assessing LC3I to LC3II conversion. Their results suggest that inhibiting SHIP1 via miR-155 improves viability in macrophages and reduces bacterial burdens71. In contrast, miR-125a could target UV radiation resistance-associated gene (UVRAG) to inhibit autophagy activation and antimicrobial responses to Mtb75. Mtb utilized miR-33 to downregulate autophagy and reshape lipid metabolism in hosts to enhance intracellular survival and persistence by influencing critical autophagy components, such as ATG5, ATG12, and UVRAG124. Mtb infection could also lead to inhibition of miR-17 and concomitant increase of its targets, myeloid cell leukemia sequence 1 (Mcl-1) and signal transducer and activator of transcription 3 (STAT3), a transcriptional activator of Mcl-1. miR-17 overexpression reduced phosphorylation of protein kinase C delta (PKCδ) and attenuated autophagy during Mtb infection125. Furthermore, BCG-challenged macrophages exhibited higher levels of miR-144-3p, which targets ATG4a and inhibits antimicrobial defense126. Collectively, these findings suggest that dysregulated miRNAs may be the result of a complex interplay between bacterial factors and autophagy components (Fig. 2).
Apoptosis
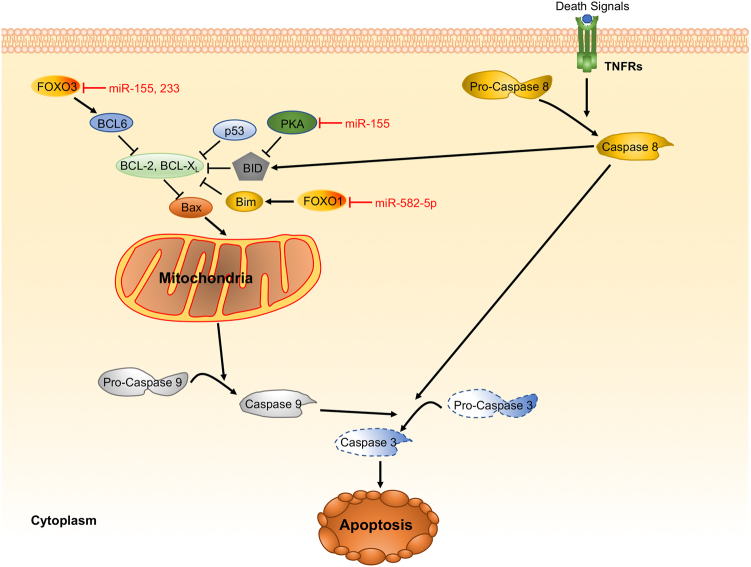
Apoptosis is considered as programmed cell death that may play a part in host defense against pathogens. Apoptosis is typically identified with cell shrinkage, DNA fragmentation, mitochondrial permeability, membrane blebbing, and particularly activated caspases (caspase 3)127. Intracellular pathogens may modulate host apoptosis signaling to facilitate their proliferation and evade host defenses. Indeed, numerous miRNAs have been reported to regulate apoptosis-related genes during bacterial infection (Fig. 3). Three highly regulated genes (Lpin1, Pmaip1, and Tspan14) associated with apoptosis/DNA damage have been validated as the targets of miR-155 in H. pylori-infected BMMs. miR-155−/− BMMs succumbed to apoptosis upon cisplatin treatment versus wild-type counterparts. Furthermore, H. pylori-infected miR-155−/− BMMs showed poly (ADP ribose) polymerase (PARP)/procaspase-3 cleavage levels similar to controls21. BCG challenge-augmented miR-155 was dependent on TLR2-associated signals. miR-155 also impacts PKA signaling axis by targeting PKI-α, which is a negative modulator of PKA. Notably, miR-155-driven PKA circuit enhanced apoptotic effector activities, thus leading to apoptosis in BCG-infected macrophages128. Huang et al. reported that miR-155 inhibited apoptosis of monocytes by targeting Forkhead Box O3 (FOXO3) using an Mtb infection model. Interestingly, miR-233 has similar functions with miR-155 by targeting the same gene129,130.
Fig. 3.
Representative miRNAs in the regulation of apoptosis. Apoptosis has been observed as a response to infection by a wide range of bacterial pathogens. Bacteria can activate several proapoptotic proteins and miRNAs to induce apoptosis. Deregulated miRNAs in infection by bacterial pathogens are involved in networks that control innate immunity and apoptosis pathways of their host cells
miR-582-5p highly expressed in monocytes was found to be increased in patients with active TB. THP-1 cells transfected with miR-582-5p mimics were much less apoptotic than the cells transfected with a negative control, suggesting miR-582-5p being an inhibitor of apoptosis, which may be through inhibition of FOXO1 expression. Via this mechanism, miR-582-5p should impact immune responses to Mtb infection131. MPT64, a secreted protein by Mtb also impeded apoptosis of RAW264.7 macrophages via a NF-κB/miR-21/BCL-2 mechanism. NF-κB was assumed to be the chief transcription factor for miR-21-mediated gene regulation. miR-21 then facilitates BCL-2 expression to block apoptosis132. These results suggest that miRNAs may participate in control of bacteria-mediated apoptosis.
Crosstalk between miRNAs and signaling pathways
As mentioned above, one miRNA may target many mRNAs, and a single mRNA may also be regulated by several or many miRNAs. The target mRNAs of one miRNA usually involve different signaling pathways, and one miRNA can be induced by different PAMPs. It has been speculated that there is a common signaling crosstalk mechanism for mounting protective immune responses against infection. For instance, it has been reported that TLR signaling links the autophagy pathway to phagocytosis and apoptosis in macrophages133,134. It appears that TLR2 and MyD88 are critical to the biogenesis of miR-125a during Mtb infection. miR-125a inhibited the induction of autophagy in macrophages by suppressing UVRAG protein expression, which is essential for the coordination of autophagic maturation and endocytic trafficking75. Wu et al. showed that SP110 nuclear body protein downregulated miR-125a in macrophages, but upregulated Bcl2-modifying factor (Bmf), an apoptosis-inducing protein. miR-125a interacts with the Bmf 3′-UTR to inhibit Bmf expression, which suppressed Mtb-induced macrophage apoptosis82. As we can see from the above description, miRNA roles in bacterial infection are mainly involved in host immune responses against pathogens, which are different from those in several other diseases, such as cancers. Indeed, the same miRNA may play a different role in different models through the same target(s). miR-146a could negatively regulate NF-κB activation by inhibiting the expression of IRAK1 and TRAF6, significantly inhibiting breast tumor growth135. It can also modulate innate immune responses via targeting IRAK1 and TRAF6 in bacterial infection models115. This functional diversity of miR-146a may be caused by crosstalk between different signaling pathways. Therefore, better understanding of the crosstalk mechanisms underlying the deregulation of miRNAs during bacterial infection is crucial for improving our understanding of immune signaling.
MiRNAs as biomarkers for bacterial infection
Diagnosis of infection is important for the control of the spread of bacterial invasion and for effective treatment of infection136. It is demonstrated that miRNAs are stable in patients’ serum. miRNAs are hard to degrade by RNases, and hence can be used as invaluable biomarkers to detect bacterial infection at earlier stages. Studies showed that 97 miRNAs expressed in a unique manner in pulmonary TB patient sera versus healthy individuals (90 increase while 7 decrease). Among these changed products, miR-361-5p, miR-889 and miR-576-3p stood out to differentiate TB patients from normal subjects or different pathogenic infections136. In another study, a larger size of subjects (326 serum samples) were enrolled to search for biomarkers for TB infection in the lung. The authors identified six different miRNA products (miR-101, miR-22, miR-29c, miR-320b miR-378, and miR-483-5p) in the serum as specific biomarkers to diagnose TB lung infection versus normal subjects. A reasonable sensitivity (~95.0%) and specificity (~91.8%) was achieved by combining the 6 miRNAs versus using single miRNA 137.
Nevertheless, studying miR-29a expression in human TB cases is still controversial. Fu et al. employed specific miRCURY LNA microarrays to compare the levels of circulating miRNAs between patients with active pulmonary TB and their healthy counterparts. The analysis of the ROC curve showed that upregulated miR-29a may distinguish TB patients from healthy controls138. In contrast, another group reported decreased expression of miR-26a, miR-29a, and miR-142-3p in whole blood in children with tuberculosis compared to healthy children with latent Mtb infection (LTBI)139. However, Awuah et al. concluded that although median miR-29a expression was slightly higher in TB patients, there was no significant difference compared with LTBI patients140. These plausible results might be due to choices of patients and starting material for miRNA quantification.
The miRNA expression profile may have several differences between children and adults. The diagnostic value of miRNA-31 in peripheral blood mononuclear cells (PBMCs) of sixty-five children with pulmonary tuberculosis has been reported. Studies have shown that miRNA-31 in pediatric TB patients exhibited less expression compared to that in normal controls. Importantly, serum miRNA-31 levels correlated inversely with production of IL6, TNF-α, NF-κB, and IFN-γ 141.
Multidrug-resistant (MDR) Mtb could result in extended, complicated disease, leading to increased treatment costs or treatment failure. A comparative miRNA analysis was conducted and indicated 142 miRNAs expressed in a different manner between an MDR Mtb strain and a sensitive Mtb strain, (48 increased while 94 decreased). Importantly, only six miRNAs were similarly expressed between the MDR and sensitive Mtb strains, whereas 108 miRNAs were only observed in the MDR Mtb strain142. Profiling miRNAs was carried out in plasma samples from cavitary pulmonary tuberculosis (CP-TB) patients, non-cavitary pulmonary tuberculosis (NCP-TB) patients and normal individuals, which revealed candidate biomarkers (miR-769-5p, miR-320a and miR-22-3p) for diagnosis of TB. Additionally, miR-320a may be useful for diagnosing drug-resistant TB143. miR-155 levels were lower in patients with MDR TB than healthy subjects. Also, this miRNA was increased in treated patients versus naive patients. miR-16 levels were the lowest in serum of MDR TB patients compared to TB-naive, TB-treated and healthy control groups144. Thus, analyzing miRNA expression patterns in MDR and drug-sensitive Mtb may reveal novel mechanisms of drug resistance in TB research.
Several other serum miRNAs are also differentially regulated upon bacterial infection as biomarkers. Serum miR-133a was the highest expressed in mice of a cecal pole ligation and puncture sepsis model, and establishing the strong correlations between miR-133a and disease severity, classical markers of inflammation and bacterial infection, and organ failure of patients with sepsis145. miR-155 and miR-197 may possess superior values in distinguishing patients with pneumonia and TB from controls146. miR-144 levels in sputum and serum were shown to be increased in TB patients, but markedly decreased following anti-Mtb treatment147. miR-155 and miR-155* preferentially expressed in PBMCs of tuberculosis patients showing Mtb-specific antigen, indicating their diagnostic potential for specific Mtb antigens148. Moreover, miR-155 was increased in both early (week 2) and late (week 11) M. bovis-infected cattle, whereas upregulation was only detected in late stages of BCG-vaccinated cattle. This suggested that miR-155 may be used as a prognostic marker for distinguishing vaccinated from controls149. miR-183 was increased in serum samples from TB patients versus healthy subjects. Interestingly, miR-183 expression was positively correlated with macrophage function, as shown by their augmented phagocytosis and enzymatic activity in the group with high serum miR-183 150.
Further studies have also profiled miRNA expression in other clinical samples. Let-7c expression decreased in samples ranging from non-atrophic gastritis to atrophic-metaplastic gastritis, intra-epithelial neoplasia, and invasive GC. It increased again significantly following H. pylori eradication14. After the identification of miRNAs and their target genes in normal gastroduodenal biopsy, H. pylori-infected gastroduodenal biopsy and H. pylori-infected gastroduodenal ulcer biopsy samples, Cheng et al. found that increase in miR-155 and miR-146b could decrease H. pylori-induced IL6 expression in gastroduodenal ulcer. This relationship between miR-155 and miR-146b and IL6 might reduce the clearance of H. pylori and contribute to ulcer development and maintenance151. Exosomes, as a rich source of miRNAs, can protect miRNAs from degradation, and have the potential to be a very promising biomarker. Sun et al. detected miRNA levels in bovine milk exosomes derived from lactating Holstein cows infected by S. aureus. They reported that bta-miR-142-5p and bta-miR-223 expressed differently as exosomal miRNAs, and may be potential biomarkers for the early diagnosis of bacterial infection, particularly for mammary glands 152.
Single-nucleotide polymorphisms (SNPs) in the processing sites of miRNAs may affect miRNA expression and function, which are involved in the pathogenesis of infectious diseases153,154. However, different ethnic groups may have distinct attributes for this type of genetic epidemiological research. For instance, the miR-499 rs3746444 T > C rather than miR-146a rs2910164 C > G likely led to increased infection incidence in the lung in Uygur Chinese. Kazak Chinese exhibited a marked different SNP frequency compared to Uygur (miR-146a C > G, but not miR-499 T > C). These same SNPs may also be related to Mtb in Tibetan people. Despite discrepant reports, the analysis of mRNA SNPs may be not a good measure for evaluating Mtb sensitivity in Southern Han Chinese 155,156.
miRNAs may serve as a valuable diagnostic marker that may be useful only for specific bacterial infections. Accumulating evidence suggests that almost all the miRNA biomarkers evaluated to date have been for the diagnosis of pulmonary tuberculosis and H. pylori-associated gastritis, which may reflect differences in the field difference due to intense interest in these bacteria. Pulmonary tuberculosis, H. pylori-associated gastritis and other specific bacterial diseases with clear induction factors would be more suitable for early diagnosis via miRNA expression profiling. However, the accuracy of detecting miRNA levels among different samples remains quite challenging. A study of whole-blood miRNA features for the effective early diagnosis of pulmonary tuberculosis revealed that evaluating certain specialized miRNAs in combination could be accomplished with reasonable sensitivities and specificities 157.
MiRNAs as therapeutic targets for bacterial infection
The revelation that miRNAs function as an important regulator in bacterial infection suggests their great application potential as novel therapeutic targets. Indeed, many miRNAs have been developed and investigated in clinical trials for several types of diseases. The first miRNA-based drugs, MRX34, entered a phase 1 trial in patients with primary liver cancer or metastatic cancer in 2013158. Miravirsen, a 15-nucleotide locked nucleic acid-modified antisense oligonucleotide that can sequester and thus inhibit miR-122, could induce significant virologic responses after subcutaneous injections in patients with chronic HCV infection159. Though there are no bacterial infection-related miRNA-based drugs evaluated in clinical research to date, miRNAs still represent a promising approach for future therapies or as immune modulators against invading pathogens. Several miRNAs with excellent immune regulation efficiency, such as miR-33124, miR-15544, miR-2965, miR-146a115, and miR-302b52, may have great developmental potential for further clinical studies.
The application of miRNA-based therapeutics still faces many hurdles before it can be translated into clinical practice for bacterial infections. The spatio-temporal expression of miRNAs and their targets at different stages during infection must first be precisely identified. If the miRNAs that can target numerous genes can be specifically designed, this could represent a significant advantage of miRNA-targeted approaches. As the multicenter phase I clinical study of MRX34 was terminated due to five immune-related serious adverse events last year, the efficacy and safety of miRNA-based drugs need to be carefully assessed160. The tissue/target specificity delivery and stability of miRNA-based drugs is another current limiting factor for satisfactory therapeutic effects. In addition to well-investigated virus and non-virus delivery systems, recent reports indicate that exosomes may be an emerging high-efficiency delivery approach 161,162.
Conclusion and perspectives
miRNA research has provided a unique angle for studying the mechanisms underlying how the innate immune system senses and responds to microbial pathogens. Over the past decade, the role of miRNAs in bacterial pathogen infection has greatly enhanced our understanding of cellular physiology and immunology. In particular, miR-155 and miR-146 are the two most well-understood miRNAs, with well-characterized roles in immunity and inflammation regulation during bacterial infection.
However, due to the complexity of bacterial infections, one miRNA may regulate several different targets in different stages during bacterial infection. The precise mechanism underling the regulatory function of miRNAs must be explored. Future studies are needed to further clarify how miRNAs with diverse targets may affect overall host responses during infection. In certain instances, the continuous development of pathological processes is accompanied by changes in the miRNA expression profile. It is necessary to distinguish that are the most important regulators and which intervention strategy is the most effective.
Bacterial pathogens have developed a variety of virulence factors to facilitate bacterial colonization in their hosts, invade deeper tissues and evade host defenses163,164. Currently, researchers have only identified bacterial infections that can cause changes in the expression of a variety of miRNA. Many miRNAs could be induced during infection with different types of bacteria. It has been noted that several aberrant miRNAs during bacterial infection are also dysregulated in other diseases. For example, levels of miR-155 are altered upon infection with P. aeruginosa, Mtb and H. pylori, and miR-155 plays a role in host immune responses23,54,71. Many reports have shown that miR-155 is involved in several other diseases, such as rheumatoid arthritis165, breast cancer166, atherosclerosis167, Crohn’s disease168, and others. The specificity of miRNA expression induced by certain bacteria or their virulence factors requires more investigation. This would be of the utmost importance for the application of unique miRNAs for diagnosis and treatment.
Furthermore, mutations within miRNAs may also alter their target selection, thereby preventing them from inhibiting tuberculosis-related genes, thus increasing host susceptibility to disease. Amila et al. investigated the genetic association of pulmonary tuberculosis with six human miRNA genes that have been predicted to interact with tuberculosis genes. However, this study did not show differences in the sequences compared with healthy individuals without antecedents of tuberculosis169. Despite the negative results in this study, larger samples are needed for clarification in future studies, especially in bacterial pathogen-susceptible or resistant populations. Even in the absence of clinical symptoms, several host-adapted bacterial pathogens (e.g., Mtb, S. typhimurium and H. pylori) are capable of maintaining infections in mammalian hosts even in the presence of inflammation, specific antimicrobial mechanisms and a robust adaptive immune response, which may be due to persistent infections170,171. However, the role of miRNAs in the fundamental genetics of bacterial persistence in the presence of immuno-surveillance has only recently begun to be clarified.
The relationship between miRNAs and bacterial pathogens and the underlying mechanisms urgently require broader investigation. Hence, the elucidation of the function of miRNAs on host–pathogen interactions may lead to the discovery of novel and effective preventive measures and the development of rational therapeutic strategies.
Acknowledgements
This project was supported by the NIH R01 AI101973-01 and P20 GM113123 to M.W.
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Xikun Zhou, Email: xikunzhou@scu.edu.cn.
Min Wu, Phone: +701 777 4875, Email: min.wu@med.und.edu.
References
- 1.Didelot X, Walker AS, Peto TE, Crook DW, Wilson DJ. Within-host evolution of bacterial pathogens. Nat. Rev. Microbiol. 2016;14:150–162. doi: 10.1038/nrmicro.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanneganti TD. Central roles of NLRs and inflammasomes in viral infection. Nat. Rev. Immunol. 2010;10:688–698. doi: 10.1038/nri2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nish S, Medzhitov R. Host defense pathways: role of redundancy and compensation in infectious disease phenotypes. Immunity. 2011;34:629–636. doi: 10.1016/j.immuni.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 7.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct. Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl Acad. Sci. USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, et al. Cellular microRNAs up-regulate transcription via interaction with promoter TATA-box motifs. RNA. 2014;20:1878–1889. doi: 10.1261/rna.045633.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat. Rev. Drug. Discov. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 12.Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136:1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polk DB, Peek RM., Jr. Helicobacter pylori: gastric cancer and beyond. Nat. Rev. Cancer. 2010;10:403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fassan M, et al. Let-7c down-regulation in Helicobacter pylori-related gastric carcinogenesis. Oncotarget. 2016;7:4915–4924. doi: 10.18632/oncotarget.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teng GG, et al. Let-7b is involved in the inflammation and immune responses associated with Helicobacter pylori infection by targeting Toll-like receptor 4. PLoS ONE. 2013;8:e56709. doi: 10.1371/journal.pone.0056709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi Y, et al. CagA mediates epigenetic regulation to attenuate let-7 expression in Helicobacter pylori-related carcinogenesis. Gut. 2013;62:1536–1546. doi: 10.1136/gutjnl-2011-301625. [DOI] [PubMed] [Google Scholar]
- 17.Tang B, et al. Compromised autophagy by MIR30B benefits the intracellular survival of Helicobacter pylori. Autophagy. 2012;8:1045–1057. doi: 10.4161/auto.20159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiga K, et al. Epigenetic silencing of miR-210 increases the proliferation of gastric epithelium during chronic Helicobacter pylori infection. Nat. Commun. 2014;5:4497. doi: 10.1038/ncomms5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang YM, et al. Helicobacter pylori-induced posttranscriptional regulation of H-K-ATPase alpha-subunit gene expression by miRNA. Am. J. Physiol. Gastrointest. Liver. Physiol. 2014;306:G606–613. doi: 10.1152/ajpgi.00333.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie G, et al. Helicobacter pylori promote B7-H1 expression by suppressing miR-152 and miR-200b in gastric cancer cells. PLoS ONE. 2017;12:e0168822. doi: 10.1371/journal.pone.0168822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch M, Mollenkopf HJ, Klemm U, Meyer TF. Induction of microRNA-155 is TLR- and type IV secretion system-dependent in macrophages and inhibits DNA-damage induced apoptosis. Proc. Natl Acad. Sci. USA. 2012;109:E1153–1162. doi: 10.1073/pnas.1116125109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oertli M, et al. MicroRNA-155 is essential for the T cell-mediated control of Helicobacter pylori infection and for the induction of chronic Gastritis and Colitis. J. Immunol. 2011;187:3578–3586. doi: 10.4049/jimmunol.1101772. [DOI] [PubMed] [Google Scholar]
- 23.Fassi Fehri L, et al. Helicobacter pylori induces miR-155 in T cells in a cAMP-Foxp3-dependent manner. PLoS ONE. 2010;5:e9500. doi: 10.1371/journal.pone.0009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao B, et al. Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. J. Infect. Dis. 2009;200:916–925. doi: 10.1086/605443. [DOI] [PubMed] [Google Scholar]
- 25.Hoces de la Guardia A, et al. Inflammatory cytokine and microRNA responses of primary human dendritic cells cultured with Helicobacter pylori strains. Front Microbiol. 2013;4:236. doi: 10.3389/fmicb.2013.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z, et al. MicroRNA-146a negatively regulates PTGS2 expression induced by Helicobacter pylori in human gastric epithelial cells. J. Gastroenterol. 2013;48:86–92. doi: 10.1007/s00535-012-0609-9. [DOI] [PubMed] [Google Scholar]
- 27.Saito Y, et al. Overexpression of miR-142-5p and miR-155 in gastric mucosa-associated lymphoid tissue (MALT) lymphoma resistant to Helicobacter pylori eradication. PLoS ONE. 2012;7:e47396. doi: 10.1371/journal.pone.0047396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lv X, et al. A multi-epitope vaccine CTB-UE relieves Helicobacter pylori-induced gastric inflammatory reaction via up-regulating microRNA-155 to inhibit Th17 response in C57/BL6 mice model. Hum. Vaccine Immunother. 2014;10:3561–3569. doi: 10.4161/hv.36096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu K, et al. MicroRNA-155-enhanced autophagy in human gastric epithelial cell in response to Helicobacter pylori. Saudi J. Gastroenterol. 2016;22:30–36. doi: 10.4103/1319-3767.173756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z, et al. Upregulated microRNA-146a negatively modulate Helicobacter pylori-induced inflammatory response in human gastric epithelial cells. Microbes Infect. 2010;12:854–863. doi: 10.1016/j.micinf.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Li N, et al. H. pylori related proinflammatory cytokines contribute to the induction of miR-146a in human gastric epithelial cells. Mol. Biol. Rep. 2012;39:4655–4661. doi: 10.1007/s11033-011-1257-5. [DOI] [PubMed] [Google Scholar]
- 32.Feng Y, et al. FoxM1 is overexpressed in Helicobacter pylori-induced gastric carcinogenesis and is negatively regulated by miR-370. Mol. Cancer Res. 2013;11:834–844. doi: 10.1158/1541-7786.MCR-13-0007. [DOI] [PubMed] [Google Scholar]
- 33.Kang DW, et al. MicroRNA-320a and microRNA-4496 attenuate Helicobacter pylori cytotoxin-associated gene A (CagA)-induced cancer-initiating potential and chemoresistance by targeting beta-catenin and ATP-binding cassette, subfamily G, member 2. J. Pathol. 2017;241:614–625. doi: 10.1002/path.4866. [DOI] [PubMed] [Google Scholar]
- 34.Zhou X, Xu G, Yin C, Jin W, Zhang G. Down-regulation of miR-203 induced by Helicobacter pylori infection promotes the proliferation and invasion of gastric cancer by targeting CASK. Oncotarget. 2014;5:11631–11640. doi: 10.18632/oncotarget.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X, Li L, Su J, Zhang G. Decreased miR-204 in H. pylori-associated gastric cancer promotes cancer cell proliferation and invasion by targeting SOX4. PLoS ONE. 2014;9:e101457. doi: 10.1371/journal.pone.0101457. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Miao L, Liu K, Xie M, Xing Y, Xi T. miR-375 inhibits Helicobacter pylori-induced gastric carcinogenesis by blocking JAK2-STAT3 signaling. Cancer Immunol. Immunother. 2014;63:699–711. doi: 10.1007/s00262-014-1550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geng Y, et al. MicroRNA-27b suppresses Helicobacter pylori-induced gastric tumorigenesis through negatively regulating Frizzled7. Oncol. Rep. 2016;35:2441–2450. doi: 10.3892/or.2016.4572. [DOI] [PubMed] [Google Scholar]
- 38.Gunn JS. Salmonella host-pathogen interactions: a special topic. Front Microbiol. 2011;2:191. doi: 10.3389/fmicb.2011.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malik-Kale P, et al. Salmonella - at home in the host cell. Front Microbiol. 2011;2:125. doi: 10.3389/fmicb.2011.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoeke L, et al. Intestinal Salmonella typhimurium infection leads to miR-29a induced caveolin 2 regulation. PLoS ONE. 2013;8:e67300. doi: 10.1371/journal.pone.0067300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ordas A, et al. MicroRNA-146 function in the innate immune transcriptome response of zebrafish embryos to Salmonella typhimurium infection. BMC Genomics. 2013;14:696. doi: 10.1186/1471-2164-14-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulte LN, Eulalio A, Mollenkopf HJ, Reinhardt R, Vogel J. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 2011;30:1977–1989. doi: 10.1038/emboj.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maudet C, et al. Functional high-throughput screening identifies the miR-15 microRNA family as cellular restriction factors for Salmonella infection. Nat. Commun. 2014;5:4718. doi: 10.1038/ncomms5718. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang T, et al. Salmonella enterica serovar enteritidis modulates intestinal epithelial miR-128 levels to decrease macrophage recruitment via macrophage colony-stimulating factor. J. Infect. Dis. 2014;209:2000–2011. doi: 10.1093/infdis/jiu006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bao H, et al. Genome-wide whole blood microRNAome and transcriptome analyses reveal miRNA-mRNA regulated host response to foodborne pathogen Salmonella infection in swine. Sci. Rep. 2015;5:12620. doi: 10.1038/srep12620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao M, et al. Regulation signature of miR-143 and miR-26 in porcine Salmonella infection identified by binding site enrichment analysis. Mol. Genet. Genomics. 2016;291:789–799. doi: 10.1007/s00438-015-1146-z. [DOI] [PubMed] [Google Scholar]
- 48.Wu G, et al. Cecal MicroRNAome response to Salmonella enterica serovar Enteritidis infection in White Leghorn Layer. BMC Genomics. 2017;18:77. doi: 10.1186/s12864-016-3413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou X, et al. Transient receptor potential channel 1 deficiency impairs host defense and proinflammatory responses to bacterial infection by regulating protein kinase cα signaling. Mol. Cell. Biol. 2015;35:2729–2739. doi: 10.1128/MCB.00256-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hirsch EB, Tam VH. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev. Pharm. Outcomes Res. 2010;10:441–451. doi: 10.1586/erp.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mun J, et al. MicroRNA-762 is upregulated in human corneal epithelial cells in response to tear fluid and Pseudomonas aeruginosa antigens and negatively regulates the expression of host defense genes encoding RNase7 and ST2. PLoS ONE. 2013;8:e57850. doi: 10.1371/journal.pone.0057850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou X, et al. MicroRNA-302b augments host defense to bacteria by regulating inflammatory responses via feedback to TLR/IRAK4 circuits. Nat. Commun. 2014;5:3619. doi: 10.1038/ncomms4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, et al. Pseudomonas aeruginosa infection augments inflammation through miR-301b repression of c-Myb-mediated immune activation and infiltration. Nat. Microbiol. 2016;1:16132. doi: 10.1038/nmicrobiol.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang K, et al. miR-155 suppresses bacterial clearance in Pseudomonas aeruginosa-induced keratitis by targeting Rheb. J. Infect. Dis. 2014;210:89–98. doi: 10.1093/infdis/jiu002. [DOI] [PubMed] [Google Scholar]
- 55.Ren Z, Ambros VR. Caenorhabditis elegans microRNAs of the let-7 family act in innate immune response circuits and confer robust developmental timing against pathogen stress. Proc. Natl Acad. Sci. USA. 2015;112:E2366–2375. doi: 10.1073/pnas.1422858112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhi L, Yu Y, Li X, Wang D, Wang D. Molecular control of innate immune response to pseudomonas aeruginosa infection by intestinal let-7 in caenorhabditis elegans. PLoS. Pathog. 2017;13:e1006152. doi: 10.1371/journal.ppat.1006152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muraleedharan CK, et al. Inactivation of the miR-183/96/182 cluster decreases the severity of pseudomonas aeruginosa-induced keratitis. Invest. Ophthalmol. Vis. Sci. 2016;57:1506–1517. doi: 10.1167/iovs.16-19134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Folkesson A, et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat. Rev. Microbiol. 2012;10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 59.Fabbri E, et al. Expression of microRNA-93 and Interleukin-8 during Pseudomonas aeruginosa-mediated induction of proinflammatory responses. Am. J. Respir. Cell Mol. Biol. 2014;50:1144–1155. doi: 10.1165/rcmb.2013-0160OC. [DOI] [PubMed] [Google Scholar]
- 60.Grootjans J, Kaser A, Kaufman RJ, Blumberg RS. The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 2016;16:469–484. doi: 10.1038/nri.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dai LL, Gao JX, Zou CG, Ma YC, Zhang K. Q. mir-233 modulates the unfolded protein response in C. elegans during Pseudomonas aeruginosa infection. PLoS. Pathog. 2015;11:e1004606. doi: 10.1371/journal.ppat.1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim JK, Kim TS, Basu J, Jo EK. MicroRNA in innate immunity and autophagy during mycobacterial infection. Cell. Microbiol. 2017;19:e12687. doi: 10.1111/cmi.12687. [DOI] [PubMed] [Google Scholar]
- 63.Dorhoi A, et al. MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J. Clin. Invest. 2013;123:4836–4848. doi: 10.1172/JCI67604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singh Y, et al. Mycobacterium tuberculosis controls microRNA-99b (miR-99b) expression in infected murine dendritic cells to modulate host immunity. J. Biol. Chem. 2013;288:5056–5061. doi: 10.1074/jbc.C112.439778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma F, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat. Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 66.Ghorpade DS, et al. Sonic hedgehog-dependent induction of microRNA 31 and microRNA 150 regulates Mycobacterium bovis BCG-driven toll-like receptor 2 signaling. Mol. Cell. Biol. 2013;33:543–556. doi: 10.1128/MCB.01108-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J, et al. MicroRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting Rheb. PLoS. Pathog. 2013;9:e1003697. doi: 10.1371/journal.ppat.1003697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J, et al. MicroRNA-155 induction by Mycobacterium bovis BCG enhances ROS production through targeting SHIP1. Mol. Immunol. 2014;62:29–36. doi: 10.1016/j.molimm.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 69.Iwai H, et al. MicroRNA-155 knockout mice are susceptible to Mycobacterium tuberculosis infection. Tuberc. (Edinb.). 2015;95:246–250. doi: 10.1016/j.tube.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 70.Kumar R, et al. Identification of a novel role of ESAT-6-dependent miR-155 induction during infection of macrophages with Mycobacterium tuberculosis. Cell. Microbiol. 2012;14:1620–1631. doi: 10.1111/j.1462-5822.2012.01827.x. [DOI] [PubMed] [Google Scholar]
- 71.Rothchild AC, et al. MiR-155-regulated molecular network orchestrates cell fate in the innate and adaptive immune response to Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA. 2016;113:E6172–E6181. doi: 10.1073/pnas.1608255113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siddle KJ, et al. Bacterial infection drives the expression dynamics of microRNAs and their isomiRs. PLoS. Genet. 2015;11:e1005064. doi: 10.1371/journal.pgen.1005064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao X, et al. Kruppel-like factor 4 regulates macrophage polarization. J. Clin. Invest. 2011;121:2736–2749. doi: 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sahu SK, et al. MicroRNA 26a (miR-26a)/KLF4 and CREB-C/EBPbeta regulate innate immune signaling, the polarization of macrophages and the trafficking of Mycobacterium tuberculosis to lysosomes during infection. PLoS. Pathog. 2017;13:e1006410. doi: 10.1371/journal.ppat.1006410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim JK, et al. MicroRNA-125a inhibits autophagy activation and antimicrobial responses during mycobacterial infection. J. Immunol. 2015;194:5355–5365. doi: 10.4049/jimmunol.1402557. [DOI] [PubMed] [Google Scholar]
- 76.Ni B, Rajaram MV, Lafuse WP, Landes MB, Schlesinger LS. Mycobacterium tuberculosis decreases human macrophage IFN-gamma responsiveness through miR-132 and miR-26a. J. Immunol. 2014;193:4537–4547. doi: 10.4049/jimmunol.1400124. [DOI] [PubMed] [Google Scholar]
- 77.Liu Z, et al. Analysis of miRNA expression profiling in human macrophages responding to Mycobacterium infection: induction of the immune regulator miR-146a. J. Infect. 2014;68:553–561. doi: 10.1016/j.jinf.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 78.Li M, et al. microRNA-146a promotes mycobacterial survival in macrophages through suppressing nitric oxide production. Sci. Rep. 2016;6:23351. doi: 10.1038/srep23351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xue X, Qiu Y, Yang HL. Immunoregulatory role of MicroRNA-21 in macrophages in response to bacillus calmette-guerin infection involves modulation of the TLR4/MyD88 signaling pathway. Cell. Physiol. Biochem. 2017;42:91–102. doi: 10.1159/000477118. [DOI] [PubMed] [Google Scholar]
- 80.Fu X, et al. MicroRNA-206 regulates the secretion of inflammatory cytokines and MMP9 expression by targeting TIMP3 in Mycobacterium tuberculosis-infected THP-1 human macrophages. Biochem. Biophys. Res. Commun. 2016;477:167–173. doi: 10.1016/j.bbrc.2016.06.038. [DOI] [PubMed] [Google Scholar]
- 81.Kumar M, et al. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-kappaB pathway. Cell Host Microbe. 2015;17:345–356. doi: 10.1016/j.chom.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 82.Wu Y, et al. The Transcriptional Foundations of Sp110-mediated Macrophage (RAW264.7) Resistance to Mycobacterium tuberculosis H37Ra. Sci. Rep. 2016;6:22041. doi: 10.1038/srep22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schnitger AK, et al. Listeria monocytogenes infection in macrophages induces vacuolar-dependent host miRNA response. PLoS ONE. 2011;6:e27435. doi: 10.1371/journal.pone.0027435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Izar B, Mannala GK, Mraheil MA, Chakraborty T, Hain T. microRNA response to Listeria monocytogenes infection in epithelial cells. Int J. Mol. Sci. 2012;13:1173–1185. doi: 10.3390/ijms13011173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lind EF, Elford AR, Ohashi PS. Micro-RNA 155 is required for optimal CD8 + T cell responses to acute viral and intracellular bacterial challenges. J. Immunol. 2013;190:1210–1216. doi: 10.4049/jimmunol.1202700. [DOI] [PubMed] [Google Scholar]
- 86.Archambaud C, et al. The intestinal microbiota interferes with the microRNA response upon oral Listeria infection. MBio. 2013;4:e00707–00713. doi: 10.1128/mBio.00707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang LJ, et al. Dermal adipocytes protect against invasive Staphylococcus aureus skin infection. Science. 2015;347:67–71. doi: 10.1126/science.1260972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Andreotti CS, et al. Staphylococcus aureus chronic intramammary infection modifies protein expression of transforming growth factor beta (TGF-beta) subfamily components during active involution. Res. Vet. Sci. 2014;96:5–14. doi: 10.1016/j.rvsc.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 89.Domingo-Gonzalez R, et al. Transforming growth factor-beta induces microRNA-29b to promote murine alveolar macrophage dysfunction after bone marrow transplantation. Am. J. Physiol. Lung. Cell. Mol. Physiol. 2015;308:L86–95. doi: 10.1152/ajplung.00283.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fang L, et al. Genome-wide transcriptional and posttranscriptional regulation of innate immune and defense responses of bovine mammary gland to staphylococcus aureus. Front Cell Infect. Microbiol. 2016;6:193. doi: 10.3389/fcimb.2016.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tanaka K, et al. MiR-142 is required for staphylococcus aureus clearance at skin wound sites via small GTPase-mediated regulation of the neutrophil actin cytoskeleton. J. Invest. Dermatol. 2017;137:931–940. doi: 10.1016/j.jid.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 92.Podsiad A, et al. MicroRNA-155 regulates host immune response to postviral bacterial pneumonia via IL-23/IL-17 pathway. Am. J. Physiol. Lung. Cell. Mol. Physiol. 2016;310:L465–475. doi: 10.1152/ajplung.00224.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee MH, et al. A postinfluenza model of Staphylococcus aureus pneumonia. J. Infect. Dis. 2010;201:508–515. doi: 10.1086/650204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cremer TJ, et al. MiR-155 induction by microbes/microbial ligands requires NF-kappaB-dependent de novo protein synthesis. Front Cell Infect. Microbiol. 2012;2:73. doi: 10.3389/fcimb.2012.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Verschoor CP, et al. MicroRNA-155 is required for clearance of Streptococcus pneumoniae from the nasopharynx. Infect. Immun. 2014;82:4824–4833. doi: 10.1128/IAI.02251-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bandyopadhyay S, Long ME, Allen LA. Differential expression of microRNAs in Francisella tularensis-infected human macrophages: miR-155-dependent downregulation of MyD88 inhibits the inflammatory response. PLoS ONE. 2014;9:e109525. doi: 10.1371/journal.pone.0109525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huck O, et al. Identification and characterization of microRNA differentially expressed in macrophages exposed to porphyromonas gingivalis infection. Infect. Immun. 2017;85:e00771–00716. doi: 10.1128/IAI.00771-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moon HG, Yang J, Zheng Y, Jin Y. miR-15a/16 regulates macrophage phagocytosis after bacterial infection. J. Immunol. 2014;193:4558–4567. doi: 10.4049/jimmunol.1401372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kalantari P, et al. miR-718 represses pro-inflammatory cytokine production through targeting PTEN. J. Biol. Chem. 2017;292:5634–5644. doi: 10.1074/jbc.M116.749325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chu Q, Sun Y, Cui J, Xu T. Inducible microRNA-214 contributes to the suppression of NFkB mediated inflammatory response via targeting MyD88 in fish. J. Biol. Chem. 2017;292:5282–5290. doi: 10.1074/jbc.M117.777078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arkatkar T, et al. Murine MicroRNA-214 regulates intracellular adhesion molecule (ICAM1) gene expression in genital Chlamydia muridarum infection. Immunology. 2015;145:534–542. doi: 10.1111/imm.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sunkavalli U, et al. Analysis of host microRNA function uncovers a role for miR-29b-2-5p in Shigella capture by filopodia. PLoS. Pathog. 2017;13:e1006327. doi: 10.1371/journal.ppat.1006327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yuk JM, Yoshimori T, Jo EK. Autophagy and bacterial infectious diseases. Exp. Mol. Med. 2012;44:99–108. doi: 10.3858/emm.2012.44.2.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang ZM, Zhang AR, Xu M, Lou J, Qiu WQ. TLR-4/miRNA-32-5p/FSTL1 signaling regulates mycobacterial survival and inflammatory responses in Mycobacterium tuberculosis-infected macrophages. Exp. Cell. Res. 2017;352:313–321. doi: 10.1016/j.yexcr.2017.02.025. [DOI] [PubMed] [Google Scholar]
- 105.Ma C, et al. microRNA-124 negatively regulates TLR signaling in alveolar macrophages in response to mycobacterial infection. Mol. Immunol. 2014;62:150–158. doi: 10.1016/j.molimm.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 106.Ma C, et al. Mycobacterium bovis BCG triggered MyD88 induces miR-124 feedback negatively regulates immune response in alveolar epithelial cells. PLoS ONE. 2014;9:e92419. doi: 10.1371/journal.pone.0092419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zou M, et al. MicroRNA-3178 ameliorates inflammation and gastric carcinogenesis promoted by Helicobacter pylori new toxin, Tip-alpha, by targeting TRAF3. Helicobacter. 2017;22:e12348. doi: 10.1111/hel.12348. [DOI] [PubMed] [Google Scholar]
- 108.Liu Y, et al. miR-223 is upregulated in monocytes from patients with tuberculosis and regulates function of monocyte-derived macrophages. Mol. Immunol. 2015;67:475–481. doi: 10.1016/j.molimm.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 109.Garg M, Potter JA, Abrahams VM. Identification of microRNAs that regulate TLR2-mediated trophoblast apoptosis and inhibition of IL6 mRNA. PLoS ONE. 2013;8:e77249. doi: 10.1371/journal.pone.0077249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qi J, et al. microRNA-210 negatively regulates LPS-induced production of proinflammatory cytokines by targeting NF-kappaB1 in murine macrophages. FEBS Lett. 2012;586:1201–1207. doi: 10.1016/j.febslet.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 111.Bazzoni F, et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl Acad. Sci. USA. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li T, et al. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat. Immunol. 2010;11:799–805. doi: 10.1038/ni.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nahid MA, Satoh M, Chan EK. Mechanistic role of microRNA-146a in endotoxin-induced differential cross-regulation of TLR signaling. J. Immunol. 2011;186:1723–1734. doi: 10.4049/jimmunol.1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vegh P, et al. MicroRNA profiling of the bovine alveolar macrophage response to Mycobacterium bovis infection suggests pathogen survival is enhanced by microRNA regulation of endocytosis and lysosome trafficking. Tuberc. (Edinb.). 2015;95:60–67. doi: 10.1016/j.tube.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 115.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Quinn EM, Wang JH, O’Callaghan G, Redmond HP. MicroRNA-146a is upregulated by and negatively regulates TLR2 signaling. PLoS ONE. 2013;8:e62232. doi: 10.1371/journal.pone.0062232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl Acad. Sci. USA. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xu G, et al. microR-142-3p down-regulates IRAK-1 in response to Mycobacterium bovis BCG infection in macrophages. Tuberc. (Edinb.). 2013;93:606–611. doi: 10.1016/j.tube.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 119.Androulidaki A, et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31:220–231. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Benakanakere MR, et al. Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes. J. Biol. Chem. 2009;284:23107–23115. doi: 10.1074/jbc.M109.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gu X, Gao Y, Mu DG, Fu EQ. MiR-23a-5p modulates mycobacterial survival and autophagy during mycobacterium tuberculosis infection through TLR2/MyD88/NF-kappaB pathway by targeting TLR2. Exp. Cell. Res. 2017;354:71–77. doi: 10.1016/j.yexcr.2017.03.039. [DOI] [PubMed] [Google Scholar]
- 122.Yang XJ, et al. Mir-30d increases intracellular survival of Helicobacter pylori through inhibition of autophagy pathway. World J. Gastroenterol. 2016;22:3978–3991. doi: 10.3748/wjg.v22.i15.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim JK, et al. MIR144* inhibits antimicrobial responses against Mycobacterium tuberculosis in human monocytes and macrophages by targeting the autophagy protein DRAM2. Autophagy. 2017;13:423–441. doi: 10.1080/15548627.2016.1241922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ouimet M, et al. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat. Immunol. 2016;17:677–686. doi: 10.1038/ni.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kumar R, et al. MicroRNA 17-5p regulates autophagy in Mycobacterium tuberculosis-infected macrophages by targeting Mcl-1 and STAT3. Cell. Microbiol. 2016;18:679–691. doi: 10.1111/cmi.12540. [DOI] [PubMed] [Google Scholar]
- 126.Guo L, et al. MicroRNA-144-3p inhibits autophagy activation and enhances Bacillus Calmette-Guerin infection by targeting ATG4a in RAW264.7 macrophage cells. PLoS ONE. 2017;12:e0179772. doi: 10.1371/journal.pone.0179772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ashida H, et al. Cell death and infection: a double-edged sword for host and pathogen survival. J. Cell. Biol. 2011;195:931–942. doi: 10.1083/jcb.201108081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ghorpade DS, Leyland R, Kurowska-Stolarska M, Patil SA, Balaji KN. MicroRNA-155 is required for Mycobacterium bovis BCG-mediated apoptosis of macrophages. Mol. Cell. Biol. 2012;32:2239–2253. doi: 10.1128/MCB.06597-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huang J, et al. MiR-155 is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO3. Mol. Med Rep. 2015;12:7102–7108. doi: 10.3892/mmr.2015.4250. [DOI] [PubMed] [Google Scholar]
- 130.Xi X, et al. MicroRNA-223 is upregulated in active tuberculosis patients and inhibits apoptosis of macrophages by targeting FOXO3. Genet Test. Mol. Biomark. 2015;19:650–656. doi: 10.1089/gtmb.2015.0090. [DOI] [PubMed] [Google Scholar]
- 131.Liu Y, Jiang J, Wang X, Zhai F, Cheng X. miR-582-5p is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO1. PLoS ONE. 2013;8:e78381. doi: 10.1371/journal.pone.0078381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang Q, Liu S, Tang Y, Liu Q, Yao Y. MPT64 protein from Mycobacterium tuberculosis inhibits apoptosis of macrophages through NF-kB-miRNA21-Bcl-2 pathway. PLoS ONE. 2014;9:e100949. doi: 10.1371/journal.pone.0100949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sanjuan MA, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 134.Zhang Y, Bliska JB. Role of Toll-like receptor signaling in the apoptotic response of macrophages to Yersinia infection. Infect. Immun. 2003;71:1513–1519. doi: 10.1128/IAI.71.3.1513-1519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu R, et al. FOXP3 controls an miR-146/NF-kappaB negative feedback loop that inhibits apoptosis in breast cancer cells. Cancer Res. 2015;75:1703–1713. doi: 10.1158/0008-5472.CAN-14-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Qi Y, et al. Altered serum microRNAs as biomarkers for the early diagnosis of pulmonary tuberculosis infection. Bmc. Infect. Dis. 2012;12:384. doi: 10.1186/1471-2334-12-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang X, et al. Screening and identification of six serum microRNAs as novel potential combination biomarkers for pulmonary tuberculosis diagnosis. PLoS ONE. 2013;8:e81076. doi: 10.1371/journal.pone.0081076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fu Y, Yi Z, Wu X, Li J, Xu F. Circulating microRNAs in patients with active pulmonary tuberculosis. J. Clin. Microbiol. 2011;49:4246–4251. doi: 10.1128/JCM.05459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kleinsteuber K, et al. Decreased expression of miR-21, miR-26a, miR-29a, and miR-142-3p in CD4(+) T cells and peripheral blood from tuberculosis patients. PLoS ONE. 2013;8:e61609. doi: 10.1371/journal.pone.0061609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Afum-Adjei Awuah A, Ueberberg B, Owusu-Dabo E, Frempong M, Jacobsen M. Dynamics of T-cell IFN-gamma and miR-29a expression during active pulmonary tuberculosis. Int. Immunol. 2014;26:579–582. doi: 10.1093/intimm/dxu068. [DOI] [PubMed] [Google Scholar]
- 141.Wang JX, Xu J, Han YF, Zhu YB, Zhang WJ. Diagnostic values of microRNA-31 in peripheral blood mononuclear cells for pediatric pulmonary tuberculosis in Chinese patients. Genet. Mol. Res. 2015;14:17235–17243. doi: 10.4238/2015.December.16.23. [DOI] [PubMed] [Google Scholar]
- 142.Ren N, et al. MicroRNA signatures from multidrug resistant Mycobacterium tuberculosis. Mol. Med Rep. 2015;12:6561–6567. doi: 10.3892/mmr.2015.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cui JY, et al. Characterization of a novel panel of plasma microRNAs that discriminates between Mycobacterium tuberculosis infection and healthy individuals. PLoS. ONE. 2017;12:e0184113. doi: 10.1371/journal.pone.0184113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wagh V, Urhekar A, Modi D. Levels of microRNA miR-16 and miR-155 are altered in serum of patients with tuberculosis and associate with responses to therapy. Tuberculosis (Edinb.). 2017;102:24–30. doi: 10.1016/j.tube.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 145.Tacke F, et al. Levels of circulating miR-133a are elevated in sepsis and predict mortality in critically ill patients. Crit. Care. Med. 2014;42:1096–1104. doi: 10.1097/CCM.0000000000000131. [DOI] [PubMed] [Google Scholar]
- 146.Abd-El-Fattah AA, Sadik NA, Shaker OG, Aboulftouh ML. Differential microRNAs expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell. Biochem. Biophys. 2013;67:875–884. doi: 10.1007/s12013-013-9575-y. [DOI] [PubMed] [Google Scholar]
- 147.Lv Y, et al. Sputum and serum microRNA-144 levels in patients with tuberculosis before and after treatment. Int. J. Infect. Dis. 2016;43:68–73. doi: 10.1016/j.ijid.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 148.Wu J, et al. Analysis of microRNA expression profiling identifies miR-155 and miR-155* as potential diagnostic markers for active tuberculosis: a preliminary study. Hum. Immunol. 2012;73:31–37. doi: 10.1016/j.humimm.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 149.Golby P, Villarreal-Ramos B, Dean G, Jones GJ, Vordermeier M. MicroRNA expression profiling of PPD-B stimulated PBMC from M. bovis-challenged unvaccinated and BCG vaccinated cattle. Vaccine. 2014;32:5839–5844. doi: 10.1016/j.vaccine.2014.07.034. [DOI] [PubMed] [Google Scholar]
- 150.Zhang C, et al. High serum miR-183 level is associated with the bioactivity of macrophage derived from tuberculosis patients. Int J. Clin. Exp. Pathol. 2015;8:655–659. [PMC free article] [PubMed] [Google Scholar]
- 151.Cheng SF, Li L, Wang L. M. miR-155 and miR-146b negatively regulates IL6 in Helicobacter pylori (cagA + ) infected gastroduodenal ulcer. Eur. Rev. Med. Pharmacol. Sci. 2015;19:607–613. [PubMed] [Google Scholar]
- 152.Sun J, et al. MicroRNA expression profiles of bovine milk exosomes in response to Staphylococcus aureus infection. BMC Genomics. 2015;16:806. doi: 10.1186/s12864-015-2044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sun G, et al. SNPs in human miRNA genes affect biogenesis and function. RNA. 2009;15:1640–1651. doi: 10.1261/rna.1560209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Moszyńska A, Gebert M, Collawn J, Bartoszewski R. SNPs in microRNA target sites and their potential role in human disease. Open Biol. 2017;7:170019. doi: 10.1098/rsob.170019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang X, et al. Association of the miR-146a, miR-149, miR-196a2 and miR-499 polymorphisms with susceptibility to pulmonary tuberculosis in the Chinese Uygur, Kazak and Southern Han populations. Bmc. Infect. Dis. 2015;15:41. doi: 10.1186/s12879-015-0771-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Li D, et al. Genetic study of two single nucleotide polymorphisms within corresponding microRNAs and susceptibility to tuberculosis in a Chinese Tibetan and Han population. Hum. Immunol. 2011;72:598–602. doi: 10.1016/j.humimm.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 157.Latorre I, et al. A novel whole-blood miRNA signature for a rapid diagnosis of pulmonary tuberculosis. Eur. Respir. J. 2015;45:1173–1176. doi: 10.1183/09031936.00221514. [DOI] [PubMed] [Google Scholar]
- 158.Bouchie A. First microRNA mimic enters clinic. Nat. Biotechnol. 2013;31:577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]
- 159.Janssen HL, et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 160.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug. Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 161.Kamerkar S, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Acunzo M, et al. Selective targeting of point-mutated KRAS through artificial microRNAs. Proc. Natl Acad. Sci. USA. 2017;114:E4203–E4212. doi: 10.1073/pnas.1620562114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ribet D, Cossart P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 2015;17:173–183. doi: 10.1016/j.micinf.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 164.Rosenberg E. The diversity of bacterial pathogenicity mechanisms. Genome Biol. 2005;6:320. doi: 10.1186/gb-2005-6-5-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Abdul-Maksoud RS, et al. Serum miR-210 and miR-155 expression levels as novel biomarkers for rheumatoid arthritis diagnosis. Br. J. Biomed. Sci. 2017;74:209–213. doi: 10.1080/09674845.2017.1343545. [DOI] [PubMed] [Google Scholar]
- 166.He XH, et al. miR-155 downregulates ErbB2 and suppresses ErbB2-induced malignant transformation of breast epithelial cells. Oncogene. 2016;35:6015–6025. doi: 10.1038/onc.2016.132. [DOI] [PubMed] [Google Scholar]
- 167.Zheng B, et al. Exosome-mediated miR-155 transfer from smooth muscle cells to endothelial cells induces endothelial injury and promotes atherosclerosis. Mol. Ther. 2017;25:1279–1294. doi: 10.1016/j.ymthe.2017.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 168.Zheng Y, et al. miR-155 regulates IL-10-producing CD24hiCD27 + B cells and impairs their function in patients with Crohn’s Disease. Front. Immunol. 2017;8:914. doi: 10.3389/fimmu.2017.00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Amila A, et al. Sequence comparison of six human microRNAs genes between tuberculosis patients and healthy individuals. Int J. Mycobacteriol. 2015;4:341–346. doi: 10.1016/j.ijmyco.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 170.Monack DM, Mueller A, Falkow S. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat. Rev. Microbiol. 2004;2:747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- 171.Monack DM. Helicobacter and salmonella persistent infection strategies. Cold Spring Harb. Perspect. Med. 2013;3:a010348. doi: 10.1101/cshperspect.a010348. [DOI] [PMC free article] [PubMed] [Google Scholar]