Abstract
Sugarcane biomass has been used for sugar, bioenergy and biomaterial production. The majority of the sugarcane biomass comes from the culm, which makes it important to understand the genetic control of biomass production in this part of the plant. A meta-transcriptome of the culm was obtained in an earlier study by using about one billion paired-end (150 bp) reads of deep RNA sequencing of samples from 20 diverse sugarcane genotypes and combining de novo assemblies from different assemblers and different settings. Although many genes could be recovered, this resulted in a large combined assembly which created the need for clustering to reduce transcript redundancy while maintaining gene content. Here, we present a comprehensive analysis of the effect of different assembly settings and clustering methods on de novo assembly, annotation and transcript profiling focusing especially on the coding transcripts from the highly polyploid sugarcane genome. The new coding sequence-based transcript clustering resulted in a better representation of transcripts compared to the earlier approach, having 121,987 contigs, which included 78,052 main and 43,935 alternative transcripts. About 73%, 67%, 61% and 10% of the transcriptome was annotated against the NCBI NR protein database, GO terms, orthologous groups and KEGG orthologies, respectively. Using this set for a differential gene expression analysis between the young and mature sugarcane culm tissues, a total of 822 transcripts were found to be differentially expressed, including key transcripts involved in sugar/fiber accumulation in sugarcane. In the context of the lack of a whole genome sequence for sugarcane, the availability of a well annotated culm-derived meta-transcriptome through deep sequencing provides useful information on coding genes specific to the sugarcane culm and will certainly contribute to understanding the process of carbon partitioning, and biomass accumulation in the sugarcane culm.
Keywords: Bioinformatics, Computational biology, Genetics, Plant biology
1. Introduction
Sugarcane is a major source of sugar (sucrose) and a key energy crop used to produce ethanol and generate electricity. Sugarcane biomass could play a very important role in supporting second generation biofuel production, reviewed in [1, 2, 3]. Developing sugarcane as a crop for a wider range of industrial use could be aided by improved understanding of the genetic and environmental control of biomass composition. Our knowledge of sugarcane genomics is hindered by the complexity of this highly polyploid crop, the lack of a full genome sequence or complete transcriptome and proteome databases. Characterization of the transcriptome will contribute directly to understanding the molecular basis of key traits and will also support the generation of a well annotated genome sequence.
Sugarcane transcriptome studies have been based on limited resources including the sorghum genome [4], sugarcane EST database [5] and Saccharum officinarum gene indices [6]. Due to the lack of a well-represented transcriptome and a reference genome, the de novo transcriptomes derived directly from the samples of each study are still considered to be the best option for representing the samples in transcriptome profiling studies. In transcriptome construction, the de novo assemblers, parameters and clustering techniques significantly affect the assembly results. For data generated from short-read technologies, i.e., those from Illumina, the widely used assemblers include Trinity [7], CLC Genomics Workbench (CLC-GWB, CLC Bio-Qiagen, Aarhus, Denmark), Velvet/OASES [8], SOAPdenovo-Trans [9] and TransAbyss [10]. Trinity, Velvet/OASES, SOAPdenovo-Trans and TransAbyss were designed for transcriptome assembly in addition to recovering the transcript isoforms; while CLC-GWB has been mostly used for genome assembly but has also been used in transcriptome assembly [11, 12], which allows flexibility in its parameters of word size and bubble size. Sugarcane transcriptomes have been assembled using mostly Illumina RNA-Seq short-read data of different tissue types and genotypes; and employing different transcriptome assemblers, including Trinity package [13, 14], and Velvet/OASES pipeline [15, 16]. Recently, we have used the long-read Iso-Seq technology for sugarcane [17] and showed recovery of more full-length transcripts compared to that from short-read Illumina technology. However, it is still costly to produce a high quality transcriptome at a sufficient depth using the long-read technologies at the moment, while short-read technologies offer lower cost per base and thereby a high depth of coverage. To date, most transcriptome assemblies in sugarcane using short-read technology were based on a single assembly or setting strategy. Various studies, (e.g. [18,11]), have shown that combining assemblies of different settings and assemblers improved the assembly and identified a greater gene content compared to the use of a single assembler or setting. This however, generates a need to cluster the resultant assembly to reduce the redundancy using tools such as CD-HIT-EST [19] or OASES [8].
The high ploidy and complex structure of the sugarcane genome suggests that every sugarcane cross may have a distinct chromosome combination and resulting gene set [20]. The gene expression in any given cross may be unique to that particular cross [21]. The sugarcane genome contains 80–130 chromosomes and up to ~14 homo(eo)logous gene copies, originating from two different progenitors [22, 23]. While the total number of genes predicted for sugarcane is about 35,000, the challenge in transcriptome assembly resides in the number of transcript isoforms resulting from the different homo(eo)logous chromosomes and alternative splicing of each of the gene families. It is still unknown how many transcript isoforms the many gene families in the sugarcane genome produce; however, this could be in the hundreds of thousands. In Arabidopsis, about 300,000 transcripts were found to result from 25,000 genes [24]. A total of ∼107,000 non-redundant sugarcane transcript isoforms have been generated [17], but the actual total number of isoforms could be much higher for the complex sugarcane genome, depending upon the genotype, developmental stage and growth condition. Recovering transcript isoforms from short-read data is a challenging task for a non-model species such as sugarcane, given that these isoforms could be present in the samples at different levels of abundance with potential to introduce errors and mis-assemblies into the resultant contigs.
Advances in sequencing technologies in recent years, allows a great amount of data to be generated. However, it is crucial to use this data to construct high quality transcriptomes representing well the genes expressed in the genotypes and samples studied [25]. Assessment of transcriptome quality needs to be standardized. It was suggested in [12] that the transcriptome quality can be assessed based on the rate of reads mapping back, recovery of widely conserved and expressed orthologs, N50 length statistics and the total number of unigenes. More importantly, erroneous, mis-assembled and chimeric contigs can be estimated and removed by several analysis tools like Detonate [26] and Transrate [27], using a contig impact score obtained from read mapping (i.e., a good contig is the one that has pairs of reads mapped to it, in the right mapping direction, with a high expression value). Additionally, more biological or real contigs can be retained by using the protein metrics obtained from programs such as TransDecoder [28] or Evidence Directed Gene Construction for Eukaryotes [29] (hereafter referred to as Evigen). BUSCO [30] and CEGMA [31] can be used to assess the recovery of the highly conserved orthologs in the transcriptome.
Meta-transcriptome assembly combines the total content of gene transcripts in a community (or of different genotypes) considered as a unique entity, at different developmental stages and conditions, in order to obtain the whole expression profile of the community [32]. This approach is an important frontier, however it requires careful validation of the new methods or workflows associated with it. The meta-transcriptome assembly strategy has been shown to have worked well for Nicotiana benthamiana [18] and for Eleusine indica [11] using assemblies of different settings from different assemblers. A sugarcane meta-transcriptome surveyed on 20 diverse genotypes derived from Illumina short-read sequencing and described in an earlier report [17], was utilized for this study.
In the current study, we evaluated the influence of different assemblers, settings and clustering approaches on the quality of transcriptome assembly through different metrics including contig statistics (number, contig average length and N50), read mapping scores (RSEM-EVAL) and comparative metrics against a sugarcane transcript database through Conditional Reciprocal Best BLAST (Transrate), BUSCO/CEGMA alignment and protein metrics. A transcript clustering approach employing the Evigen tool, based only on the coding fraction, was used to generate a more usable sugarcane culm-derived transcript set for transcript profiling analysis, compared to the initial de novo set generated in our earlier report [17]. Further characterization including an improved annotation and a gene expression analysis were performed to evaluate the resultant meta-transcriptome assembly specific to the sugarcane culm, representing sugarcane varieties of different genetic backgrounds and tissues of different developmental stages. The newly clustered transcript set reported here, together with the PacBio long-read transcriptome (SUGIT) [17], will provide useful information on coding genes specific to the sugarcane culm and contribute toward understanding the process of carbon partitioning, and biomass accumulation in the sugarcane culm.
2. Materials and methods
2.1. Samples collection, RNA-Seq and read data processing
This study was based on 20 sugarcane genotypes of diverse genetic background (provided in Table S1, which was adapted from [17, 33] with additional information regarding the parental genotypes and cultivar types). For each of the 20 genotypes, one top and one bottom internodal sample was collected, resulting in a final sample set of 40. The RNA-Seq and read data processing were previously described [17]. In brief, about 3 μg of RNA from each sample was used for library preparation (Illumina TruSeq stranded with Ribo-Zero Plant Library Prep Kit for total RNA library), indexed and sequenced to provide 2 × 150 bp paired-end (PE) reads, using an Illumina HiSeq4000 instrument at the Translational Research Institute, The University of Queensland, Australia. A total of 1,509,867,086 PE reads was generated. The read quality score and adapter remaining in the reads were assessed by FastQC [34] and trimmed using CLC-GWB v9.0. Only PE reads with a quality score of ≤0.001 (equivalent to Phred Q30 or the accuracy of the base calling of 99.9%), ≤2 of ambiguous nucleotides, and a length of ≥75 bp were retained. The rRNA content in the data was checked by mapping reads (length fraction 0.9, similarity fraction 0.9, in CLC-GWB) against a set of rRNA sequences extracted from the sugarcane chloroplast genome (rrn16 and rrn23) [35], mitochondrial genome (rrn18 and rrn26) [36]; and sugarcane cytoplasmic rRNA genes (RPS4, RPL17, RPS24 and RPS10) from the SoGI database [6]. Table S2 showed the reads mapping onto these selected genes estimated by using data from top and bottom internodal samples of one of the genotypes (QN05-1509). Reads showing homology to the sugarcane chloroplast genome and sorghum mitochondrial genome were removed using BBDuk, BBmap v36.02 [37], with a k-mer of 31. Prior to de novo assembly, the total clean trimmed read data set (1,015,845,414 PE reads) from 20 genotypes was concatenated into one interleaved file for downstream analysis including read digital normalization by using the perl script insilico_read_normalization.pl from Trinity package [38] and BBnorm tool [37].
2.2. Influence of settings and assemblers on the quality of de novo assembly output
Four assemblers including Trinity r2013-08-14 [7], CLC-GWB v9.0, Velvet/OASES v1.2.10 [8] and SOAPdenovo-Trans v1.03 [9] were employed as described in [17]. Additionally, different settings of word size (15–64) and bubble size (50–5000) were used in CLC-GWB to study the effect of these settings on the assembly quality statistics including contigs number, N50, contig average length; mapping and comparative metrics (details described in the next section). Three packages CD-HIT-EST ver4.6 [19], OASES and CAP3 [39] were employed in redundancy reduction of the assembly. The parameters of “-s 0.95 -c 0.95 -n 10” were used in CD-HIT-EST to cluster those contigs of 95% identity and of at least 95% length of the longest representative contigs in the cluster. The parameters “-merge yes -cov_cutoff 1 -edgeFractionCutoff 0.01 -min_trans_lgth 300” were used in OASES for contig merging, error correction and length filtering. The overlap percent identity cutoff “-p 95” and other default parameters were used in CAP3 to assemble contigs of defined identity into longer sequences.
2.3. Transcriptome quality assessment
A combination of different metrics was used in assessing the quality of the transcriptome assemblies, including N50 length statistics, rate of reads mapping back, recovery of widely conserved and expressed orthologs, full-length count and coding potentials. The RSEM-EVAL scores obtained from Detonate v1.11 [26] were employed to assess the quality of the assemblies. This package offers a novel metric based on a reference-free probabilistic model for quality assessment of de novo assemblies, using the assembly and the read data it is derived from. The SUGIT database [17] was used to estimate the true transcript length distribution for sugarcane. A subset containing ∼59 million normalized PE reads of data from 20 genotypes was used to generate the RSEM-EVAL score based on the evidence that reads mapped to the contigs in Detonate analysis. Additionally, Conditional Reciprocal Best BLAST (CRBB) [40] by BLAST+ v2.2.29 from Transrate v1.0.3 [27] was utilized by aligning the contigs against the reference to count the number of CRBB hits against four transcript reference databases including sorghum transcripts [41], SUGIT, SUCEST [5, 42] and Saccharum officinarum Gene Indices (SoGI) [6]. Results in Table S3 suggested that the full-length SUGIT database had more CRBB hits and hence was chosen for further analyses in this study.
The transcriptome assembly completeness was assessed by CEGMA [31] and BUSCO [30]. Full-length transcript counting was done by BLASTX homology search (BLAST+ v2.3.0, e-value = −20, -max_target_seqs 1) against the UniProt Viridiplantae database [43] and running perl script analyze_blastPlus_topHit_coverage.pl from the Trinity package. Coding potential was analyzed using Evigen to obtain other protein metrics including number of primary/alternative transcripts and the average length of the largest 1000 proteins. We included two transcriptome datasets from [13] and SoGI as the reference group.
2.4. Transcript annotation
The final newly-clustered transcriptome assembly from this study was compared against the UniProt Viridiplantae protein database using BLASTX (BLAST+ v2.3.0, e-value = −10), sorghum transcripts, SoGI, and SUCEST and SUGIT using BLASTN (BLAST+ v2.3.0, e-value = −10). The functional annotation of the final transcript set was carried out in Blast2GO v4.0.2 [44] with default parameters on the BLASTX result against the NCBI non-redundant (NR) protein database with 100 hits (e-value = −10). The MapMan v3.5.1R2 program [45, 46] was used in visualizing the annotation, by employing the mapping files generated by Mercator sequence annotator [45]. The Gene Ontology (GO) terms were extracted and plotted using the program WEGO [47] for three categories, biological process, molecular function and cellular component. Eukaryotic orthologous groups of the transcriptome were identified by OrthoMCL 5 [48] with default settings using BLASTP of translated protein sequences against OrthoMCL proteins with an e-value = −5, and 50% match.
2.5. Differential expression analysis
The pipeline for differentially expressed (DE) transcript identification was adapted from the Trinity v2.2.0 package [7], employing R program v3.2.0 [49] and R packages including Bioconductor v3.4 [50], DESeq2 package [51], limma [52], ctc [53] Biobase [54], cluster 2.0.4 [55], ape [56] and gplots [57]. To calculate the transcript expression, the clean RNA-Seq reads (quality score ≤0.01 or Phred score ≥20 to retain more reads in each sample, ≤2 ambiguous nucleotides, and length of ≥75 bp) from the top and bottom internodal tissue samples of three selected genotypes (QC02-402, Q200 and KQB08-32953) were aligned against the transcriptome with Bowtie v2.2.7 [58] with the following parameters “--no-mixed --no-discordant --gbar 1000 --end-to-end -k 200”. A sorted alignment file in BAM format was generated by SAMtools-1.3.1 [59] and used for the program RSEM [60] in estimating the transcript abundance in raw read counts for statistical models in differential expression analysis (counts.matrix files). The transcript expression was normalized as fragments per kilobase of feature sequence per million fragments mapped (FPKM) [61] and transcripts per million transcripts (TPM) [62]. Cross-sample normalization was carried out by Trimmed Mean of M-values (TMM) to obtain TMM-normalized FPKM values [63]. The transcript differential expression analysis was performed on the matrix of raw read counts using a perl script run_DE_analysis.pl in Trinity which employs a negative binomial model in DESeq2 package. DE transcripts were identified, extracted and clustered by running the script analyze_diff_expr.pl on the TMM-normalized value matrix. The cut-off for DE transcripts was at a false discovery rate (FDR) adjusted p-value ≤0.05 and a fold-change ≥2. The up-regulated and down-regulated transcripts were analyzed by MapMan v3.5.1R2.
2.6. Data analysis
Basic assembly statistics were determined by QUAST (Quality Assessment For Genome Assemblies) [64]. Venn diagrams were created by the online tool InteractiVenn [65]. All analyses in a Linux environment were conducted at the High Performance Computer clusters (Euramoo, Flashlite and Tinaroo) at the Research Computing Center, The University of Queensland, Australia [66]. All analyses in CLC-GWB were run on a QAAFI CLC Genomics Server, the University of Queensland, Australia. Other analyses were conducted in Microsoft Excel 2013 including XL Toolbox NG v7.3.12 [67] and RStudio v0.9.8/R v3.1.2.
3. Results and discussion
3.1. Read digital normalization
Despite the short read-length, RNA-Seq using Illumina sequencing technology has been utilized widely in transcriptome assembly thanks to the greater depth of coverage and a low error rate, compared to other sequencing platforms [68]. The total number of raw reads generated for this study was 1,509,867,086, with a pair distance estimated to be 64–302 bp, of which, 1,015,845,414 reads survived after quality and length trimming, having a quality of Phred Q30 and above [17]. The aim of having good depth of sequencing was to include more gene content and better transcriptome completeness, yet it was challenging for the data processing and computational steps, since this required high performance computing facilities for transcriptome construction. Not all transcripts express at the same level, and as shown in [12], the dynamic transcript abundance in the samples could result in an incomplete transcriptome assembly, often by fragmentation of contigs or failure in assembly of contigs. Sampling reads could help to reduce the size of the data, but at the same time, this causes considerable loss in gene content as it would in turn proportionally reduce the sequencing depth and affect the lowly expressed transcripts. Highly expressed transcripts can be reduced by experimental normalization employing a duplex specific nuclease enzyme (for examples, see [12, 17]) or digital normalizations, and therefore simplify the assembly algorithm. Digital normalization can be performed by khmer [69], BBnorm [37] or Trinity normalization [38]. The digital normalization technique resized the read data by reducing the over-representation of highly expressed transcripts (through highly abundant k-mers) to a level defined by the user (termed as maximum coverage, MC), and retained the reads that were originally from the less expressed transcripts which were below the user-defined cut-off. This ensured that the gene content remained the same as in the original data but the analysis required less computational resources and this sped up the assembly process.
Apart from the total read dataset (non-normalized reads), this study was based on four different normalized read datasets of different MC, having 59,054,880 reads (6%) at a MC50, 213,165,230 reads (21%) at a MC2000 (by Trinity in silico normalization); and 378,337,000 reads (∼37%) at MC10,000 (by BBnorm), as described previously [17].
3.2. Influence of settings on the quality of de novo assembly output
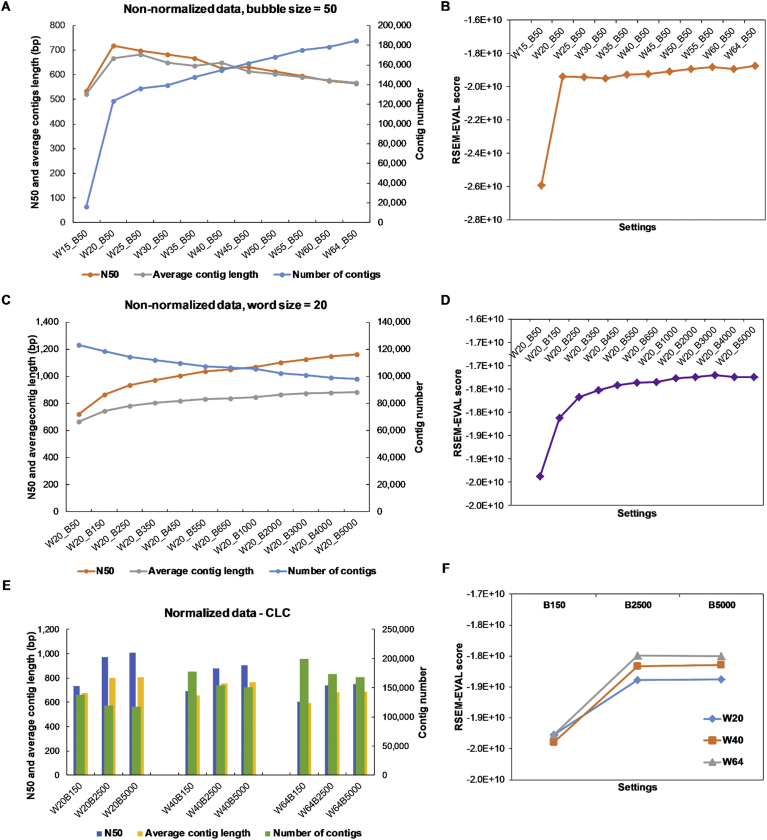
Changes in the settings (k-mer/word size, bubble size) affected the transcript statistics and quality metrics. We tested the effect of word size and bubble size on the assemblies using those from CLC-GWB, since this assembler allowed both these parameters to be changed. As shown in the Fig. 1A, an increase of the word size from 15 to 64 (at a fixed bubble size of 50, hereafter, W denotes word size and B for bubble size), generated more transcripts, while the assembly N50 and average contig length were reduced. The lowest contig number obtained was at W15_B50, which could be attributed to the word size being too short to resolve the repetitive content and complexity of the sugarcane transcriptome. The reads from different transcripts might have been collapsed into one due to their high similarity when the reads were broken down into short fragments at a word size of 15 bp. Table 1 and Fig. 1B show that as the word size increased from 15 to 64, the Detonate RSEM-EVAL score, the number of CRBB hits against the SUGIT database and percentage (%) of good contigs increased. The result reported for setting W15_B50 was in agreement with that for contig statistics, which had the lowest figures amongst the tested settings (RSEM-EVAL score of −2.59 × 1010, CRBB hits of 9,089, number of SUGIT sequences with CRBB of 8,300 and 82.4% good contigs). As the word size increased from 20 to 64, the RSEM-EVAL scores were slightly increased (−1.95 × 1010 to −1.87 × 1010), while that of assembly contigs with CRBB hit against SUGIT database was increased from 55,894 to 91,486; the number of SUGIT sequences with a CRBB hit was increased from 33,773 to 43,405; and the % of good contigs was increased from 93.2 to 97%. Assembly contig length and N50 can be manually increased by using different assemblers or adjusting the assembly settings, however, these metrics do not always reflect the transcriptome assembly quality [70,71]. The influence of word size on the contig assemblies has been studied in previous studies on different assemblers including CLC-GWB, Velvet, OASES, Bridger and SOAP [11,70]. Our results are in agreement with those in [70], in which a higher N50 and lower contig number were obtained for a lower k-mer size. Taking this together with the Detonate and Transrate results, it could be that for this dataset of PE reads 2 × 150 bp, when only the single setting was used, a larger word size could generate more contigs of lower N50 and average length, but with improved mapping and comparative metrics. A large word size may respond more sensitively to the differences in transcript abundance, i.e., reads from different transcripts isoforms of different expression levels, while a smaller word size may tend to assemble reads from different transcripts isoforms into the same contig, and hence, reduce the number of contigs in the assembly.
Fig. 1.
Effect of word size and bubble size on de novo assembly, performed in CLC-GWB. (A) Effect of word size on contig assembly. (B) Effect of word size on RSEM-EVAL score of assembly. (C) Effect of bubble size on contig assembly. (D) Effect of bubble size on RSEM-EVAL score of assembly. (E) Contig assembly in response to changes in word size and bubble size. (F) Assembly RSEM-EVAL score in response to changes in word size and bubble size. W denotes word size and B denotes bubble size.
Table 1.
Effect of word size and bubble size on assembly quality measured by Detonate and Transrate.
| Assembly | RSEM_EVAL scorea | SUGIT transcripts |
Good contigsd | % Good contigs | ||
|---|---|---|---|---|---|---|
| CRBB hitsb | N refs with CRBBc | |||||
| Word size | W15_B50 | -25,925,250,958 | 9,089 | 8,300 | 13,242 | 82.42 |
| W20_B50 | -19,380,072,750 | 55,894 | 33,773 | 114,829 | 93.23 | |
| W25_B50 | -19,421,340,895 | 57,372 | 34,013 | 128,286 | 94.07 | |
| W30_B50 | -19,496,141,905 | 59,888 | 34,706 | 131,210 | 94.39 | |
| W35_B50 | -19,281,907,606 | 67,973 | 37,144 | 140,335 | 94.99 | |
| W40_B50 | -19,215,617,161 | 72,865 | 38,402 | 147,726 | 95.37 | |
| W45_B50 | -19,097,783,732 | 76,624 | 39,741 | 154,954 | 95.71 | |
| W50_B50 | -18,927,399,593 | 80,304 | 40,781 | 161,660 | 96.05 | |
| W55_B50 | -18,835,828,725 | 84,991 | 42,022 | 168,938 | 96.43 | |
| W60_B50 | -18,936,543,261 | 86,758 | 42,152 | 172,140 | 96.69 | |
| W64_B50 | -18,759,903,834 | 91,486 | 43,405 | 178,809 | 96.96 | |
| Bubble size | W20_B50 | -19,380,072,750 | 55,894 | 33,773 | 114,829 | 93.23 |
| W20_B150 | -18,123,264,583 | 54,469 | 33,434 | 110,251 | 93.13 | |
| W20_B250 | -17,676,552,772 | 52,661 | 32,806 | 106,413 | 92.96 | |
| W20_B350 | -17,523,013,646 | 51,425 | 32,345 | 103,694 | 92.81 | |
| W20_B450 | -17,418,564,960 | 50,285 | 31,884 | 101,551 | 92.77 | |
| W20_B550 | -17,368,072,051 | 49,086 | 31,446 | 99,462 | 92.63 | |
| W20_B650 | -17,346,490,818 | 48,440 | 31,061 | 98,611 | 92.67 | |
| W20_B1000 | -17,274,641,924 | 47,491 | 30,905 | 97,653 | 92.55 | |
| W20_B2000 | -17,241,940,468 | 45,698 | 30,354 | 94,482 | 92.36 | |
| W20_B3000 | -17,208,740,562 | 45,491 | 30,241 | 92,987 | 92.17 | |
| W20_B4000 | -17,240,401,414 | 44,233 | 29,750 | 91,388 | 92.15 | |
| W20_B5000 | -17,250,943,232 | 43,935 | 29,550 | 90,434 | 92.18 | |
The higher the score, the better the assembly.
Number of contigs in assembly with a Conditional Reciprocal Best BLAST (CRBB) hit.
Number of sequences (out of 107,598 sequences in the SUGIT database) with a CRBB hit.
Contig with a positive RSEM-EVAL impact score.
Fig. 1C indicates that when the bubble size was increased from 50 to 5000, a lower number of contigs of a longer N50 and average contig length was obtained. The RSEM-EVAL scores obtained ranged from −1.94 × 1010 to −1.72 × 1010, and showed similar results to those from the settings of W20_B1000 to W20_B5000 (Fig. 1D); while the number of CRBB hits and number of good contigs were reduced due to a lower total contig number at a higher bubble size. The number of contigs with CRBB hit was reduced from 55,894 to 43,935; SUGIT sequences hits dropped from 33,773 to 29,550; and good contigs from 93.2% to 92.2%. This could be due to the fact that the longer bubble size resolved the conflict of bases (which could be biologically true), and extended the contigs compared to a shorter bubble size.
From the above results, we performed another analysis, using a set of normalized data (MC50, ∼59 million PE reads), at three different word sizes, 20, 40 and 64, in combination with three different bubble sizes of 150, 2500 and 5000. Results presented in Fig. 1E were consistent with the previous separate observations using the non-normalized read data (Fig. 1A and C). As the word size increased, more contigs were generated but the N50 and the average contig length were reduced. When the bubble size was increased, fewer contigs were generated with a longer N50 and average contig length. In all cases, there were more differences between assemblies obtained from bubble sizes of 150 and 2500 than from bubble sizes of 2500 and 5000. As a majority of the transcripts observed in the sugarcane cDNA library were <3000 bp in length (see Figure S7 in [17]), it could be that at the bubble size of 2500, the contig length obtained was longer as more read conflicts were resolved in the majority of transcripts, while at a bubble size of 5000, only the conflicts in reads from those transcripts in the range of 2500–5000 were further resolved. It was also shown that changes in bubble size at a word size of 20 (small) or 64 (large) affected the assembly results more (N50, average contig length and contig number) than that at a medium word size (40). The RSEM-EVAL scores were increased from B150 to B2500 and did not show much difference between the B2500 and B5000 (Fig. 1F and Table S4). These results suggest that, if only contig number, N50 and average contig length were considered, a small word size combined with a large bubble size resulted in the best assembly with lower contig number of a longer N50 and average length. However, the results from Detonate and Transrate suggested otherwise, a medium to larger word size (i.e. 25–64) combined with a medium to large bubble size (1000–5000) would be better in obtaining improved quality score and comparative metrics. Increasing bubble size in the lower range (i.e., B50–B1000) could affect contig length statistics and quality score more significantly than in a higher range (i.e., B1000–B5000). A larger bubble size, however, tends to incorporate more mis-assembled and chimeric transcripts (CLC-GWB manual), and could explain the increase in contig size and the reduction of the contig number.
3.3. Different assemblers and transcript assembly output
Different assemblers employ different algorithms in contig construction, therefore, even when using similar parameters, they produce varied assemblies [70, 72, 73]. The analyses reported here were based on four different de novo assemblies assembled by four assemblers, Trinity, CLC-GWB, Velvet/OASES and SOAPdenovo-Trans, previously described in [17]; and are referred to as Trinity-assembly, CLC-assembly, OASES-assembly and SOAP-assembly, respectively. A wide range of settings (k-mer size and bubble size) and the usage of different assemblers were applied to maximize the gene content and incorporate different transcripts into the transcriptome assembly, as suggested by several studies, i.e., [11, 18]. The contig number, N50, cumulative length and length distribution in the assemblies varied depending upon assemblers. The total contig number from the Trinity-assembly was 431,255 (N50: 2,194 bp), while that of the CLC-GWB assembly, OASES-assembly and SOAP-assembly were 508,239 (N50: 1,014 bp), 798,345 (N50: 516 bp) and 289,705 (N50: 674 bp), respectively, as reported earlier [17]. The Trinity-assembly had the highest N50 amongst the assemblies, while the OASES-assembly, despite having more contigs, had a shorter contig N50 length in general. In this study, these contig sets were pooled together for clustering step using different clustering packages. A more updated quality assessment of these four assemblies (referred to as single-assembler derived assemblies), is reported below, taking the protein metrics by Evigen and mapping metrics through Detonate and Transrate packages into account.
3.4. Assembly clustering and redundancy reduction
Application of three strategies to reduce the redundancy of the pooled assembly, employing CD-HIT-EST [74], OASES and CAP3 [39] programs, resulted in the total number of contigs being significantly reduced. The reduced assemblies in the three cases were referred to as the CDHIT-clustered assembly, Oases-clustered assembly and CAP3-assembled assembly, respectively. The summary statistics for the three clustered assemblies are presented in the Table 2, including only contigs in the range of 300 bp to 10 kb. The CDHIT-clustered assembly had 906,566 contigs with an N50 of 1,671 bp, while the OASES-clustered assembly had more contigs (1,383,279 contigs) with a shorter N50 of 1,331 and CAP3-assembled assembly had less contigs (839,331 contigs) with a longer N50 of 1,758 due to the overlapping contigs merging together during scaffolding. Compared to CD-HIT-EST and OASES, the assembly and scaffolding by the CAP3 program helped to reduce the number of contigs, however, it introduced an average of 92 ambiguous bases (Ns) per 100 kb (0.092%) into the sequences through the scaffolding process. All clustered assemblies had about the same GC content of ∼43.6%. It is important to note that, the CDHIT-clustered assembly was used as a representative of the de novo assembly strategy for comparison against the SUGIT transcriptome database, showing that the de novo assembly using Illumina short reads incorporated more gene content compared to the PacBio long-read derived transcriptome [17].
Table 2.
Comparison of three clustered assemblies used in this study.
| Assembly | CDHIT-clustered assembly* | Oases-clustered assembly | CAP3-assembled assembly |
|---|---|---|---|
| Contigs ≥300 bp | 906,566 | 1,383,279 | 839,331 |
| Contigs ≥1000 bp | 294,867 | 410,658 | 295,282 |
| Contigs ≥2000 bp | 130,095 | 155,453 | 131,443 |
| Contigs ≥3000 bp | 57,437 | 66,584 | 58,414 |
| Contigs ≥4000 bp | 23,416 | 27,110 | 24,080 |
| Contigs ≥5000 bp | 9,227 | 10,731 | 9,625 |
| Total length (≥300 bp) | 966,867,516 | 1,392,306,487 | 940,125,432 |
| Total length (≥1000 bp) | 646,818,455 | 842,812,564 | 651,587,956 |
| Total length (≥2000 bp) | 412,768,843 | 489,235,645 | 418,538,133 |
| Total length (≥3000 bp) | 235,893,013 | 273,427,809 | 240,741,264 |
| Total length (≥4000 bp) | 119,115,268 | 137,864,255 | 122,879,692 |
| Total length (≥5000 bp) | 56,369,155 | 65,423,551 | 58,916,716 |
| Total contigs | 906,566 | 1,383,279 | 839,331 |
| Largest contig(bp) | 9,990 | 9,991 | 9,990 |
| Total length (bp) | 966,867,516 | 1,392,306,487 | 940,125,432 |
| GC (%) | 43.67 | 43.61 | 43.67 |
| N50 | 1,671 | 1,331 | 1,758 |
| N75 | 745 | 691 | 812 |
| L50 | 168,723 | 282,937 | 158,900 |
| L75 | 385,929 | 654,771 | 354,745 |
| Ambiguous bases (N) per 100 kb | 0 | 0 | 92 |
*Adapted from [17].
3.5. Transcriptome completeness based upon CEGMA/BUSCO alignment
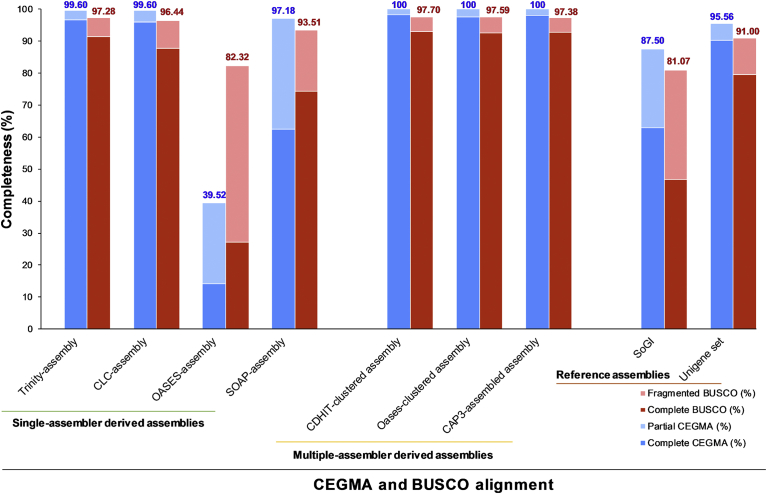
In this comparison, three groups of datasets, including the single assembler-derived assemblies (Trinity-assembly, CLC-assembly, OASES-assembly and SOAP-assembly), the clustered assemblies (CDHIT-clustered assembly, Oases-clustered assembly and CAP3-assembled assembly) and the reference group (SoGI database [6] and a unigene set from [13]) were included. The CEGMA and BUSCO alignments showed that, in all cases, the clustered assemblies exhibited a higher completeness level than those from single assembler-derived assemblies, and the reference datasets (Fig. 2, Tables 3 and 4). The three clustered assemblies had 97.6–98.4% CEGMA (when only the complete CEG proteins were counted) and 100% CEGMA (including all complete and partial CEG proteins). The single assembler-derived assemblies had ∼14.1–96.8% CEGMA when only the complete CEG proteins were counted and 39.5–99.6% CEGMA when all complete and partial CEG proteins were counted. Amongst the single assembler-derived assemblies, the Trinity-assembly performed the best, incorporating 96.8% complete CEGMA, followed by CLC-assembly (96.0%), and SOAP-assembly (62.5%), while the OASES-assembly incorporated only 14.1% due to the short contigs in the assembly. In the reference group, SoGI dataset had 62.9% CEGMA alignment (87.5% including partial CEGMA alignment), while the unigene set had 90.3% CEGMA alignment (95.6% including partial CEGMAs), respectively. There were no missing CEGMA in the clustered-assemblies, while there was 0.4–60.5% missing CEGMA in the single assembler-derived assemblies (with 0.4–2.8% of Trinity, CLC and SOAP-assembly, while the OASES-assembly had a large 60.5% missing CEGMA) and 4.4–12.5% missing CEGMA in the reference group. Similarly, in the BUSCO alignment against 956 conserved proteins, the clustered-assemblies were shown to have higher completeness levels, by having up to ∼93% (or 97.4–97.7% including fragmented BUSCOs), compared to that of the single-assembler derived assemblies (27.2–91.5% and 82.3–97.3%, respectively), and that of the reference group (46.7–79.6% complete BUSCO proteins and 81.1–91% including complete and fragmented BUSCO proteins).
Fig. 2.
Transcriptome completeness based upon CEGMA and BUSCO alignment.
Table 3.
CEGMA alignment for assembly completeness between single assemblies, clustered assemblies and reference assemblies.
| Assembly | Contig count | # CEGs Protein | Complete CEGs count | % Completeness | Partial CEGS | % Partials | Missing CEGs | % Missing | Total CEGs | % Total complete and partial CEGs |
|---|---|---|---|---|---|---|---|---|---|---|
| Trinity-assembly | 431,255 | 248 | 240 | 96.77 | 7 | 2.82 | 1* | 0.40 | 247 | 99.60 |
| CLC-assembly | 508,239 | 248 | 238 | 95.97 | 9 | 3.63 | 1** | 0.40 | 247 | 99.60 |
| OASES-assembly | 798,345 | 248 | 35 | 14.11 | 63 | 25.40 | 150 | 60.48 | 98 | 39.52 |
| SOAP-assembly | 289,705 | 248 | 155 | 62.50 | 86 | 34.68 | 7 | 2.82 | 241 | 97.18 |
| CDHIT-clustered | 906,566 | 248 | 244 | 98.39 | 4 | 1.61 | 0 | 0.00 | 248 | 100.00 |
| Oases-clustered | 1,383,279 | 248 | 242 | 97.58 | 6 | 2.42 | 0 | 0.00 | 248 | 100.00 |
| CAP3-assembled | 839,331 | 248 | 243 | 97.98 | 5 | 2.02 | 0 | 0.00 | 248 | 100.00 |
| SoGI database | 121,342 | 248 | 156 | 62.90 | 61 | 24.60 | 31 | 12.50 | 217 | 87.50 |
| Unigene set | 72,269 | 248 | 224 | 90.32 | 13 | 5.24 | 11 | 4.44 | 237 | 95.56 |
Missing CEGs: *KOG0434 **KOG1088.
Table 4.
BUSCO alignment for assembly completeness between single assemblies, clustered assemblies and reference assemblies.
| Assembly | BUSCO Notation Assessment |
||||||
|---|---|---|---|---|---|---|---|
| Total BUSCO groups searched | Total complete (%) | Single-copy BUSCOs (%) | Duplicated BUSCOs (%) | Fragmented BUSCOs (%) | Missing BUSCOs (%) | % Total complete and fragmented BUSCOs | |
| Trinity-assembly | 956 | 91.53 | 21.03 | 70.50 | 5.75 | 2.72 | 97.28 |
| CLC-assembly | 956 | 87.87 | 22.07 | 65.79 | 8.58 | 3.56 | 96.44 |
| OASES-assembly | 956 | 27.20 | 13.18 | 14.02 | 55.13 | 17.68 | 82.32 |
| SOAP-assembly | 956 | 74.37 | 35.15 | 39.23 | 19.14 | 6.49 | 93.51 |
| CDHIT-clustered | 956 | 92.99 | 10.04 | 82.95 | 4.71 | 2.30 | 97.70 |
| Oases-clustered | 956 | 92.68 | 3.35 | 89.33 | 4.92 | 2.41 | 97.59 |
| CAP3-assembled | 956 | 92.78 | 11.72 | 81.07 | 4.60 | 2.62 | 97.38 |
| SoGI database | 956 | 46.65 | 26.67 | 19.98 | 34.41 | 18.93 | 81.07 |
| Unigene set | 956 | 79.60 | 63.81 | 15.79 | 11.40 | 9.00 | 91.00 |
Amongst all the compared assemblies, the OASES-assembly, SOAP-assembly and SoGI had the largest proportion of partial/fragmented alignment, having 25.4% CEGMA/55.1% BUSCO, 34.7% CEGMA/19.1% BUSCO and 24.6% CEGMA/34.4% BUSCO, respectively. The OASES-assembly and SoGI database had the highest level of missing proteins, 60.5% CEGMA/17.7% BUSCO and 12.5% CEGMA/18.9% BUSCO, respectively. In the BUSCO alignment, the clustered-assemblies were shown to have the highest duplication level. This could be due to the fact that the clustered-assemblies were derived from a pooled assembly. In addition to the true biological transcript isoforms, these assemblers and settings might have assembled the same transcripts or transcript isoforms into many contigs of different lengths that were retained by the clustering process. The OASES-assembly had more fragmented contigs which could explain a low CEGMA completeness (required longer alignment length compared to BUSCO). The SoGI contains sugarcane gene indices and ESTs (fragmented mRNAs) collected from many experiments of various tissues, which could contribute to the low completeness and more fragmented BUSCOs. This database represented ∼90% of the predicted sugarcane genes in the forms of short ESTs and assembled tentative contigs. All in all, pooling and clustering contigs from multiple assemblies improved the protein metric assessment. This is in agreement with previous reports, such as that of [18], in which the authors concluded that although the method required high computational and storage capabilities, the de novo assembly was more complete, representing the samples from which it was derived, better than that from individual assemblies alone. This may be particularly useful for many polyploid crop species, such as sugarcane.
3.6. Detonate RSEM-EVAL score and Transrate metrics
Using these novel metrics which take the mapping of reads against the contigs into account in assessing the assembly quality, the RSEM-EVAL score and CRBB hits against the SUGIT database were obtained (Tables 5 and S5). The higher the RSEM-EVAL score, the better the assembly is considered, even though this score is always negative [26]. In the group of single assembler-derived assemblies, the score ranged from −1.997 × 1010 to −1.41 × 1010. The assemblies were ordered based on their RSEM-EVAL from the highest to the lowest, as follows: CLC-assembly > SOAP-assembly > Trinity-assembly > OASES-assembly. Amongst the clustered assemblies, the range was from −1.61 × 1010 to −1.43 × 1010, in which the order was CDHIT-clustered assembly > CAP3-assembled assembly > Oases-clustered assembly. Amongst the reference group, the SoGI and unigene set had RSEM-EVAL score of −2.05 × 1010 and −1.83 × 1010, respectively. These reference sets were not derived directly from the reads used in this study, therefore, lower RSEM-EVAL scores are expected. The result suggests that the low score of the OASES-assembly could be due to the high number of fragmented contigs that resulted in broken read pairs in mapping. The CLC-assembly and the CDHIT-clustered assembly were found to have the highest scores of all assemblies compared. The number of sequences with a hit against the SUGIT database corresponded to the number of contigs in each of the assemblies. The clustered assemblies had more sequences from SUGIT database hits in general, with the OASES-clustered assembly having the highest number of SUGIT sequence hits, however, it had the lowest % of good contigs.
Table 5.
Quality assessment of assemblies by Detonate and Transrate.
| Assembly | RSEM_EVAL scorea | SUGIT transcripts |
Contigs with positive impact scored | %Contigs with positive impact score | |
|---|---|---|---|---|---|
| CRBB hitsb | N refs with CRBBc | ||||
| Trinity-assembly | -15,240,221,881 | 229,152 | 44,765 | 274,889 | 63.74 |
| CLC-assembly | -14,103,274,074 | 257,035 | 59,991 | 384,245 | 75.60 |
| OASES-assembly | -19,974,094,466 | 376,082 | 61,612 | 459,890 | 57.61 |
| SOAP-assembly | -14,863,304,548 | 182,813 | 54,837 | 273,789 | 94.51 |
| CD-HIT-clustered | -14,369,832,453 | 465,467 | 68,603 | 511,561 | 56.43 |
| Oases-clustered | -16,058,124,749 | 655,184 | 70,611 | 549,043 | 39.69 |
| CAP3-assembled | -14,615,684,149 | 436,219 | 67,299 | 464,628 | 55.36 |
| SoGI database | -20,480,838,298 | 85,621 | 41,261 | 75,426 | 62.16 |
| Unigene set | -18,324,896,664 | 28,403 | 23,193 | 59,130 | 81.82 |
The higher the score, the better the assembly.
Number of contigs in assembly with a Conditional Reciprocal Best BLAST (CRBB) hit with the SUGIT database.
Number of sequences in the SUGIT database with a CRBB hit.
Contig with a positive RSEM_EVAL impact score.
3.7. Full-length transcript counting against the Viridiplantae proteins
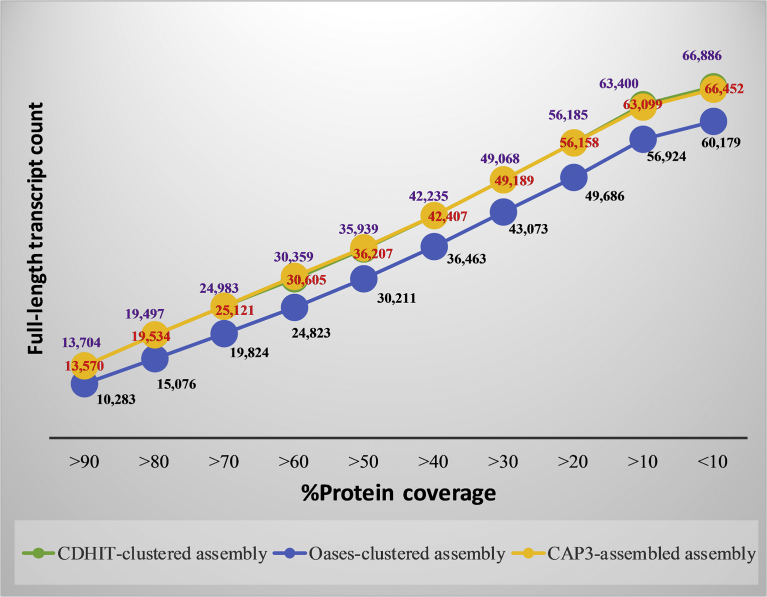
Since it was shown in the previous analyses that the clustered assemblies performed better in general (in CEGMA and BUSCO protein alignments, Detonate and Transrate assessments), only the clustered assemblies were used in this full-length transcript counting. The full-length transcript counting was done by comparing the assemblies against the UniProt Viridiplantae protein database. The number of transcripts appearing to be full-length (covering at least 90% of Viridiplantae proteins) or nearly full-length (covering at least 70% of Viridiplantae proteins) was counted and compared (Fig. 3). Among the three clustered assemblies, the CDHIT-clustered assembly included most full-length transcripts which covered at least 90% of the known proteins from the Viridiplantae database. A total of 13,704 transcript counts was reported for the CDHIT-clustered assembly [17], compared to 10,283 and 13,570 counts for the OASES-clustered assembly and the CAP3-assembled assembly, respectively. At a 70% cut-off it was 24,983, 19,824 and 25,121 for the CDHIT-clustered assembly, the OASES-clustered assembly and the CAP3-assembled assembly, respectively. The CDHIT-clustered assembly and CAP3-assembled assembly had a higher number of full-length transcripts compared to the OASES-clustered assembly, due to the similar approach in retaining/extending the contigs that differed from the way that OASES pipeline worked. CAP3, however, performed scaffolding by introducing ambiguous bases into the contigs, which could be the reason for the protein homology search being lower than for the CDHIT-clustered assembly at a cut-off of 90% and higher at 70%.
Fig. 3.
Full-length transcript count against the Viridiplantae proteins. Purple, blue and red colors represent values for CDHIT-clustered assembly, Oases-clustered assembly and CAP3-assembled assembly, respectively.
3.8. Potential coding transcripts and protein prediction of transcriptome
The results in Table 6 show that among three final assemblies, the OASES-clustered assembly had the highest number of predicted transcripts and average length of 1,000 largest proteins, hereafter, referred to as AP-1000 (94,398 transcripts including main and alternative, and AP-1000 of 304 aa), compared to that of CDHIT-clustered assembly (83,041 contigs and 298 aa [17]) and CAP3-assembled assembly (73,885 contigs and 300 aa). Both SoGI and unigene sets gave a lower number of predicted transcripts (41,042 and 13,205, respectively), which could be due to the lower duplication level/isoforms or short transcripts in these assemblies that was reflected in the lower fraction of the predicted alternative transcripts. In relation to the main transcripts, the CDHIT-clustered assembly had the highest number of main transcripts among the three (56,766 compared to 40,617 in the OASES-clustered assembly and 53,691 in the CAP3-assembled assembly), while the OASES-clustered assembly had the highest number of alternative transcripts among the three (53,781 compared to 26,275 in CDHIT-clustered assembly and 20,194 in CAP3-assembled assembly). This shows the effect of using different approaches to processing transcript contigs, since the three datasets resulted from the same initial pooled assembly by using three different tools. It was found that Evigen performed better on only the coding fraction of the assembly, which is discussed in the next section.
Table 6.
Potential coding and transcript prediction based on the Evigen pipeline.
| Assembly | CDHIT-clustered assembly | Oases-clustered assembly | CAP3-assembled assembly | SoGI database | Unigene set |
|---|---|---|---|---|---|
| Main transcripts | 56,766 | 40,617 | 53,691 | 32,013 | 13,205 |
| Alternate transcripts | 26,275 | 53,781 | 20,194 | 9,029 | – |
| Total (main + alternate) | 83,041 | 94,398 | 73,885 | 41,042 | 13,205 |
| Minimum length (aa) | 64 | 64 | 64 | 42 | 64 |
| Maximum length (aa) | 616 | 580 | 616 | 534 | 620 |
| Average length (aa) | 139.3 | 138.4 | 141.2 | 165.2 | 155.2 |
| AP-1000** | 298 | 304 | 300 | 287 | 298 |
**Average length of 1000 largest proteins (length expressed as amino acid, aa).
Considering together the transcript prediction and the CEGMA/BUSCO alignment in the previous sections, the CDHIT-clustered assembly was chosen for down-stream processing, as it had a good transcript length prediction and better CEGMA/BUSCO alignment, more full-length transcripts matching with the Viridiplantae protein database and better RSEM-EVAL score as well as Transrate comparative metrics.
3.9. Clustered de novo assembly, which assembler contributes more?
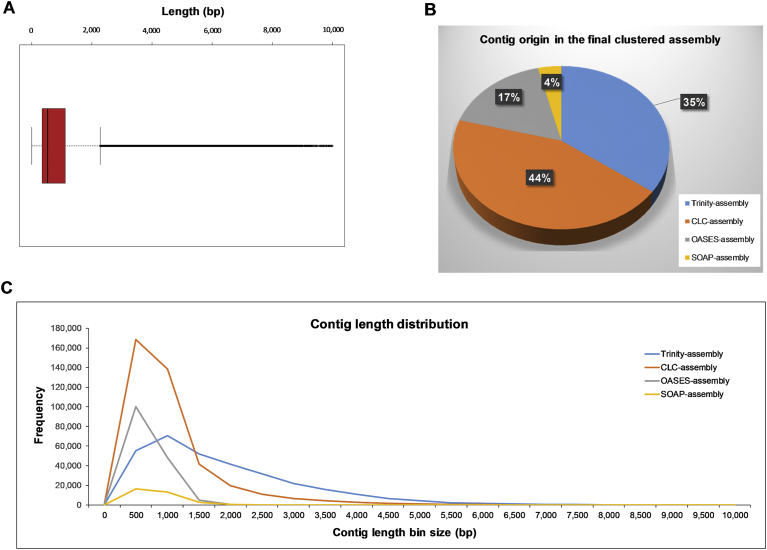
As mentioned earlier, the CDHIT-clustered assembly had 906,566 contigs, of ∼967 Mb, and having an N50 of 1,671 bp (Fig. 4A). Investigating the contig composition in this assembly, it was found that 44% of the contigs originated from the CLC-assembly, while 35%, 17% and 4% were from Trinity-assembly, OASES-assembly and SOAP-assembly, respectively (Fig. 4B). The larger number of transcripts derived from the CLC-assembly could be the result of more settings conducted using the CLC-WB and the longer contigs it produced. The lower number of contigs derived from the OASES-assembly (despite it having the highest contig number before clustering) and especially from the SOAP-assembly, could be due to the shorter contigs obtained in these assemblies compared to those from CLC-GWB or Trinity. This resulted in the shorter contigs getting clustered and being removed by CD-HIT at a similarity setting of 95%. The CLC-assembly contributed more contigs to the final assembly, but Trinity contributed more contigs with longer length, especially those that had a length >1,500 bp (Fig. 4C). This CDHIT-clustered assembly was used for further generation and characterization of the final de novo assembly by different coding-based clustering approaches, including comparison with the initial clustered transcript set from the earlier report [17].
Fig. 4.
Length distribution and expression of transcripts from the CDHIT-clustered assembly. (A) Boxplot of contig length of the CDHIT-clustered assembly. (B) Percentage of contig composition based on the assembly origins. (C) Length distribution of the contigs in the CDHIT-clustered assembly based on their assembly origins.
Transcriptome de novo assembly, particularly from short read data, is a challenging task in higher plants due to the fact that the plant genome contains thousands of genes, and the alternatively spliced transcripts from each gene [75]. More challenging is that the actual number of transcripts that are expressed in a certain situation does not correspond to the number of genes or known transcripts of that species. The total number and nature of transcripts expressed in one condition is different from that expressed under another condition. RNA-Seq has been used intensively for transcriptome de novo assembly for many plant species, i.e., wheat [76], tobacco [18], and sugarcane [13, 14, 15, 16, 77]. In sugarcane, most transcriptome studies have been based on a single-assembler approach employing Trinity or Velvet/OASES, which could be limited by the range of transcripts that the specific assembler is designed to capture. In this study, we have presented further quality assessment of the assembly including CEGMA/BUSCO completeness alignment, RSEM-EVAL score, CRBB hits, full-length transcript counts, coding transcript prediction and protein metrics, supporting the conclusion that the strategy using multi-assemblers/multiple settings improved the transcript de novo assembly. The sugarcane meta-transcriptome was derived from combining several individual culms by including several samples of diverse genetic backgrounds (for a wider representation of variety-specific genes), in combination with multiple settings and assemblers (for capturing transcript isoforms). The results are in agreement with studies on assembly of the transcriptomes of diploid species [11] and especially of those from polyploid species, including those for allo-tetraploid Nicotiana benthamiana [18], tetraploid peanut [73] and hexaploid wheat [76].
As the Evigen pipeline clusters transcripts based on their protein sequences, we attempted to run the program on only the coding fraction of the CDHIT-clustered assembly which was first retained by using the Portrait package [78]. The Portrait-retained set of 535,295 transcript contigs (59.05% of the total) was then clustered by Evigen, resulting in an improved assembly compared to that clustered by Evigen on the total CDHIT-clustered assembly, as described earlier [17]. This new Evigen-clustered set had 121,987 transcript contigs; which accounted for about 6% of the total pooled transcripts, and ∼13.46% of the CDHIT-clustered assembly (Tables 7 and S5); and showed an RSEM-EVAL score of −1.74 × 1010, had 55,093 sequences (∼46%) with a CRBB hit against 22,317 sequences in the SUGIT database. A total of 99,680 contigs in this transcriptome (∼82% of the final set) were validated by mapping against a set of ∼59 million normalized PE reads, and categorized as “good contigs with a positive impact score”, which had both PE reads mapped to, in the correct orientation. Considering that the remaining 22,307 contigs (∼18% of the final set, classified as bad contigs with negative impact score) could be biologically functional transcripts (they exhibited ORFs and were categorized as biologically real by Evigen) but may have been expressed at a low level that might have not been validated by the subset of normalized reads, we retained all 121,987 transcripts for downstream analysis, and referred to this as the final de novo assembly. This final set was composed of 78,052 main transcripts and 43,935 alternative transcripts with an N50 of 1,669 bp, an improved AP-1000 metric of 1,372 aa, and a CEGMA completeness level of 96.4%.
Table 7.
Summary statistics of the final de novo assembly.
| De novo assembly summary | |
|---|---|
| Total length of sequence (bp) | 128,307,893 |
| Total number of sequences | 121,987 |
| N25 (bp) | 3,049 |
| N50 (bp) | 1,669 |
| N75 (bp) | 692 |
| Total GC count (bp) | 61,352,407 |
| GC %: | 47.82 |
| RSEM-EVAL score | -17,384,096,424 |
| Contigs with positive impact score | 99,680 |
| %Contigs with positive impact score | 81.71 |
| CRBB hits | 55,903 |
| N refs with CRBB* | 22,317 |
*Number of SUGIT sequences with hit.
3.10. Comparative analysis against other databases
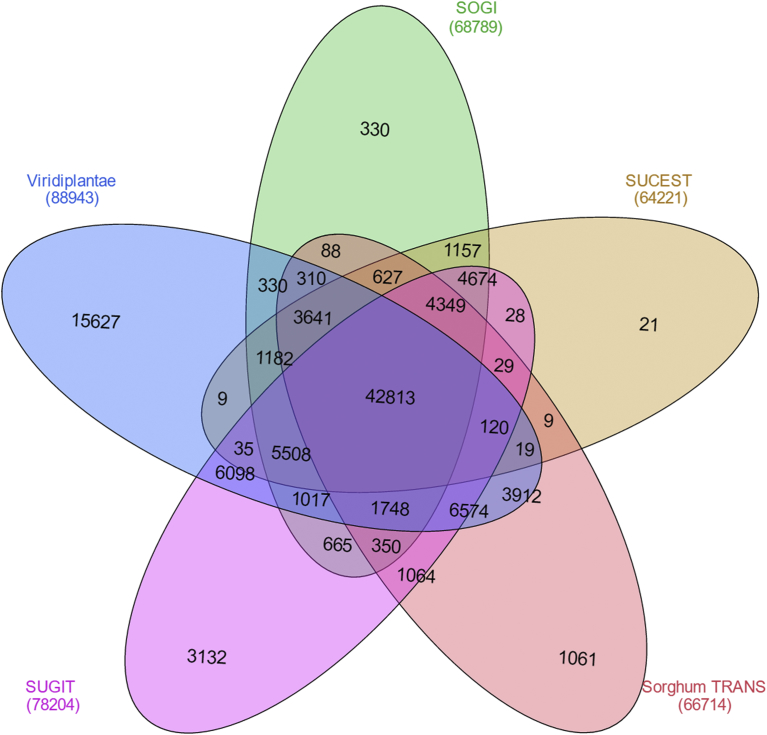
Of 121,987 sequences in the final de novo assembly, 88, 943 contigs (72.9%) matched 38,887 entries from the Viridiplantae protein database, 66,714 sequences (54.7%) matched the sorghum transcripts, 68,789 contigs (56.4%) matched the SoGI database, 64,221 contigs (52.7%) matched the SUCEST dataset and 78,204 contigs (64.1%) matched the SUGIT database. Taken together, a total of 106,527 (87.3% of the final set) matched one of the five databases, as presented in Table 8, and Venn diagram in Fig. 5. We found that 42,813 common contigs matched against all 5 databases used in the BLAST analysis. There were more transcripts that were unique to Viridiplantae (15,627 sequences), which is composed of protein sequences from all plant species. There were 1,061 sequences that matched uniquely against the sorghum transcripts and 3,132 sequences unique to the SUGIT database. These unique sequences could be novel for sugarcane since these were not present in the SoGI or SUCEST databases. The full-length transcript count of the final de novo assembly against the SUGIT and Viridiplantae protein databases were 7,282 and 9,722 (at >90% coverage) and 12,468 and 16,671 (at >70% coverage), respectively (Table S6).
Table 8.
Blast summary of the final de novo assembly.
| Database | De novo contigs matched | % de novo assembly | Database entries matched |
|---|---|---|---|
| Viridiplantae protein | 88,943 | 72.91 | 38,887 |
| Sorghum transcripts | 66,714 | 54.69 | 22,922 |
| SoGI database | 68,789 | 56.39 | 32,467 |
| SUCEST database | 64,221 | 52.65 | 23,284 |
| SUGIT database | 78,204 | 64.11 | 33,488 |
| Total sequences matched | 106,527 | 87.33 | – |
Fig. 5.
Comparative analysis of the final de novo assembly against the Viridiplantae protein database, sorghum and sugarcane transcript database.
3.11. Transcript functional annotation
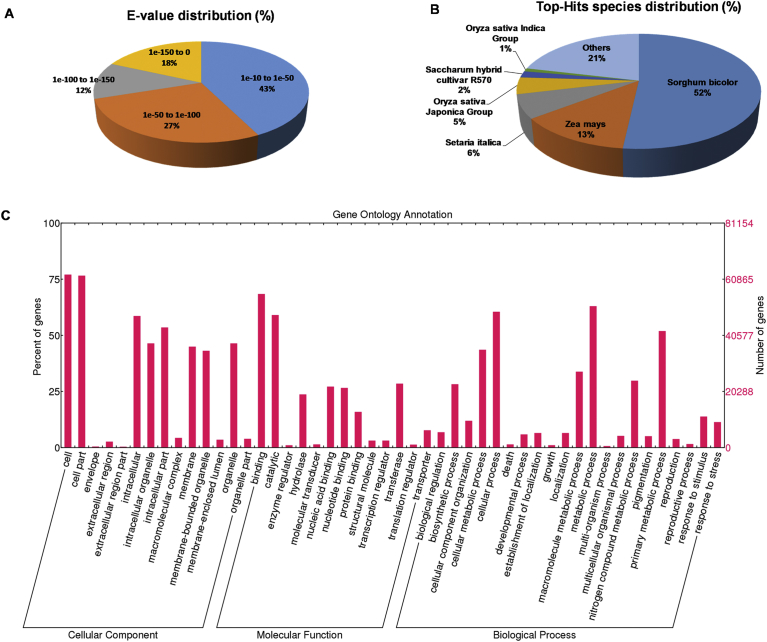
The functional annotation of the final de novo assembly was done using the results from BLASTX against the NCBI non-redundant (NR) protein database (100 hits, e-value = −10). A summary of the annotation results is presented in Fig. 6, including sequence length distribution of the final set, data distribution, e-value distribution of the blast hits, and the species distribution. Of a total 121,987 sequences, 92,861 sequences (76.1%) matched the NCBI NR protein database. The majority of e-values ranged from 1e-10 to 1e-50 (43%), followed by 27% hits with e-values from −50 to −100, 18% hits with e-values from 1e-150 to 0, and 12% hits with e-value of 1e-100 to 1e-150 (Fig. 6A). The majority of hits (52%) were attributed to Sorghum bicolor, the most closely related species to sugarcane, while 13% of the top hits were from Zea mays, 6% from Setaria italica, 5% from Oryza sativa Japonica group, 2% from Saccharum hybrid cultivar R570 and 1% from Oryza sativa Indica group (Fig. 6B). In general, most of the top hits were attributed to the grass family, and cultivar R570 was the most represented cultivar amongst sugarcane genotypes. Fig. 6C shows GO terms distribution for 81,154 transcripts of the final de novo assembly annotated by Blast2GO [44] and plotted by WEGO [47]. The most abundant GO terms were cell/cell part, intracellular, intracellular part (cellular component); binding, catalytic and transferase (molecular function); cellular process, metabolic process and primary metabolic process (biological process). Of the total predicted main transcripts, 8,089 transcripts (∼10.4%) were annotated against 3,251 KOs in the KEGG metabolic pathway. A total of 74,618 transcripts were assigned to 11,853 orthologous groups by the program OrthMCL5. The list of orthologous groups is provided in the Table S7.
Fig. 6.
Summary of functional annotation of the final assembly. (A) E-value distribution. (B) Top-hit species distribution. (C) Gene ontology terms annotation.
3.12. Transcript isoform estimation
In relation to number of transcript isoforms in the final de novo assembly, selected genes were used to estimate the number of transcript isoforms predicted based on the annotation result (Table S8). It is shown that there were 11 transcripts belonging to cellulose synthase (CesA) and CesA-like that appeared to be full-length transcripts, covering at least 90% of the transcript in the SUGIT database (and 16 transcripts covering at least 70% of full-length SUGIT transcripts). A total of two transcripts of cinnamyl alcohol dehydrogenase (CAD), three transcripts as 4-coumarate-CoA ligase 1 (4CL 1), one transcript as caffeoyl-CoA O-methyltransferase (CCoAOMT), nine transcripts as cinnamoyl CoA reductase (CCR) and three transcripts as phenylalanine lyase (PAL) were found covering at least 90% of the SUGIT full-length transcripts. When only 70% full-length SUGIT transcripts were considered, there were four, one, seven, two, 11 and eight transcripts found, respectively for CAD, Caffeic acid O-3-methyltransferase (COMT), 4CL 1, CCoAOMT, CCR and PAL, repspectively. Protein-wise, 18 CesA/CesA-like, three CAD, one COMT, five 4CL 1, two CCoAOMT, 11 CCR and four PAL transcripts covered at least 70% of Viridiplantae proteins.
3.13. Transcript differential expression analysis
To evaluate the usability of the final de novo transcriptome assembly, an experiment was set up to investigate the differential expression of transcripts between the young and mature tissues of the sugarcane culm. Three genotypes (QC02-402, Q200 and KQB08-32953) were selected for testing the transcript expression between the young and mature tissues in this analysis, in which, for each genotype, one top internodal and one bottom internodal tissue sample was used. An average mapping back rate of RNA-Seq read data against the final de novo assembly was ∼71% (Table S9) suggesting this is suitable for transcript profiling analysis. It is noteworthy that this final set contains only coding transcripts, while the RNA-Seq data had reads originally from coding and also non-coding RNAs. The assemblies before Evigen clustering (CDHIT-clustered assembly and Portrait-retained set) had 96–98% and 95–97% reads mapping back, respectively, however they also had 94–97% and 92–96% reads that mapped more than 1 times, indicating the high transcript redundancy.
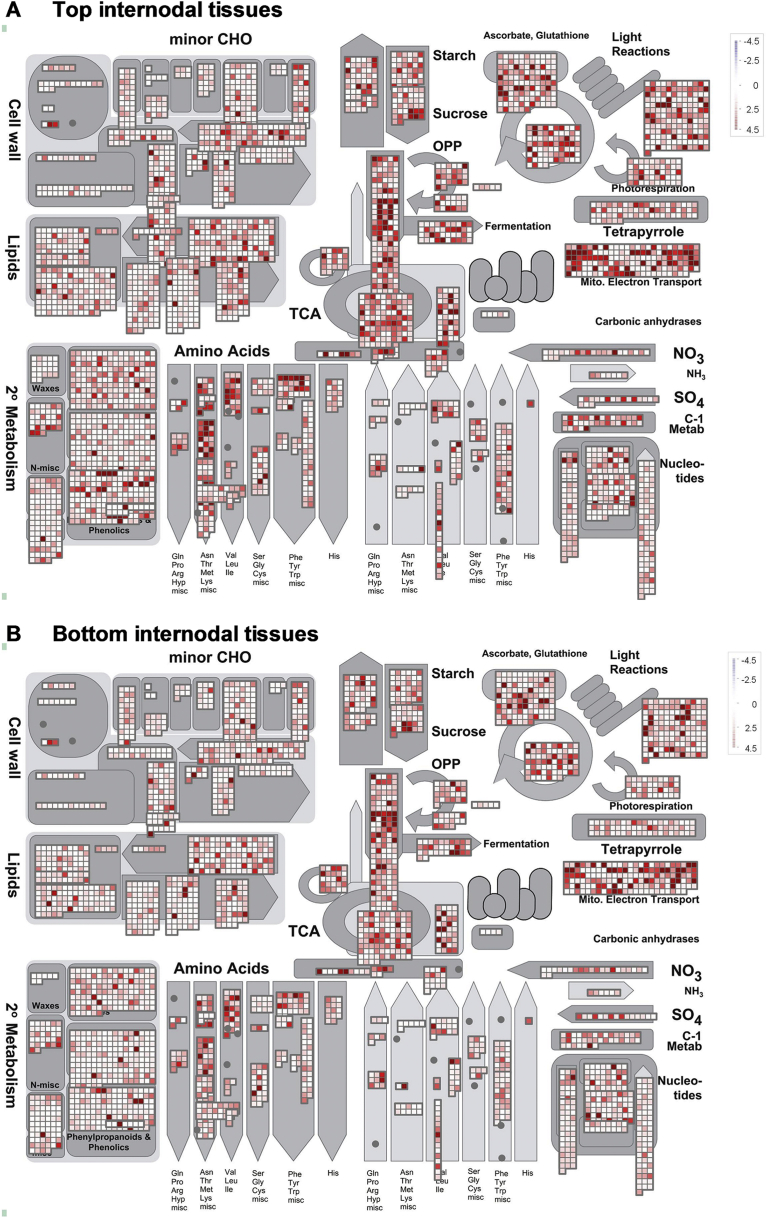
Fig. 7 shows the result analysed by the MapMan Image Annotator module for the top and bottom internodal samples from the three genotypes. In this analysis, we included only transcripts with a log2(TMM-normalised FPKM +1)> 0.3 for visualization and comparison between the two groups of samples. It was observed that the top internodal samples had a higher expression level of the transcript compared to the bottom internodal samples. This is expected as the top internode represents the young and growing tissues where the metabolism is active whereas in the bottom internodal tissues some of the metabolic processes might have ceased or slowed down.
Fig. 7.
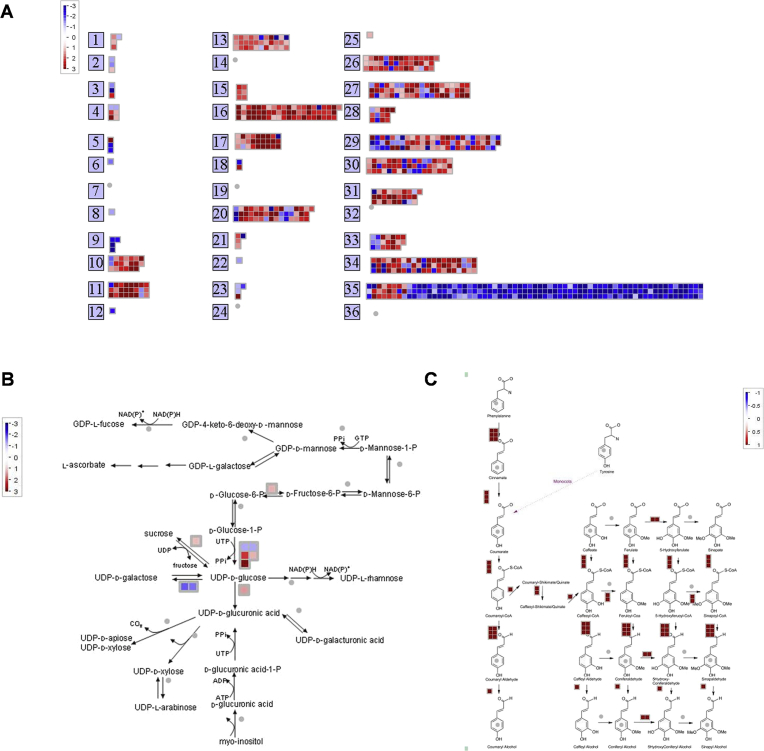
Transcript expression measured by RNA-Seq in the top and bottom internodal tissues analysed by the MapMan Image Annotator module. (A) Top internodal tissue samples. (B) Bottom internodal tissue samples. Log2(TMM-normalised FPKM +1)> 0.3 was used. 2o Metabolism denotes Secondary Metabolism.
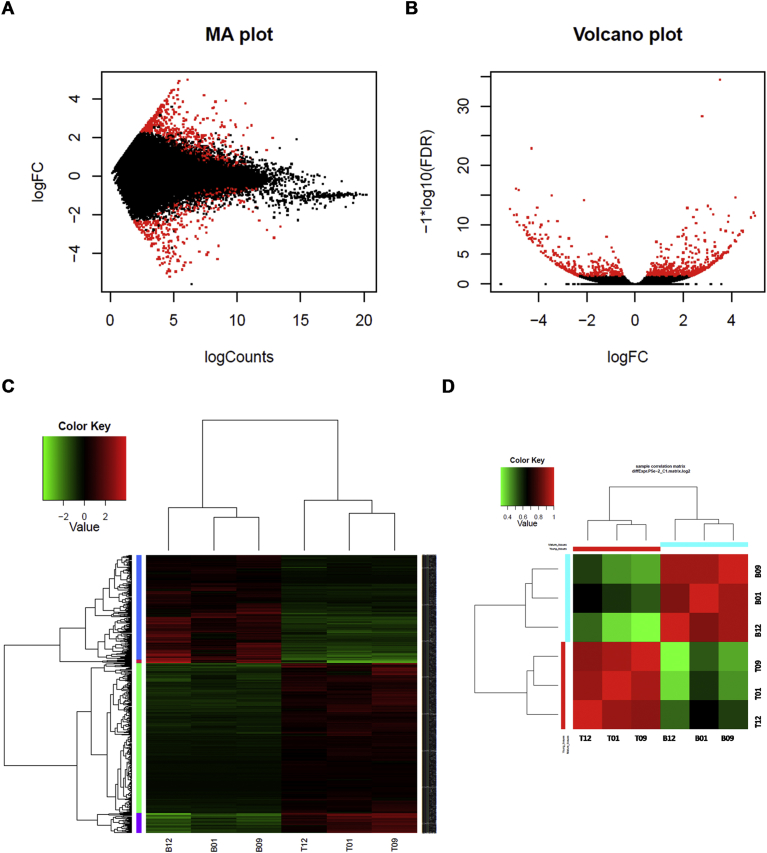
When top and bottom internodal tissues were compared in the differential expression analysis using the DESeq2 package at a FDR-corrected p-value ≤0.05 and fold change ≥2, it was found that a total of 822 transcripts were differentially expressed (DE). This included 504 down-regulated transcripts and 318 up-regulated transcripts in the bottom internodal tissues (or up-regulated and down-regulated in the top tissues, respectively, details provided in Tables S10 and S11). It is important to mention that the transcripts/genes “down-regulated” in the bottom internode (mature tissue) are actually transcripts/genes that are down-regulated during maturation and those “up-regulated” transcripts/genes in the mature tissue are up-regulated during maturation. Summary of the differential expression analysis includes the MA (Fig. 8A) and volcano plots where the DE transcripts are highlighted (Fig. 8B), clustered DE transcripts between the top and bottom internodal tissues (Fig. 8C), and sample correlation (Fig. 8D). The identified DE transcripts and functional classifications are illustrated in Fig. 9A, including transcripts related to carbohydrates (CHO)/cell wall metabolism (function classifications 1, 2 and 10) and phenylpropanoid pathway (in secondary metabolism category, functional classification 16) which includes the monolignol metabolism. Notably, the identified DE transcripts involved in major/minor CHO metabolism include: fructose-bisphosphate aldolase, chloroplast precursor (EC 4.1.2.13) (ALDP), sucrose phosphate synthase 3F (SPS3F), cytosolic fructose-1,6-bisphosphatase (EC 3.1.3.11), NADP-dependent oxidoreductase phosphofructokinase 3 (PFK3), and glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12). Transcripts related to cell wall precursor metabolism (Fig. 9B) include: UDP-D-glucose/UDP-D-galactose 4-epimerase 2 (UGE2), sucrose synthase 2 (EC 2.4.1.13), UDP-glucose 6-dehydrogenase (EC 1.1.1.22), CesA (EC 2.4.1.12) and CesA-like, COBRA-like 5 protein precursor (protein BRITTLE CULM1), IRX9 gene and pectin lyase-like superfamily protein. Transcripts involved in the lignin biosynthesis (Fig. 9C) include: PAL (EC 4.3.1.5), CAD (EC 1.1.1.195), cinnamic acid 4-hydroxylase (C4H, EC 1.14.13.11), 4CL 2 (EC 6.2.1.12), hydroxycinnamoyl-Coenzyme A shikimate/quinate hydroxycinnamoyl transferase (HCT, EC 2.3.1.106), coumarate 3-hydroxylase (C3H, E.C. 1.14.14.1), CCoAOMT (EC 2.1.1.104), CCR1 (EC 1.2.1.44) and ferulate 5-hydroxylase (F5H, EC 1.14.13). This result is consistent with the earlier reports on DE transcripts in the immature, maturing and mature internodes of the sugarcane plant [79, 80] and also in our other studies using the SUGIT database [81]. The down-regulation of some of the transcripts including the COBRA-like 5 protein precursor, CAD, CCR, CesA, CesA-like C5 and IRX9 during maturation was validated by quantitative real-time PCR (qPCR) [81] on RNA samples from two out of three genotypes used in this study (QC02-401 and Q200).
Fig. 8.
Differential expression analysis of transcripts between the top and bottom internodal tissue samples. (A) MA plot. (B) Volcano plot. (C) Up- and down-regulated transcripts between the top and bottom internodal tissues. (D) Comparison between samples in each groups of tissues. T denotes top tissue, and B denotes bottom tissue. 01, 09 and 12 are codes for genotypes QC02-402, Q200 and KQB08-32953, respectively.
Fig. 9.
Differentially expressed transcripts between the top and bottom tissues. (A) Summary of identified differentially expressed transcripts annotated as different MapMan functional classifications. (B) Cell-wall precursor metabolism. (C) Secondary metabolism – phenylpropanoid pathway. Log2FC was used in generating this figure, red color denotes the up-regulated transcript while blue color denotes the down-regulated transcripts in top internodal tissue (or down-regulated/up-regulated in the bottom tissues, respectively). Each heatmap is representative of one of the identified differentially expressed transcripts.
4. Conclusion
Due to the tissue- /genotype-specificity of the sugarcane transcriptome and in the context of sugarcane lacking a reference genome, it is widely agreed that the best transcriptome to be used for transcript expression profiling is the one that is assembled directly from the samples. To aid the construction of a culm-derived meta-transcriptome, a large scale deep RNA sequencing of 20 genotypes of diverse genetic backgrounds had been performed, aiming to cover a wide range of transcripts that are expressed in the culm. In this current study, further analyses of the effect of different settings, assemblers and processing methods on assembly output through different quality assessment was performed and discussed. The separation of the coding fraction of the transcriptome prior to protein-based transcript prediction resulted in a more usable set of 121,987 transcripts being retained. The updated annotation showed that 76% and 73% of this dataset matched the NCBI NR protein and the Viridiplantae protein databases, respectively, while the rest could contain potentially novel genes in sugarcane. About 67% of the transcriptome was annotated against the GO terms; while ∼61% and ∼10% (taking only main transcripts) were assigned to 11,853 orthologous groups and 3,251 KOs in the KEGG metabolic pathway, respectively. A total of 822 transcripts were differentially expressed (DE), including 504 down-regulated transcripts and 318 up-regulated transcripts during maturation of the sugarcane plant. Among these, there were important transcripts involved in fiber and sugar accumulation in the sugarcane culms. This study provides useful information on coding genes specific to the sugarcane culm from a diverse set of genotypes that was not previously available and the transcript set identified will facilitate further gene expression studies in the sugarcane culm, especially in understanding the processes of carbon partitioning and biomass accumulation in the sugarcane culm.
Declarations
Author contribution statement
Nam V. Hoang, Angelo Furtado: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Prathima P. Thirugnanasambandam: Analyzed and interpreted the data; Wrote the paper.
Frederick C. Botha: Conceived and designed the experiments; Wrote the paper.
Robert J. Henry: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by the Queensland Government and Sugar Research Australia (SRA, grant number LP160100939). Nam V. Hoang was supported by the Australian Agency for International Development (AusAID) through an Australian Awards Scholarship. The funders had no role in the design of the study, collection, analysis, and interpretation of data, nor in writing the manuscript.
Competing interest statement
The authors declare no conflict of interest.
Additional information
RNA-Seq read data associated with this study has been submitted as sequence read archive (SRA) in NCBI with the BioProject ID PRJNA356226, BioSample SAMN06323325, and accessions from SRR5258946 to SRR5259025, as mentioned previously in [17]. The final transcriptome assembly derived from this work can be accessed in Figshare under the DOI 10.6084/m9.figshare.5103838 or at https://doi.org/10.6084/m9.figshare.5103838.
Acknowledgements
We thank SRA staff from Brandon Station, Burdekin, Queensland, Australia for helping with the sample collecting and processing and Ravi Nirmal for helping with sample collection and transport.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Hotta C. The biotechnology roadmap for sugarcane improvement. Trop. Plant Biol. 2010;3:75–87. [Google Scholar]
- 2.de Souza A.P., Grandis A., Leite D.C.C., Buckeridge M.S. Sugarcane as a bioenergy source: history, performance, and perspectives for second-generation bioethanol. Bioenerg. Res. 2014;7:24–35. [Google Scholar]
- 3.Hoang N.V., Furtado A., Botha F.C., Simmons B.A., Henry R.J. Potential for genetic improvement of sugarcane as a source of biomass for biofuels. Front Bioeng Biotechnol. 2015;3:182. doi: 10.3389/fbioe.2015.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paterson A. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 5.SUCEST-FUN Database. 2015. http://sucest-fun.org [Google Scholar]
- 6.SoGI . 2017. Saccharum officinarum Gene Indices.ftp://occams.dfci.harvard.edu/pub/bio/tgi/data/Saccharum_officinarum/ [Google Scholar]
- 7.Grabherr M.G. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29 doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulz M.H., Zerbino D.R., Vingron M., Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie Y. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics. 2014;30:1660–1666. doi: 10.1093/bioinformatics/btu077. [DOI] [PubMed] [Google Scholar]
- 10.Birol I. De novo transcriptome assembly with ABySS. Bioinformatics. 2009;25 doi: 10.1093/bioinformatics/btp367. [DOI] [PubMed] [Google Scholar]
- 11.Chen S., McElroy J.S., Dane F., Peatman E. Optimizing transcriptome assemblies for Eleusine indica leaf and seedling by combining multiple assemblies from three de novo assemblers. Plant Genome. 2015;8 doi: 10.3835/plantgenome2014.10.0064. [DOI] [PubMed] [Google Scholar]
- 12.Honaas L.A. Selecting superior de novo transcriptome assemblies: lessons learned by leveraging the best plant genome. PLoS One. 2016;11 doi: 10.1371/journal.pone.0146062. e0146062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardoso-Silva C.B. De novo assembly and transcriptome analysis of contrasting sugarcane varieties. PLoS One. 2014;9 doi: 10.1371/journal.pone.0088462. e88462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M. De novo analysis of transcriptome reveals genes associated with leaf abscission in sugarcane (Saccharum officinarum L.) BMC Genom. 2016;17:195. doi: 10.1186/s12864-016-2552-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vicentini R. Large-scale transcriptome analysis of two sugarcane genotypes contrasting for lignin content. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134909. e0134909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santa Brigida A.B. Sugarcane transcriptome analysis in response to infection caused by Acidovorax avenae subsp. avenae. PLoS One. 2016;11 doi: 10.1371/journal.pone.0166473. e0166473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoang N.V. A survey of the complex transcriptome from the highly polyploid sugarcane genome using full-length isoform sequencing and de novo assembly from short read sequencing. BMC Genom. 2017;18:395. doi: 10.1186/s12864-017-3757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakasugi K., Crowhurst R., Bally J., Waterhouse P. Combining transcriptome assemblies from multiple de novo assemblers in the allo-tetraploid plant Nicotiana benthamiana. PLoS One. 2014;9 doi: 10.1371/journal.pone.0091776. e91776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22 doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 20.Matsuoka S. Energy cane: its concept, development, characteristics, and prospects. Adv. Bot. 2014;2014:13. [Google Scholar]
- 21.Grivet L., Arruda P. Sugarcane genomics: depicting the complex genome of an important tropical crop. Curr. Opin. Plant Biol. 2002;5:122–127. doi: 10.1016/s1369-5266(02)00234-0. [DOI] [PubMed] [Google Scholar]
- 22.Vettore A.L. Analysis and functional annotation of an expressed sequence tag collection for tropical crop sugarcane. Genome Res. 2003;13:2725–2735. doi: 10.1101/gr.1532103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Souza G.M. The sugarcane genome challenge: strategies for sequencing a highly complex genome. Trop. Plant Biol. 2011;4:145–156. [Google Scholar]
- 24.Hilson P. Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res. 2004;14:2176–2189. doi: 10.1101/gr.2544504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang R. A high quality Arabidopsis transcriptome for accurate transcript-level analysis of alternative splicing. Nucleic Acids Res. 2017;45:5061–5073. doi: 10.1093/nar/gkx267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B. Evaluation of de novo transcriptome assemblies from RNA-Seq data. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith-Unna R., Boursnell C., Patro R., Hibberd J.M., Kelly S. TransRate: reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 2016;26:1134–1144. doi: 10.1101/gr.196469.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.TransDecoder. 2016. https://transdecoder.github.io/ [Google Scholar]
- 29.Evidence Directed Gene Construction for Eukaryotes. 2016. http://arthropods.eugenes.org/EvidentialGene/about/EvidentialGene_trassembly_pipe.html [Google Scholar]
- 30.Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 31.Parra G., Bradnam K., Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 32.Parro V., Paz M.M. In: Encyclopedia of Astrobiology. Amils Ricardo., editor. Springer Berlin Heidelberg; 2014. pp. 1–3. [Google Scholar]
- 33.Hoang N.V. High-throughput profiling of the fiber and sugar composition of sugarcane biomass. Bioenerg. Res. 2016;10:400–416. [Google Scholar]
- 34.Andrews S. 2010. FastQC a Quality Control Tool for High Throughput Sequence Data.http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 35.Hoang N.V., Furtado A., McQualter R.B., Henry R.J. Next generation sequencing of total DNA from sugarcane provides no evidence for chloroplast heteroplasmy. New Negat. Plant Sci. 2015;1–2:33–45. [Google Scholar]
- 36.Shearman J.R. The two chromosomes of the mitochondrial genome of a sugarcane cultivar: assembly and recombination analysis using long PacBio reads. Sci. Rep. 2016;6:31533. doi: 10.1038/srep31533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.BBMap. 2016. http://sourceforge.net/projects/bbmap/ [Google Scholar]
- 38.Haas B.J. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8 doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang X., Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9 doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aubry S., Kelly S., Kümpers B.M.C., Smith-Unna R.D., Hibberd J.M. Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004365. e1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phytozome. 2016. https://phytozome.jgi.doe.gov/pz/portal.html [Google Scholar]
- 42.Vettore A.L., Silva F. R. d., Kemper E.L., Arruda P. The libraries that made SUCEST. Genet. Mol. Biol. 2001;24:1–7. [Google Scholar]
- 43.UniProt. 2016. http://www.uniprot.org/taxonomy/33090 [Google Scholar]
- 44.Conesa A., Gotz S. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008;2008:619832. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lohse M. Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 2014;37:1250–1258. doi: 10.1111/pce.12231. [DOI] [PubMed] [Google Scholar]
- 46.Usadel B. A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant Cell Environ. 2009;32:1211–1229. doi: 10.1111/j.1365-3040.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- 47.Ye J. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L., Stoeckert C.J., Jr., Roos D.S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Team, R. C. 2013. R: a Language and Environment for Statistical Computing.http://www.R-project.org/ [Google Scholar]
- 50.Gentleman R.C. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5 doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ritchie M.E. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv007. e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lucas A., Jasson S. 1985. Using amap and ctc Packages for Huge Clustering; p. 58. The Newsletter of the R Project Volume 6/5, December 2006 1. [Google Scholar]
- 54.Huber W. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maechler M., Rousseeuw P., Struyf A., Hubert M., Hornik K. 2012. Cluster: Cluster Analysis Basics and Extensions; p. 56. R package version 1. [Google Scholar]
- 56.Paradis E., Claude J., Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 57.Warnes G.R. 2009. gplots: various R programming tools for plotting data. R package version 2. [Google Scholar]
- 58.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li B., Dewey C.N. R.S.E.M: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:1–16. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trapnell C. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wagner G.P., Kin K., Lynch V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theo. Biosci. = Theor. Biowiss. 2012;131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 63.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11 doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.InteractiVenn. 2017. http://www.interactivenn.net/ [Google Scholar]
- 66.Research Computing Centre, 2016. http://www.rcc.uq.edu.au. (Accessed 2016).
- 67.Kraus D. Consolidated data analysis and presentation using an open-source add-in for the Microsoft Excel® spreadsheet software. Med. Writ. 2014;23:25–28. [Google Scholar]
- 68.Quail M. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genom. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crusoe M.R. The khmer software package: enabling efficient nucleotide sequence analysis. F1000Research. 2015;4:900. doi: 10.12688/f1000research.6924.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rana S.B., Zadlock F.J.I.V., Zhang Z., Murphy W.R., Bentivegna C.S. Comparison of de novo transcriptome assemblers and k-mer strategies using the Killifish, Fundulus heteroclitus. PLoS One. 2016;11 doi: 10.1371/journal.pone.0153104. e0153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O'Neil S.T., Emrich S.J. Assessingde novotranscriptome assembly metrics for consistency and utility. BMC Genom. 2013;14 doi: 10.1186/1471-2164-14-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He B. Optimal assembly strategies of transcriptome related to ploidies of eukaryotic organisms. BMC Genom. 2015;16:1–10. doi: 10.1186/s12864-014-1192-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chopra R. Comparisons of de novo transcriptome assemblers in diploid and polyploid species using peanut (Arachis spp.) RNA-Seq data. PLoS One. 2015;9 doi: 10.1371/journal.pone.0115055. e115055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reddy A.S., Marquez Y., Kalyna M., Barta A. Complexity of the alternative splicing landscape in plants. Plant Cell. 2013;25:3657–3683. doi: 10.1105/tpc.113.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duan J., Xia C., Zhao G., Jia J., Kong X. Optimizing de novo common wheat transcriptome assembly using short-read RNA-Seq data. BMC Genom. 2012;13 doi: 10.1186/1471-2164-13-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dharshini S. De novo sequencing and transcriptome analysis of a low temperature tolerant Saccharum spontaneum clone IND 00-1037. J. Biotechnol. 2016;231:280–294. doi: 10.1016/j.jbiotec.2016.05.036. [DOI] [PubMed] [Google Scholar]
- 78.Arrial R.T., Togawa R.C., Brigido Mde M. Screening non-coding RNAs in transcriptomes from neglected species using PORTRAIT: case study of the pathogenic fungus Paracoccidioides brasiliensis. BMC Bioinf. 2009;10:239. doi: 10.1186/1471-2105-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Casu R. Identification of a novel sugar transporter homologue strongly expressed in maturing stem vascular tissues of sugarcane by expressed sequence tag and microarray analysis. Plant Mol. Biol. 2003;52:371–386. doi: 10.1023/a:1023957214644. [DOI] [PubMed] [Google Scholar]
- 80.Casu R.E. Tissue-specific transcriptome analysis within the maturing sugarcane stalk reveals spatial regulation in the expression of cellulose synthase and sucrose transporter gene families. Plant Mol. Biol. 2015 doi: 10.1007/s11103-015-0388-9. [DOI] [PubMed] [Google Scholar]
- 81.Hoang N.V., Furtado A., O’Keeffe A.J., Botha F.C., Henry R.J. Association of gene expression with biomass content and composition in sugarcane. PLoS One. 2017;12 doi: 10.1371/journal.pone.0183417. e0183417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.