Abstract
Probiotics are live microorganisms that provide health benefits to humans. Some lactic acid bacteria (LAB) are probiotic organisms used in the production of fermented foods, such as yogurt, cheese, and pickles. Given their widespread consumption, it is important to understand the physiological state of LAB in foods such as yogurt. However, this analysis is complicated, as it is difficult to separate the LAB from milk components such as solid curds, which prevent cell separation by dilution or centrifugation. In this study, we successfully separated viable LAB from yogurt by density gradient centrifugation. The recovery rate was >90 %, and separation was performed until the stationary phase. Recovered cells were observable by microscopy, meaning that morphological changes and cell viability could be directly detected at the single-cell level. The results indicate that viable LAB can be easily purified from fermented milk. We expect that this method will be a useful tool for the analysis of various aspects of probiotic cells, including their enzyme activity and protein expression.
Keywords: Food analysis, Microbiology
1. Introduction
Some lactic acid bacteria (LAB) are frequently included in yogurt and have beneficial effects on humans, including the improvement of gastrointestinal disorders and the prevention of certain allergies (Nagpal et al., 2012). However, few studies have focused on how specific LAB characteristics cause these effects. LAB can be easily isolated from yogurt (Tabasco et al., 2007); however, the isolated cells must be obtained through culture with adequate selective medium, making their characteristics quite different from those of LAB in milk products. LAB in yogurt are mixed with solid curds, making it difficult to gently separate the cells. Therefore, a simple method for cell isolation from fermented milk is necessary to determine the activity of LAB in fermented milk. For example, measurement of enzyme activity or protein expression of LAB in fermented milk is necessary to isolate LAB in the same physiological state as that in milk. Furthermore, the isolation of a large amount of LAB cells in a short time would be better.
Separation of LAB from yogurt can be accomplished by homogenizing and dissolving curds using alkaline conditions and lysis treatment with proteases or surfactants (Gunasekera et al., 2002). After these processes, the cells are collected by centrifugation. However, this method would be lethal to LAB, resulting in low viability of the separated cells. The immunomagnetic separation method is currently a popular choice for separation of microbial cells from milk (Ertas et al., 2013; Lim et al., 2016; Luciani et al., 2016). This method uses magnetic beads coated by antibodies specific for the target cells. Cells bound to the beads are removed from milk components by magnetic action. However, this method is complicated and expensive. There are some problems with analyzing the behavior of LAB in milk. For example, the observation of morphological change of LAB in milk could not be easily done. Though it would be the basic analysis, the presence of the card makes it difficult. The gene expression of LAB in milk has been investigated by microarray analysis (Azcarate-Peril et al., 2009). mRNA extraction may be hampered by large amounts of milk components. The impact of solid milk components must be resolved to achieve fast separation of cells under mild conditions.
Therefore, we have developed a means to isolate LAB by density gradient centrifugation (DGC) using Percoll. DGC is a useful technique for separating cells of different densities that has been applied to the separation of bacteria from soil (Liu et al., 2010) and aquatic environments (Garrison and Bochdansky, 2015), and to the separation of subpopulations of pure cultured cells (Nishino et al., 2003). Percoll consists of colloidal silica particles. Percoll was used because of its non-toxicity, low viscosity, and ease of preparation at the desired osmolarity and pH (Pertoft, 2000). The ability to easily isolate viable LAB will be valuable for the study of their characteristics in fermented milk.
2. Material and methods
2.1. Bacteria and culture conditions
The LAB Lactococcus lactis ssp. lactis NBRC100933 was cultured at 30 °C in bromocresol purple (BCP) medium. The BCP medium contained 0.5 % polypepton (Japan Pharmaceutical Company, Japan), 0.25 % yeast extract (Difco, USA), 0.1 % glucose, 0.1 % Tween 80, 0.01 % L-cysteine, and 0.006 % bromocresol purple. The pH of the BCP medium was adjusted to 7.0 with NaOH. Cells from overnight cultures were inoculated into skim milk medium [10 % reconstituted skim milk supplemented with 0.1 % yeast extract (Difco) and 1 % glucose] and grown at 30 °C. Cultured cells were harvested at various growth phases. Commercial fermented milk was purchased and used within its prescribed shelf life. Some details on the nature and the composition of these commercial products are found at Table 2.
Table 2.
Application of the DGC method to commercially fermented milk.
| Fermented milk |
aCFUs |
Total cell counts | Recovery rate | |
|---|---|---|---|---|
| before DGC | after DGC | |||
| A | 5.4×108 ± 1.9×107 | 4.9×108 ± 2.6×107 | 3.7×108 ± 1.3×107 | 90.13 ± 1.65 |
| B | 5.3×108 ± 6.5×107 | 4.6×108 ± 4.5×107 | 2.7×108 ± 7.2×107 | 87.27 ± 2.18 |
| C | 2.3×108 ± 1.4×107 | 1.9×108 ± 1.8×107 | 6.7×108 ± 2.0×108 | 82.53 ± 8.87 |
| D | 4.8×108 ± 3.6×107 | 4.2×108 ± 4.2×107 | 5.8×108 ± 1.0×107 | 87.40 ± 2.48 |
A: Plain yogurt containing sugar syrup, pectin, flavor, and tea extract, B: Apple yogurt containing juice, soy milk, flavor, acidifier, and stabilizers (polysaccharides), C: Blueberry yogurt containing fruit pulp, vegetable oil, sugar syrup, flavor, acidifier, and stabilizers (polysaccharides and gelatin), D: Yogurt made with raw milk.
All values are the mean ± standard deviation of triplicates.
CFUs = colony forming units per mL.
2.2. Density gradient centrifugation (DGC)
A 1 mL sample of fermented milk was diluted with 9 mL of phosphate-buffered saline (PBS; pH 6.8). Unless otherwise stated, the diluted cell suspension (1 mL) was layered on top of 10 mL of a Percoll gradient working solution, which contained 3 mL of Percoll (GE Healthcare UK Ltd.) and 7 mL of PBS. The final Percoll concentration was 30 % (vol/vol). The mixture was centrifuged at 2 610 × g for 30 min at 25 °C in a swing-bucket centrifuge (Kubota Model 2410, Tokyo, Japan) to pellet the separated cells, which were resuspended in 1 mL of PBS.
2.3. Determination of recovery rate and culturability
Separated cell suspensions were diluted with saline to equal cell concentrations to determine the recovery rate and culturability of intact and centrifuged samples. Total cell counts (TCs) were obtained by epifluorescence microscopy (Olympus CX-31, Tokyo, Japan) after staining with 4,6-diamidino-2-phenylindole (DAPI; Wako, Osaka, Japan). Plate counts were determined by counting the number of CFUs on BCP agar. The recovery rate was calculated using the following equation: Recovery rate = (CFUs in 1 mL of sedimented cell suspension after DGC/CFUs in 1 mL of diluted cell suspension before DGC) × 100. Culturability was expressed as (CFUs after DGC/TCs after DGC) × 100.
3. Results and discussion
Consumption of LAB in yogurt is beneficial to human health. However, the physiological characteristics of LAB are not well understood, as viable LAB cannot be easily separated from milk components. We investigated the separation of LAB from fermented milk by DGC using Percoll.
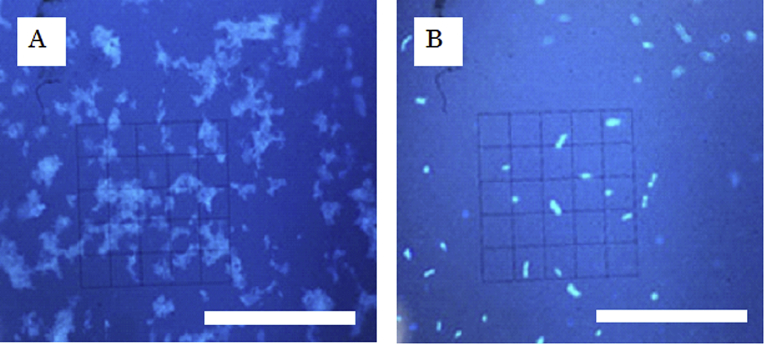
Fig. 1 shows microscopic images before and after LAB separation from fermented milk. Before separation, the milk components formed large aggregates with the microbial cells, even when blended with the PBS buffer, making it difficult to discriminate LAB from abiotic substances (Fig. 1A). After separation, LAB were clearly observable (Fig. 1B), enabling morphological analysis and quantitation.
Fig. 1.
Separation of LAB from fermented milk by DGC (bar, 50 μm). A. Before DGC separation, aggregated substances were mixtures of LAB and milk components. B. After DGC separation, LAB cells were clearly observable.
Table 1 shows the recovery rates during various growth phases. The recovery rate was >90 % during the exponential (4 h) and stationary (24 h) phases, but decreased to approximately 25 % during the death phase (168 h). This decrease was confirmed during the late stationary phase (96 h). The culturability of the separated cells showed similar values during the exponential and stationary phases, and drastically decreased during the death phase.
Table 1.
Recovery rate and culturability during various growth phases.
| Growth phase |
aCFUs |
Total cell counts | Recovery rate | Culturability | |
|---|---|---|---|---|---|
| before DGC | after DGC | ||||
| Exponential | 4.8×108 ± 7.4×107 | 4.7×108 ± 5.4×107 | 4.1×108 ± 1.6×107 | 98.87 ± 4.97 | 114.76 ± 8.80 |
| Stationary | 1.7×109 ± 1.4×108 | 1.6×109 ± 6.1×107 | 1.9×109 ± 1.1×108 | 94.66 ± 5.34 | 85.51 ± 7.69 |
| Late stationary | 4.5×108 ± 2.9×107 | 2.4×108 ± 4.4×107 | 1.9×109 ± 2.0×107 | 53.02 ± 11.33 | 12.29 ± 2.41 |
| Death | 3.1×106 ± 2.7×106 | 8.4×105 ± 7.4×105 | 1.6×109 ± 2.0×107 | 27.91 ± 6.60 | 0.05 ± 0.05 |
All values are the mean ± standard deviation of triplicates.
CFUs = colony-forming units per mL.
We also tested the method with commercially fermented milk products. Table 2 shows the results for various yogurts. The recovery rate was >80 % in all samples, indicating that this method is applicable to commercial yogurt.
LAB were easily collected from yogurt by this method, which can be performed in only 40 minutes and is inexpensive. DGC is usually performed by ultracentrifugation, but this separation involves centrifugation with a swing-bucket rotor at low speed. The separated LAB actively proliferated, and their colony-forming ability was nearly equal to that of non-treated LAB. As LAB can be separated from yogurt in a short period, the state of the separated LAB should closely resemble the state of LAB in milk. The simplicity of this method may be the key to obtaining accurate results that reflect the state of LAB in fermented milk products.
Studies on the characteristics of LAB in yogurt have typically analyzed isolated colonies on plates. These results would not reflect the conditions in milk, as these cells are cultured in medium. Flow cytometry has also been applied (Gunasekera et al., 2000), but has some limitations. First, the recovered cell numbers are small, which is a disadvantage for analyses that require many cells (e.g., determination of enzyme activity or enzyme purification or proteomic analysis). Second, the cell sorting process, as well as the fluorescent staining required, may stress the cells, reducing their viability and changing their state. The DGC method was capable of collecting >80 % of the LAB in the samples, in an active, unstressed state.
Pretreatment with the DGC method may be effective for molecular biology approaches, as has been reported for similar approaches (Mohania et al., 2008, Stefanis et al., 2016). For example, LAB protein expression analysis was previously performed by microarray using mRNA extracted directly from fermented milk (Azcarate-Peril et al., 2009), but this extraction method resulted in low mRNA recovery owing to the presence of a large quantity of milk components. Separation by our method prior to mRNA extraction could improve the mRNA yield dramatically, resulting in more expansive and accurate results. The enzyme activity of LAB in the yogurt could also be more easily measured by this method. Generally, it is difficult to distinguish between LAB enzymes and milk casein in yogurt. Separation of these enzymes from casein would make enzymatic analysis easier and more reliable.
The viability of the separated cells remained >90 % during the exponential and stationary phases with laboratory-fermented milk samples, and >80 % with commercial yogurts, demonstrating the broad utility of this method. This method could be used with other matrices such as probiotic juice or cheese. However, the recovery rate of viable cells decreased in the death phase, even though the total cells (live and dead cells) were recovered successfully. This tendency could not be resolved by adjusting the centrifugation conditions or the composition of the Percoll gradient working solution. We previously reported that the buoyant cell density became lighter in stationary phase Vibrio parahaemolyticus (Nishino et al., 2003); we ascertained that the same phenomenon occurred in LAB (data not shown). Therefore, this decrease in the recovery rate of viable cells in the death phase may be caused by the buoyant cell density change. In addition, the loss of culturability suggested that the separation operation might have stressed the cells after the late stationary phase.
In conclusion, we have developed an easy, fast, and inexpensive method for separation of LAB from yogurt. This method will be useful in the elucidation of the roles of LAB in fermented milk.
Declarations
Author contribution statement
Tomohiko Nishino: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Yusuke Matsuda, Yuna Yamazaki: Performed the experiments; Analyzed and interpreted the data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We thank Ms. Y. Aihara for technical assistance.
References
- Azcarate-Peril M.A., Tallon R., Klaenhammer T.R. Temporal gene expression and probiotic attributes of Lactobacillus acidophilus during growth in milk. J. Dairy Sci. 2009;92:870–886. doi: 10.3168/jds.2008-1457. [DOI] [PubMed] [Google Scholar]
- Ertas N., Gonulalan Z., Yildirim Y. Detection of Escherichia coli O157:H7 using immunomagnetic separation and mPCR in Turkish foods of animal origin. Lett. Appl. Microbiol. 2013;57:373–379. doi: 10.1111/lam.12124. [DOI] [PubMed] [Google Scholar]
- Garrison C.E., Bochdansky A.B. A simple separation method for downstream biochemical analysis of aquatic microbes. J. Microbiol. Methods. 2015;111:78–86. doi: 10.1016/j.mimet.2015.01.025. [DOI] [PubMed] [Google Scholar]
- Gunasekera T.S., Attfield P.V., Veal D.A. A flow cytometry method for rapid detection and enumeration of total bacteria in milk. Appl. Environ. Microbiol. 2000;66:1228–1232. doi: 10.1128/aem.66.3.1228-1232.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera T.S., Sørensen A., Attfield P.V. Inducible gene expression by nonculturable bacteria in milk after pasteurization. Appl. Environ. Microbiol. 2002;68:1988–1993. doi: 10.1128/AEM.68.4.1988-1993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M.C., Lee G.H., Huynh D.T.N. Biological preparation of highly effective immunomagnetic beads for the separation, concentration, and detection of pathogenic bacteria in milk. Colloids Surf. B Biointerfaces. 2016;145:854–861. doi: 10.1016/j.colsurfb.2016.05.077. [DOI] [PubMed] [Google Scholar]
- Liu J., Li J., Feng L. An improved method for extracting bacteria from soil for high molecular weight DNA recovery and BAC library construction. J. Microbiol. 2010;48:728–733. doi: 10.1007/s12275-010-0139-1. [DOI] [PubMed] [Google Scholar]
- Luciani M., Di Febo T., Zilli K. Rapid detection and isolation of Escherichia coli O104:H4 from milk using monoclonal antibody-coated magnetic beads. Front. Microbiol. 2016;15:942. doi: 10.3389/fmicb.2016.00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohania D., Nagpal R., Kumar M. Molecular approaches for identification and characterization of lactic acid bacteria. J. Dig. Dis. 2008;9:190–198. doi: 10.1111/j.1751-2980.2008.00345.x. [DOI] [PubMed] [Google Scholar]
- Nagpal R., Kumar A., Kumar M. Probiotics, their health benefits and applications for developing healthier foods: a review. FEMS Microbiol. Lett. 2012;334:1–15. doi: 10.1111/j.1574-6968.2012.02593.x. [DOI] [PubMed] [Google Scholar]
- Nishino T., Nayak B.B., Kogure K. Density-dependent sorting of physiologically different cells of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 2003;69:3569–3572. doi: 10.1128/AEM.69.6.3569-3572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertoft H. Fractionation of cells and subcellular particles with Percoll. J. Biochem. Biophys. Methods. 2000;44:1–30. doi: 10.1016/s0165-022x(00)00066-x. [DOI] [PubMed] [Google Scholar]
- Stefanis C., Mantzourani I., Plessas S. Reviewing classical and molecular techniques regarding profiling of probiotic character of microorganisms. Curr. Res. Nutr. Food Sci. 2016;4:27–47. [Google Scholar]
- Tabasco R., Paarup T., Janer C. Selective enumeration and identification of mixed cultures of Streptococcus thermophilus, Lactobacillus delbrueckii subsp. bulgaricus, L. acidophilus, L. paracasei subsp. paracasei and Bifidobacterium lactis in fermented milk. Int. Dairy J. 2007;17:1107–1114. [Google Scholar]