Abstract
Membrane proteins are functionally regulated by the composition of the surrounding lipid bilayer. The late endosomal compartment is a central site for the generation of ceramide, a bioactive sphingolipid, which regulates responses to cell stress. The molecular interactions between ceramide and late endosomal transmembrane proteins are unknown. Here, we uncover in atomistic detail the ceramide interaction of Lysosome Associated Protein Transmembrane 4B (LAPTM4B), implicated in ceramide-dependent cell death and autophagy, and its functional relevance in lysosomal nutrient signaling. The ceramide-mediated regulation of LAPTM4B depends on a sphingolipid interaction motif and an adjacent aspartate residue in the protein’s third transmembrane (TM3) helix. The interaction motif provides the preferred contact points for ceramide while the neighboring membrane-embedded acidic residue confers flexibility that is subject to ceramide-induced conformational changes, reducing TM3 bending. This facilitates the interaction between LAPTM4B and the amino acid transporter heavy chain 4F2hc, thereby controlling mTORC signaling. These findings provide mechanistic insights into how transmembrane proteins sense and respond to ceramide.
Short abstract
A ceramide-regulated element in the late endosomal protein LAPTM4B controls its interaction with the amino acid transporter heavy chain 4F2hc.
Introduction
It has been estimated that 20–30% of genes encode membrane-spanning proteins,1 and their proper function depends on favorable interactions with the lipids surrounding them.2 Sphingolipids in particular constitute one of the key lipid classes able to modulate protein activity. They can influence membrane-spanning proteins by modulating physical membrane properties such as fluidity or membrane thickness.3 They can also act as bona fide ligands4,5 or govern membrane protein behavior through direct molecular interactions as annular lipids.6 Overall, annular lipids regulate the function and conformation of transmembrane (TM) proteins by optimizing membrane thickness to match the length of the hydrophobic domain of the protein, and also by specific interactions with structural features of membrane-spanning domains.6−9 Further, sphingolipids foster the stability of cellular membranes and may segregate into functional domains with cholesterol,10,11 thereby promoting protein function.
The functional significance of interactions between sphingolipids and TM domains was corroborated by the discovery of a specific C18-sphingomyelin interaction motif (VXXTLXXIY) in the single membrane-spanning vesicular transport protein p24.12 Thereafter, additional putative sphingolipid-binding motifs conforming to the relaxed motif ([V/I/T/L]XX[[V/I/T/L][V/I/T/L]XX[V/I/T/L][F/W/Y]) were identified in over 600 proteins, four of which were experimentally verified to show sphingolipid binding.13 However, for these proteins, nothing is known regarding their regulation by sphingolipids. Over 10 proteins that contain sphingolipid interaction motifs localize to endosomal/lysosomal compartments12,13 that represent central organelles in sphingolipid degradation. Sphingomyelin is the most abundant sphingolipid in mammals, and its immediate catabolic product ceramide contributes to the pathology of several diseases including cancer14 and obesity.15 Ceramides have been shown to affect the composition of membrane domains16 and to regulate cell signaling by promoting receptor clustering.17 However, the mechanisms underlying these phenomena are poorly understood, and bona fide sphingolipid interaction motifs that preferentially interact with membrane ceramide have not been identified.
We recently showed that the Lysosome Associated Protein Transmembrane 4B (LAPTM4B) interacts with ceramide and promotes its clearance from late endosomal compartments, thereby regulating cell sensitivity to chemotherapy-induced cell death.18 LAPTM4B was originally identified as a highly upregulated transcript in hepatocellular carcinoma,19 and has been shown to associate with poor outcome in breast cancers20 and acute myeloid leukemia.21 Studies have also indicated LAPTM4B to be involved in the regulation of mTORC signaling and autophagy.22−26 LAPTM4B was shown to promote the recruitment of the amino acid transporter 4F2hc/LAT1 to lysosomes, thereby enhancing lysosomal leucine uptake and stimulating mTORC1 activity.22 Interestingly, ceramide can induce the internalization of nutrient transport proteins, including 4F2hc.27,28
Here, by combining atomistic computer simulations with biochemical and cell biological experiments, we identify a functional ceramide-regulated unit in the third TM domain of LAPTM4B and show that it consists of a sphingolipid-binding motif and an adjacent aspartate residue. We provide evidence that the interaction of LAPTM4B with ceramide plays a role in the 4F2hc-mediated regulation of mTORC1. Introducing mutations in the ceramide interaction motif leads to altered association between LAPTM4B and 4F2hc and dysregulated mTORC1 signaling. Based on our results, we propose that ceramide controls the interaction of LAPTM4B with 4F2hc and thereby enhances downstream nutrient signaling.
Results
Identification of the Ceramide Interaction Site of LAPTM4B
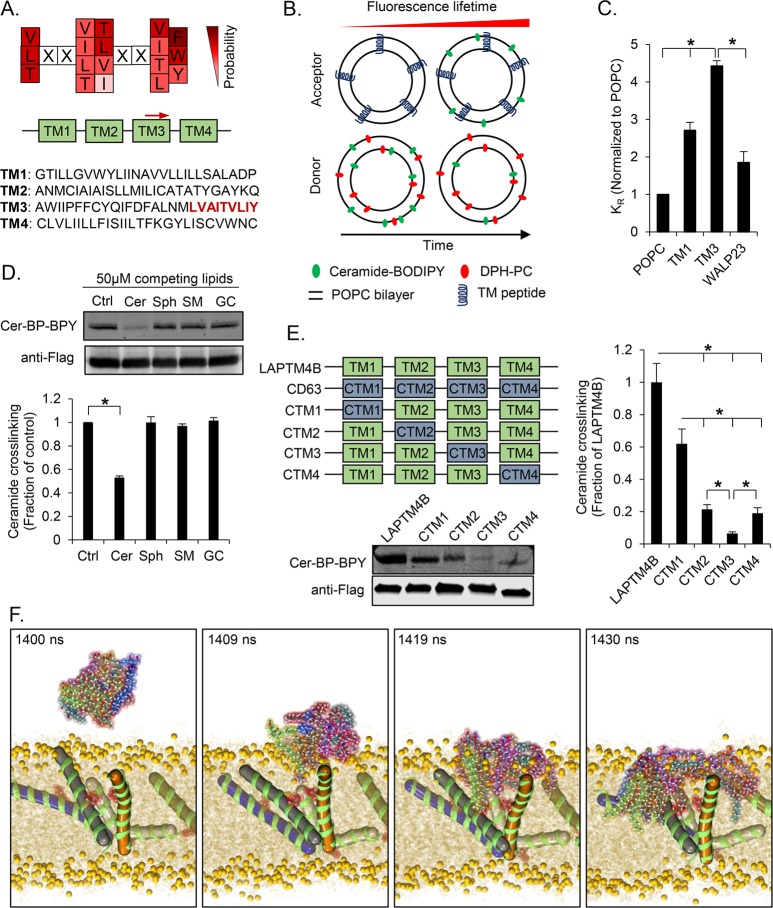
We have previously shown that LAPTM4B binds a cross-linkable ceramide with high specificity.18 Now, we aimed to identify the specific structural features that are important in this interaction. Analysis of the LAPTM4B primary structure suggested that it contains four transmembrane domains (TM1–TM4) each 23 amino acids long (predicted by TMHMM server 2.0; http://www.cbs.dtu.dk/services/TMHMM/). Further examination revealed that LAPTM4B contains two amino acid stretches that match the postulated relaxed sphingolipid-binding motif (Figure 1A),12,13 one located in TM3 and the other one partially overlapping with the predicted TM1. To assess whether these TM domains play a role in the interaction between ceramide and LAPTM4B, we established an in vitro lipid transfer assay that measures the association of a fluorescent ceramide probe with large unilamellar vesicles (LUVs) containing synthetic LAPTM4B-derived TM peptides (Figure 1B, Figure S1A,B). In this assay, ceramide–BODIPY displayed a substantially higher association with TM3-LUVs compared to the control LUVs containing WALP23 (Figure 1C, Figure S1C,D). In contrast, LUVs hosting TM1 showed only a marginally higher uptake of the ceramide probe compared to WALP23-LUVs (Figure 1C) or TM2-LUVs (Figure S1D), which lack the consensus sphingolipid-binding sequence. These results suggest that the consensus sphingolipid-binding motif in TM3 may be functional in the ceramide interaction.
Figure 1.
LAPTM4B contains a functional sphingolipid-binding motif. (A) Schematic representation of a conserved sphingolipid-binding signature (top panel). All four LAPTM4B transmembrane domains (TMs) are shown (bottom panel). The position of the putative relaxed sphingolipid-binding motif is indicated by a red arrow, and its sequence is highlighted in red. (B) The association of ceramide–BODIPY with LAPTM4B-TM-containing acceptor LUVs was studied by a FRET-based in vitro lipid transfer assay. DPH-PC served as FRET donor and ceramide–BODIPY as FRET acceptor. (C) Quantification of ceramide–BODIPY affinity for TM peptide-containing LUVs from n = 3 experiments, mean ± SEM, p* < 0.05. LUVs without peptides (POPC) and LUVs containing WALP23 served as controls. (D) Cross-linking of Cer-BP-BPY to LAPTM4B in the presence of 50 μM competing lipids (ceramide, Cer; sphingosine, Sph; sphingomyelin, SM; glucosylceramide, GC) was assessed by immunoprecipitation, in-gel fluorescence imaging of cross-linked Cer-BP-BPY, and immunoblotting of precipitated Flag-tagged LAPTM4B. Top panel, representative experiment; bottom panel, quantification of cross-linked Cer-BP-BPY for competing lipids compared to control (n = 3 experiments, mean ± SEM, p* < 0.05). (E) Schematic representation of LAPTM4B, CD63, and LAPTM4B TM domain mutants. Cross-linking of Cer-BP-BPY to LAPTM4B WT and mutants was assessed as described in part D. Bottom panel, representative experiment; right panel, quantification of Cer-BP-BPY cross-linking to TM mutants compared to LAPTM4B (n = 3 experiments, mean ± SEM, p* < 0.05). (F) Intake of self-assembled ceramide micelles into TM3 peptide-containing membranes by atomistic simulations.
TM3 Is Necessary for Ceramide Binding to LAPTM4B in Cells
Next, we addressed whether the in vitro lipid transfer experiments reflect the behavior of LAPTM4B in the cellular context. For this purpose, we generated a LAPTM4B knockout cell line (Figure S2A,B), and expressed wild type (WT) or mutant LAPTM4B constructs on the null background (Figure S2C,D). The previously characterized cross-linkable probe ceramide-BP-BPY18 (Figure S2E) was used as a reporter for the ceramide–protein interaction. The cross-linking of ceramide-BP-BPY to LAPTM4B was competed by an excess of C6-ceramide, but not by the immediate metabolites sphingosine, C6-sphingomyelin, or C6-glucosylceramide, suggesting that the probe specifically reports interaction of ceramide with LAPTM4B (Figure 1D). Initially, four LAPTM4B chimeras were generated with single TM domains exchanged for the corresponding domain from the tetraspanin CD63 used as a control protein (mutant constructs CTM1–CTM4, Figure 1E and Figure S2). Exchanging TM2, TM3, or TM4 resulted in a reduced interaction with cross-linkable ceramide, suggesting that these domains cooperate for efficient ceramide binding in the holoprotein (Figure 1E). In agreement with the in vitro experiments, the TM3-specific LAPTM4B mutant (CTM3) displayed the lowest level of ceramide-BP-BPY cross-linking (Figure 1E), suggesting that this TM domain is of particular importance for the ceramide interaction with LAPTM4B also in cells.
Atomistic Dissection of TM3–Ceramide Interaction
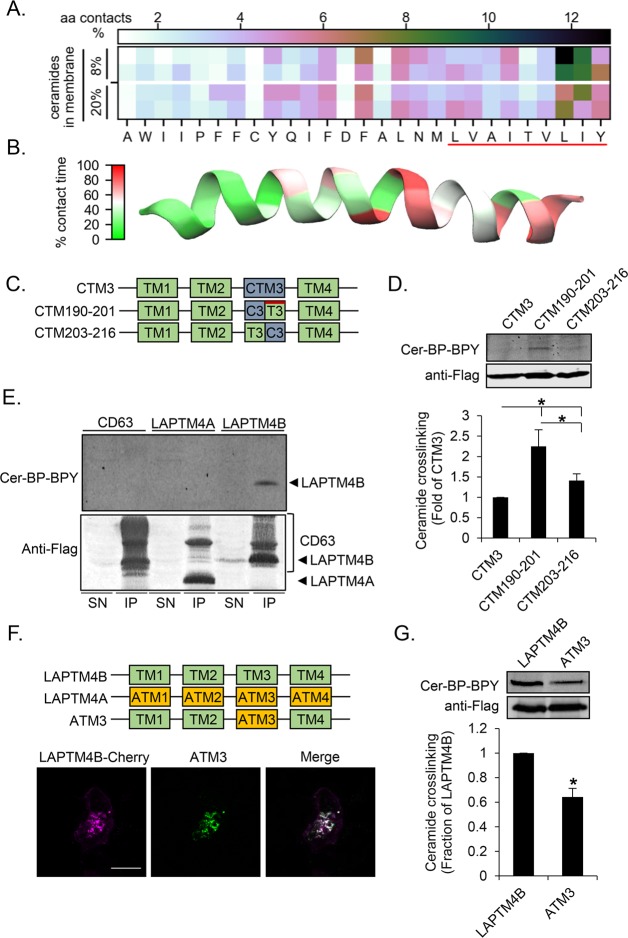
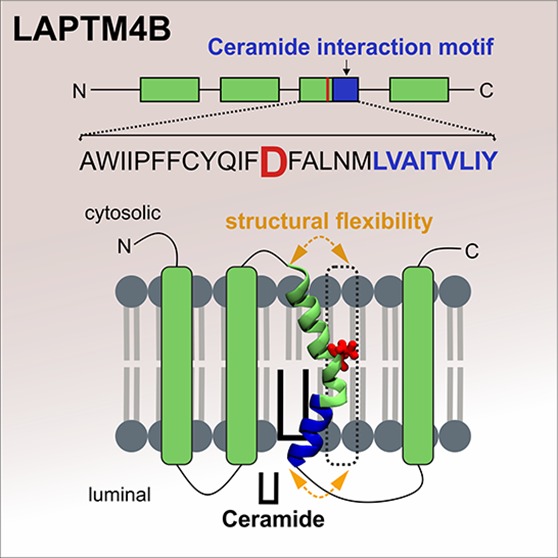
To elucidate how LAPTM4B-TM3 interacts with ceramide, we next modeled the interactions by atomistic simulations. POPC membrane systems with or without transmembrane peptides were set up to mimic the composition of the LUVs in the biochemical assay (Figure 1B, Tables S1–S3). C16-ceramides (N-palmitoyl-D-erythro-sphingosine) were then included as monomers in the aqueous phase. The ceramides rapidly formed micelles in the aqueous phase, and did not subsequently insert into peptide-free or POPC-WALP23 bilayers during 5500 and 5000 ns, respectively (Table S3). In contrast, membranes containing LAPTM4B-TM3 internalized micellar ceramide in the time range 190–1415 ns in four separate simulations (Figure 1F, Movie S1). These simulations revealed the molecular mechanism for the interaction of ceramides with LAPTM4B-TM3-containing membranes. First, TM3 was found to be flexible around the central aspartate residue (D202) which caused shielded hydrophobic groups to become exposed to the aqueous phase (Figure 1F, Table S4), thereby facilitating the incorporation of ceramide into the lipid bilayer. Second, simulated systems where ceramides were introduced randomly and symmetrically within the membrane indicated that ceramide predominantly interacts with TM3 in the region that constitutes the postulated sphingolipid-binding motif (Figure 2A,B).
Figure 2.
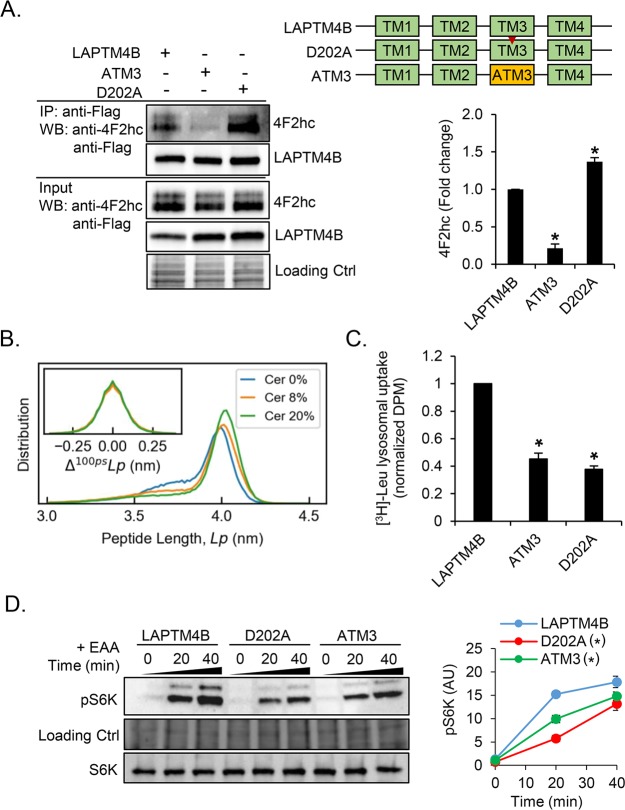
The LAPTM4B sphingolipid-binding motif in TM3 is necessary for the interaction with cross-linkable ceramide. (A) Ceramide atom contacts per residue with LAPTM4B-TM3 in membranes containing 8 or 20 mol % ceramide. The red line highlights the sphingolipid interaction motif. (B) Heat map of TM3 showing percent of the simulation time that each residue has ≥5 contacts with ceramide in a membrane containing 8 mol % ceramide. (C) Schematic representation of LAPTM4B TM3 mutants. The red bar indicates the sphingolipid interaction motif. (D) Cross-linking of Cer-BP-BPY to CTM3, CTM190-201, and CTM203-216 mutants was assessed by immunoprecipitation, in-gel fluorescence imaging of cross-linked Cer-BP-BPY, and immunoblotting of precipitated Flag-tagged LAPTM4B mutant proteins. Top panel, representative experiment; bottom panel, quantifications of cross-linked Cer-BP-BPY to half-TM mutants compared to CTM3 (n = 3 experiments, mean ± SEM, p* < 0.05). (E) Cross-linking of Cer-BP-BPY to LAPTM4B WT, and the control proteins CD63 and LAPTM4A, was assessed as described in part D. SN, supernatant; IP, immunoprecipitate. (F) Schematic representation of LAPTM4B, LAPTM4A, and ATM3 mutants. The cellular localization of LAPTM4B and ATM3 in A431 cells was visualized by immunofluorescence microscopy. Scale bar 20 μm. (G) Cross-linking of Cer-BP-BPY to LAPTM4B and the ATM3 mutant was assessed as described in part D. Top panel, representative experiment; bottom panel, quantification of Cer-BP-BPY cross-linked to ATM3 compared to LAPTM4B (n = 3 experiments, mean ± SEM, p* < 0.05).
Ceramide Interaction Motif Promotes Ceramide Binding to LAPTM4B in Cells
To assess the functionality of the identified ceramide interaction motif in TM3, we reinserted the wild type LAPTM4B sequences in either the cytosolic or luminal leaflet-spanning parts of the non-ceramide-binding CTM3 mutant (CTM190-201 and CTM203-216, respectively, see Figure 2C and Figure S2C,D). Reintroducing the wild type sphingolipid-binding motif in the late endosomal luminal membrane leaflet (CTM190-201) substantially rescued the ceramide interaction, while reintroducing the wild type sequence in the cytosolic leaflet (CTM203-216) had a weaker effect (Figure 2D). As the mutant proteins CTM3, CTM190-301, and CTM203-216 predominantly localized to the endoplasmic reticulum (Figure S3), we next generated a ceramide interaction domain mutant that retained the same localization as the wild type protein. For this purpose, we used LAPTM4A, a LAPTM4B paralog that shows negligible binding to cross-linkable ceramide (Figure 2E). LAPTM4A and LAPTM4B display 89% similarity over the length of TM3, with the nonconservative mutations residing in the ceramide interaction motif (Figure S2C,D). A mutant LAPTM4B protein containing the third TM domain from LAPTM4A (ATM3) colocalized with the wild type protein (Figure 2F) and displayed reduced interaction with cross-linkable ceramide (Figure 2G). Taken together, these results indicate that efficient LAPTM4B interaction with cross-linkable ceramide in cells requires an intact sphingolipid-binding motif in TM3.
Aspartate 202 in TM3 Is Necessary for the Ceramide Interaction with LAPTM4B in Cells
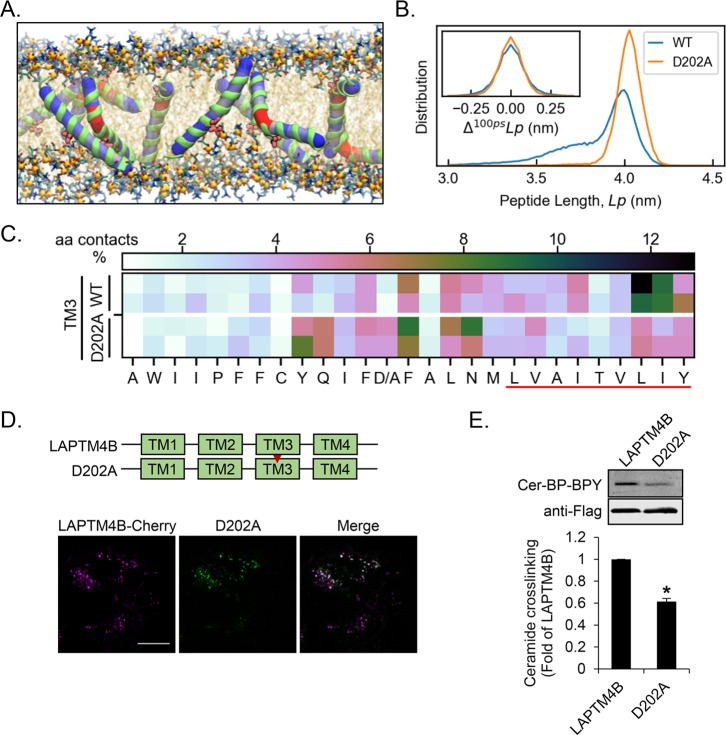
A striking observation from the atomistic simulations was the presence of a flexible helix region surrounding the central aspartate of TM3 (Figure 3A, see also Figure S1E). It has been shown that the acidic amino acids aspartate and glutamate can affect the membrane positioning of hydrophobic α-helices.29,30 Interestingly, the sphingolipid-binding motif in p2412 is accompanied by a membrane-embedded acidic glutamate residue. This feature is absent from the structurally similar but non-sphingolipid-binding protein p23.12 We therefore considered that acidic residues in TM domains might play an important role in protein–sphingolipid interactions.
Figure 3.
Aspartate 202 in TM3 of LAPTM4B is central for the ceramide interaction. (A) LAPTM4B-derived TM3 can bend in the region surrounding the central aspartate shown in red (van der Waals representation). (B) The D202A mutation reduces the flexibility of TM3 (increased peptide length) and dynamics (change in peptide length per 100 ps interval) compared to the WT. (C) Ceramide interaction with WT-TM3 and the D202A mutant. The WT simulation data are the same as in Figure 2A and included for comparison. The red line highlights the sphingolipid-binding motif. (D) Schematic representation of LAPTM4B WT and the D202A point mutant. The cellular localization of LAPTM4B and the D202A mutant in A431 cells was visualized by immunofluorescence microscopy. Scale bar 20 μm. (E) Cross-linking of Cer-BP-BPY to LAPTM4B and D202A mutant was assessed by immunoprecipitation, in-gel fluorescence imaging of cross-linked Cer-BP-BPY, and immunoblotting of precipitated Flag-tagged LAPTM4B. Top panel, representative experiment; bottom panel, quantification of cross-linked Cer-BP-BPY for D202A compared to LAPTM4B (n = 3 experiments, mean ± SEM, p* < 0.05).
In atomistic simulations, exchange of the central aspartate for the more hydrophobic alanine (D202A) reduced the dynamic flexibility of TM3 (Figure 3B). Compared to the wild type TM3-containing bilayers, the TM3-D202A membranes were more ordered, and displayed fewer contacts between hydrophobic groups and the aqueous phase (Tables S4–S9). This was accompanied by a tendency for delayed ceramide incorporation into TM3-D202A-containing bilayers (ranging from 1000 to 2970 ns) compared to the wild type TM3 bilayers (190–1415 ns). In the D202A mutant, the contact points with membrane-inserted ceramide were shifted toward the center of the peptide (Figure 3C). However, the luminal leaflet-spanning portion of TM3-D202A, which contains the sphingolipid-binding motif, was still the favored region for ceramide interaction. To investigate the functional role of the aspartate residue for ceramide binding in cells, we generated an aspartate to alanine point mutant D202A. The D202A mutant colocalized with wild type LAPTM4B in endosomes (Figure 3D), yet it displayed a significantly reduced interaction with cross-linkable ceramide (Figure 3E). Taken together, D202 and the sphingolipid-binding motif in the luminal membrane leaflet are critical for ceramide interaction. The kink in the TM helix induced by D202 may facilitate ceramide access to TM3 and may promote ceramide intake, e.g., from neighboring membranes.
Ceramide Facilitates the Interaction between LAPTM4B and 4F2hc in Late Endosomes
To gain insight into the nanoenvironment in which LAPTM4B operates in cells, we performed a mass spectrometric screen to identify specific LAPTM4B interacting proteins. Proteins identified from at least five peptides in the LAPTM4B samples, and that were simultaneously enriched by more than 3-fold compared to the negative control (coprecipitation with CD63), were considered as specific hits. This yielded 18 LAPTM4B interacting proteins (Table S10). Nine of the identified interaction partners belong to the transporter or solute carrier protein families, suggesting a function for LAPTM4B in transmembrane transport processes. The top hit in our screen was the leucine transporter heavy chain 4F2hc/SLC3A2/CD98hc, supporting the observations by Milkereit et al.22 Co-immunoprecipitation experiments confirmed that 4F2hc interacts specifically with LAPTM4B, but not with the paralog LAPTM4A or with CD63 (Figure S4). To investigate if the 4F2hc-LAPTM4B interaction is regulated by ceramide, we treated cells with bacterial sphingomyelinase (bSMase) that hydrolyzes sphingomyelin to ceramide. This enhanced the interaction between LAPTM4B and 4F2hc (Figure 4A, Figure S4), but did not cause 4F2hc to interact with LAPTM4A or CD63, suggesting that the interaction is specific (Figure S4).
Figure 4.
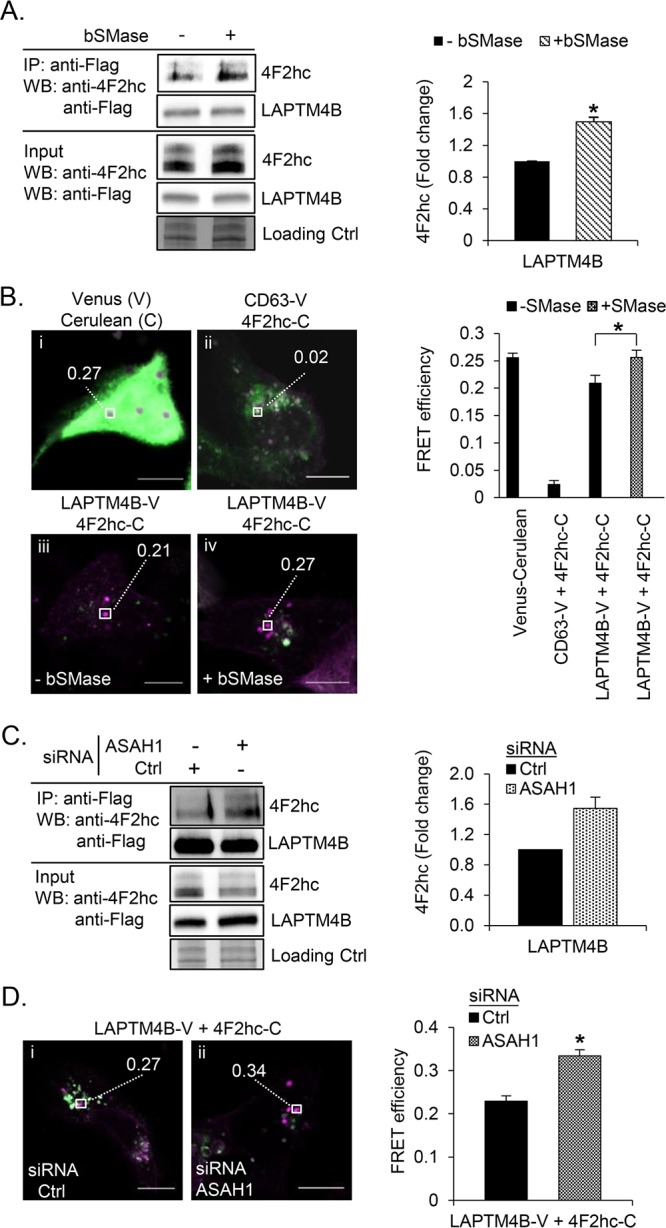
Ceramide modulates the interaction between LAPTM4B and 4F2hc. (A) LAPTM4B-3xFlag expressing cells were treated with bSMase (50 mU/mL, 30 min), and the interaction of LAPTM4B with endogenous 4F2hc was assessed by Co-immunoprecipitation and Western blotting. Left panel, representative experiment; right panel, quantification of n = 3 experiments, mean ± SEM, p* < 0.05. (B) A431 cells were treated with bSMase (50 mU/mL, 30 min), and the interaction of LAPTM4B-V (green) and 4F2hc-C (magenta) was assessed by FRET. Left panel, representative overlay images of cells co-overexpressing the indicated FRET pairs; right panel, quantification of n = 5 independent FRET experiments, mean ± SEM, p* < 0.05 [endosomes per condition: (i) Venus–Cerulean, n = 36; (ii) CD63-V/4F2hc-C, n = 38; (iii) LAPTM4B-V/4F2hc-C without bSMase, n = 51; and (iv) LAPTM4B-V/4F2hc-C with bSMase, n = 43]. Scale bar: 10 μm. (C) The interaction of LAPTM4B with endogenous 4F2hc in ASAH1-silenced LAPTM4B-3xFlag expressing cells was assessed by Co-immunoprecipitation followed by Western blotting. Left panel, representative experiment; right panel, quantification of n = 3 experiments, mean ± SEM, p* < 0.05. (D) ASAH1 was silenced in cells, and the interaction of LAPTM4B-V with 4F2hc-C was assessed by FRET. Left panel, representative FRET images; right panel, quantification of n = 3 independent experiments, mean ± SEM, p* < 0.05 [endosomes per condition: (i) LAPTM4B-V/4F2hc-C Ctrl siRNA, n = 55; and (ii) LAPTM4B-V/4F2hc-C ASAH1 siRNA, n = 60].
To determine the subcellular localization of the LAPTM4B interaction with 4F2hc we utilized fluorescence resonance energy transfer (FRET) in cells expressing LAPTM4B-venus (acceptor fluorophore) and 4F2hc-cerulean (donor fluorophore). Donor and acceptor fluorophores fused by a short amino acid linker were used as a positive control, and cells coexpressing 4F2hc-cerulean and CD63-venus were used as a negative control. FRET between the LAPTM4B and 4F2hc constructs was observed in punctate intracellular structures, in line with the observation that overexpressed LAPTM4B recruits 4F2hc to lysosomes.22 Treating the cells with bSMase further increased the FRET efficiency in these organelles (Figure 4B). Since 4F2hc functions as the heavy chain of the leucine transporter 4F2hc/LAT1, we tested whether leucine uptake would be affected by LAPTM4B knockout or overexpression. Interestingly, while 4F2hc depletion reduced the Na+-independent leucine uptake by ∼50%, LAPTM4B knockout or overexpression did not affect leucine uptake from the extracellular environment (Figure S5). These observations are in line with the idea that the functionally relevant interaction between LAPTM4B and 4F2hc takes place in the late endosomal compartment rather than on the plasma membrane.22
To further address how late endosomal ceramide affects the interaction between LAPTM4B and 4F2hc, we attenuated ceramide degradation in this compartment by depleting cells of acid ceramidase (ASAH1). Co-immunoprecipitation (Figure 4C) and FRET experiments (Figure 4D) showed that ASAH1-depleted cells have increased interaction between LAPTM4B and 4F2hc compared to controls. These findings support a role for late endosomal ceramide in regulating the interaction of LAPTM4B and 4F2hc.
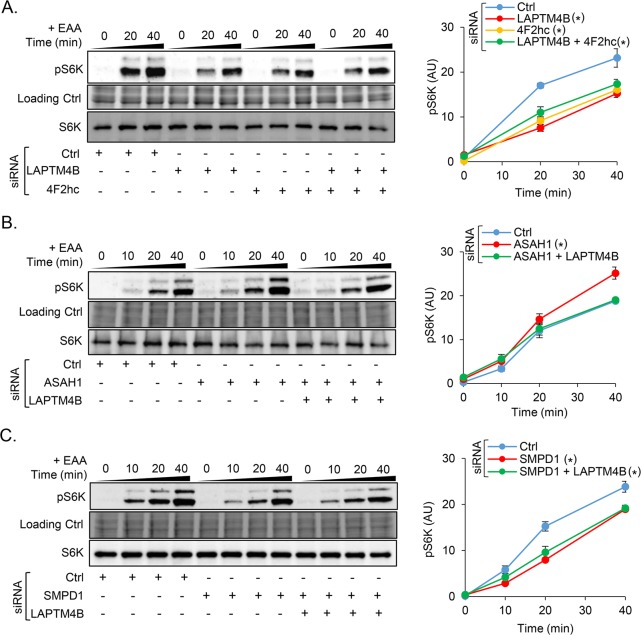
LAPTM4B Promotes S6K Phosphorylation in a Ceramide- and 4F2hc-Dependent Manner
LAPTM4B has been shown to regulate mTORC1 signaling by recruiting 4F2hc to lysosomes.22 In line with this, we observed decreased phosphorylation of the mTORC1 substrate S6K in LAPTM4B knockout cells (Figure S6A). This effect was reversed by reintroducing LAPTM4B (Figure S6B) indicating that it is specific. The LAPTM4B-dependent regulation of S6K was also observed by gene silencing; cells treated with siRNA, targeting either LAPTM4B or 4F2hc, displayed reduced phosphorylation of S6K (Figure 5A). Codepleting cells of LAPTM4B and 4F2hc did not have an additional inhibitory effect on S6K phosphorylation (Figure 5A), suggesting that LAPTM4B and 4F2hc function in the same pathway. Next, we silenced ASAH1 to address whether ceramide generated in the late endosomal compartments regulates S6K via LAPTM4B. Cells depleted of ASAH1 displayed increased levels of phospho-S6K, and this effect was reversed in cells codepleted of ASAH1 and LAPTM4B (Figure 5B). Moreover, depleting cells of acid sphingomyelinase (SMPD1) to reduce late endosomal ceramide levels also caused a reduction in S6K phosphorylation (Figure 5C), strengthening the notion that late endosomal ceramide is involved in this signaling pathway. Codepleting cells of SMPD1 and LAPTM4B did not have an additional inhibitory effect on S6K phosphorylation, suggesting a shared pathway (Figure 5C). Similar effects were observed in HeLa cells depleted of SMPD1 and LAPTM4B (Figure S6C). This implies that an analogous regulation also operates in other cancer cell models.
Figure 5.
Ceramide-enhanced phosphorylation of S6K is dependent on LAPTM4B and 4F2hc. (A) A431 cells were treated with siRNAs against LAPTM4B- and/or 4F2hc, (B) ASAH1- or/and LAPTM4B, and (C) SMPD1- or/and LAPTM4B, respectively. Cells were starved (1 h with EBSS), and S6K/pS6K expression was assessed by Western blotting upon refeeding with stimulation medium for the indicated times. Left panels, show representative experiments; right panels, quantifications of n ≥ 3 experiments, mean ± SEM. Differences are significant for p* < 0.05 compared to the control.
We next assessed whether the LAPTM4B ATM3 and D202A mutants show differences in their interaction with 4F2hc as compared to the wild type protein. The ATM3 mutant which lacks the ceramide interaction motif showed a severely attenuated interaction with 4F2hc (Figure 6A), supporting a role for ceramide in driving the hetero-oligomerization of LAPTM4B and 4F2hc. Unexpectedly, the D202A point mutant displayed a significantly enhanced interaction with 4F2hc compared to the wild type LAPTM4B protein (Figure 6A). Since the atomistic simulations showed that the D202A mutation abrogates the kinked conformation of TM3 (Figure 3B), we considered that reduced TM3 bending may promote the interaction between LAPTM4B and 4F2hc. In a similar way, high ceramide concentration might restrict TM3-WT flexibility to promote the interaction with 4F2hc. This idea was supported by additional atomistic simulations showing that addition of ceramide in the bilayer reduces the flexibility of TM3-WT (Figure 6B), analogously to the D202A mutation (see Figure 3B).
Figure 6.
The ceramide interaction motif in LAPTM4B-TM3 affects binding to 4F2hc, lysosomal leucine uptake, and S6K phosphorylation. (A) Schematic representation of LAPTM4B WT, the D202A point mutant, and ATM3 (top right panel). The interaction of LAPTM4B WT, D202A, or ATM3 with endogenous 4F2hc was assessed by Co-immunoprecipitation followed by Western blotting. Left panel, representative experiment; right bottom panel, quantification of n = 3 experiments, mean ± SEM, differences are significant for p* < 0.05 compared to LAPTM4B. (B) Including ceramide in the membrane (0%, 8%, or 20% Cer) in atomistic simulations reduces the flexibility of TM3 as judged by increased peptide length. Inset shows peptide dynamics as change in length per 100 ps interval. (C) LAPTM4B, ATM3, and D202A stable cells were transfected with LAMP1-mGFP, amino acid starved (1 h with EBSS), and stimulated with EBSS-containing MEM amino acids and [3H]-leucine. Lysosomes were isolated by GFP-Trap immunoprecipitation, and radioactivity was quantified by scintillation counting. All values represent mean ± SEM (n ≥ 3), and differences are significant for p* < 0.05 compared to LAPTM4B. (D) A431 cells expressing LAPTM4B, D202A, or ATM3 were starved (1 h with EBSS), and S6K/pS6K expression was assessed by Western blotting upon refeeding with stimulation medium for the indicated times. Left panel, representative experiment; right panel, quantification of n = 3 experiments, mean ± SEM, differences are significant for p* < 0.05 compared to LAPTM4B.
To further investigate the functional role of the ceramide-regulated element in LAPTM4B, we measured lysosomal leucine and S6K phosphorylation in the ATM3 and D202A mutant cell lines. The ATM3 cells displayed substantially reduced lysosomal [3H]-leucine levels (Figure 6C), and a concurrently attenuated S6K phosphorylation (Figure 6D). These results are in line with the finding that LAPTM4B promotes mTORC activation by recruiting the leucine transporter 4F2hc/LAT1 to lysosomes.22 ASAH1 depletion did not enhance the S6K phosphorylation in ATM3 cells (Figure S7), suggesting that the sphingolipid-binding motif is required for mTORC activation by late endosomal ceramide. Interestingly, although the D202A mutant showed enhanced interaction with 4F2hc, the cells displayed reduced lysosomal leucine uptake (Figure 6C), and a lower S6K phosphorylation compared to LAPTM4B-WT cells (Figure 6D). This speaks for the importance of the LAPTM4B–ceramide interaction in the process. Taken together, these results suggest that reducing TM3 bending is sufficient to enhance the interaction between LAPTM4B and 4F2hc. However, LAPTM4B needs to interact with both 4F2hc and ceramide to promote mTORC activation.
Discussion
We recently identified LAPTM4B as a ceramide interacting protein that regulates cell sensitivity to chemotherapeutic agents.18 In the present study, we combined atomistic simulations with biochemical and cell biological experiments to pinpoint in atomistic detail the structural features that are central for the ceramide interaction of LAPTM4B, as well as characterized its functional relevance for LAPTM4B interaction with the leucine transporter heavy chain and downstream nutrient signaling from lysosomes.
We identified a functional ceramide-regulated element in the third TM domain of LAPTM4B, consisting of a ceramide interaction motif and a membrane-embedded aspartate, which regulates ceramide access to the interaction site. While several TM domains contribute to the interaction with cross-linkable ceramide in cells, the luminal half of TM3 containing the sphingolipid interaction motif is the most critical determinant. It is therefore likely that LAPTM4B senses ceramide generated in late endosomes as a result of sphingolipid degradation. In isolated TM3 peptides, the contacts of ceramide with the interaction motif are highly dynamic. However, as several TM domains contribute to the interaction in the full protein, ceramide might form a more stable complex with LAPTM4B in cells. As such, the present study provides the first functional characterization of a sphingolipid-binding motif in a multi-membrane-spanning protein. A common characteristic between LAPTM4B and the single membrane span p24 is that sphingolipid binding regulates the assembly of functional protein complexes: sphingomyelin binding promotes the homodimerization of p24 and regulates vesicular trafficking,12 while ceramide binding to LAPTM4B promotes its interaction with 4F2hc and tunes the response to nutrient signaling. Moreover, the observed LAPTM4B ceramide binding may be linked to our previous findings of LAPTM4B-mediated late endosomal ceramide clearance.18 Besides binding, LAPTM4B (or one of its interacting proteins) might promote ceramide translocation or control the trafficking and metabolic fate of ceramide-rich domains.
A key finding of our study is that the membrane-embedded aspartate residue provides TM3 with flexibility that is subject to ceramide-induced conformational changes. Ceramide-enriched lipid domains have been associated with receptor clustering and activation,31−33 but changes in protein conformation related to this are poorly understood. Studies on supported lipid bilayers have shown that thick ceramide-enriched domains compartmentalize liquid ordered (Lo) phase proteins and decrease their diffusion rate within the plane of the membrane.34−36 This “trapping” of signaling proteins in ceramide-rich domains has been suggested to bring them into close contact and to stabilize their interactions.36,37 We found that the LAPTM4B-4F2hc interaction in cells can be enhanced either by increasing cellular ceramides or by replacing the centrally positioned aspartate in TM3 by the more hydrophobic alanine. The atomistic simulations showed that TM3 adopts an extended conformation in the presence of membrane ceramide, and that this effect can be mimicked by exchanging aspartate for alanine. This suggests that reducing TM3 bending is sufficient for triggering the interaction with 4F2hc.
The hydrophobic lengths of the predicted LAPTM4B transmembrane segments are typical for proteins residing in thick membrane compartments such as the plasma membrane or endosomes.38 α-Helical TM domains respond to hydrophobic mismatch mainly by tilting or bending in the membrane,39 and annular lipids can to some degree compensate for hydrophobic mismatch by compressing or stretching their acyl chains.40 In our simulations, TM3-WT did not markedly affect the thickness of the POPC bilayer (P–P distance 3.90 nm with TM3-WT and 3.89 nm without). In contrast, TM3-D202A increased the membrane thickness to 3.97 nm, which is comparable to the effect of including 8 mol % ceramide in the membrane (4.00 nm). This suggests that wild type TM3 is accommodated in the POPC membrane by bending, while TM3-D202A forces the stretching of lipid acyl chains to provide a matching hydrophobic thickness. We propose that TM3 adopts a kinked structure that is incompatible with 4F2hc interaction in thin membranes. Instead, in thick ceramide-enriched domains TM3 adopts an elongated and less flexible structure, which promotes the interaction with 4F2hc. Interestingly, although the D202A mutant displays higher interaction with 4F2hc, this was not sufficient to rescue lysosomal leucine levels or enhance S6K phosphorylation. Considering that the D202A mutant shows reduced interaction with cross-linkable ceramide it is likely that a combination of ceramide binding and reduced TM3 flexibility are necessary for efficient LAPTM4B/4F2hc-dependent downstream signaling.
Acidic amino acids are underrepresented within TM helices, with an estimated 1.4% incidence in the membrane core of human proteins.41 Remarkably, the originally identified sphingolipid-binding motif in p2412 is accompanied by an adjacent acidic residue, and the GPI-attachment protein 1 (GAA1) that displayed the highest interaction with a cross-linkable sphingolipid probe in the follow-up study13 also contains an acidic glutamate residue near its sphingolipid-binding motif. Moreover, examination of the identified four membrane-spanning proteins expressing relaxed sphingolipid-binding motifs13 reveals that 42% (13 of 31) of them are associated with TM-embedded acidic amino acids. These data argue that the principle revealed in this study for LAPTM4B may be more general: Membrane-embedded acidic residues in the vicinity of a sphingolipid-binding motif provide transmembrane domains with structural flexibility that is amenable to regulation by sphingolipids, thereby affecting protein interactions in response to lipid binding.
Acknowledgments
We acknowledge the technical support by the Biomedicum Imaging Unit, University of Helsinki, and the Proteomic Unit, Institute of Biotechnology, University of Helsinki. This work was supported by the Academy of Finland (Grants 303771 and 266092 to T.B., and the Center of Excellence Project (307415) (I.V., E.I.)), the European Research Council (Advanced Grant GROWDED-PRO-LIPIDS (290974) (I.V.)), and the Sigrid Juselius Foundation (T.B., E.I., I.V.). T.B. was supported by the Magnus Ehrnrooth Foundation, the Ruth and Nils-Erik Stenbäck Foundation, the Liv och Hälsa Foundation, and the University of Helsinki three-year research grants. K.Z. was supported by the ILS doctoral programme at the University of Helsinki. H.M.-S. acknowledges support from the Czech Science Foundation (Grant 208/12/G016). We also thank CSC—IT Center for Science (Espoo, Finland) for computing resources.
Glossary
Abbreviations
- ASAH1
acid ceramidase
- bSMase
bacterial sphingomyelinase
- Cer
ceramide
- LAT1
CD98 light chain
- Lo
liquid ordered
- LUV
large unilamellar vesicle
- SM
sphingomyelin
- SMPD1
acid sphingomyelinase
- TM
transmembrane domain
- 4F2hc
CD98 heavy chain
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.7b00582.
Additional detailed descriptions of materials, methods, and figures including behavior of LAPTM4B transmembrane domain peptides in LUVs; genomic modifications of LAPTM4B knockout, wild type, and mutant cell lines; cellular localization of the LAPTM4B mutant constructs; interaction of endogenous 4F2hc with LAPTM4B and control proteins; LAPTM4B not affecting 4F2hc-dependent leucine uptake into cells; LAPTM4B affecting mTORC1 activity; and interaction of ceramide with LAPTM4B affecting mTORC1 activity (PDF)
Tables S1–S3: description of the simulated systems (XLSX)
Tables S4–S9: fundamental properties of the simulated membranes, and their incorporation of micellar ceramide (XLSX)
Table S10: full results of the proteomic screen (XLSX)
Tables S11–S13: list of used DNA constructs, oligos, and siRNAs (XLSX)
Movie S1: entry of micellar ceramide into a TM3-containing POPC membrane (AVI)
Author Contributions
○ Co-first authorship: K.Z. and A.D. Conceptualization: T.B., K.Z., A.D., H. M.-S., E.I., I.V. Experimental design: T.B., K.Z., A.D., H.M.-S., T.N., Y.A.K., S.L. Performed experiments: K.Z., A.D., H.M.-S., T.B., T.N., Y.A.K., S.L. Wrote the manuscript: T.B., A.D., K.Z., H.M.-S., E.I., I.V. Funding acquisition: T.B., E.I., I.V. Supervision: T.B., E.I., I.V.
The authors declare no competing financial interest.
Supplementary Material
References
- Krogh A.; Larsson B.; von Heijne G.; Sonnhammer E. L. L. Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete Genomes. J. Mol. Biol. 2001, 305, 567–580. 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- van Meer G.; Voelker D. R.; Feigenson G. W. Membrane Lipids: Where They Are and How They Behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis J. C. M.; Menon A. K. Lipid Landscapes and Pipelines in Membrane Homeostasis. Nature 2014, 510, 48–57. 10.1038/nature13474. [DOI] [PubMed] [Google Scholar]
- Hanson M. A.; Roth C. B.; Jo E.; Griffith M. T.; Scott F. L.; Reinhart G.; Desale H.; Clemons B.; Cahalan S. M.; Schuerer S. C.; et al. Crystal Structure of a Lipid G Protein-Coupled Receptor. Science 2012, 335, 851–855. 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-J. J.; Van Brocklyn J. R.; Thangada S.; Liu C. H.; Hand A. R.; Menzeleev R.; Spiegel S.; Hla T. Sphingosine-1-Phosphate as a Ligand for the G Protein-Coupled Receptor EDG-1. Science 1998, 279, 1552–1555. 10.1126/science.279.5356.1552. [DOI] [PubMed] [Google Scholar]
- Lee A. G. Biological Membranes: The Importance of Molecular Detail. Trends Biochem. Sci. 2011, 36, 493–500. 10.1016/j.tibs.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Landreh M.; Marty M. T.; Gault J.; Robinson C. V. A Sliding Selectivity Scale for Lipid Binding to Membrane Proteins. Curr. Opin. Struct. Biol. 2016, 39, 54–60. 10.1016/j.sbi.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras F.-X.; Ernst A. M.; Wieland F.; Brugger B. Specificity of Intramembrane Protein-Lipid Interactions. Cold Spring Harbor Perspect. Biol. 2011, 3, a004705–a004705. 10.1101/cshperspect.a004705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guixà-González R.; Javanainen M.; Gómez-Soler M.; Cordobilla B.; Domingo J. C.; Sanz F.; Pastor M.; Ciruela F.; Martinez-Seara H.; Selent J. Membrane Omega-3 Fatty Acids Modulate the Oligomerisation Kinetics of Adenosine A2A and Dopamine D2 Receptors. Sci. Rep. 2016, 6, 19839. 10.1038/srep19839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K.; Ikonen E. Functional Rafts in Cell Membranes. Nature 1997, 387, 569–572. 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Simons K.; Sampaio J. L. Membrane Organization and Lipid Rafts. Cold Spring Harbor Perspect. Biol. 2011, 3, a004697. 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras F.-X.; Ernst A. M.; Haberkant P.; Björkholm P.; Lindahl E.; Gönen B.; Tischer C.; Elofsson A.; von Heijne G.; Thiele C.; et al. Molecular Recognition of a Single Sphingolipid Species by a Protein’s Transmembrane Domain. Nature 2012, 481, 525–529. 10.1038/nature10742. [DOI] [PubMed] [Google Scholar]
- Björkholm P.; Ernst A. M.; Hacke M.; Wieland F.; Brügger B.; von Heijne G. Identification of Novel Sphingolipid-Binding Motifs in Mammalian Membrane Proteins. Biochim. Biophys. Acta, Biomembr. 2014, 1838, 2066–2070. 10.1016/j.bbamem.2014.04.026. [DOI] [PubMed] [Google Scholar]
- Zeidan Y. H.; Jenkins R. W.; Korman J. B.; Liu X.; Obeid L. M.; Norris J. S.; Hannun Y. A. Molecular Targeting of Acid Ceramidase: Implications to Cancer Therapy. Curr. Drug Targets 2008, 9, 653–661. 10.2174/138945008785132358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaurasia B.; Summers S. A. Ceramides – Lipotoxic Inducers of Metabolic Disorders. Trends Endocrinol. Metab. 2015, 26, 538–550. 10.1016/j.tem.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Megha; London E. Ceramide Selectively Displaces Cholesterol from Ordered Lipid Domains (Rafts) Implications for Lipid Raft Structure and Function. J. Biol. Chem. 2004, 279, 9997–10004. 10.1074/jbc.M309992200. [DOI] [PubMed] [Google Scholar]
- Bollinger C. R.; Teichgräber V.; Gulbins E. Ceramide-Enriched Membrane Domains. Biochim. Biophys. Acta, Mol. Cell Res. 2005, 1746, 284–294. 10.1016/j.bbamcr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Blom T.; Li S.; Dichlberger A.; Bäck N.; Kim Y. A. Y. A.; Loizides-Mangold U.; Riezman H.; Bittman R.; Ikonen E. LAPTM4B Facilitates Late Endosomal Ceramide Export to Control Cell Death Pathways. Nat. Chem. Biol. 2015, 11, 799–806. 10.1038/nchembio.1889. [DOI] [PubMed] [Google Scholar]
- Shao G.-Z.; Zhou R.-L.; Zhang Q.-Y.; Zhang Y.; Liu J.-J.; Rui J.-A.; Wei X.; Ye D.-X. Molecular Cloning and Characterization of LAPTM4B, a Novel Gene Upregulated in Hepatocellular Carcinoma. Oncogene 2003, 22, 5060–5069. 10.1038/sj.onc.1206832. [DOI] [PubMed] [Google Scholar]
- Li Y.; Zou L.; Li Q.; Haibe-Kains B.; Tian R.; Li Y.; Desmedt C.; Sotiriou C.; Szallasi Z.; Iglehart J. D.; et al. Amplification of LAPTM4B and YWHAZ Contributes to Chemotherapy Resistance and Recurrence of Breast Cancer. Nat. Med. 2010, 16, 214–218. 10.1038/nm.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. W. K.; Mitchell A.; Kennedy J. A.; Chen W. C.; McLeod J.; Ibrahimova N.; Arruda A.; Popescu A.; Gupta V.; Schimmer A. D.; et al. A 17-Gene Stemness Score for Rapid Determination of Risk in Acute Leukaemia. Nature 2016, 540, 433–437. 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- Milkereit R.; Persaud A.; Vanoaica L.; Guetg A.; Verrey F.; Rotin D. LAPTM4b Recruits the LAT1–4F2hc Leu Transporter to Lysosomes and Promotes mTORC1 Activation. Nat. Commun. 2015, 6, 7250. 10.1038/ncomms8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X.; Sun Y.; Thapa N.; Liao Y.; Hedman A. C.; Anderson R. A. LAPTM4B Is a PtdIns(4,5)P2 Effector That Regulates EGFR Signaling, Lysosomal Sorting, and Degradation. EMBO J. 2015, 34, 475–490. 10.15252/embj.201489425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X.; Thapa N.; Sun Y.; Anderson R. A. A. A Kinase-Independent Role for EGF Receptor in Autophagy Initiation. Cell 2015, 160, 145–160. 10.1016/j.cell.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergarajauregui S.; Martina J. a; Puertollano R. LAPTMs Regulate Lysosomal Function and Interact with Mucolipin 1: New Clues for Understanding Mucolipidosis Type IV. J. Cell Sci. 2011, 124, 459–468. 10.1242/jcs.076240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Zhang Q.; Tian R.; Wang Q.; Zhao J. J.; Iglehart J. D.; Wang Z. C.; Richardson A. L. Lysosomal Transmembrane Protein LAPTM4B Promotes Autophagy and Tolerance to Metabolic Stress in Cancer Cells. Cancer Res. 2011, 71, 7481–7489. 10.1158/0008-5472.CAN-11-0940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther G. G.; Peralta E. R.; Rosales K. R.; Wong S. Y.; Siskind L. J.; Edinger A. L. Ceramide Starves Cells to Death by Downregulating Nutrient Transporter Proteins. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 17402–17407. 10.1073/pnas.0802781105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther G. G.; Liu G.; Ramirez M. U.; McMonigle R. J.; Kim S. M.; McCracken A. N.; Joo Y.; Ushach I.; Nguyen N. L.; Edinger A. L. Loss of TSC2 Confers Resistance to Ceramide and Nutrient Deprivation. Oncogene 2014, 33, 1776–1787. 10.1038/onc.2013.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo G. A.; London E. Position and Ionization State of Asp in the Core of Membrane-Inserted Alpha Helices Control Both the Equilibrium between Transmembrane and Nontransmembrane Helix Topography and Transmembrane Helix Positioning. Biochemistry 2004, 43, 8794–8806. 10.1021/bi049696p. [DOI] [PubMed] [Google Scholar]
- Monné M.; Nilsson I.; Johansson M.; Elmhed N.; von Heijne G. Positively and Negatively Charged Residues Have Different Effects on the Position in the Membrane of a Model Transmembrane Helix. J. Mol. Biol. 1998, 284, 1177–1183. 10.1006/jmbi.1998.2218. [DOI] [PubMed] [Google Scholar]
- Grassme H.; Jendrossek V.; Bock J.; Riehle A.; Gulbins E. Ceramide-Rich Membrane Rafts Mediate CD40 Clustering. J. Immunol. 2002, 168, 298–307. 10.4049/jimmunol.168.1.298. [DOI] [PubMed] [Google Scholar]
- Detre C.; Kiss E.; Varga Z.; Ludányi K.; Pászty K.; Enyedi Á.; Kövesdi D.; Panyi G.; Rajnavölgyi É.; Matkó J. Death or Survival: Membrane Ceramide Controls the Fate and Activation of Antigen-Specific T-Cells Depending on Signal Strength and Duration. Cell. Signalling 2006, 18, 294–306. 10.1016/j.cellsig.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Morad S. A. F.; Cabot M. C. Ceramide-Orchestrated Signalling in Cancer Cells. Nat. Rev. Cancer 2013, 13, 51–65. 10.1038/nrc3398. [DOI] [PubMed] [Google Scholar]
- Carrer D. C.; Kummer E.; Chwastek G.; Chiantia S.; Schwille P. Asymmetry Determines the Effects of Natural Ceramides on Model Membranes. Soft Matter 2009, 5, 3279. 10.1039/b901883b. [DOI] [Google Scholar]
- Carter Ramirez D. M.; Kim Y. A.; Bittman R.; Johnston L. J. Lipid Phase Separation and Protein-Ganglioside Clustering in Supported Bilayers Are Induced by Photorelease of Ceramide. Soft Matter 2013, 9, 4890–4899. 10.1039/c3sm50240f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiantia S.; Ries J.; Chwastek G.; Carrer D.; Li Z.; Bittman R.; Schwille P. Role of Ceramide in Membrane Protein Organization Investigated by Combined AFM and FCS. Biochim. Biophys. Acta, Biomembr. 2008, 1778, 1356–1364. 10.1016/j.bbamem.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Castro B. M.; Prieto M.; Silva L. C. Ceramide: A Simple Sphingolipid with Unique Biophysical Properties. Prog. Lipid Res. 2014, 54, 53–67. 10.1016/j.plipres.2014.01.004. [DOI] [PubMed] [Google Scholar]
- Sharpe H. J.; Stevens T. J.; Munro S. A Comprehensive Comparison of Transmembrane Domains Reveals Organelle-Specific Properties. Cell 2010, 142, 158–169. 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt A.; Killian J. A. Orientation and Dynamics of Transmembrane Peptides: The Power of Simple Models. Eur. Biophys. J. 2010, 39, 609–621. 10.1007/s00249-009-0567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. G. How Lipids Affect the Activities of Integral Membrane Proteins. Biochim. Biophys. Acta, Biomembr. 2004, 1666, 62–87. 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Illergård K.; Kauko A.; Elofsson A. Why Are Polar Residues within the Membrane Core Evolutionary Conserved?. Proteins: Struct., Funct., Genet. 2011, 79, 79–91. 10.1002/prot.22859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.