SUMMARY
Despite decades of effort, Helicobacter pylori infections remain difficult to treat. Over half of the world's population is infected by H. pylori, which is a major cause of duodenal and gastric ulcers as well as gastric cancer. During chronic infection, H. pylori localizes within the gastric mucosal layer, including deep within invaginations called glands; thanks to its impressive ability to survive despite the harsh acidic environment, it can persist for the host's lifetime. This ability to survive and persist in the stomach is associated with urease production, chemotactic motility, and the ability to adapt to the fluctuating environment. Additionally, biofilm formation has recently been suggested to play a role in colonization. Biofilms are surface-associated communities of bacteria that are embedded in a hydrated matrix of extracellular polymeric substances. Biofilms pose a substantial health risk and are key contributors to many chronic and recurrent infections. This link between biofilm-associated bacteria and chronic infections likely results from an increased tolerance to conventional antibiotic treatments as well as immune system action. The role of this biofilm mode in antimicrobial treatment failure and H. pylori survival has yet to be determined. Furthermore, relatively little is known about the H. pylori biofilm structure or the genes associated with this mode of growth. In this review, therefore, we aim to highlight recent findings concerning H. pylori biofilms and the molecular mechanism of their formation. Additionally, we discuss the potential roles of biofilms in the failure of antibiotic treatment and in infection recurrence.
KEYWORDS: Helicobacter pylori, biofilm, regulation, gastritis, antibiotics
INTRODUCTION
The observation that bacteria reside within communities is centuries old. In fact, Antonie van Leeuwenhoek commented on the presence of “animalcules” in plaque on teeth in 1623, but the biofilm theory was not developed until 1978 (1). Many names were subsequently used in early studies of the bacterial communities that we now know as biofilms, which can be defined as adherent aggregates of microorganisms encased with an extracellular polymeric substance (EPS) (2–4). Biofilms present one of the most successful modes of life; the importance of the microbial biofilm is demonstrated by its presence in the fossil record dating back over 3 million years (4, 5). Today, biofilms have been identified in a myriad of ecological niches, and they are increasingly being recognized for their importance in human health; up to 80% of microbial infections in humans involve biofilm growth (3, 6). Indeed, biofilms are often associated with chronic infectious diseases since they provide an additional level of protection against environmental, host, and antimicrobial assaults (reviewed in references 4 and 6–8). There is a growing body of evidence that indicates that the bacterium Helicobacter pylori can establish a biofilm.
H. pylori is considered one of the most remarkable human pathogens. More than 10,000 years of coexistence and current colonization estimates at nearly half of the human population distinguish H. pylori as one of the most prevalent global pathogens as well as the most common bacterial infection (9). H. pylori infects the gastric epithelium and will persist as a chronic infection unless treated. Rates of infection with this bacterium are strongly associated with socioeconomic status and hygiene conditions. As such, the prevalence of H. pylori varies from more than 80% in developing countries to less than 40% in industrialized countries (10). While H. pylori colonization is often associated with asymptomatic infections, H. pylori is responsible for gastric diseases such as chronic gastritis, peptic and duodenal ulcers, and gastric cancers (11–13). Indeed, H. pylori is the number one risk factor for the development of gastric adenocarcinoma, which occurs in approximately 1 to 2% of infected individuals (14, 15).
Since its discovery in the 1980s, studies of H. pylori have focused on the planktonic, free-floating, nonattached mode of growth; however, recent evidence suggests that H. pylori can also grow in a surface-attached biofilm mode. Studies are beginning to elucidate H. pylori biofilm development both in vitro and in vivo (16–23). While the examination of H. pylori biofilms is relatively new, research in this particular field has been slowly gaining momentum over the past few years. Given the chronicity associated with H. pylori infection, it is perhaps not surprising that this bacterium has been observed in biofilms.
En masse, the available data support further consideration of the potential role of H. pylori biofilms in gastric infections. These considerations are especially relevant given that (i) H. pylori is notorious for being recalcitrant to antibiotic treatment and the immune response (24–27) and (ii) biofilms can impact the efficacy of antibiotic treatment and clearance by the host immune response. Indeed, recent U.S. estimates have found that 20 to 25% of infected individuals are not cured by current therapy (28). In this review, we provide insights into current research on the composition of H. pylori biofilms and associated genetic factors. Additionally, we discuss how biofilms could be associated with the failure of antibiotic treatment and infection recurrence.
DEMONSTRATION OF H. PYLORI BIOFILMS
In Vitro Models
Although the role of H. pylori biofilms is unknown, researchers are beginning to develop in vitro models to investigate the biofilm phenotype. As such, several studies have demonstrated the ability of H. pylori to produce a biofilm in vitro (18, 29, 30) (Fig. 1). Indeed, one group demonstrated biofilm production by clinical, laboratory, and mouse-adapted strains at the air-liquid interface on glass coverslips after 3 to 5 days of shaking culture in brucella broth (BB) supplemented with 7% fetal calf serum (FCS) (30). These biofilms were composed mainly of coccoid bacteria within a three-dimensional architecture of stacked layers and holes (30). In comparison, Yonezawa et al. demonstrated that H. pylori Sydney strain 1 (SS1), a mouse-colonizing strain used extensively in H. pylori research, formed relatively little biofilm biomass compared to those formed by ATCC 43579 and other clinical isolates (31). Scanning electron microscopy (SEM) analysis revealed that a biofilm formed by strain TK1402 consisted of thick layers of cells with a bacillary morphology (31). Conversely, biofilms formed by other strains (e.g., SS1) exhibited thin layers of amorphous cells mixed with some autolyzed cells (31). The difference in morphological forms associated with H. pylori biofilms is intriguing. The two dominant morphologies adopted by H. pylori cells include a bacillary-spiral form and a coccoid form. Both forms have been observed in vitro (32–34) and in vivo (35, 36). The exact nature of the coccoid form is not fully understood, and little is known about this form in Helicobacter pylori. Furthermore, there is currently a split in the H. pylori field concerning the significance of the coccoid form; some consider these cells to be dead/degraded, and others consider these cells to represent a viable but nonculturable (VBNC) form (32, 37).
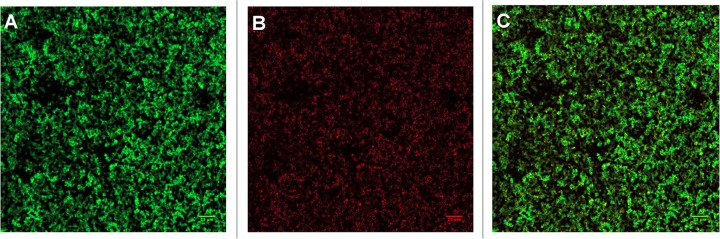
FIG 1.
Confocal laser scanning microscopy image of an H. pylori G27 biofilm stained with the FilmTracer Live/Dead biofilm viability kit. The biofilm was formed in 96-well plates after 3 days of growth using brucella broth medium supplemented with 10% FBS. (A) Live cells; (B) dead or compromised cells; (C) merge.
In other species, VBNC cells are metabolically less active, are capable of long-term survival, and resuscitate under suitable conditions (37, 38). In general, various stress signals induce some bacteria to enter the VBNC state. These signals include low pH, osmolarity, nutrient starvation, and antibiotic exposure (30, 39–42). Similarly, in H. pylori, the coccoid form is known to dominate after prolonged culture, under starvation conditions, and after exposure to antimicrobials (32, 37).
The observed differences in biofilm-associated cell morphology may be due to differences in growth conditions employed in different studies. Indeed, growth conditions are important factors that have been shown to influence biofilm formation of other species (43–45). Similarly, variations in the compositions of liquid media appear to have a significant influence on H. pylori biofilm formation (20). The fastidious nature of H. pylori is well known and was in fact a major hurdle during the initial isolation of the bacterium (46). H. pylori is microaerophilic and thrives under conditions that include a carbon dioxide-enriched (5 to 10%) atmosphere, high humidity, and a temperature of 35°C to 37°C (46). In addition, cultures require rich medium that is supplemented with specific factors to promote optimal growth. Common H. pylori liquid media begin with a complex base, such as BB, brain heart infusion (BHI) broth, or Ham's F-12 medium, which are then supplemented with serum and additional carbon sources (e.g., fetal bovine serum [FBS] or 2,6-dimethyl-β-cyclodextrin [46, 47]). One study assessed the effects of four types of common liquid culture media on H. pylori biofilm formation: BB supplemented with 2% FBS, BHI broth supplemented with 2% FBS, and Ham's F-12 medium supplemented with or without 2% FBS. As expected, significant variation in biofilm production by both H. pylori ATCC 43629 and clinical strain 9/10 was observed (20). Interestingly, Ham's F-12 medium without serum optimized biofilm formation despite slowing bacterial growth (20). Mechanistically, this is not fully understood but may involve enhanced EPS production (20). Future studies will be required to understand the role of various medium types in H. pylori biofilm formation.
A similar study found that the presence of FBS appears to negatively impact H. pylori biofilm development (48). One mechanism by which this response could occur is via the disruption of surface adherence, which is a crucial step in biofilm formation. Indeed, Williams et al. observed that serum negatively affects H. pylori adherence to surfaces (48). That group demonstrated that lower concentrations (≤1%) of serum were needed to optimally promote H. pylori adherence to abiotic surfaces and to inhibit swimming (48). Similarly, Shao et al. demonstrated that the removal of serum induced bacterial aggregation and, thus, biofilm formation (49). These results are intriguing since serum is commonly used to supplement H. pylori liquid media to promote planktonic growth; it is thought to provide essential growth factors and to reduce fatty acid toxicity (50). Overall, even if the use of complex and rich media for in vitro biofilm growth does not necessarily reflect the natural environment that H. pylori encounters in vivo, the optimization of growth media and conditions that promote biofilm formation will be helpful for future studies to help gain a better understanding of H. pylori biofilms and their potential role in gastric infection.
An additional difference that cannot be overlooked when comparing data from H. pylori studies is the differences in strains being studied. Without a side-by-side comparison of strains in various media, it is difficult to tease out whether differences in the morphologies of H. pylori within biofilms are due to medium differences or due to inherent differences betweens strains. Given the well-documented heterogeneity observed among H. pylori isolates (51, 52), it is perhaps not surprising that biofilm formation varies between strains. Indeed, several groups have compared multiple strains and observed differences in their abilities to form biofilms (30, 31, 34). For example, among the clinical strains assessed by Yonezawa et al., an isolate (TK1402) from a Japanese patient with duodenal and gastric ulcers showed a significantly higher level of biofilm biomass than those of the other tested strains (31). However, a correlation between their abilities to form a biofilm and gastric ulcers requires further investigation.
Extragastric Biofilms
Few reports have documented the presence of H. pylori in the environment (53–56). The fastidious nature of H. pylori, the specificity of the gastric niche, and the tendency of H. pylori to transition to the coccoid form when stressed all seem contrary to the long-term survivability of extragastric H. pylori. However, given that biofilm formation can occur as a response to environmental stressors (4, 7), perhaps H. pylori biofilm formation is part of ex vivo survival. Indeed, a majority of the early H. pylori biofilm studies focused on the possibility that H. pylori forms a biofilm as a mechanism to survive in aquatic reservoirs (57–59). The presence of H. pylori in water-associated biofilms has been demonstrated by using PCR (29, 58), peptide DNA probes (60), and fluorescence in situ hybridization (FISH) (56, 61); however, despite positive indications using all of these methodologies, the gold standard is still culture. Two groups reported positive H. pylori cultures from wastewater (56, 62). Although those two groups did not comment on biofilm formation, a separate study presented in vitro evidence that suggests that interactions with specific bacteria (for example, Mycobacterium chelonae) within a multispecies biofilm may contribute to H. pylori viability (63). Together, these findings offer precedent to support the exploration of the ability of H. pylori to survive in multispecies biofilms. Additional proposed extragastric environments for H. pylori include dental plaque (reviewed in reference 64) and biofilms associated with vegetables (65–67). The existence of live extragastric H. pylori is not definitive; however, the potential involvement of biofilms as a means to foster a more favorable colonization niche is worth further exploration.
Gastric Biofilms
Biofilms are often associated with chronic infectious diseases since they provide an additional level of protection against environmental, host, and antimicrobial assaults (4, 6, 7, 68, 69). Indeed, the ability of H. pylori to form biofilms may contribute to the longevity of this infection. Carron et al. and Coticchia et al. provided the first evidence of biofilm formation by H. pylori during colonization of the human gastric mucosa (16, 17). Using biopsy specimens and SEM analyses, they demonstrated the presence of dense layers of what appeared to be biofilm-associated bacteria at the mucosal surface of H. pylori-positive patients (16, 17). The SEM images from both studies were consistent with a community of organisms embedded in the EPS (16, 17). H. pylori-negative patients, in contrast, had smooth mucosa with little evidence of a bacterial community. Biofilm-like growth was seen in over 97.3% of the analyzed mucosal surfaces in H. pylori-positive patients, compared to only 1.64% in H. pylori-negative patients (17). Recently, Attaran et al. demonstrated H. pylori biofilm formation in a mouse model (36). Through the use of SEM and immunofluorescence with a polyclonal anti-H. pylori-specific antibody, those investigators found that clinical isolates colonize the mouse gastric mucosa in a manner consistent with a biofilm. This included the presence of an amorphous extracellular matrix at the mucosal surface (36). SEM revealed a dominant proportion of H. pylori biofilm cells to be in the coccoid form, which is consistent with previous observations (17, 35). However, it remains unclear whether the biofilm matrix in vivo consists of bacterial self-produced polymeric substances, a host-derived matrix (e.g., collagen, laminin, and fibronectin), or both. Further studies will be needed to determine the contribution of the host extracellular matrix.
In addition to colonizing the surface of epithelial cells and the mucus gel layer, the presence of H. pylori within the glands of the murine and human gastrointestinal tracts has been observed (70–72). These glands were suggested to be protective niches that could support the growth of H. pylori. At the height of infection, around 50% of glands are occupied, and some of them could contain more than 200 bacteria (70, 72). Interestingly, aggregates of H. pylori found in the gastric glands may be consistent with it being in the biofilm growth mode (Fig. 2). Studies to evaluate H. pylori biofilm development in the gastric glands and the potential contribution of this growth mode to colonization and persistence are warranted.
FIG 2.
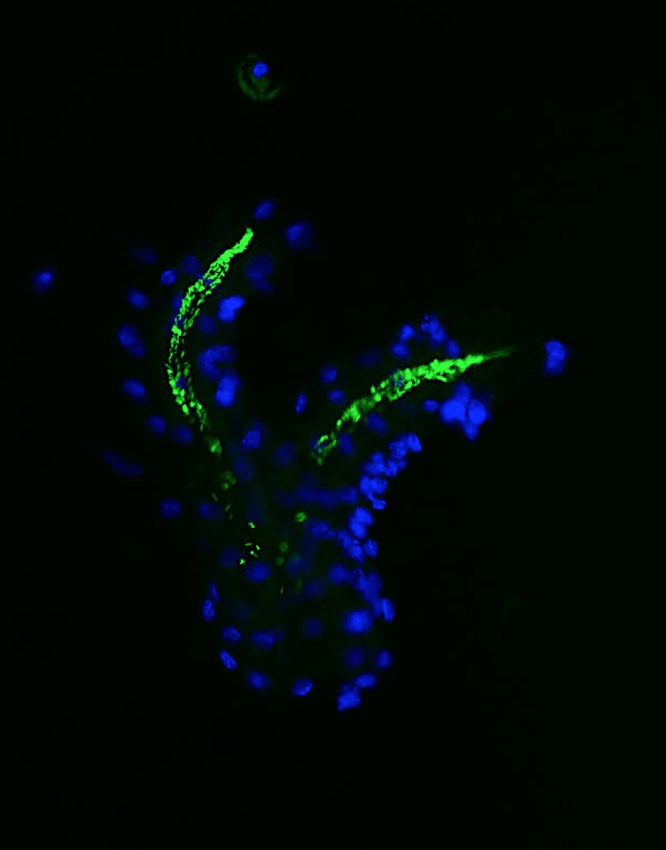
Gastric gland colonization by H. pylori. H. pylori SS1 was visualized by green fluorescent protein (GFP) expression (green), while gland cells were stained by using DNA Hoechst fluorescent dye (blue).
MOLECULAR MECHANISMS OF H. PYLORI BIOFILM FORMATION
Omic Approaches
In support of the theory that biofilms form by similar mechanisms regardless of the bacterial species, the genes demonstrated to be important for biofilm formation tend to fall into a few categories: adherence, quorum sensing, cell wall synthesis, the stress response, carbohydrate metabolism, and cell division (73). Genomics, transcriptomics, and proteomics are high-throughput strategies that have been implemented to identify factors associated with H. pylori biofilms (Table 1). One genomic study compared the sequences of wild-type strain J99 and 31 H. pylori clinical isolates that each had a range of biofilm-forming abilities as a means to identify genes enriched in those strains that were high biofilm producers (34). The strains were classified as low, moderate, or high biofilm producers based on crystal violet staining intensity. By studying the genetic differences between strains, those authors were able to identify several genes that appear to be associated with biofilm formation. Indeed, three hypothetical genes (K74_10375, K747_09130, and K747_06625) were significantly correlated with biofilm formation (34). K747_06625 is predicted to contain a homing endonuclease and a ParB-like domain, the latter of which is associated with biofilm formation in some bacteria (74, 75). Additionally, four functional genes, coding for a flagellar protein (jhp_1117), an alpha-(1,3)-fucosyltransferase (fucT), an outer membrane protein (OMP) (encoded by homD), and a cytotoxin-associated gene pathogenicity island (cag PAI) protein (CagA), were also associated with biofilm formation in H. pylori (34). The role of the cag PAI proteins in H. pylori biofilms was further investigated by the creation of deletion mutations in cagA and the entire cag PAI. Both mutations resulted in a significant decrease in biofilm biomass compared to that of the wild type (34).
TABLE 1.
Some genes associated with biofilm formation by H. pylori
| Gene | Gene product | Approach(es) | Reference(s) |
|---|---|---|---|
| K74_10375 | Hypothetical gene | Genomica | 34 |
| K747_09130 | Hypothetical gene | ||
| K747_06625 | Hypothetical gene | ||
| fucT | Alpha-(1,3)-fucosyltransferase | ||
| homD | Outer membrane protein | ||
| cagA | Cytotoxin-associated gene pathogenicity island | ||
| cag26-cagA | Cytotoxin-associated gene pathogenicity island | Proteomicb | 49 |
| cag24-cagD | Cytotoxin-associated gene pathogenicity island | ||
| cagE | Cytotoxin-associated gene pathogenicity island | Mutagenesis | 30 |
| napA | Neutrophil-activating protein | Proteomic,b mutagenesis | 77 |
| luxS | Quorum sensing | Mutagenesisd | 30, 78 |
| tlpB | Chemoreceptor | Mutagenesis,d transcriptomicc | 78 |
| aibA | Periplasmic binding proteins | ||
| aibB | Periplasmic binding proteins | ||
| arsR | Regulatory response | Mutagenesis,d proteomicb | 23, 49 |
| alpB | Outer membrane protein | Mutagenesisd | 99 |
Whole-genome sequencing of these 32 clinical strains was performed on the Illumina MiSeq platform.
Two-dimensional gel electrophoresis (2-DE) coupled with the identification of proteins through peptide mass fingerprinting (PMF) by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS).
Real-time quantitative reverse transcription–PCR (RT-qPCR).
Phenotype confirmed by molecular complementation.
Other work similarly supported the importance of the cag PAI proteins. Using a proteomic approach, Shao et al. observed that two Cag proteins, Cag26/CagA and Cag24/CagD, were highly expressed in H. pylori biofilms that had been induced by serum starvation compared to planktonic H. pylori cultured under the same conditions (49). Similarly, a study also showed that the creation of isogenic strains lacking CagE altered biofilm formation by H. pylori, further supporting the role of the Cag PAI in biofilm formation (30). These results are particularly relevant to pathogenesis, since infections with cag PAI-containing strains are often associated with an increased risk for gastric cancer (76).
The proteomics study by Shao et al. described above also revealed 35 other proteins that showed altered expressions between biofilm and planktonic H. pylori cultures (49). These proteins are associated with various biological functions, including motility, virulence, signal transduction, and regulation (49). These observations strongly support the facts that biofilm differs from the planktonic mode of growth and that H. pylori biofilm cells harbor a distinct proteomic profile. Another group utilized a proteomic analysis to identify biofilm-associated proteins (77). In that study, NapA, a secreted neutrophil-activating exoprotein, was associated with H. pylori biofilms (77). NapA expression was significantly upregulated during biofilm formation, and a napA-deficient mutant strain produced a reduced biomass and less cell aggregation than did wild-type strains (77).
Targeted Studies of Candidate Genes Associated with Biofilms
In addition to the omics approaches described above, other groups have taken a more targeted approach. Cole et al. evaluated genes known to be associated with biofilm formation in other bacterial species for a potential role in H. pylori biofilm development (30). Of the potential biofilm-associated genes, only the loss of luxS, a quorum-sensing gene, had a significant effect on biofilm formation (30). The contribution of LuxS is perhaps not surprising given that H. pylori has only one known type of quorum-sensing system, autoinducer 2 (AI-2), which is synthesized by LuxS. In comparison, H. pylori has multiple ways to sense AI-2. One method is via the chemoreceptor TlpB, which senses AI-2 as a chemorepellent when AI-2 is bound to either the AibA or AibB periplasmic binding protein (78, 79). LuxS-deficient H. pylori mutant strains do not produce AI-2 and exhibit increased adherence and biofilm biomass compared to their wild-type counterparts (30, 78). H. pylori biofilms from strains lacking AI-2 chemoreception (ΔtlpB, ΔaibA, or ΔaibB) showed enhanced biomass, large microcolonies, cell clustering, and an increased number of attached bacteria (78). Thus, it seems that AI-2 sensing via the chemotaxis system decreases biofilm formation. While this role is in contrast to what is seen in many bacterial species, given that another role of AI-2 sensing is to enhance motility, flagellar gene expression, and morphogenesis (80, 81), perhaps a role in decreased biofilm formation is not surprising.
Regulatory Proteins Associated with H. pylori Biofilm Formation
Preliminary investigations into the role of regulatory proteins in H. pylori biofilm formation have also been conducted. Recently, the expressions of 16 transcriptional regulators in H. pylori 26695 were analyzed under various conditions, including in cells found in biofilms (82). Compared with planktonic cells, biofilm-grown cells showed increased expression levels of all investigated transcriptional factors, with the exception of the heat shock regulator HrcA. Biofilm-induced transcription factors included the response regulator ArsR, two sigma factors (RpoD and FliD), and the metalloregulators Fur, NikR, and CrdR (82). Although the roles that these transcriptional factors play in biofilm formation are largely unknown, the data suggest that a large and complex regulatory network likely controls gene transcription during H. pylori biofilm growth. Two regulatory proteins that have been investigated in more detail are the response regulators ArsR (HP0166) and CrdR (HP1365) (23, 30). While the loss of CrdR from 26695 resulted in no difference in biofilm formation (30), mutation of ArsR to a nonphosphorylatable form or deletion of its cognate sensor kinase ArsS in G27 resulted in a dramatic increase in biofilm formation (23). ArsRS-deficient mutant strains showed increases in cell aggregation and biofilm biomass, which were visible by SEM (23). The ArsRS system is known to regulate OMPs; therefore, one possibility is that the adherence and/or autoaggregation of bacteria is linked to the expression of particular OMPs at the cell surface. These traits in turn control biofilm formation. ArsR was also identified in the proteomic analysis conducted by Shao et al. (49). Numerous candidate genes have now been proposed to play a role in H. pylori biofilm formation. However, more extensive studies, including the evaluation of genes in multiple strain backgrounds, are necessary to truly understand how these genes contribute to biofilm formation.
COMPOSITION OF THE BIOFILM MATRIX
As stated in the introduction, biofilms can be defined as adherent aggregates of microorganisms encased in an EPS (83). Several groups have begun to characterize the EPS of the H. pylori biofilm. The earliest studies focused on polysaccharide components (18). When H. pylori ATCC 43504 (NCTC 11637) was grown in a continuous culture using a glass fermentor, a biofilm formed at the air-liquid interface. The biofilm materials from the wall of the glass culture vessel were dissolved in saturated phenol, and the monosaccharides of both crude and partially purified biofilm material were analyzed by gas-liquid chromatography (18). This method revealed the presence of carbohydrates with a high carbon-to-nitrogen ratio (18). Although the precise composition and structure of the polysaccharide were not identified, several components of H. pylori lipopolysaccharide (LPS), such as C14 and C16 lipids, N-acetylglucosamine, fucose, glucose, galactose, and glycomannoheptose, were identified (18). LPS-like material has been described as a component of the biofilm matrix in other bacteria and suggested to play a role in biofilm formation (84–86).
In a similar study that used nuclear magnetic resonance (NMR), the biofilm matrix of H. pylori strain ATCC 43504 was shown to contain mannose-related proteoglycans (proteomannans) and 1,3- or 1,4-mannosyl linkages (77). Proteomannans consist of a beta-1,3/1,6-glucan structure attached to a surface of complex proteoglycans. Interestingly, these same types of molecules also compose the inner layer of the Candida albicans cell envelope (87) and have been shown to have immunomodulatory properties (88).
Extracellular DNA (eDNA) has also been found as part of the biofilm matrix of several bacterial species and is thought to contribute to biofilm structure, genetic exchange, and bacterial variability (8, 89–91). Similar to other species, eDNA has been observed as a component of the H. pylori biofilm matrix; however, in the H. pylori biofilm, the role of eDNA is not fully understood (92). DNase I treatment of a preformed biofilm did not disperse an H. pylori biofilm, suggesting that eDNA does not play a role in biofilm architecture or that the eDNA might be protected by other molecules (92). In a follow-up study, it was observed that during biofilm formation by strain ATCC 43629, eDNA was detected in outer membrane vesicles (OMVs) from both biofilm (bOMV) and planktonic (pOMV) fractions (19). OMVs are 20- to 500-nm-diameter bilayer structures that contain phospholipids, proteins, LPS, and DNA (93, 94). OMVs were shown to be part of the biofilm matrix in several Gram-negative bacterial species (95, 96), including that of H. pylori (97). Furthermore, the presence of OMVs was observed in biofilms from the strong-biofilm-forming H. pylori strain TK1402 but not from weak biofilm producers. This suggests that OMVs might promote H. pylori biofilm formation (97, 98). Together, these findings support the hypothesis that eDNA may be protected by OMVs and may avoid DNase I treatment (92). Interestingly, larger amounts of eDNA are associated with bOMVs than with pOMVs (19). However, eDNA was also found at the pOMV membrane surface and the H. pylori cell wall, suggesting that H. pylori may just generally be covered with eDNA. Free eDNA, as well as that associated with OMVs, could potentially be utilized by H. pylori for genetic recombination. Sampling of this DNA by the bacteria could contribute to genetic variability and possibly enhance bacterial survival (89–91). Thus, the precise role of OMVs and eDNA in biofilm formation is still not clear (19).
In addition to eDNA, proteins such as the AlpB OMP have been shown to be associated with OMVs and are thought to contribute to biofilm formation (99). Indeed, a study compared protein profiles of OMVs obtained from a wild-type strong-biofilm-forming strain (H. pylori TK1402) to those from isogenic biofilm-defective mutant strains. Those authors observed weaker expression of a 52-kDa protein in the biofilm-defective mutant strains than in the wild-type profile; this protein was identified as AlpB (99). To confirm the role of AlpB in biofilm formation, an alpB-deficient strain was further evaluated for biofilm formation. The alpB mutant strain exhibited a defect in biofilm formation, suggesting that AlpB may play a role in cell attachment, cell-to-cell aggregation, and, thus, biofilm development (99). It is not currently known, however, whether AlpB needs to be in OMVs to exert this effect.
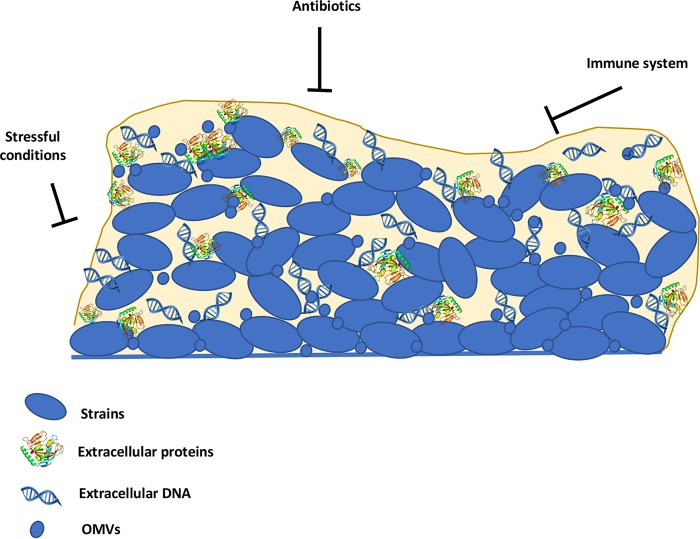
In summary, current data suggest that the H. pylori biofilm matrix contains at least proteomannans, LPS-related structures, eDNA, proteins, and OMVs (Fig. 3). Elucidation of the role of each of these components and their importance in the biofilm matrix will be noteworthy future studies.
FIG 3.
Schematic representation of an H. pylori biofilm. Extracellular polymeric substances in H. pylori, composed of extracellular DNA, extracellular proteins, and OMVs, may act as a shield to protect the bacterial community from immune cells and antimicrobials.
IMMUNE EVASION AND TREATMENT FAILURE
Studies of H. pylori Biofilms and Antibiotic Resistance
While there are still many unanswered questions regarding H. pylori biofilm formation, the implications of biofilm-associated infections are well documented for other species (3, 83, 100). The possibility that H. pylori may adopt a biofilm mode of growth during colonization of the gastric mucosa and gastric glands could have a profound impact on antimicrobial treatment. For multiple bacteria, biofilms are known to render infections of multiple bacteria more difficult to eradicate with antimicrobial therapy (3, 8, 83, 100, 101). Bacterial biofilms resist antimicrobial treatments by using a transient and noninheritable mechanism called tolerance, which is related to the physiological state of the biofilm cells and the physical barrier formed by the extracellular matrix (8, 100, 101). In addition to antibiotic tolerance, biofilms often represent a reservoir of genetic diversity by promoting genetic exchange between different subpopulations. Several studies have documented the increased dissemination of antibiotic resistance genes in biofilms through horizontal gene transfer, integrative conjugative elements, and natural transformation (102–104). Additionally, bacterial cells within biofilms undergo several stressful conditions, including major nutrient starvation, which could increase the mutation frequency and the emergence of antibiotic-resistant mutant strains (8, 69, 105, 106).
Biofilm-associated H. pylori was shown to be more resistant in vitro to clarithromycin, which is one of the common antibiotics used to treat H. pylori infections (107). Specifically, the MIC increased by 16-fold and the minimum bactericidal concentration (MBC) increased by up to 4-fold in biofilm cells compared to planktonic ones (107). H. pylori biofilm cells also showed an increased mutation rate, which could promote clarithromycin-resistant mutations (107).
The contribution of H. pylori biofilms to the spread of antibiotic resistance has not been fully explored; however, the emergence of resistance to clarithromycin, levofloxacin, and metronidazole urges us to develop new strategies that take into account not only bacterial resistance but also the mode of growth.
While the field of H. pylori biofilm research is fairly new, groups have already begun to explore alternative therapeutic approaches that may target and eradicate biofilms. The use of the mucolytic, thiol-containing compound N-acetylcysteine (NAC) has shown promise in both humans and mice (21, 108). In early studies that looked at H. pylori-infected mice, daily treatment with NAC (120 mg for 14 days) successfully reduced the H. pylori load by almost 1 log compared to the nontreated group. Pretreatment with NAC (40 mg/day) significantly reduced the H. pylori load but did not completely prevent colonization (108). Furthermore, NAC was also shown to augment the activity of clarithromycin and lansoprazole, a proton pump inhibitor (PPI) dual therapy. H. pylori-positive patients that received both dual therapy and NAC showed significantly better eradication of H. pylori than did those on dual therapy only (109). The mechanism by which NAC functions in the treatment of H. pylori is not clear; however, subsequent studies showed that NAC was effective at dispersing preexistent in vitro H. pylori biofilms and at preventing their formation (21). NAC is believed to function by disrupting disulfide bonds that cross-link glycoproteins in the mucus. While the exact proteins targeted by NAC are not known, it is possible that NAC targets the proteins of the biofilm matrix and leads to their destruction. In a clinical trial, NAC was demonstrated to similarly disperse H. pylori biofilms during in vivo infections (21). Indeed, subjects with a history of at least 4 H. pylori eradication failures who were treated with NAC before receiving a traditional antibiotic regimen demonstrated better clearance of H. pylori (65%) than did the non-NAC group (20%) (21). The exact molecular mechanisms underlying the reported therapeutic effect of NAC are not clear, since this molecule seems to be able to target both H. pylori biofilms and the gastric mucus. Further studies are required to decipher NAC's activity against H. pylori and its biofilms and to help assess its efficacy for possible future antimicrobial therapies.
Studies of H. pylori Biofilms and Immune Evasion
In addition to enhanced defenses against therapeutic interventions, biofilms also defend against the host immune system. Gaddy et al. demonstrated that exposure to subinhibitory concentrations of the host antimicrobial protein calprotectin (CP) resulted in altered lipid A structures, which led to a decrease in surface hydrophobicity and an increase in biofilm formation (22). CP is produced by neutrophils and other myeloid cells (22, 110–113). One function of CP is to sequester essential metals, including manganese and zinc; this sequestration contributes to a process known as nutritional immunity (112). Interestingly, chemical chelation of zinc from H. pylori cultures produced biofilm results similar to those seen with the addition of CP. These findings suggest that one mechanism that H. pylori may utilize to elude the host response is the alteration of its cell surface and subsequent biofilm formation.
Another mechanism of immune interference associated with H. pylori biofilms may be via the proteomannans associated with the matrix. In C. albicans, these mannose-related proteoglycans purportedly display several immunomodulatory roles. These roles include the suppression of B and T lymphocytes (88) and the promotion of mast cell degranulation, a process that releases several mediators (e.g., histamine, serotonin, serine protease, and proteoglycans) that affect the gastrointestinal mucosal barrier (114). Interestingly, the pathogenesis of H. pylori-infected gastritis has been associated with mast cell degranulation (115, 116) and regulatory T cells that actively suppress the T cell response (117–119).
Together, these preliminary data suggest that H. pylori found within a biofilm may be protected from both antimicrobials and the immune system; however, the role of H. pylori biofilms in antimicrobial resistance and clearance by the immune system in vivo remains to be elucidated.
CONCLUSION
H. pylori-associated diseases are a scourge for human health and still raise major health concerns worldwide. The increase in antimicrobial resistance, the high infection rates, and the ability of H. pylori to subvert the host immune response have made this pathogen one of the most successful human pathogens. Cures against H. pylori infection frequently fail, and consequently, multidrug-resistant strains have emerged and become a very serious problem. Evidence suggests that H. pylori can form biofilms and that these structures may have a role in persistence, survivability, and/or recalcitrance to antimicrobial treatment within the host or environment. Thus, these structures require further study. However, there are many challenges that researchers must keep in mind to create informative in vitro H. pylori biofilm models. These challenges include culture conditions, the culture system, and differences between strains. While these challenges may seem daunting, it is important to remember that similar problems have been faced in other biofilm fields, and awareness of these issues will aid in the design of experiments and the interpretation of their results. Hopefully, the recent data that indicate that H. pylori bacteria within a biofilm are distinct from planktonic bacteria will inspire future studies and research to demystify the unexplored role of biofilms in H. pylori infection and, thus, improve the efficacy of future eradication therapies.
ACKNOWLEDGMENTS
We thank Philip Domenico, Shuai Hu, and Carmen Schwechheimer for their helpful suggestions and comments.
The work described here was supported by National Institutes of Health National Institute of Allergy and Infectious Diseases (NIAID) grants RO1AI116946 (to K.M.O.) and R21AI121517 (to D.S.M.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the Department of Defense, the Uniformed Services University, or the NIH.
APPENDIX 1
OPEN QUESTIONS
What conditions or signals drive H. pylori to produce a biofilm?
What are the steps of biofilm formation for H. pylori? What genes are required?
Do biofilms play a role in chronic disease and the persistence of H. pylori?
What role(s) does H. pylori biofilms play in the pathogenesis of gastritis, peptic ulcers, and gastric cancer?
Are biofilms present at any specific points in time or locations during infections?
If H. pylori biofilms form in vivo, do they modulate the immune response?
Does H. pylori multiply within a biofilm?
Do H. pylori biofilms play a role in antibiotic therapy failure?
Are cells in H. pylori biofilms dormant or antibiotic tolerant?
APPENDIX 2
H. PYLORI TREATMENTS
Triple therapy for H. pylori infection remains the first-line therapy in areas with low clarithromycin resistance (<15%) and consists of a proton pump inhibitor (PPI) (e.g., omeprazole, esomeprazole, or lansoprazole), clarithromycin, and amoxicillin for a duration that ranges from 7 to 14 days (120, 121). However, over the past decade, success rates have fallen to unacceptable levels (≤80%) (122). Resistance to clarithromycin has been proposed as the main factor for eradication therapy failure (121, 123).
As a second-line therapy for patients in whom first-line therapies have failed, a bismuth-containing quadruple-therapy regimen is usually recommended (120, 121, 124–126). Bismuth-containing quadruple therapy consists of a PPI, bismuth subsalicylate, metronidazole, and tetracycline for a duration of 10 to 14 days. Bismuth-based regimens offer an effective option as rescue therapy, with cure rates being relatively higher than those with first-line therapies (121, 122, 124). Bismuth is a nontoxic heavy metal with broad-spectrum bactericidal activity (127). Its action against H. pylori remains unclear, but like many heavy metals, it may play a role in iron sequestration (127). Similar effects have been observed when H. pylori was exposed to either bismuth compounds or iron chelators, which suggests that bismuth may act mainly through an iron deprivation mechanism to inhibit H. pylori growth (128). Interestingly, previous reports have demonstrated the efficacy of bismuth against several biofilms (129–131). While the effect of bismuth on H. pylori biofilms has not been studied, it is possible that bismuth-containing therapies act through the action of bismuth against biofilms. In such a scenario, bismuth might decrease the H. pylori biomass and thus enhance the effect of the antibiotics. The eradication of H. pylori is still one of the most difficult challenges in gastroenterology. One route to explore is whether antibiofilm compounds would enhance treatment outcomes.
REFERENCES
- 1.Costerton JW, Geesey GG, Cheng KJ. 1978. How bacteria stick. Sci Am 238:86–95. doi: 10.1038/scientificamerican0178-86. [DOI] [PubMed] [Google Scholar]
- 2.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 3.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 4.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 5.Westall F. 2005. Evolution. Life on the early Earth: a sedimentary view. Science 308:366–367. doi: 10.1126/science.1107227. [DOI] [PubMed] [Google Scholar]
- 6.Lebeaux D, Ghigo JM, Beloin C. 2014. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev 78:510–543. doi: 10.1128/MMBR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 8.Hathroubi S, Mekni MA, Domenico P, Nguyen D, Jacques M. 2017. Biofilms: microbial shelters against antibiotics. Microb Drug Resist 23:147–156. doi: 10.1089/mdr.2016.0087. [DOI] [PubMed] [Google Scholar]
- 9.Polk DB, Peek RM Jr. 2010. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer 10:403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ierardi E, Giorgio F, Losurdo G, Di Leo A, Principi M. 2013. How antibiotic resistances could change Helicobacter pylori treatment: a matter of geography? World J Gastroenterol 19:8168–8180. doi: 10.3748/wjg.v19.i45.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marshall BJ, Warren JR. 1983. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet i:1273–1275. [PubMed] [Google Scholar]
- 12.International Agency for Research on Cancer, WHO. 1994. Schistosomes, liver flukes and Helicobacter pylori. IARC Monogr Eval Carcinog Risks Hum 61:1–241. [PMC free article] [PubMed] [Google Scholar]
- 13.Kusters JG, van Vliet AH, Kuipers EJ. 2006. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev 19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkin DM. 2006. The global health burden of infection-associated cancers in the year 2002. Int J Cancer 118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 15.McColl KEL. 2010. Helicobacter pylori infection. N Engl J Med 362:1597–1604. doi: 10.1056/NEJMcp1001110. [DOI] [PubMed] [Google Scholar]
- 16.Carron MA, Tran VR, Sugawa C, Coticchia JM. 2006. Identification of Helicobacter pylori biofilms in human gastric mucosa. J Gastrointest Surg 10:712–717. doi: 10.1016/j.gassur.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 17.Coticchia JM, Sugawa C, Tran VR, Gurrola J, Kowalski E, Carron MA. 2006. Presence and density of Helicobacter pylori biofilms in human gastric mucosa in patients with peptic ulcer disease. J Gastrointest Surg 10:883–889. doi: 10.1016/j.gassur.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Stark RM, Gerwig GJ, Pitman RS, Potts LF, Williams NA, Greenman J, Weinzweig IP, Hirst TR, Millar MR. 1999. Biofilm formation by Helicobacter pylori. Lett Appl Microbiol 28:121–126. doi: 10.1046/j.1365-2672.1999.00481.x. [DOI] [PubMed] [Google Scholar]
- 19.Grande R, Di Marcantonio MC, Robuffo I, Pompilio A, Celia C, Di Marzio L, Paolino D, Codagnone M, Muraro R, Stoodley P, Hall-Stoodley L, Mincione G. 2015. Helicobacter pylori ATCC 43629/NCTC 11639 outer membrane vesicles (OMVs) from biofilm and planktonic phase associated with extracellular DNA (eDNA). Front Microbiol 6:1369. doi: 10.3389/fmicb.2015.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bessa LJ, Grande R, Di Iorio D, Di Giulio M, Di Campli E, Cellini L. 2013. Helicobacter pylori free-living and biofilm modes of growth: behavior in response to different culture media. APMIS 121:549–560. doi: 10.1111/apm.12020. [DOI] [PubMed] [Google Scholar]
- 21.Cammarota G, Branca G, Ardito F, Sanguinetti M, Ianiro G, Cianci R, Torelli R, Masala G, Gasbarrini A, Fadda G, Landolfi R, Gasbarrini G. 2010. Biofilm demolition and antibiotic treatment to eradicate resistant Helicobacter pylori: a clinical trial. Clin Gastroenterol Hepatol 8:817.e3–820.e3. doi: 10.1016/j.cgh.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 22.Gaddy JA, Radin JN, Cullen TW, Chazin WJ, Skaar EP, Trent MS, Algood HMS. 2015. Helicobacter pylori resists the antimicrobial activity of calprotectin via lipid A modification and associated biofilm formation. mBio 6:e01349-15. doi: 10.1128/mBio.01349-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Servetas SL, Carpenter BM, Haley KP, Gilbreath JJ, Gaddy JA, Merrell DS. 2016. Characterization of key Helicobacter pylori regulators identifies a role for ArsRS in biofilm formation. J Bacteriol 198:2536–2548. doi: 10.1128/JB.00324-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glupczynski Y, Megraud F, Lopez-Brea M, Andersen LP. 2001. European multicentre survey of in vitro antimicrobial resistance in Helicobacter pylori. Eur J Clin Microbiol Infect Dis 20:820–823. doi: 10.1007/s100960100611. [DOI] [PubMed] [Google Scholar]
- 25.Megraud F, Coenen S, Versporten A, Kist M, Lopez-Brea M, Hirschl AM, Andersen LP, Goossens H, Glupczynski Y, Study Group Participants. 2013. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut 62:34–42. doi: 10.1136/gutjnl-2012-302254. [DOI] [PubMed] [Google Scholar]
- 26.Smith SM, O'Morain C, McNamara D. 2014. Antimicrobial susceptibility testing for Helicobacter pylori in times of increasing antibiotic resistance. World J Gastroenterol 20:9912–9921. doi: 10.3748/wjg.v20.i29.9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones KR, Cha JH, Merrell DS. 2008. Who's winning the war? Molecular mechanisms of antibiotic resistance in Helicobacter pylori. Curr Drug Ther 3:190–203. doi: 10.2174/157488508785747899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vakil N, Vaira D. 2013. Treatment for H. pylori infection: new challenges with antimicrobial resistance. J Clin Gastroenterol 47:383–388. doi: 10.1097/MCG.0b013e318277577b. [DOI] [PubMed] [Google Scholar]
- 29.Mackay WG, Gribbon LT, Barer MR, Reid DC. 1998. Biofilms in drinking water systems: a possible reservoir for Helicobacter pylori. J Appl Microbiol 85(Suppl 1):52S–59S. doi: 10.1111/j.1365-2672.1998.tb05283.x. [DOI] [PubMed] [Google Scholar]
- 30.Cole SP, Harwood J, Lee R, She R, Guiney DG. 2004. Characterization of monospecies biofilm formation by Helicobacter pylori. J Bacteriol 186:3124–3132. doi: 10.1128/JB.186.10.3124-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yonezawa H, Osaki T, Kurata S, Zaman C, Hanawa T, Kamiya S. 2010. Assessment of in vitro biofilm formation by Helicobacter pylori. J Gastroenterol Hepatol 25(Suppl 1):S90–S94. doi: 10.1111/j.1440-1746.2009.06213.x. [DOI] [PubMed] [Google Scholar]
- 32.Cellini L, Allocati N, Di Campli E, Dainelli B. 1994. Helicobacter pylori: a fickle germ. Microbiol Immunol 38:25–30. doi: 10.1111/j.1348-0421.1994.tb01740.x. [DOI] [PubMed] [Google Scholar]
- 33.Konishi K, Saito N, Shoji E, Takeda H, Kato M, Asaka M, Ooi HK. 2007. Helicobacter pylori: longer survival in deep ground water and sea water than in a nutrient-rich environment. APMIS 115:1285–1291. doi: 10.1111/j.1600-0643.2007.00594.x. [DOI] [PubMed] [Google Scholar]
- 34.Wong EH, Ng CG, Chua EG, Tay AC, Peters F, Marshall BJ, Ho B, Goh KL, Vadivelu J, Loke MF. 2016. Comparative genomics revealed multiple Helicobacter pylori genes associated with biofilm formation in vitro. PLoS One 11:e0166835. doi: 10.1371/journal.pone.0166835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cammarota G, Sanguinetti M, Gallo A, Posteraro B. 2012. Review article: biofilm formation by Helicobacter pylori as a target for eradication of resistant infection. Aliment Pharmacol Ther 36:222–230. doi: 10.1111/j.1365-2036.2012.05165.x. [DOI] [PubMed] [Google Scholar]
- 36.Attaran B, Falsafi T, Moghaddam AN. 2016. Study of biofilm formation in C57Bl/6J mice by clinical isolates of Helicobacter pylori. Saudi J Gastroenterol 22:161–168. doi: 10.4103/1319-3767.178529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kusters JG, Gerrits MM, Van Strijp JA, Vandenbroucke-Grauls CM. 1997. Coccoid forms of Helicobacter pylori are the morphologic manifestation of cell death. Infect Immun 65:3672–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizoguchi H, Fujioka T, Nasu M. 1999. Evidence for viability of coccoid forms of Helicobacter pylori. J Gastroenterol 34(Suppl 11):S32–S36. [PubMed] [Google Scholar]
- 39.Cook KL, Bolster CH. 2007. Survival of Campylobacter jejuni and Escherichia coli in groundwater during prolonged starvation at low temperatures. J Appl Microbiol 103:573–583. doi: 10.1111/j.1365-2672.2006.03285.x. [DOI] [PubMed] [Google Scholar]
- 40.Cunningham E, O'Byrne C, Oliver JD. 2009. Effect of weak acids on Listeria monocytogenes survival: evidence for a viable but nonculturable state in response to low pH. Food Control 20:1141–1144. doi: 10.1016/j.foodcont.2009.03.005. [DOI] [Google Scholar]
- 41.Li L, Mendis N, Trigui H, Oliver JD, Faucher SP.. 2014. The importance of the viable but non-culturable state in human bacterial pathogens. Front Microbiol 5:258. doi: 10.3389/fmicb.2014.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magajna BA, Schraft H. 2015. Campylobacter jejuni biofilm cells become viable but non-culturable (VBNC) in low nutrient conditions at 4 degrees C more quickly than their planktonic counterparts. Food Control 50:45–50. doi: 10.1016/j.foodcont.2014.08.022. [DOI] [Google Scholar]
- 43.Cherifi T, Jacques M, Quessy S, Fravalo P. 2017. Impact of nutrient restriction on the structure of Listeria monocytogenes biofilm grown in a microfluidic system. Front Microbiol 8:864. doi: 10.3389/fmicb.2017.00864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uhlich GA, Chen CY, Cottrell BJ, Nguyen LH. 2014. Growth media and temperature effects on biofilm formation by serotype O157:H7 and non-O157 Shiga toxin-producing Escherichia coli. FEMS Microbiol Lett 354:133–141. doi: 10.1111/1574-6968.12439. [DOI] [PubMed] [Google Scholar]
- 45.Ghanbari A, Dehghany J, Schwebs T, Musken M, Haussler S, Meyer-Hermann M. 2016. Inoculation density and nutrient level determine the formation of mushroom-shaped structures in Pseudomonas aeruginosa biofilms. Sci Rep 6:32097. doi: 10.1038/srep32097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevenson TH, Castillo A, Lucia LM, Acuff GR. 2000. Growth of Helicobacter pylori in various liquid and plating media. Lett Appl Microbiol 30:192–196. doi: 10.1046/j.1472-765x.2000.00699.x. [DOI] [PubMed] [Google Scholar]
- 47.Albertson N, Wenngren I, Sjostrom JE. 1998. Growth and survival of Helicobacter pylori in defined medium and susceptibility to Brij 78. J Clin Microbiol 36:1232–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams JC, McInnis KA, Testerman TL. 2008. Adherence of Helicobacter pylori to abiotic surfaces is influenced by serum. Appl Environ Microbiol 74:1255–1258. doi: 10.1128/AEM.01958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shao C, Sun Y, Wang N, Yu H, Zhou Y, Chen C, Jia J. 2013. Changes of proteome components of Helicobacter pylori biofilms induced by serum starvation. Mol Med Rep 8:1761–1766. doi: 10.3892/mmr.2013.1712. [DOI] [PubMed] [Google Scholar]
- 50.Hazell SL, Markesich DC, Evans DJ, Evans DG, Graham DY. 1989. Influence of media supplements on growth and survival of Campylobacter pylori. Eur J Clin Microbiol Infect Dis 8:597–602. doi: 10.1007/BF01968136. [DOI] [PubMed] [Google Scholar]
- 51.Owen RJ, Taylor DE, Wang G, van Doorn LJ. 2001. Heterogeneity and subtyping, p 363–378. In Mobley HLT, Mendz GL, Hazell SL (ed), Helicobacter pylori: physiology and genetics. ASM Press, Washington, DC. [Google Scholar]
- 52.Hennig EE, Mernaugh R, Edl J, Cao P, Cover TL. 2004. Heterogeneity among Helicobacter pylori strains in expression of the outer membrane protein BabA. Infect Immun 72:3429–3435. doi: 10.1128/IAI.72.6.3429-3435.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moreno-Mesonero L, Moreno Y, Alonso JL, Ferrus MA. 2016. DVC-FISH and PMA-qPCR techniques to assess the survival of Helicobacter pylori inside Acanthamoeba castellanii. Res Microbiol 167:29–34. doi: 10.1016/j.resmic.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 54.Moreno-Mesonero L, Moreno Y, Alonso JL, Ferrus MA. 2017. Detection of viable Helicobacter pylori inside free-living amoebae in wastewater and drinking water samples from eastern Spain. Environ Microbiol 19:4103–4112. doi: 10.1111/1462-2920.13856. [DOI] [PubMed] [Google Scholar]
- 55.Santiago P, Moreno Y, Ferrus MA. 2015. Identification of viable Helicobacter pylori in drinking water supplies by cultural and molecular techniques. Helicobacter 20:252–259. doi: 10.1111/hel.12205. [DOI] [PubMed] [Google Scholar]
- 56.Moreno Y, Ferrus MA. 2012. Specific detection of cultivable Helicobacter pylori cells from wastewater treatment plants. Helicobacter 17:327–332. doi: 10.1111/j.1523-5378.2012.00961.x. [DOI] [PubMed] [Google Scholar]
- 57.Garcia A, Salas-Jara MJ, Herrera C, Gonzalez C. 2014. Biofilm and Helicobacter pylori: from environment to human host. World J Gastroenterol 20:5632–5638. doi: 10.3748/wjg.v20.i19.5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park SR, Mackay WG, Reid DC. 2001. Helicobacter sp. recovered from drinking water biofilm sampled from a water distribution system. Water Res 35:1624–1626. doi: 10.1016/S0043-1354(00)00582-0. [DOI] [PubMed] [Google Scholar]
- 59.Cellini L, Del Vecchio A, Di Candia M, Di Campli E, Favaro M, Donelli G. 2004. Detection of free and plankton-associated Helicobacter pylori in seawater. J Appl Microbiol 97:285–292. doi: 10.1111/j.1365-2672.2004.02307.x. [DOI] [PubMed] [Google Scholar]
- 60.Azevedo NF, Vieira MJ, Keevil CW. 2003. Establishment of a continuous model system to study Helicobacter pylori survival in potable water biofilms. Water Sci Technol 47(5):155–160. [PubMed] [Google Scholar]
- 61.Giao MS, Azevedo NF, Wilks SA, Vieira MJ, Keevil CW. 2008. Persistence of Helicobacter pylori in heterotrophic drinking-water biofilms. Appl Environ Microbiol 74:5898–5904. doi: 10.1128/AEM.00827-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu Y, Redlinger TE, Avitia R, Galindo A, Goodman K. 2002. Isolation and genotyping of Helicobacter pylori from untreated municipal wastewater. Appl Environ Microbiol 68:1436–1439. doi: 10.1128/AEM.68.3.1436-1439.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giao MS, Azevedo NF, Wilks SA, Vieira MJ, Keevil CW. 2011. Interaction of Legionella pneumophila and Helicobacter pylori with bacterial species isolated from drinking water biofilms. BMC Microbiol 11:57. doi: 10.1186/1471-2180-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andersen LP, Rasmussen L. 2009. Helicobacter pylori—coccoid forms and biofilm formation. FEMS Immunol Med Microbiol 56:112–115. doi: 10.1111/j.1574-695X.2009.00556.x. [DOI] [PubMed] [Google Scholar]
- 65.Ng CG, Loke MF, Goh KL, Vadivelu J, Ho B. 2017. Biofilm formation enhances Helicobacter pylori survivability in vegetables. Food Microbiol 62:68–76. doi: 10.1016/j.fm.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 66.Atapoor S, Safarpoor Dehkordi F, Rahimi E. 2014. Detection of Helicobacter pylori in various types of vegetables and salads. Jundishapur J Microbiol 7:e10013. doi: 10.5812/jjm.10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yahaghi E, Khamesipour F, Mashayekhi F, Safarpoor Dehkordi F, Sakhaei MH, Masoudimanesh M, Khameneie MK. 2014. Helicobacter pylori in vegetables and salads: genotyping and antimicrobial resistance properties. Biomed Res Int 2014:757941. doi: 10.1155/2014/757941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hathroubi S, Beaudry F, Provost C, Martelet L, Segura M, Gagnon CA, Jacques M. 2016. Impact of Actinobacillus pleuropneumoniae biofilm mode of growth on the lipid A structures and stimulation of immune cells. Innate Immun 22:353–362. doi: 10.1177/1753425916649676. [DOI] [PubMed] [Google Scholar]
- 69.Dorr T, Vulic M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol 8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keilberg D, Zavros Y, Shepherd B, Salama NR, Ottemann KM. 2016. Spatial and temporal shifts in bacterial biogeography and gland occupation during the development of a chronic infection. mBio 7:e01705-16. doi: 10.1128/mBio.01705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Howitt MR, Lee JY, Lertsethtakarn P, Vogelmann R, Joubert LM, Ottemann KM, Amieva MR. 2011. ChePep controls Helicobacter pylori infection of the gastric glands and chemotaxis in the Epsilonproteobacteria. mBio 2:e00098-11. doi: 10.1128/mBio.00098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sigal M, Rothenberg ME, Logan CY, Lee JY, Honaker RW, Cooper RL, Passarelli B, Camorlinga M, Bouley DM, Alvarez G, Nusse R, Torres J, Amieva MR. 2015. Helicobacter pylori activates and expands Lgr5(+) stem cells through direct colonization of the gastric glands. Gastroenterology 148:1392.e21–1404.e21. doi: 10.1053/j.gastro.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 73.Jefferson KK. 2004. What drives bacteria to produce a biofilm? FEMS Microbiol Lett 236:163–173. doi: 10.1111/j.1574-6968.2004.tb09643.x. [DOI] [PubMed] [Google Scholar]
- 74.Mashimo C, Kamitani H, Nambu T, Yamane K, Yamanaka T, Sugimori-Shinozuka C, Tatami T, Inoue J, Kamei M, Morita S, Leung KP, Fukushima H. 2013. Identification of the genes involved in the biofilm-like structures on Actinomyces oris K20, a clinical isolate from an apical lesion. J Endod 39:44–48. doi: 10.1016/j.joen.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 75.Bartosik AA, Glabski K, Jecz P, Mikulska S, Fogtman A, Koblowska M, Jagura-Burdzy G. 2014. Transcriptional profiling of ParA and ParB mutants in actively dividing cells of an opportunistic human pathogen Pseudomonas aeruginosa. PLoS One 9:e87276. doi: 10.1371/journal.pone.0087276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bourzac KM, Satkamp LA, Guillemin K. 2006. The Helicobacter pylori cag pathogenicity island protein CagN is a bacterial membrane-associated protein that is processed at its C terminus. Infect Immun 74:2537–2543. doi: 10.1128/IAI.74.5.2537-2543.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang FL, Hassanbhai AM, Chen HY, Huang ZY, Lin TL, Wu SH, Ho B. 2011. Proteomannans in biofilm of Helicobacter pylori ATCC 43504. Helicobacter 16:89–98. doi: 10.1111/j.1523-5378.2010.00815.x. [DOI] [PubMed] [Google Scholar]
- 78.Anderson JK, Huang JY, Wreden C, Sweeney EG, Goers J, Remington SJ, Guillemin K. 2015. Chemorepulsion from the quorum signal autoinducer-2 promotes Helicobacter pylori biofilm dispersal. mBio 6:e00379-15. doi: 10.1128/mBio.00379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rader BA, Wreden C, Hicks KG, Sweeney EG, Ottemann KM, Guillemin K. 2011. Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiology 157:2445–2455. doi: 10.1099/mic.0.049353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rader BA, Campagna SR, Semmelhack MF, Bassler BL, Guillemin K. 2007. The quorum-sensing molecule autoinducer 2 regulates motility and flagellar morphogenesis in Helicobacter pylori. J Bacteriol 189:6109–6117. doi: 10.1128/JB.00246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Osaki T, Hanawa T, Manzoku T, Fukuda M, Kawakami H, Suzuki H, Yamaguchi H, Yan X, Taguchi H, Kurata S, Kamiya S. 2006. Mutation of luxS affects motility and infectivity of Helicobacter pylori in gastric mucosa of a Mongolian gerbil model. J Med Microbiol 55:1477–1485. doi: 10.1099/jmm.0.46660-0. [DOI] [PubMed] [Google Scholar]
- 82.De la Cruz MA, Ares MA, von Bargen K, Panunzi LG, Martinez-Cruz J, Valdez-Salazar HA, Jimenez-Galicia C, Torres J. 2017. Gene expression profiling of transcription factors of Helicobacter pylori under different environmental conditions. Front Microbiol 8:615. doi: 10.3389/fmicb.2017.00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. 2016. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 84.Coulon C, Vinogradov E, Filloux A, Sadovskaya I. 2010. Chemical analysis of cellular and extracellular carbohydrates of a biofilm-forming strain Pseudomonas aeruginosa PA14. PLoS One 5:e14220. doi: 10.1371/journal.pone.0014220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mann EE, Wozniak DJ. 2012. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev 36:893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hathroubi S, Hancock MA, Bosse JT, Langford PR, Tremblay YD, Labrie J, Jacques M. 2015. Surface polysaccharide mutants reveal that absence of O antigen reduces biofilm formation of Actinobacillus pleuropneumoniae. Infect Immun 84:127–137. doi: 10.1128/IAI.00912-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Raa J. 2015. Immune modulation by non-digestible and non-absorbable beta-1,3/1,6-glucan. Microb Ecol Health Dis 26:27824. doi: 10.3402/mehd.v26.27824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dinadayala P, Kaur D, Berg S, Amin AG, Vissa VD, Chatterjee D, Brennan PJ, Crick DC. 2006. Genetic basis for the synthesis of the immunomodulatory mannose caps of lipoarabinomannan in Mycobacterium tuberculosis. J Biol Chem 281:20027–20035. doi: 10.1074/jbc.M603395200. [DOI] [PubMed] [Google Scholar]
- 89.Harmsen M, Lappann M, Knochel S, Molin S. 2010. Role of extracellular DNA during biofilm formation by Listeria monocytogenes. Appl Environ Microbiol 76:2271–2279. doi: 10.1128/AEM.02361-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hathroubi S, Fontaine-Gosselin SE, Tremblay YD, Labrie J, Jacques M. 2015. Sub-inhibitory concentrations of penicillin G induce biofilm formation by field isolates of Actinobacillus pleuropneumoniae. Vet Microbiol 179:277–286. doi: 10.1016/j.vetmic.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 91.Jakubovics NS, Burgess JG. 2015. Extracellular DNA in oral microbial biofilms. Microbes Infect 17:531–537. doi: 10.1016/j.micinf.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 92.Grande R, Di Giulio M, Bessa LJ, Di Campli E, Baffoni M, Guarnieri S, Cellini L. 2011. Extracellular DNA in Helicobacter pylori biofilm: a backstairs rumour. J Appl Microbiol 110:490–498. doi: 10.1111/j.1365-2672.2010.04911.x. [DOI] [PubMed] [Google Scholar]
- 93.Mashburn LM, Whiteley M. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 94.Mashburn-Warren LM, Whiteley M. 2006. Special delivery: vesicle trafficking in prokaryotes. Mol Microbiol 61:839–846. doi: 10.1111/j.1365-2958.2006.05272.x. [DOI] [PubMed] [Google Scholar]
- 95.Schooling SR, Hubley A, Beveridge TJ. 2009. Interactions of DNA with biofilm-derived membrane vesicles. J Bacteriol 191:4097–4102. doi: 10.1128/JB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang W, Chanda W, Zhong M. 2015. The relationship between biofilm and outer membrane vesicles: a novel therapy overview. FEMS Microbiol Lett 362:fnv117. doi: 10.1093/femsle/fnv117. [DOI] [PubMed] [Google Scholar]
- 97.Yonezawa H, Osaki T, Kurata S, Fukuda M, Kawakami H, Ochiai K, Hanawa T, Kamiya S. 2009. Outer membrane vesicles of Helicobacter pylori TK1402 are involved in biofilm formation. BMC Microbiol 9:197. doi: 10.1186/1471-2180-9-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yonezawa H, Osaki T, Woo T, Kurata S, Zaman C, Hojo F, Hanawa T, Kato S, Kamiya S. 2011. Analysis of outer membrane vesicle protein involved in biofilm formation of Helicobacter pylori. Anaerobe 17:388–390. doi: 10.1016/j.anaerobe.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 99.Yonezawa H, Osaki T, Fukutomi T, Hanawa T, Kurata S, Zaman C, Hojo F, Kamiya S. 2017. Diversification of the AlpB outer membrane protein of Helicobacter pylori affects biofilm formation and cellular adhesion. J Bacteriol 199:e00729-16. doi: 10.1128/JB.00729-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Olsen I. 2015. Biofilm-specific antibiotic tolerance and resistance. Eur J Clin Microbiol Infect Dis 34:877–886. doi: 10.1007/s10096-015-2323-z. [DOI] [PubMed] [Google Scholar]
- 101.Stewart PS. 2015. Antimicrobial tolerance in biofilms. Microbiol Spectr 3(3):MB-0010-2014. doi: 10.1128/microbiolspec.MB-0010-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Madsen JS, Burmolle M, Hansen LH, Sorensen SJ. 2012. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol 65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 103.Bae J, Oh E, Jeon B. 2014. Enhanced transmission of antibiotic resistance in Campylobacter jejuni biofilms by natural transformation. Antimicrob Agents Chemother 58:7573–7575. doi: 10.1128/AAC.04066-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Merod RT, Wuertz S. 2014. Extracellular polymeric substance architecture influences natural genetic transformation of Acinetobacter baylyi in biofilms. Appl Environ Microbiol 80:7752–7757. doi: 10.1128/AEM.01984-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bernier SP, Lebeaux D, DeFrancesco AS, Valomon A, Soubigou G, Coppee JY, Ghigo JM, Beloin C. 2013. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet 9:e1003144. doi: 10.1371/journal.pgen.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yonezawa H, Osaki T, Hanawa T, Kurata S, Ochiai K, Kamiya S. 2013. Impact of Helicobacter pylori biofilm formation on clarithromycin susceptibility and generation of resistance mutations. PLoS One 8:e73301. doi: 10.1371/journal.pone.0073301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huynh HQ, Couper RT, Tran CD, Moore L, Kelso R, Butler RN. 2004. N-Acetylcysteine, a novel treatment for Helicobacter pylori infection. Dig Dis Sci 49:1853–1861. doi: 10.1007/s10620-004-9583-2. [DOI] [PubMed] [Google Scholar]
- 109.Gurbuz AK, Ozel AM, Ozturk R, Yildirim S, Yazgan Y, Demirturk L. 2005. Effect of N-acetyl cysteine on Helicobacter pylori. South Med J 98:1095–1097. doi: 10.1097/01.smj.0000182486.39913.da. [DOI] [PubMed] [Google Scholar]
- 110.Clohessy PA, Golden BE. 1995. Calprotectin-mediated zinc chelation as a biostatic mechanism in host-defense. Scand J Immunol 42:551–556. doi: 10.1111/j.1365-3083.1995.tb03695.x. [DOI] [PubMed] [Google Scholar]
- 111.van Zanten SJ, Kolesnikow T, Leung V, O'Rourke JL, Lee A. 2003. Gastric transitional zones, areas where Helicobacter treatment fails: results of a treatment trial using the Sydney strain mouse model. Antimicrob Agents Chemother 47:2249–2255. doi: 10.1128/AAC.47.7.2249-2255.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gaddy JA, Radin JN, Loh JT, Piazuelo MB, Kehl-Fie TE, Delgado AG, Ilca FT, Peek RM, Cover TL, Chazin WJ, Skaar EP, Algood HMS. 2014. The host protein calprotectin modulates the Helicobacter pylori cag type IV secretion system via zinc sequestration. PLoS Pathog 10:e1004450. doi: 10.1371/journal.ppat.1004450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zackular JP, Chazin WJ, Skaar EP. 2015. Nutritional immunity: S100 proteins at the host-pathogen interface. J Biol Chem 290:18991–18998. doi: 10.1074/jbc.R115.645085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sakurai A, Yamaguchi N, Sonoyama K. 2012. Cell wall polysaccharides of Candida albicans induce mast cell degranulation in the gut. Biosci Microbiota Food Health 31:67–70. doi: 10.12938/bmfh.31.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yamamoto J, Watanabe S, Hirose M, Osada T, Ra C, Sato N. 1999. Role of mast cells as a trigger of inflammation in Helicobacter pylori infection. J Physiol Pharmacol 50:17–23. [PubMed] [Google Scholar]
- 116.Nakajima S, Bamba N, Hattori T. 2004. Histological aspects and role of mast cells in Helicobacter pylori-infected gastritis. Aliment Pharmacol Ther 20(Suppl 1):S165–S170. doi: 10.1111/j.1365-2036.2004.01974.x. [DOI] [PubMed] [Google Scholar]
- 117.Lundgren A, Suri-Payer E, Enarsson K, Svennerholm AM, Lundin BS. 2003. Helicobacter pylori-specific CD4+ CD25high regulatory T cells suppress memory T-cell responses to H. pylori in infected individuals. Infect Immun 71:1755–1762. doi: 10.1128/IAI.71.4.1755-1762.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Enarsson K, Lundgren A, Kindlund B, Hermansson M, Roncador G, Banham AH, Lundin BS, Quiding-Jarbrink M. 2006. Function and recruitment of mucosal regulatory T cells in human chronic Helicobacter pylori infection and gastric adenocarcinoma. Clin Immunol 121:358–368. doi: 10.1016/j.clim.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 119.Harris PR, Wright SW, Serrano C, Riera F, Duarte I, Torres J, Pena A, Rollan A, Viviani P, Guiraldes E, Schmitz JM, Lorenz RG, Novak L, Smythies LE, Smith PD. 2008. Helicobacter pylori gastritis in children is associated with a regulatory T-cell response. Gastroenterology 134:491–499. doi: 10.1053/j.gastro.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 120.Urgesi R, Cianci R, Riccioni ME. 2012. Update on triple therapy for eradication of Helicobacter pylori: current status of the art. Clin Exp Gastroenterol 5:151–157. doi: 10.2147/CEG.S25416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chey WD, Leontiadis GI, Howden CW, Moss SF. 2017. ACG clinical guideline: treatment of Helicobacter pylori infection. Am J Gastroenterol 112:212–239. doi: 10.1038/ajg.2016.563. [DOI] [PubMed] [Google Scholar]
- 122.Molina-Infante J, Gisbert JP. 2014. Optimizing clarithromycin-containing therapy for Helicobacter pylori in the era of antibiotic resistance. World J Gastroenterol 20:10338–10347. doi: 10.3748/wjg.v20.i30.10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Park JY, Dunbar KB, Mitui M, Arnold CA, Lam-Himlin DM, Valasek MA, Thung I, Okwara C, Coss E, Cryer B, Doern CD. 2016. Helicobacter pylori clarithromycin resistance and treatment failure are common in the USA. Dig Dis Sci 61:2373–2380. doi: 10.1007/s10620-016-4091-8. [DOI] [PubMed] [Google Scholar]
- 124.Gisbert JP, Romano M, Gravina AG, Solis-Munoz P, Bermejo F, Molina-Infante J, Castro-Fernandez M, Ortuno J, Lucendo AJ, Herranz M, Modolell I, Del Castillo F, Gomez J, Barrio J, Velayos B, Gomez B, Dominguez JL, Miranda A, Martorano M, Algaba A, Pabon M, Angueira T, Fernandez-Salazar L, Federico A, Marin AC, McNicholl AG. 2015. Helicobacter pylori second-line rescue therapy with levofloxacin- and bismuth-containing quadruple therapy, after failure of standard triple or non-bismuth quadruple treatments. Aliment Pharmacol Ther 41:768–775. doi: 10.1111/apt.13128. [DOI] [PubMed] [Google Scholar]
- 125.Fiorini G, Saracino IM, Zullo A, Gatta L, Pavoni M, Vaira D. 2017. Rescue therapy with bismuth quadruple regimen in patients with Helicobacter pylori-resistant strains. Helicobacter 22:e12448. doi: 10.1111/hel.12448. [DOI] [PubMed] [Google Scholar]
- 126.Megraud F. 2012. The challenge of Helicobacter pylori resistance to antibiotics: the comeback of bismuth-based quadruple therapy. Therap Adv Gastroenterol 5:103–109. doi: 10.1177/1756283X11432492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Domenico P, Reich J, Madonia W, Cunha BA. 1996. Resistance to bismuth among gram-negative bacteria is dependent upon iron and its uptake. J Antimicrob Chemother 38:1031–1040. doi: 10.1093/jac/38.6.1031. [DOI] [PubMed] [Google Scholar]
- 128.Bland MV, Ismail S, Heinemann JA, Keenan JI. 2004. The action of bismuth against Helicobacter pylori mimics but is not caused by intracellular iron deprivation. Antimicrob Agents Chemother 48:1983–1988. doi: 10.1128/AAC.48.6.1983-1988.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Folsom JP, Baker B, Stewart PS. 2011. In vitro efficacy of bismuth thiols against biofilms formed by bacteria isolated from human chronic wounds. J Appl Microbiol 111:989–996. doi: 10.1111/j.1365-2672.2011.05110.x. [DOI] [PubMed] [Google Scholar]
- 130.Domenico P, Baldassarri L, Schoch PE, Kaehler K, Sasatsu M, Cunha BA. 2001. Activities of bismuth thiols against staphylococci and staphylococcal biofilms. Antimicrob Agents Chemother 45:1417–1421. doi: 10.1128/AAC.45.5.1417-1421.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Huang CT, Stewart PS. 1999. Reduction of polysaccharide production in Pseudomonas aeruginosa biofilms by bismuth dimercaprol (BisBAL) treatment. J Antimicrob Chemother 44:601–605. doi: 10.1093/jac/44.5.601. [DOI] [PubMed] [Google Scholar]