Abstract
A large number of arthropod-borne viruses are endemic to East Africa. As a part of the process of undertaking a systematic characterization of the mosquito fauna of Uganda, we examined mosquitoes collected from 2008 through early 2012 for known and novel viruses. In all, 8,288 mosquito pools containing 157,554 mosquitoes were tested. Twenty-nine isolations of 11 different viruses were made from mosquitoes of nine distinct species and from pools identified only to genus Culex. Identified viruses were from family Togaviridae, alphaviruses Sindbis and Babanki viruses; family Rhabdoviridae, hapaviruses Mossuril and Kamese viruses; family Flaviviridae, flaviviruses West Nile and Usutu viruses; family Phenuiviridae, phlebovirus Arumowot virus; and family Peribunyaviridae, orthobunyaviruses Witwatersrand, Pongola, and Germiston viruses. In addition, a novel orthobunyavirus, provisionally named Mburo virus, was isolated from Coquillettidia metallica (Theobald). This is the first report of Babanki, Arumowot, and Mossuril virus isolation from Uganda.
Keywords: arbovirus, mosquito, Uganda
Arthropod-borne pathogens have contributed significantly to the emergence and reemergence of infectious diseases in recent decades (Rosenberg 2015). Of particular concern to global health are areas of the world that pose a high risk for emerging infectious disease (EID) events due to growing human populations, changes in land use altering the interface between humans and wildlife, increasing global travel and commerce, and climate change. Based on these and other criteria, East Africa has been identified as a region of high risk for EID events in the future (Jones et al. 2008). These events are frequently attributed to zoonotic viruses circulating in wildlife that infect humans via transmission by arthropods, with zoonoses from wildlife representing over 60% of all EIDs (Jones et al. 2008). Uganda, in particular, has been the site of multiple EID events, with >20 new arthropod-borne viruses (arboviruses) having been isolated there in the past 70 yr (Karabatsos 1985).
To assess the current level of known and novel arbovirus circulation in Uganda, the U.S. Centers for Disease Control and Prevention (CDC) and the Uganda Virus Research Institute (UVRI) initiated a collaborative survey beginning in 2008 with the goals of collecting and describing the current mosquito fauna of Uganda and screening for arboviruses circulating in mosquitoes, rodents, bats, and other small mammals. Previous publications have reported results from this survey, including descriptions of the mosquitoes collected, prevalence of antibodies to arboviruses in vertebrate sera, identification of host blood from engorged mosquitoes, and descriptions of viruses isolated from bird and bat hosts (Mutebi et al. 2012; Crabtree et al. 2013; Kading et al. 2013a,b; Ledermann et al. 2014). We report here the results of surveillance for arboviruses in mosquitoes collected from 21 sites in Uganda between November 2008 and February 2012.
Twenty nine (29) virus isolates were obtained from 8,288 mosquito pools (1,57,554 mosquitoes), comprising 11 arboviruses from four virus families. Isolates belong to the families Togaviridae (genus Alphavirus, n = 8), Peribunyaviridae (genus Orthobunyavirus, n = 13), Phenuiviridae (genus Phlebovirus, n = 2), Flaviviridae (genus Flavivirus, n = 3), and Rhabdoviridae (genus Hapavirus, n = 3). We report the first isolation of Arumowot virus, Mossuril virus, and Babanki virus from Uganda and additionally describe one novel virus, provisionally named Mburo virus, identified to be genetically most closely related to viruses in the Tete serogroup of the Orthobunyavirus genus, Peribunyaviridae.
Materials and Methods
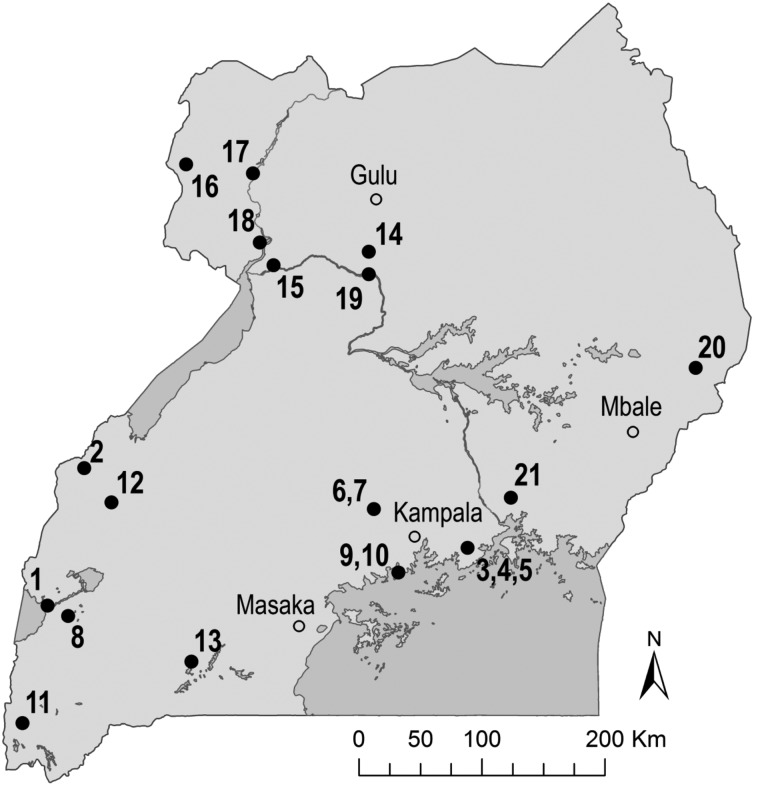
Mosquitoes were collected using CO2-baited CDC light traps and CDC gravid traps as described elsewhere (Godsey et al. 2012, Mutebi et al. 2012). Collections were made at 21 sites throughout Uganda between November 2008 and February 2012 (Fig. 1). Mosquitoes were identified to species using available keys (Edwards 1941, Gillies and DeMeillon 1968, Gillett 1972, Gillies and Coetzee 1987, Jupp 1996), and pooled by trap date, location, and type as well as species, sex, gravidity, and bloodmeal presence (≤25 mosquitoes per pool).
Fig. 1.
Map of Uganda showing mosquito collection sites. Collection sites (•) as well as cities and other locales (○) are indicated. Study site numbers correspond to Table 2.
The infection status of mosquito pools was determined by trituration of mosquitoes in 2-ml conical microcentrifuge tubes with 1.3 ml BA-1 medium (1× Medium 199 with Earle’s salts, 1% bovine albumin, 100 U/ml penicillin, 100 mg/ml streptomycin, 1.5 µg/ml gentamycin, and 1.5 µl/ml amphotericin B) and one copper bead per tube using a Qiagen Tissuelyser (Qiagen, Valencia, CA). Triturated mosquito preparations were clarified by centrifugation at 9,000 rpm at 4 °C for 20 min. Clarified homogenate supernatants were stored at −80 °C. Mosquito homogenates were tested for infectious virus by plaque titration on Vero cells in six-well plates as previously described using 100 µl of each clarified mosquito pool supernatant and with second overlays applied at 4 d postinfection (Miller et al. 1989). Plates were observed for plaques for 15 d following infection. Plate wells in which plaques were observed were harvested as previously described and viruses were amplified by infecting T-25 flasks of Vero cells with 25 µl of each plaque isolate (Crabtree et al. 2009). Flasks were observed and supernatants harvested when cytopathic effects were evident.
Viral RNA was extracted from amplified virus supernatants and identification of virus isolates was attempted by reverse transcription-polymerase chain reaction (RT-PCR) and partial Sanger dideoxy sequencing as previously described using arbovirus group-specific primers for alphaviruses, orthobunyaviruses, and flaviviruses (Kuno et al. 1998, Powers et al. 2001, Bryant et al. 2005, Lambert and Lanciotti 2008). Partial nucleic acid sequences were compared with those in the GenBank database using the BLAST program to identify isolates. Isolates that were negative by group-specific RT-PCR were tested by immunofluorescence assay (IFA) using National Institutes of Health polyvalent mouse hyperimmune ascitic fluids (grouping fluids) as previously described, to identify an arbovirus serogroup to which the isolates might be related (Sang et al. 2006). Positive IFA results were then used to direct virus-specific RT-PCR using primers designed from published related virus sequences. In cases where IFA was also negative, or where positive IFA followed by virus-specific RT-PCR did not produce positive results, isolates were sequenced using next-generation sequencing (NGS) on an Ion Torrent Personal Genome Machine (Life Technologies) as described elsewhere (Kading et al. 2013b, Ledermann et al. 2014). Genetic relatedness of assembled sequences derived from NGS was identified by using the BLAST program at the National Center for Biotechnology Information. Sequences were verified and sequence gaps filled in by using Sanger dideoxy sequencing as necessary. Phylogenetic relationships among amino acid sequences were determined by maximum likelihood analysis using the LG + G + I + F model on MEGA6 software (Tamura et al. 2013).
Results
Twenty-nine virus isolations were made from pools of Aedes (Aedimorphus) tarsalis (Newstead) (1 isolate), Coquillettidia (Cq.) fuscopennata (Theobald) (1), Cq. (Cq.) metallica (Theobald) (1), Culex (Cx.) antennatus (Becker) (1), Culex (Cx.) neavei Theobald (2), Culex (Cx.) perfuscus (Edwards) (8), Culex (Eumelanimyia) rubinotus Theobald (10), Culex (Cx.) univittatus Theobald (1), Culex species (3), and Mansonia (Mansonoides) africana (Theobald) (1) (Table 1). Viruses were isolated from 8 of the 21 collection sites included in the survey: Mweya, Queen Elizabeth National Park (QENP) (1 isolate), Sempaya, Semliki National Park (SNP) (8), Nkokonjeru (1), Luwawa Forest Reserve (LFR) (2), Segganga (10), Kibale National Park (KNP) (1), Lake Mburo National Park (LMNP) (3), and Jinja (3) (Fig. 1; Table 2). All viruses were isolated from mosquitoes collected using CDC light traps.
Table 1.
Number of isolates of each virus by collection site
| Collection site | Togaviridae |
Peribunyaviridae |
Phenuivirus | Flaviviridae |
Rhabdoviridae |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alphavirus |
Orthobunyavirus |
Phlebovirus | Flavivirus |
Hapavirus |
|||||||
| Species | Babanki | Sindbis | Pongola | Germiston | Mburo | Witwatersrand | Arumowot | West Nile | Usutu | Mossuril | Kamese |
| Mweya, Queen Elizabeth National Park | |||||||||||
| Culex neavei | – | – | – | – | – | – | – | 1 | – | – | |
| Sempaya, Semliki National Park | |||||||||||
| Culex perfuscus | 5 | – | – | – | – | – | – | – | – | 2 | 1 |
| Nkokonjeru, Kikusa | |||||||||||
| Culex species | 1 | – | – | – | – | – | – | – | – | – | – |
| Luwawa Forest Reserve, Nansomba Village | |||||||||||
| Aedes tarsalis | – | – | 1 | – | – | – | – | – | – | – | – |
| Coquillettidia fuscopennata | – | – | 1 | – | – | – | – | – | – | – | – |
| Segganga, Luvunvu Village | |||||||||||
| Culex rubinotus | – | – | – | 4 | – | 6 | – | – | – | – | – |
| Kibale National Park | |||||||||||
| Culex species | – | – | – | – | – | – | – | – | 1 | – | – |
| Lake Mburo National Park | |||||||||||
| Culex neavei | – | 1 | – | – | – | – | – | – | – | – | – |
| Mansonia africana | – | 1 | – | – | – | – | – | – | – | – | – |
| Coquillettidia metallica | – | – | – | – | 1 | – | – | – | – | – | – |
| Jinja, Kakira Sugar Works | |||||||||||
| Culex antennatus | – | – | – | – | – | – | 1 | – | – | – | – |
| Culex species | – | – | – | – | – | – | 1 | – | – | – | – |
| Culex univitattus | – | – | – | – | – | – | – | – | 1 | – | – |
Table 2.
Mosquito collection sites and numbers tested
| Site no. | Site location | Latitude/Longitude | Collection dates | No. of mosquitoes tested | No. of pools tested |
|---|---|---|---|---|---|
| 1 | Mweya, Queen Elizabeth National Park | 0° 11′ S, 29° 54′ E | Nov. 2008 | 8,540 | 377 |
| June 2009 | 13,355 | 615 | |||
| Jan. 2010 | 6,630 | 326 | |||
| 2 | Sempaya, Semliki National Park | 0° 49′ N, 30° 10′ E | Nov. 2008 | 4,259 | 236 |
| June 2009 | 5,177 | 267 | |||
| Jan. 2010 | 1,418 | 114 | |||
| 3 | Busitwe, Nkokonjeru | 0° 14′ N, 32° 57′ E | Feb. 2009 | 3,928 | 229 |
| 4 | Kikusa, Nkokonjeru | 0° 14′ N, 32° 58′ E | Dec. 2008 | 275 | 15 |
| Feb. 2009 | 1,447 | 80 | |||
| 5 | Kyambogo, Nkokonjeru | 0° 13′ N, 32° 58′ E | Dec. 2008 | 1,468 | 88 |
| Jan. 2009 | 1,471 | 76 | |||
| 6 | Nansomba Village, Luwawa Forest Reserve, Segganga | 0° 31′ N, 32° 18′ E | Feb. 2009 | 4,188 | 240 |
| 7 | Lununvu Village, Segganga | 0° 32′ N, 32° 17′ E | Feb. 2009 | 9,449 | 445 |
| 8 | Maramagambo, QENP | 0° 16′ S, 30° 03′ E | June 2009 | 488 | 45 |
| Jan. 2010 | 890 | 96 | |||
| 9 | Kitubulu Forest Reserve, Entebbe | 0° 07′ N, 32° 47′ E | June 2009 | 1,807 | 86 |
| Jan. 2010 | 12,145 | 563 | |||
| 10 | Uganda Virus Research Institute, Entebbe | 0° 07′ N, 32° 46′ E | Jan. 2010 | 21 | 13 |
| 11 | Bwindi Impenetrable Forest National Park | 01° 03′ S, 29° 43′ E | Jan. 2010 | 586 | 53 |
| June 2010 | 943 | 79 | |||
| 12 | Kibale National Park | 0° 34′ N, 30° 22′ E | June 2010 | 2,851 | 166 |
| 13 | Lake Mburo National Park | 0° 36′ S, 30° 57′ E | June 2010 | 20,809 | 1,016 |
| 14 | Chobe, Murchison Falls National Park | 2° 24′ N, 32° 13′ E | Jan. 2011 | 3,658 | 191 |
| June 2011 | 7,451 | 407 | |||
| 15 | Paraa, Murchison Falls National Park | 2° 18′ N, 31° 33′ E | Jan. 2011 | 7,476 | 368 |
| June 2011 | 1,906 | 183 | |||
| 16 | Sunguru, Arua | 3° 01′N, 30° 48'E | Jan. 2011 | 317 | 46 |
| June 2011 | 466 | 74 | |||
| 17 | Rhino Camp | 2° 58′ N, 31° 24′ E | Jan. 2011 | 18,927 | 841 |
| 18 | Pakwach | 2° 28′ N, 31° 27′ E | June 2011 | 16 | 9 |
| 19 | Karuma | 2° 15′ N, 32° 14′ E | June 2011 | 84 | 7 |
| 20 | Sipi, Mt. Elgon National Park | 1° 33′ N, 34° 38′ E | Jan. 2012 | 3,928 | 352 |
| 21 | Jinja, Kakira Sugar Works | 0° 36′ N, 33° 17′ E | Feb. 2012 | 11,180 | 585 |
| Total | 157,554 | 8,288 |
Eleven different viruses were identified in this study (Table 1). Sindbis virus (SINV; 97.6% BLAST identity) was isolated from one pool each of Cx. neavei and Ma. africana, both from the LMNP collections (June 2010). Babanki virus (BBKV; 98–99.8% BLAST identities) was isolated from six mosquito pools: five pools of Cx. perfuscus and one pool of Culex species collected at Sempaya, SNP (November 2008 and June 2009) and Kikusa, Nkokonjeru (February 2009), respectively. Pongola virus (PGAV; 97% BLAST identity) was isolated from one pool each of Ae. tarsalis and Cq. fuscopennata, both collected at Nansomba Village, LFR (March 2009). Germiston virus (GERV; 92.7–93.4% BLAST identities) was isolated from four pools of Cx. rubinotus collected at Luvunvu village, Segganga (February 2009). A novel orthobunyavirus, provisionally named Mburo virus (MBUV), was isolated from a pool of Cq. metallica collected at the LMNP site (February 2012). Arumowot virus (AMTV; 97.3% BLAST identity) was isolated from one pool each of Cx. antennatus and Culex species, both collected at Jinja (February 2012). Six isolates of Witwatersrand virus (WITV; 80% nucleotide BLAST identity) were obtained from Cx. rubinotus collected at Luvunvu village, Segganga (February 2009). We isolated West Nile virus (WNV; 98.6% BLAST identity) from a pool of Cx. neavei collected at Mweya, QENP (June 2009), and Usutu virus (USUV; 98% BLAST identity) from one pool each of Cx. univittatus and Culex species, collected at Jinja (February 2012) and KNP (June 2010), respectively. We made two isolates of Mossuril virus (MOSV; 93–96% BLAST identities) and one of Kamese virus (KAMV; 98.2% BLAST identity), all from pools of Cx. perfuscus collected at Sempaya, SNP (June 2009). Nucleic acid sequences from these isolates were obtained by using RT-PCR, with primers designed from published sequence of the KAMV polymerase gene (KAMV-184F, TATGAAAAGTGGAACAATCATC; KAMV-404R, CCTTTCTGGCCGTCCCAAC).
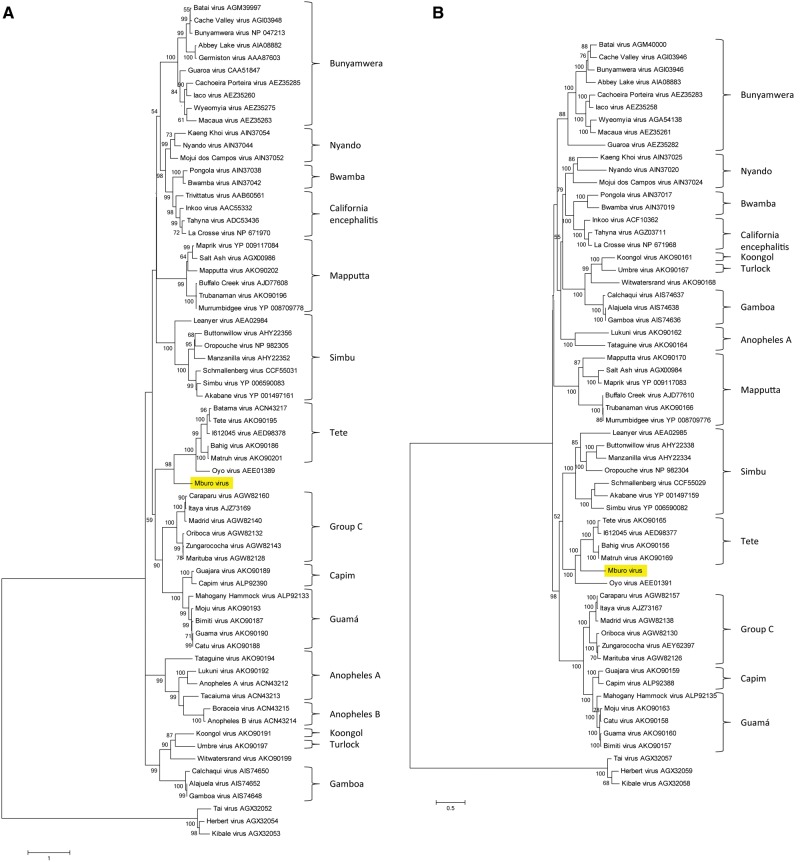
As mentioned above, a novel orthobunyavirus, provisionally named Mburo virus, was isolated from a pool of Cq. metallica mosquitoes collected at the LMNP site. To characterize this new virus, we used IFA with National Institutes of Health grouping fluids. The IFA results suggested the virus was serologically related to the orthobunyaviruses (data not shown). Next-generation sequencing was then used to obtain partial sequences from the S and L segments of the virus. BLAST comparison of the 248 amino acid nucleocapsid sequence and the 2,262 amino acid RNA-dependent RNA polymerase (RdRP) sequence suggested that the virus was most similar to viruses in the Tete serogroup and the closely related Oyo virus (OYOV) of the Orthobunyavirus genus, family Peribunyaviridae (nucleocapsid: >90% coverage, 42–46% identity to Tete serogroup, 45% identity to OYOV; RdRP: 99% coverage, 62–63% identity to Tete serogroup, 55% identity to OYOV). Amino acid sequence maximum likelihood alignment with other orthobunyaviruses confirmed this relationship (Fig. 2).
Fig. 2.
Amino acid sequence-based phylogenies of orthobunyavirus nucleocapsid and RNA-dependent RNA polymerase. Full-length amino acid sequences of nucleocapsid (A) and RNA-dependent RNA polymerase (B) were obtained from GenBank and aligned with the derived sequences of Mburo virus (highlighted; GenBank MF074140 and MF074141, respectively). Phylogeny was determined by maximum likelihood analysis using the LG + G + I + F model. The trees were rooted using three members of the Herbevirus genus, family Peribunyaviridae (Marklewitz et al. 2013). Serogroups are shown at the right for reference.
Discussion
Arboviruses comprise a diverse group of human and animal pathogens that may cause mild to severe illness and result in significant public health and economic consequences. The number of EID events in recent decades that are attributable to zoonotic arboviruses calls for increased regular surveillance to predict and control future emergence or reemergence of viruses causing infectious diseases. This study was designed to gather baseline information regarding the circulation of arboviruses throughout Uganda by sampling potential arbovirus vector mosquitoes that may be part of zoonotic cycles. Because many EID events have resulted as a consequence of increased human population and land use, collection sites for this study were selected to allow surveillance at the interface of wildlife areas and human habitation as well as in villages and undisturbed habitats.
We report the isolation of 11 arboviruses, including one novel virus and three viruses never before isolated from Uganda. Six of these viruses—BBKV, SINV, PGAV, GERV, WNV, and USUV—have been isolated from humans and associated with febrile illness, often accompanied by rash and arthritis. West Nile virus has also been shown to cause more severe disease including CNS symptoms (Davis et al. 2006). Of the known viruses isolated in this study, three—BBKV, AMTV, and MOS—have not been reported previously from Uganda. Additionally, five of the known arboviruses we isolated were obtained from mosquitoes of species not previously associated with these viruses: PGAV from Cq. fuscopennata, WNV from Cx. neavei, USUV from Cx. univitattus, and MOSV and KAMV from Cx. perfuscus.
The six isolates that were identified as Witwatersrand virus were compared with GenBank sequences at both the nucleotide and amino acid levels. At the nucleotide level, BLAST identities were 80% compared with the single Witwatersrand virus represented in GenBank that was isolated in 1958. However, amino acid comparisons yielded BLAST identities of 94%, indicating that although changes had occurred in the nucleotide sequence over the 50+ yr between these isolations, the amino acid sequence was highly conserved.
We report herein the isolation of a novel virus that we have provisionally named Mburo virus (MBUV). This virus is genetically most closely related to viruses in the Tete serogroup and OYOV of the Orthobunyavirus genus, family Peribunyaviridae. Several viruses of the Tete serogroup are among the relatively small number of orthobunyaviruses that are known to be transmitted by or have been isolated from ticks. Further, many members of this serogroup have been repeatedly isolated from birds (Shchetinin et al. 2015). It is therefore not surprising that MBUV was isolated from Cq. metallica, as Coquillettidia species are known to be ornithophilic (Williams et al. 1958, Mukwaya 1972, Rickenbach et al. 1974, Chandler and Highton 1976, Beier et al. 1990, Crabtree et al. 2013). In a previous study in Uganda, all identified bloodmeals from Coquillettidia species mosquitoes were exclusively from birds, except those from Cq. fuscopennata. Additionally, using the methods of Crabtree et al. (2013), we identified the bloodmeals of six Cq. metallica collected in Uganda, five of which were birds (M.B.C., unpublished data). The single reported isolation of OYOV was made from the blood of a pig in 1964 (R.B. Tesh, personal communication). No arthropod association for OYOV has yet been documented. Mburo virus may also be related to Sedlec virus (SEDV; Hubalek et al. 1990). Sedlec virus was not included in our analyses owing to the limited sequence information available. However, based on partial sequence analysis, the L segment of SEDV appeared to be similar to OYOV and the Tete serogroup, but distinct from MBUV (data not shown), while the S segment was more closely related to the Simbu serogroup–Leanyer virus clade (Bakonyi et al. 2013). Further study is needed to determine whether MBUV may also be tick-borne and what its vertebrate host range might be. Regrettably, owing to a freezer malfunction, the isolate was lost and additional characterization will not be possible until the virus is isolated again in the future.
Although a relatively low number of isolates were obtained from these mosquito collections given the large number of mosquitoes tested, the number was not unexpected when compared with reports of other arbovirus surveillance studies in the region. In 2009, Crabtree, et al. reported the isolation of 27 virus strains from 2,651 mosquito pools collected during an outbreak of Rift Valley fever virus (RVFV) in Kenya (2006–2007; Crabtree et al. 2009). Environmental conditions during this period were conducive to a very high abundance of mosquito vectors, and as a result, increased interaction between mosquitoes and humans or nonhuman hosts, thereby supporting increased virus transmission (Nguku et al. 2010). Similarly, in a study by Ochieng et al. (2013) conducted in Kenya from 2007 to 2012, mosquito collections were made only during wet seasons when mosquito population density was high, again resulting in increased host and vector contact supporting increased virus transmission. During that study, 83 virus isolates were obtained from 15,890 mosquito pools, many of which were floodwater Aedes. In our study, collections were made at most sites during multiple seasons, both dry and wet, and as a result, mosquito population density was quite low for some collections. Additionally, some collection sites were close to bodies of water (lakes, rivers, and swamps), resulting in collection of a high number of Coquillettidia mosquitoes which are not known to vector many arboviruses (Karabatsos 1985, Mutebi et al. 2012). In addition to the effect of habitat on the composition of mosquito collections, trapping methods also play an important role. In the study mentioned above by Ochieng et al. mosquitoes were collected using both CO2-baited CDC light traps and human bait collections, resulting in collection of a higher number of anthropophilic mosquitoes which may be more likely to transmit viruses to humans. Owing to study design constraints, our survey did not include human-baited collections.
Our collections also comprised many naïve or newly emerged specimens as indicated by 1) the presence in some collections of relatively higher numbers of males which emerge before females, 2) the low proportion of engorged specimens collected (0.74%), and 3) the collection of a relatively small proportion of gravid specimens (0.85%). These younger mosquitoes would be less likely to have taken a first bloodmeal and therefore would not have had the opportunity to acquire a virus from a host.
Comparison of viruses isolated in our study with those of the two Kenya studies mentioned above suggests that similar viruses are circulating in both countries. Viruses isolated in all three studies include SINV, BBKV, PGAV, and WNV. In our study, the greatest number of viruses isolated were from the family Peribunyaviridae (n = 13), followed by the Togaviridae (n = 8); in both Kenya studies, the family most represented in the virus isolations was the Togaviridae, followed by Peribunyaviridae and Flaviviridae (Crabtree et al. 2009) or Flaviviridae and Peribunyaviridae (Ochieng et al. 2013).
The results of this surveillance study are important if we are to detect, prevent, and control outbreaks of emerging and reemerging infectious diseases caused by arboviruses. The number and identity of isolated viruses that cause febrile illness will suggest directions for human diagnostic testing to determine arboviral agents causing febrile illnesses.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Acknowledgments
We thank the Uganda Wildlife Authority (UWA) for granting us access to Semliki National Park (SNP), Queen Elizabeth National Park (QENP), and Kibale National Park (KNP) and the park rangers at SNP, QENP, and KNP for their assistance and for use of UWA facilities. We also thank T. Okello and M. Driciru for use of laboratory facilities at QENP; F. Ssenfunka, J.-B. Lwanga, S. Wakaalo, J. Mugga, G. Kyazze, and D. Ssemwogerere of UVRI, and K. Saxton-Shaw and J. Lederman of CDC Ft. Collins for field assistance during this project, and K. Boegler of CDC Ft. Collins for assistance creating the site map. Lastly, we thank R. Rosenberg for his assistance and support during this project. This study was supported in part by funds from USAID Emerging Pandemic Threat Program.
References Cited
- Bakonyi T., Kolodziejek J., Rudolf I., Bercic R., Nowotny N., Hubalek Z.. 2013. Partial genetic characterization of Sedlec virus (Orthobunyavirus, Bunyaviridae). Infect. Genetics Evol. J. Mol. Epidemiol. Evol. Genetics Infect. Dis. 19: 244–249. [DOI] [PubMed] [Google Scholar]
- Beier J. C., Odago W. O., Onyango F. K., Asiago C. M., Koech D. K., Roberts C. R.. 1990. Relative abundance and blood feeding behavior of nocturnally active culicine mosquitoes in western Kenya. J. Am. Mosq. Control Assoc. 6: 207–212. [PubMed] [Google Scholar]
- Bryant J. E., Crabtree M. B., Nam V. S., Yen N. T., Duc H. M., Miller B. R.. 2005. Isolation of arboviruses from mosquitoes collected in northern Vietnam. Am. J. Trop. Med. Hyg. 73: 470–473. [PubMed] [Google Scholar]
- Chandler J. A., Highton R. B.. 1976. Studies on some ornithophilic mosquitoes (Diptera, Culicidae) of the Kano Plain, Kenya. Bull. Entomol. Res. 66: 133–143. [Google Scholar]
- Crabtree M., Sang R., Lutomiah J., Richardson J., Miller B.. 2009. Arbovirus surveillance of mosquitoes collected at sites of active Rift Valley fever virus transmission: Kenya, 2006–2007. J. Med. Entomol. 46: 961–964. [DOI] [PubMed] [Google Scholar]
- Crabtree M. B., Kading R. C., Mutebi J. P., Lutwama J. J., Miller B. R.. 2013. Identification of host blood from engorged mosquitoes collected in western Uganda using cytochrome oxidase I gene sequences. J. Wildl. Dis. 49: 611–626. [DOI] [PubMed] [Google Scholar]
- Davis L. E., DeBiasi R., Goade D. E., Haaland K. Y., Harrington J. A., Harnar J. B., Pergam S. A., King M. K., DeMasters B. K., Tyler K. L.. 2006. West Nile virus neuroinvasive disease. Ann. Neurol. 60: 286–300. [DOI] [PubMed] [Google Scholar]
- Edwards F. W. 1941. Mosquitoes of the Ethiopian Region III. Culcine adults and pupae. London British Museum of Natural History, London, United Kingdom. [Google Scholar]
- Gillett J. D. 1972. Common African mosquitos and their medical importance. William Heinemann Medical Books Ltd, London, United Kingdom. [Google Scholar]
- Gillies M. T., DeMeillon B.. 1968. The Anophelinae of Africa South of the Sahara (Ethiopian Zoogeographical region). South African Institute for Medical Research, Johannesburg, South Africa. [Google Scholar]
- Gillies M. T., Coetzee M.. 1987. A supplement to the Anophelinae of Africa South of the Sahara (Afrotropical Region). The South African Institute for Medical Research, Johannesburg, South Africa. [Google Scholar]
- Godsey M. S. Jr., Burkhalter K., Young G., Delorey M., Smith K., Townsend J., Levy C., Mutebi J. P.. 2012. Entomologic investigations during an outbreak of West Nile virus disease in Maricopa County, Arizona, 2010. Am. J. Trop. Med. Hyg. 87: 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubalek Z., Juricova Z., Halouzka J., Butenko A. M., Kondrasina N. G., Guscina E. A., Morozova T. N.. 1990. Isolation and characterization of Sedlec virus, a new bunyavirus from birds. Acta Virol. 34: 339–345. [PubMed] [Google Scholar]
- Jones K. E., Patel N. G., Levy M. A., Storeygard A., Balk D., Gittleman J. L., Daszak P.. 2008. Global trends in emerging infectious diseases. Nature 451: 990–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupp P. G. 1996. Mosquitoes of Southern Africa. Ekogilde, South Africa. [Google Scholar]
- Kading R. C., Borland E. M., Cranfield M., Powers A. M.. 2013a. Prevalence of antibodies to alphaviruses and flaviviruses in free-ranging game animals and nonhuman primates in the greater Congo basin. J. Wildl. Dis. 49: 587–599. [DOI] [PubMed] [Google Scholar]
- Kading R. C., Gilbert A. T., Mossel E. C., Crabtree M. B., Kuzmin I. V., Niezgoda M., Agwanda B., Markotter W., Weil M. R., Montgomery J. M., et al. 2013b. Isolation and molecular characterization of Fikirini rhabdovirus, a novel virus from a Kenyan bat. J. Gen. Virol. 94: 2393–2398. [DOI] [PubMed] [Google Scholar]
- Karabatsos N. 1985. International catalogue of arboviruses. American Society of Tropical Medicine and Hygiene, San Antonio, TX. [Google Scholar]
- Kuno G., Chang G. J., Tsuchiya K. R., Karabatsos N., Cropp C. B.. 1998. Phylogeny of the genus Flavivirus. J. Virol. 72: 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert A. J., Lanciotti R. S.. 2008. Molecular characterization of medically important viruses of the genus Orthobunyavirus. J. Gen. Virol. 89: 2580–2585. [DOI] [PubMed] [Google Scholar]
- Ledermann J. P., Zeidner N., Borland E. M., Mutebi J. P., Lanciotti R. S., Miller B. R., Lutwama J. J., Tendo J. M., Andama V., Powers A. M.. 2014. Sunguru virus: a novel virus in the family Rhabdoviridae isolated from a chicken in north-western Uganda. J. Gen. Virol. 95: 1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklewitz M., Zirkel F., Rwego I. B., Heidemann H., Trippner P., Kurth A., Kallies R., Briese T., Lipkin W. I., Drosten C.. et al. 2013. Discovery of a unique novel clade of mosquito-associated bunyaviruses. J. Virol. 87: 12850–12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B. R., Mitchell C. J., Ballinger M. E.. 1989. Replication, tissue tropisms and transmission of yellow fever virus in Aedes albopictus. Trans. R. Soc. Trop. Med. Hyg. 83: 252–255. [DOI] [PubMed] [Google Scholar]
- Mukwaya L. G. 1972. Host preference of Mansonia (Coquillettidia) spp. in Uganda, with special reference to M. metallica (Theo.) (Dipt., Culicidae). Bull. Entomol. Res. 62: 87–90. [Google Scholar]
- Mutebi J. P., Crabtree M. B., Kading R. C., Powers A. M., Lutwama J. J., Miller B. R.. 2012. Mosquitoes of western Uganda. J. Med. Entomol. 49: 1289–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguku P. M., Sharif S. K., Mutonga D., Amwayi S., Omolo J., Mohammed O., Farnon E. C., Gould L. H., Lederman E., Rao C., et al. 2010. An investigation of a major outbreak of Rift Valley fever in Kenya: 2006-2007. Am. J. Trop. Med. Hyg. 83: 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochieng C., Lutomiah J., Makio A., Koka H., Chepkorir E., Yalwala S., Mutisya J., Musila L., Khamadi S., Richardson J., et al. 2013. Mosquito-borne arbovirus surveillance at selected sites in diverse ecological zones of Kenya; 2007–2012. Virol. J. 10: 140.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers A. M., Brault A. C., Shirako Y., Strauss E. G., Kang W., Strauss J. H., Weaver S. C.. 2001. Evolutionary relationships and systematics of the alphaviruses. J. Virol. 75: 10118–10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickenbach A., Boreham P. F., Weitz B., Germain M., Eouzan J. P.. 1974. Etude des preferences trophiques des moustiques (Diptera, Culicidae) de la region de Younde (Cameroun) par la methode des tests d precipitines. Cahiers ORSTOM Serie Entomologie Medicale et Parasitologie XII: 179–189. [Google Scholar]
- Rosenberg R. 2015. Detecting the emergence of novel, zoonotic viruses pathogenic to humans. Cell. Mol. Life Sci. 72: 1115–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang R., Onyango C., Gachoya J., Mabinda E., Konongoi S., Ofula V., Dunster L., Okoth F., Coldren R., Tesh R., et al. 2006. Tickborne arbovirus surveillance in market livestock, Nairobi, Kenya. Emerg. Infect. Dis. 12: 1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchetinin A. M., Lvov D. K., Deriabin P. G., Botikov A. G., Gitelman A. K., Kuhn J. H., Alkhovsky S. V.. 2015. Genetic and phylogenetic characterization of Tataguine and Witwatersrand Viruses and other Orthobunyaviruses of the Anopheles A, Capim, Guama, Koongol, Mapputta, Tete, and Turlock Serogroups. Viruses 7: 5987–6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M. C., Weitz B., McClelland G.A.H.. 1958. Natural hosts of some species of Taeniorhynchus Lynch Arribalzaga (Diptera, Culicidae) collected in Uganda, as determined by the precipitin test. Ann. Trop. Med. Parasitol. 52: 186–190. [DOI] [PubMed] [Google Scholar]