Abstract
Objective:
Carbapenems are effective agents to treat multidrug-resistant (MDR) strains of bacteria, including Pseudomonas aeruginosa. However, there is a potential threat of emergence of carbapenem-resistant P. aeruginosa (CRPA). The aim of this study was to determine antibiotic susceptibility patterns and metallo-beta-lactamase (MBL)-mediated resistance in clinical P. aeruginosa isolates.
Materials and Methods:
Different clinical specimens were subjected to conventional culture-based identification of P. aeruginosa. Antimicrobial susceptibility patterns and MBL production were evaluated using the Kirby-Bauer and combined double-disk synergy test methods, respectively. Multiplex polymerase chain reaction was performed to investigate the presence of the blaIMP, blaVIM, blaNDM, blaSPM, and blaSIM genes.
Results:
A total of 71 clinical P. aeruginosa isolates were recovered, of which 28.17% were identified as CRPA. The most active antibiotics were colistin and polymyxin B (92.96% susceptibility to each). A total of 35% and 50% of CRPA isolates were MDR and extensively drug-resistant (XDR), respectively. MBL activity was shown in 20% of CRPA. A total of 90%, 40%, and 5% of CRPA isolates harbored the blaIMP, blaVIM, and blaNDM genes, respectively. No correlation was found between the MBL-encoding genes of P. aeruginosa and patient characteristics.
Conclusion:
Although the prevalence of CRPA in our therapeutic centers was relatively low, this rate of carbapenem resistance reflects a threat limiting treatment choices. A high prevalence of MDR/XDR phenotypes among the MBL-producer isolates suggests the need for continuous assessment of antimicrobial susceptibility and surveillance of antibiotic prescription. In addition, infection control measures are needed to prevent further dissemination of these organisms.
KEYWORDS: Carbapenem, Metallo-β-lactamase genes, Pseudomonas aeruginosa
INTRODUCTION
Pseudomonas aeruginosa, a member of the ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter species) group of pathogens, is responsible for life-threatening nosocomial infections, especially in critically ill and immunocompromised patients and is known by potential drug resistance mechanisms [1,2]. Multidrug-resistant P. aeruginosa (MDR-PA) is a matter of great concern as it not only causes severe and fatal infections but also increases the length of hospital stay, resulting in increased treatment costs [3]. Carbapenems are effective antibiotics against MDR-PA infections [4]. However, their use in the management of infections is threatened by the development of carbapenem-resistant P. aeruginosa (CRPA) strains [4,5]. Resistance to the carbapenems in P. aeruginosa is often caused by impermeability through alteration or loss of the porin OprD [6], increased expression of an efflux pump [7], or the production of class B metallo-β-lactamases (MBLs) [4].
MBLs that have been detected primarily in P. aeruginosa [8] are categorized in class B in the structural classification scheme and group 3 in the functional classification system [9]. Various types of MBLs, such as the IMP, VIM, SIM, SPM, GIM, and NDM families have been described in P. aeruginosa worldwide [4]. Class B carbapenemases can hydrolyze most β-lactam group antibiotics except for monobactams (aztreonam). They are inhibited in vitro by metal chelators, such as ethylenediaminetetetraacetic acid (EDTA), while clavulanic acid has no activity against these enzymes [9].
MBL-encoding genes are frequently located in mobile or transferable genetic structures, such as plasmids, transposons and integrons, which can contribute to their spread in different bacteria, including Acinetobacter species and Enterobacteriaceae [4]. We are several years away from implementation of a therapeutic inhibitor of MBLs, and hence, their continuous spread is becoming an important challenge for antibiotic therapy [10].
The present study was performed to characterize the prevalence of carbapenem resistance, and MBL production among P. aeruginosa isolates from hospitalized patients in Gorgan, in northern Iran. Herein, we describe the susceptibility profiles and class B carbapenemase genes carried by the isolates, as it might help in the worldwide comparison of resistance mechanisms of MDR Gram-negative bacilli.
MATERIALS AND METHODS
Study population and species identification
This project was approved by the Ethics Committee of Golestan University of Medical Sciences with the ethical code number IR. GUMS. REC.1395.87. Informed consent was signed by all patients who were hospitalized at four therapeutic centers, including 5th Azar, Sayyad Shirazi, Ayatollah Taleghani, and Hakim Jurjani hospitals. From February 2016 to July 2017, different biological materials, including urine, respiratory tract samples, blood, eye swabs, wound swabs, ear swabs, and a heart biopsy were obtained. All specimens were cultured on MacConkey agar and nonselective blood agar media (Merck, Darmstadt, Germany). P. aeruginosa isolates were identified by standard microbiological and biochemical methods, such as colony morphology and pigmentation, Gram staining, oxidase, catalase, motility, indole, citrate, and oxidative-fermentative tests, and growth at 42°C. All isolates were then stored in tryptic soy broth stock (Merck, Darmstadt, Germany) with 15% glycerol at −80°C until used.
Antimicrobial susceptibility profiling
Antibiotic susceptibility profiles of all isolates were determined using the Kirby-Bauer diffusion testing method on Mueller–Hinton agar plates (Merck, Darmstadt, Germany), according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [11]. The antibiotic disks (MAST, Merseyside, UK) used were as follows: imipenem (10 μg), meropenem (10 μg), ceftazidime (30 μg), piperacillin-tazobactam (100 μg/10 μg), ciprofloxacin (5 μg), levofloxacin (5 μg), gentamicin (10 μg), amikacin (30 μg), tobramycin (10 μg), polymyxin B (300 U), and colistin (10 μg). Quality control testing was performed using the recommended organisms, P. aeruginosa ATCC 27853 and E. coli ATCC 35218. To characterize patterns of multiple drug resistance, isolates were defined as MDR-PA, extensively drug-resistant-PA (XDR-PA), and pandrug-resistant-PA (PDR-PA) according to Magiorakos et al. [12] as follows: MDR-PA, if isolates were resistant to ≥1 agent in ≥3 antimicrobial groups; XDR-PA, if isolates were resistant to ≥1 agent in all but ≤2 groups; PDR-PA, if isolates were resistant to all antimicrobials studied. Furthermore, all isolates that met criteria of resistance to any carbapenem (imipenem or meropenem) tested, as characterized by the CLSI [11], were considered CRPA.
Phenotypic detection of metallo-beta-lactamase
CRPA cultures were evaluated for MBL production using the combined double-disk synergy test (CDDST) [13]. In brief, 100 mM EDTA solution (Merck, Darmstadt, Germany) as an MBL inhibitor (iMBL) was made by dissolving 3.72 g of disodium EDTA.2H2O in 100 mL of distilled water. The pH was adjusted to 8.0 by NaOH and sterilized by autoclaving. A 0.5 McFarland suspension of the organism was inoculated onto Mueller–Hinton agar plates. Two disks each of imipenem (10 μg) and ceftazidime (30 μg) antimicrobials were placed on the inoculated Mueller–Hinton plates at a distance of 2 cm. For both β-lactam agents, 8 μL of iMBL solution was directly added to one of the disks. After an overnight incubation period at 37°C, an increase in the inhibition zone ≥8 mm around the combined disk compared to that the corresponding antimicrobial disk alone was considered positive for an MBL [13]. An MBL-producing strain of P. aeruginosa was used as the positive control.
Detection of metallo-beta-lactamase determinants
All CRPA isolates were examined for MBL-encoding genes, including blaIMP, blaVIM, blaNDM, blaSPM, and blaSIM, by a multiplex polymerase chain reaction (PCR) assay using specific primers [Table 1][14]. A DNA template was prepared using the boiling method. In brief, a few colonies of an overnight culture were suspended in 200 μL of sterile distilled water. The bacterial suspension was boiled for 15 min and centrifuged at 12000 rmp for 10 min, and the supernatant was transferred to a fresh Eppendorf tube as a template. PCR amplification was performed in 25 μL reaction mixture with 2x Master Mix Red (Ampliqon, Herlev, Denmark) containing 1 × PCR buffer ([Tris-HCl, pH 8.5], [NH4]2S04, 4 mM MgCl2, 0.2% Tween20), 0.2 U/μL Taq DNA polymerase, 2 mM MgCl2, and 0.4 mM of each deoxynucleotide, 0.2 μM of each primer (Macrogen, Seoul, Korea), 10 ng template DNA, and sterile distilled water. PCR was performed using the following thermal cycling protocol: initial denaturation at 94°C for 10 min, followed by 36 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 40 s, extension at 72°C for 50 s and a final extension at 72°C for 5 min. PCR products were electrophoresed on 1.5% agarose gel stained with DNA safe stain (SinaClon, Tehran, Iran), then photographed under an ultraviolet transilluminator. Sequencing of the PCR amplicons was performed with anABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA), and then, the nucleotide sequences were compared with known sequences at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/).
Table 1.
Oligonucleotide primers used to amplify metallo-β-lactamase genes
| Gene | Primer (5’→3’) | Amplicon size (bp) |
|---|---|---|
| blaIMP | F: GGAATAGAGTGGCTTAAYTCTC R: GGTTTAAYAAAACAACCACC |
232 |
| blaVIM | F: GATGGTGTTTGGTCGCATA R: CGAATGCGCAGCACCAG |
390 |
| blaSPM | F: AAAATCTGGGTACGCAAACG R: ACATTATCCGCTGGAACAGG |
271 |
| blaSIM | F: TACAAGGGATTCGGCATCG R: TAATGGCCTGTTCCCATGTG |
570 |
| blaNDM | F: GGTTTGGCGATCTGGTTTTC R: CGGAATGGCTCATCACGATC |
621 |
Data analysis
SPSS software version 18.0 for Windows (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Categorical and continuous quantitative variables were analyzed using Pearson's Chi-squared and independent samples t-tests, respectively. Statistical significance in this study was a P < 0.05.
RESULTS
Bacterial isolates
A total of 71 nonrepetitive P. aeruginosa isolates were recovered, of which 37 (52.12%) were from males and 34 (47.88%) were from females. The age distribution of patients was as follows: <1 year old (n = 13, 18.31%), 1–15 years old (n = 15, 21.12%), 16–35 years old (n = 7, 9.86%), 36–55 years old (n = 9, 12.67%), and > 55 years old (n = 27, 38.03%). The origins of isolates were urine (n = 31, 43.66%), the respiratory tract (n = 28, 39.43%), blood (n = 5, 7.04%), eye swabs (n = 3, 4.22%), wound wabs (n = 2, 2.81%), ear swab (n = 1, 1.41%), and heart biopsy (n = 1, 1.41%).
Antimicrobial susceptibility profile
According to results from the antibiotic susceptibility test, 20 (28.17%) and 51 (71.83%) isolates were identified as CRPA and carbapenem-susceptible (CSPA), respectively. CRPA isolates were recovered from the respiratory tract (n = 10, 50%), urine (n = 5, 25%), blood (n = 3, 15%), and one wound and one ear swab (n = 1, 5% each). The resistance rates for β-lactams, including imipenem, meropenem, ceftazidime, and piperacillin/tazobactam were 26.76% (n = 19), 18.31% (n = 13), 22.53% (n = 16), and 18.31% (n = 13), respectively. The resistance rates for non-β-lactam antimicrobials, including ciprofloxacin, levofloxacin, gentamicin, amikacin, tobramycin, polymyxin B, and colistin were 25.35% (n = 18), 29.57% (n = 21), 21.12% (n = 15), 15.49% (n = 11), 19.72% (n = 14), 7.04% (n = 5), and 7.04% (n = 5), respectively. Polymyxin B and colistin showed the highest in vitro activity against the isolates, with a susceptibility rate 92.96% (n = 66) for each; while the highest resistance rate was seen against levofloxacin (29.57%, n = 21).
Except for polymyxin B and colistin, the degrees of resistance for all antibiotics tested were significantly higher in the CRPA group than the CSPA group [Table 2]. Although a significant percentage of both CSPA and CRPA isolates showed susceptibility to polymyxin B (90.2% vs. 100%) and colistin (92.15% vs. 95%), CSPA isolates were significantly more susceptible to the remaining antibiotics tested. Based on the criteria mentioned earlier, 11.26% (n = 8) and 15.49% (n = 11) were identified as the MDR-PA and XDR-PA phenotype, respectively. The percentages of MDR-PA and XDR-PA among the CRPA were 35% and 50%, respectively; while the prevalence of MDR-PA and XDR-PA was low in the CSPA group (1.96% each).
Table 2.
Comparison of susceptibility of carbapenem-resistant Pseudomonas aeruginosa and carbapenem-susceptible Pseudomonas aeruginosa isolates to antibiotics tested
| Antimicrobial agent | Number (%) of resistant isolates | P | Number (%) of susceptible isolates | P | ||
|---|---|---|---|---|---|---|
| CRPA (n=20) | CSPA (n=51) | CRPA (n=20) | CSPA (n=51) | |||
| CAZ | 12 (60) | 4 (7.84) | 0.0001 | 8 (40) | 46 (90.2) | 0.0001 |
| PTZ | 10 (50) | 3 (5.88) | 0.001 | 8 (40) | 46 (90.2) | 0.0001 |
| CIP | 12 (60) | 6 (11.76) | 0.0001 | 7 (35) | 44 (86.27) | 0.0001 |
| LEV | 13 (65) | 8 (15.68) | 0.0001 | 5 (25) | 43 (84.31) | <0.0001 |
| GM | 12 (60) | 3 (5.88) | 0.0001 | 8 (40) | 47 (92.15) | 0.0001 |
| AK | 8 (40) | 3 (5.88) | 0.007 | 12 (60) | 48 (94.11) | 0.007 |
| TN | 11 (55) | 3 (5.88) | 0.0001 | 9 (45) | 48 (94.11) | 0.0001 |
| PB | 0 | 5 (9.8) | 0.9 | 20 (100) | 46 (90.2) | 0.9 |
| CO | 1 (5) | 4 (7.84) | 1 | 19 (95) | 47 (92.15) | 1 |
CAZ: Ceftazidime, PTZ: Piperacillin/tazobactam, CIP: Ciprofloxacin, LEV: Levofloxacin, GM: Gentamicin, AK: Amikacin, TN: Tobramicin, PB: Polymyxin B, CO: Colistin, CRPA: Carbapenem-resistant PA, CSPA: Carbapenem-susceptible PA, PA: Pseudomonas aeruginosa
Metallo-beta-lactamase activity and related genes
Of the 20 CRPA isolates, 4 (20%) were MBL-positive with the CDDST [Figure 1], and all showed multi-resistance patterns. Overall, 19 (95%) of CRPA isolates were found to carry MBL-encoding determinants [Figure 2]. The presence of MBL-encoding genes in CRPA isolates was not correlated with patient characteristics, including clinical specimen type, hospital ward, age, or gender (P > 0.06) [Table 3]. MBL genes detected were blaIMP(n = 18, 90%), blaVIM(n = 8, 40%), and blaNDM(n = 1, 5%). The blaIMP gene co-existed with blaVIM in eight (40%) isolates. In addition, blaIMP co-existed with the blaNDM gene in only one (5%) CRPA isolate. Sequencing data confirmed the presence of the blaIMP-1, blaVIM-2, and blaNDM-1v ariants in isolates. No amplicon was obtained with primers targeting blaSPM, and blaSIM. All 4 isolates with MBL activity in the CDDST were positive for MBL genes, of which 1 (25%), 1 (25%), and 2 (50%) isolates carried the blaIMP, blaVIM, and both genes, respectively. Furthermore, 15 (75%) of the CRPA which lacked MBL activity were positive for particular genes. Only one CRPA isolate was negative for MBL activity and any of the genes tested.
Figure 1.
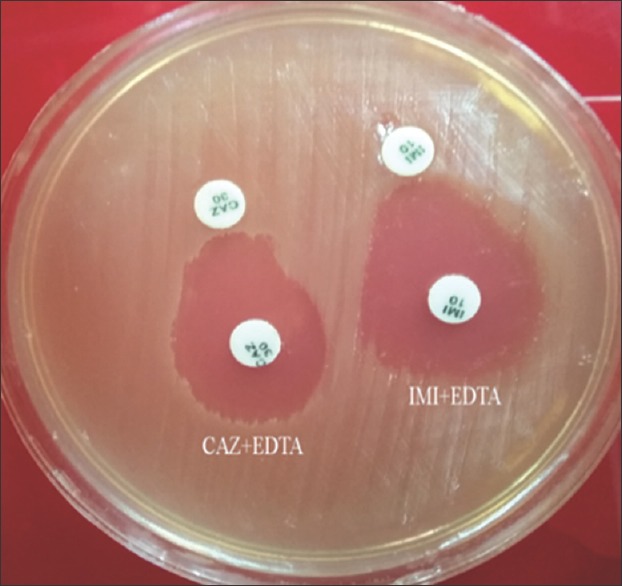
Phenotypic combined double-disk synergy test to detect metallo-beta-lactamase production in carbapenem-resistant P. aeruginosa isolates. Imipenem and imipenem + EDTA disks and ceftazidime and ceftazidime + EDTA disks were used to detect metallo-beta-lactamase -producers. A zone diameter difference between the disks alone and the disks plus EDTA of ≥8 mm was interpreted as metallo-beta-lactamase production
Figure 2.
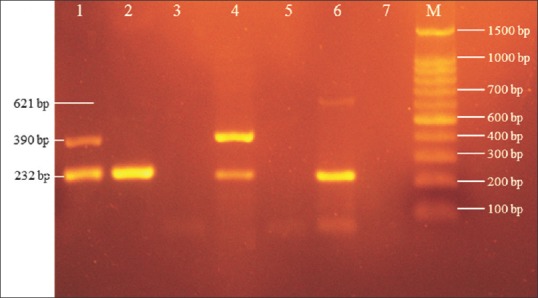
Agarose gel electrophoresis of multiplex PCR products. Lanes 1 and 4, isolates with blaIMP (232 bp) and blaVIM (390 bp) genes; Lane 2, isolate with the blaIMP (232 bp) gene; Lane 6, isolates with blaIMP (232 bp) and blaNDM (621 bp) genes; Lanes 3 and 5, isolates without any genes studied; Lane 7, negative control (distilled water); M, 100 bp DNA ladder
Table 3.
Characteristics of carbapenem-resistant Pseudomonas aeruginosa isolated from hospitalized patients
| Isolate | Patient characteristics | Resistance pattern | MDR | XDR | CDDST | MBL genes | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (year) | Gender | Hospital ward | Isolation material | IMP | VIM | NDM | SIM | SPM | |||||
| PA2 | 26 | Female | Emergency room | Blood | IMI, MEM, CAZ, PTZ | + | - | - | + | + | - | - | - |
| PA3 | 78 | Female | Internal medicine | Respiratory tract | IMI, CAZ, PTZ, LEV | - | + | + | - | + | - | - | - |
| PA6 | 83 | Female | ICU | Respiratory tract | IMI, GM, CAZ, PTZ, CIP, LEV | - | + | - | + | + | - | - | - |
| PA7 | 71 | Female | ICU | Respiratory tract | IMI, MEM, GM, AK, TN, PTZ, CIP, LEV | - | + | - | + | + | - | - | - |
| PA9 | 63 | Male | Internal medicine | Respiratory tract | IMI, MEM, GM, TN, CAZ, CIP, LEV | - | + | - | + | - | + | - | - |
| PA11 | 79 | Male | Outpataint | Urine | IMI, MEM, GM, TN, CIP, LEV | + | - | - | + | - | - | - | - |
| PA13 | 79 | Male | Emergency room | Respiratory tract | IMI, MEM, GM, AK, TN, CIP | + | - | - | + | - | - | - | - |
| PA15 | 20 | Female | Internal medicine | Respiratory tract | IMI, MEM, GM, AK, TN, CAZ, CIP, LEV | - | + | + | + | - | - | - | - |
| PA20 | 9 | Male | Infectious diseases | Ear swab | IMI | - | - | - | + | - | - | - | - |
| PA21 | 52 | Female | Outpataint | Urine | IMI | - | - | - | + | + | - | - | - |
| PA22 | 25 | Male | Outpataint | Urine | IMI, GM, AK, CIP, LEV | + | - | - | + | - | - | - | - |
| PA25 | 68 | Male | ICU | Respiratory tract | IMI, MEM, GM, CIP, LEV | + | - | + | + | + | - | - | - |
| PA31 | 1 | Female | Outpataint | Urine | MEM | - | - | - | + | - | - | - | - |
| PA33 | 68 | Male | Infectious diseases | Respiratory tract | IMI, MEM, GM, AK, TN, CAZ, PTZ, CIP, LEV | - | + | - | + | - | - | - | - |
| PA44 | 7 | Female | Emergency room | Wound swab | IMI, MEM, GM, AK, TN, CAZ, PTZ, CIP | - | + | + | + | + | - | - | - |
| PA45 | 81 | Female | Outpataint | Urine | IMI, CIP, LEV, CO | + | - | - | + | - | - | - | - |
| PA48 | 1 | Female | PICU | Blood | IMI, MEM, CAZ, PTZ | + | - | - | + | + | - | - | - |
| PA52 | 46 | Male | Nephrology | Blood | IMI, MEM, GM, AK, TN, CAZ, PTZ, CIP, LEV | - | + | - | - | - | - | - | - |
| PA55 | 1 | Female | ICU | Respiratory tract | IMI, MEM, CAZ, PTZ, LEV | - | + | - | + | - | - | - | - |
| PA71 | 72 | Female | Emergency room | Respiratory tract | IMI, GM, AK, TN, CAZ, PTZ, CIP, LEV | - | + | - | + | - | - | - | - |
IMI: Imipenem, MEM: Meropenem, CAZ: Ceftazidime, PTZ: Piperacillin/tazobactam, CIP: Ciprofloxacin, LEV: Levofloxacin, GM: Gentamicin, AK: Amikacin, TN: Tobramicin, CO: Colistin, PA: Pseudomonas aeruginosa, MDR: Multidrug-resistant, XDR: Extensively drug-resistant, CDDST: Combined double-disk synergy test, MBL: Metallo-β-lactamase, ICU: Intensive Care Unit, PICU: Pediatric ICU, +: Yes, -: No
DISCUSSION
Antimicrobial resistance is an unpleasant event with a rising impact on patient safety, especially in Intensive Care Units (ICUs). Acutely, ill patients are prone to colonization and infection by antibiotic-resistant bacteria due to long-term hospitalization repeated exposure to antimicrobial agents and implementation of invasive therapeutic procedures. This serious array of risk factors leads to a vicious cycle of increased infection rates, the increased necessity for broad-spectrum antibiotics, reduced antimicrobial efficacy, and increased selection of drug resistance [15].
Carrying acquired MBL determinants enables bacteria to become resistant to broad-spectrum β-lactam antibiotics, including carbapenems, which are often the “last resort” agents for Gram-negative pathogens causing hospital-associated infections. Furthermore, MBL-producing pathogens usually develop to the multi-resistant phenotype, because they originate nosocomially and acquired MBL-related genes typically cluster with other drug resistance determinants in the variable region of multi-resistance integrons [16]. These reasons explain why infections caused by MBL producers can pose a great challenge for antibiotic chemotherapy.
In this study, we described clinical carbapenem-resistant MDR/XDR P. aeruginosa isolates, which are capable of producing MBL type carbapenemase. Although the prevalence of CRPA in the present study was relatively low (28.17%), this rate of carbapenem resistance reflects a threat limiting treatment choices in our therapeutic centers. The ratio of CRPA differs by geographic region, kind of infection and specimen source, and selective pressure due to antibiotics [17]. In a recent study in Algeria, Meradji et al. [18] found that 18.75% of clinical P. aeruginosa isolates were resistant to carbapenems. In our country, there are different rates of resistance to carbapenems in P. aeruginosa. Mirsalehian et al. [19] reported a 100% rate of carbapenem-resistant and MDR-PA isolates from burn patients in Tehran, the capital of Iran. Furthermore, Farajzadeh Sheikh et al. [20] showed that among P. aeruginosa isolates from Ahvaz in Southwest Iran, 58.7% and 31.8% were resistant to imipenem and meropenem, respectively, and 44.4% showed multiple-drug resistance. However, lower prevalences of CRPA isolates were reported previously by Tarhani et al. [21], and Forozsh et al. [22]. In addition, we found a significantly higher multi-resistance rate in the CRPA group compared with the CSPA group, and the most effective antibiotics against both groups were polymyxin B and colistin. These findings indicate that polymyxins have increasingly become the last resort treatment for multi-resistant P. aeruginosa infections.
Various phenotypic methods, such as the MBL Etest, double-disk synergy test (DDST), and CDSST have been suggested for screening for MBL producing P. aeruginosa. Although both the DDST and CDDST are simple to perform and cheaper than the MBL Etest, they have shown incompatible results depending on the methodology used, β-lactam substrates, iMBLs, and the bacterial genus tested [13,20,23,24]. In the present study, 20% of all CRPA isolates were positive for MBL production on the phenotypic screening test, whereas 95% were positive for MBL-encoding genes. The reported low rates of MBL activity among the CRPA could be because detection of carbapenemase activity in clinical isolates is challenging work. In addition, discordant results for MBL detection between the phenotypic and genotypic methods were reported in Inacio et al.'s study [25], where 30% of P. aeruginosa were MBL producers with the Etest and DD methods using several iMBLs, whereas 87.5% were positive for the blaSPM-1, and blaVIM-1 genes. In this manner, these MBL producers might act as silent reservoirs of such resistance determinants, with the ability to spread [26].
Overall, blaIMP and blaVIM are the most prevalent genes encoding MBLs detected in many regions, including Iran [27,28]. However, our study is in contrast to the findings obtained by Hakemi Vala et al. [29] and Bejestani et al. [30] in Tehran, Iran, who reported no blaVIM producing-P. aeruginosa from clinical materials. There are few studies reporting the incidence of blaNDM positive P. aeruginosa in our country. Most recently, Haghi et al. [31] found two clinical MDR-PA harboring the blaNDM-1 in Zanjan, Iran. Although only one CRPA isolate was identified to carry this gene in the present study, it is of concern as blaNDM producers can spread rapidly and this finding implies that numerous new blaNDM cases will be found in the near future [32]. In addition, we found one non-MBL isolate negative for all genes tested. Therefore, the carbapenem resistance ability of the latter isolate may be mediated by other resistance mechanisms, such as loss of the oprD outer membrane protein-encoding gene, up-regulated efflux pumps, and production of other carbapenemases [33]. Further investigation is needed by our research group to address as many of these resistance mechanisms as possible.
Some correlation between the MBL-encoding genes of P. aeruginosa and patient characteristics has been demonstrated [34,35,36]. In a study in Brazil, Lucena et al. [34] found that ICU stay and urinary tract infections serve as significant risk factors for MBL-positive P. aeruginosa infections. Conversely, in two individual studies, Shirani et al. [37] and Zavascki et al. [38] found no significant correlations between MBL-positive P. aeruginosa and clinical characteristics of patients. Similarly, no specific characteristics of patients, including age, gender, hospitalization ward, and source of the isolate were associated with a higher frequency of MBL-positive P. aeruginosa infections in our study.
Herein, we demonstrated carbapenem-resistant MDR/XDR P. aeruginosa isolates with carbapenemase activity. IMP- and VIM-producing P. aeruginosa were the MBL-producing strains present at our therapeutic centers. The emergence of carbapenem resistance reflects a threat limiting treatment choices and suggests the need for ongoing epidemiological and antimicrobial susceptibility studies and longitudinal surveillance of antibiotic prescription. The continuous presence of such problematic organisms and their intra-hospital and inter-hospital spread is a matter of concern and should be taken into consideration by hospital infection control teams.
Financial support and sponsorship
This study was supported financially by Golestan University of Medical Sciences and Health Services grant no. 950429096.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
The authors would like to thank all colleagues who helped us throughout the project.
REFERENCES
- 1.Rice LB. Progress and challenges in implementing the research on ESKAPE pathogens. Infect Control Hosp Epidemiol. 2010;31(Suppl 1):S7–10. doi: 10.1086/655995. [DOI] [PubMed] [Google Scholar]
- 2.Ardebili A, Lari AR, Beheshti M, Lari ER. Association between mutations in GyrA and ParC genes of Acinetobacter baumannii clinical isolates and ciprofloxacin resistance. Iran J Basic Med Sci. 2015;18:623–6. [PMC free article] [PubMed] [Google Scholar]
- 3.Davane M, Suryawanshi N, Pichare A, Nagoba BS. Pseudomonas aeruginosa from hospital environment. J Microbiol Infect Dis. 2014;4:42–3. [Google Scholar]
- 4.Kateete DP, Nakanjako R, Namugenyi J, Erume J, Joloba ML, Najjuka CF, et al. Carbapenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii at Mulago hospital in Kampala, Uganda (2007-2009) Springerplus. 2016;5:1308. doi: 10.1186/s40064-016-2986-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liakopoulos A, Mavroidi A, Katsifas EA, Theodosiou A, Karagouni AD, Miriagou V, et al. Carbapenemase-producing Pseudomonas aeruginosa from central greece: Molecular epidemiology and genetic analysis of class I integrons. BMC Infect Dis. 2013;13:505. doi: 10.1186/1471-2334-13-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rojo-Bezares B, Cavalié L, Dubois D, Oswald E, Torres C, Sáenz Y, et al. Characterization of carbapenem resistance mechanisms and integrons in Pseudomonas aeruginosa strains from blood samples in a French hospital. J Med Microbiol. 2016;65:311–9. doi: 10.1099/jmm.0.000225. [DOI] [PubMed] [Google Scholar]
- 7.Lee JY, Ko KS. OprD mutations and inactivation, expression of efflux pumps and ampC, and metallo-β-lactamases in carbapenem-resistant Pseudomonas aeruginosa isolates from South Korea. Int J Antimicrob Agents. 2012;40:168–72. doi: 10.1016/j.ijantimicag.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Pitout JD, Gregson DB, Poirel L, McClure JA, Le P, Church DL. Detection of Pseudomonas aeruginosa producing metallo-β-lactamases in a large centralized laboratory. J Clin Microbiol. 2005;43:3129–35. doi: 10.1128/JCM.43.7.3129-3135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zavascki AP, Carvalhaes CG, Picão RC, Gales AC. Multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii: Resistance mechanisms and implications for therapy. Expert Rev Anti Infect Ther. 2010;8:71–93. doi: 10.1586/eri.09.108. [DOI] [PubMed] [Google Scholar]
- 10.Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-beta-lactamases: The quiet before the storm? Clin Microbiol Rev. 2005;18:306–25. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. CLSI document. 26th ed. Wayne, PA: 2016. Performance standards for antimicrobial susceptibility testing; pp. M100–S21. [Google Scholar]
- 12.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 13.Picão RC, Andrade SS, Nicoletti AG, Campana EH, Moraes GC, Mendes RE, et al. Metallo-beta-lactamase detection: Comparative evaluation of double-disk synergy versus combined disk tests for IMP-, GIM-, SIM-, SPM-, or VIM-producing isolates. J Clin Microbiol. 2008;46:2028–37. doi: 10.1128/JCM.00818-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119–23. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 15.De Francesco MA, Ravizzola G, Peroni L, Bonfanti C, Manca N. Prevalence of multidrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa in an Italian hospital. J Infect Public Health. 2013;6:179–85. doi: 10.1016/j.jiph.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Lagatolla C, Tonin EA, Monti-Bragadin C, Dolzani L, Gombac F, Bearzi C, et al. Endemic carbapenem-resistant Pseudomonas aeruginosa with acquired metallo-beta-lactamase determinants in European hospital. Emerg Infect Dis. 2004;10:535–8. doi: 10.3201/eid1003.020799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrow BJ, Pillar CM, Deane J, Sahm DF, Lynch AS, Flamm RK, et al. Activities of carbapenem and comparator agents against contemporary US Pseudomonas aeruginosa isolates from the CAPITAL surveillance program. Diagn Microbiol Infect Dis. 2013;75:412–6. doi: 10.1016/j.diagmicrobio.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Meradji S, Barguigua A, Zerouali K, Mazouz D, Chettibi H, Elmdaghri N, et al. Epidemiology of carbapenem non-susceptible Pseudomonas aeruginosa isolates in Eastern Algeria. Antimicrob Resist Infect Control. 2015;4:27. doi: 10.1186/s13756-015-0067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirsalehian A, Kalantar-Neyestanaki D, Taherikalani M, Jabalameli F, Emaneini M. Determination of carbapenem resistance mechanism in clinical isolates of Pseudomonas aeruginosa isolated from burn patients, in Tehran, Iran. J Epidemiol Glob Health. 2017;7:155–9. doi: 10.1016/j.jegh.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farajzadeh Sheikh A, Rostami S, Jolodar A, Tabatabaiefar MA, Khorvash F, Saki A, et al. Detection of metallo-beta lactamases among carbapenem-resistant Pseudomonas aeruginosa. Jundishapur J Microbiol. 2014;7:e12289. doi: 10.5812/jjm.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarhani M, Hakemi-Vala M, Hashemi A, Nowroozi J, Khanbababee G. Detection of metallo-β-Lactamases and Klebsiella pneumonia carbapenemases in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Arch Pediatr Infect Dis. 2016;4:e35905. [Google Scholar]
- 22.Forozsh FM, Irajian G, Moslehi TZ, Fazeli H, Salehi M, Rezania S, et al. Drug resistance pattern of Pseudomonas aeruginosa strains isolated from cystic fibrosis patients at Isfahan AL Zahra hospital, Iran (2009-2010) Iran J Microbiol. 2012;4:94–7. [PMC free article] [PubMed] [Google Scholar]
- 23.Galani I, Rekatsina PD, Hatzaki D, Plachouras D, Souli M, Giamarellou H, et al. Evaluation of different laboratory tests for the detection of metallo-beta-lactamase production in Enterobacteriaceae. J Antimicrob Chemother. 2008;61:548–53. doi: 10.1093/jac/dkm535. [DOI] [PubMed] [Google Scholar]
- 24.Lee K, Yong D, Yum JH, Lim YS, Bolmström A, Qwärnström A, et al. Evaluation of etest MBL for detection of blaIMP-1 and blaVIM-2 allele-positive clinical isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2005;43:942–4. doi: 10.1128/JCM.43.2.942-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inacio HS, Bomfim MR, França RO, Farias LM, Carvalho MA, Serufo JC, et al. Phenotypic and genotypic diversity of multidrug-resistant Pseudomonas aeruginosa isolates from bloodstream infections recovered in the hospitals of Belo Horizonte, Brazil. Chemotherapy. 2014;60:54–62. doi: 10.1159/000365726. [DOI] [PubMed] [Google Scholar]
- 26.Picão RC, Carrara-Marroni FE, Gales AC, Venâncio EJ, Xavier DE, Tognim MC, et al. Metallo-β-lactamase-production in meropenem-susceptible Pseudomonas aeruginosa isolates: Risk for silent spread. Mem Inst Oswaldo Cruz. 2012;107:747–51. doi: 10.1590/s0074-02762012000600007. [DOI] [PubMed] [Google Scholar]
- 27.Neyestanaki DK, Mirsalehian A, Rezagholizadeh F, Jabalameli F, Taherikalani M, Emaneini M, et al. Determination of extended spectrum beta-lactamases, metallo-beta-lactamases and ampC-beta-lactamases among carbapenem resistant Pseudomonas aeruginosa isolated from burn patients. Burns. 2014;40:1556–61. doi: 10.1016/j.burns.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Peymani A, Naserpour Farivar T, Mohammadi Ghanbarlou M, Najafipour R. Dissemination of Pseudomonas aeruginosa producing bla IMP-1 and bla VIM-1 in Qazvin and Alborz educational hospitals, Iran. Iran J Microbiol. 2015;7:302–9. [PMC free article] [PubMed] [Google Scholar]
- 29.Hakemi Vala M, Hallajzadeh M, Hashemi A, Goudarzi H, Tarhani M, Sattarzadeh Tabrizi M, et al. Detection of ambler class A, B and D ß-lactamases among Pseudomonas aeruginosa and Acinetobacter baumannii clinical isolates from burn patients. Ann Burns Fire Disasters. 2014;27:8–13. [PMC free article] [PubMed] [Google Scholar]
- 30.Bejestani FB, Hakemi Vala M, Momtaheni R, Bejestani OB, Gholami M. The frequency of imp and vim genes among Pseudomonas aeruginosa isolates from children's medical center of Tehran. Arch Clin Infect Dis. 2015;10:e20991. [Google Scholar]
- 31.Haghi F, Keramati N, Hemmati F, Zeighami H. Distribution of integrons and gene cassettes among metallo-β-lactamase producing Pseudomonas aeruginosa clinical isolates. Infect Epidemiol Med. 2017;3:36–40. [Google Scholar]
- 32.Manenzhe RI, Zar HJ, Nicol MP, Kaba M. The spread of carbapenemase-producing bacteria in Africa: A systematic review. J Antimicrob Chemother. 2015;70:23–40. doi: 10.1093/jac/dku356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar V, Sen MR, Nigam C, Gahlot R, Kumari S. Burden of different beta-lactamase classes among clinical isolates of ampC-producing Pseudomonas aeruginosa in burn patients: A prospective study. Indian J Crit Care Med. 2012;16:136–40. doi: 10.4103/0972-5229.102077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucena A, Dalla Costa LM, Nogueira KS, Matos AP, Gales AC, Paganini MC, et al. Nosocomial infections with metallo-beta-lactamase-producing Pseudomonas aeruginosa: Molecular epidemiology, risk factors, clinical features and outcomes. J Hosp Infect. 2014;87:234–40. doi: 10.1016/j.jhin.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 35.Hirakata Y, Yamaguchi T, Nakano M, Izumikawa K, Mine M, Aoki S, et al. Clinical and bacteriological characteristics of IMP-type metallo-beta-lactamase-producing Pseudomonas aeruginosa. Clin Infect Dis. 2003;37:26–32. doi: 10.1086/375594. [DOI] [PubMed] [Google Scholar]
- 36.Van der Bij AK, Van Mansfeld R, Peirano G, Goessens WH, Severin JA, Pitout JD, et al. First outbreak of VIM-2 metallo-β-lactamase-producing Pseudomonas aeruginosa in the Netherlands: Microbiology, epidemiology and clinical outcomes. Int J Antimicrob Agents. 2011;37:513–8. doi: 10.1016/j.ijantimicag.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 37.Shirani K, Ataei B, Roshandel F. Antibiotic resistance pattern and evaluation of metallo-beta lactamase genes (VIM and IMP) in Pseudomonas aeruginosa strains producing MBL enzyme, isolated from patients with secondary immunodeficiency. Adv Biomed Res. 2016;5:124. doi: 10.4103/2277-9175.186986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zavascki AP, Barth AL, Gaspareto PB, Gonçalves AL, Moro AL, Fernandes JF, et al. Risk factors for nosocomial infections due to Pseudomonas aeruginosa producing metallo-beta-lactamase in two tertiary-care teaching hospitals. J Antimicrob Chemother. 2006;58:882–5. doi: 10.1093/jac/dkl327. [DOI] [PubMed] [Google Scholar]


