Abstract
Background: Breast cancer is the highest incidence of tumor in women, which seriously threaten women's health. The occurrence and progression of breast cancer is linked to inactivation or downregulation of tumor suppressors, and activation or upregulation of oncogenes. However, the mechanism of PAK7 involving in the occurrence and progression of breast cancer is not yet fully understood.
Methods: PAK7 expression was analyzed by RT-qPCR and immunohistochemistry and correlated with clinicopatholgical parameters in breast cancer tissue microarray. The effects of PAK7 on breast cancer cells were detected by CCK-8 assay, colon formation assay, wound healing and transwell assays, and flow cytometry. The relationship between PAK7 and Wnt/β-catenin signaling pathway was determined by western blotting, TOP/FOP flash, co-Immunoprecipitation and co-localization assays.
Results: PAK7 expression was significantly increased in breast cancer tissues and positively correlated with pathological differentiation and TNM stage of breast cancer. Overexpression of PAK7 could significantly promote proliferation and migration of breast cancer cells, and inhibit apoptosis. In contrast, PAK7 knockdown significantly inhibited the proliferation and migration of breast cancer cells and promoted apoptosis. In addition, PAK7 could activate Wnt/β-catenin signaling pathway in breast cancer cells. Further study found that PAK7 could directly bind to GSK3β and β-catenin, and regulate β-catenin degradation by phosphorylating GSK3β.
Conclusions: Our study demonstrated that PAK7, as an oncogene, involved in breast cancer progression by activating the Wnt/β-catenin signaling pathway, suggesting that the potential applicability of PAK7 as a target for breast cancer treatment.
Keywords: P21-activated kinase 7, Wnt/β-catenin signaling pathway, cell proliferation, cell migration, breast cancer
Introduction
Breast cancer is a major killer of modern women's health. In 2012, global estimated new cases of breast cancer reached 1,676,600, accounting for 25.2% of all female cancer patients. At the same time, global estimated deaths reached 521,900, accounting for 14.7% of all female cancer patients 1. Breast cancer is one of the most important malignant tumors that endanger the life and health of resident, and the incidence has been increasing in the last ten years in China 2. Several decades effort have revealed that the development and progression of breast cancer is linked to inactivation or downregulation of tumor suppressors, and activation or upregulation of oncogenes, leading to oncogenic signaling activation such as Wnt/β-catenin 3. To explore these genes regulating which signaling pathway and its mechanisms are of great concern to us, as it may provide clues for the clinical treatment of breast cancer.
P21-activated kinases (PAKs) are serine/threonine protein kinases that are important regulators of many oncogenic signaling pathways 4. Numerous studies have found that increased PAKs expression or activation of mutations leads to enhanced cell proliferation and migration, and also evasion of apoptosis, which are important mechanisms of PAKs to promote tumorigenesis 5. The mammalian PAK family consists of 6 members, which include two subgroups: group I (PAK 1-3) and group II (PAK 4-6). Because of their structural similarity, these family proteins also have some similarities in function 4. PAK activity was significantly increased in many tumors and positively correlated with advanced grade and decreased survival 4, 5.
PAK7, also known as PAK5, is the last identified member of PAK family 6-8, which is still poorly understood in tumor progression. PAK7 was originally found in brain in which promotes the formation of filopodia in nerve cells 9. However, recent studies have found that the expression level of PAK7 significantly increased in colorectal cancer 10, 11, ovarian cancer 12, gastric cancer 13-15, Neuroglioma 16, 17, Hepatocellular carcinoma 18, 19, Pancreatic cancer 20, 21, Osteosarcoma 22, and breast cancer 23-25, leading to increased proliferation and migration of tumor cells and inhibition of apoptosis. More importantly, PAK7 expression was positively correlated with the malignancy of colorectal cancer and ovarian cancer, suggesting that PAK7 indeed played an important role in tumor progression. Current studies have shown that PAK7 plays an important role in tumorigenesis, invasion and metastasis 26, which is mainly related to its mechanism of regulating cytoskeleton, inhibiting apoptosis and promoting proliferation 5. The mechanism of PAK7 involving in the progression of breast cancer was poorly understood. One study showed that PAK7 could phosphorylate GATA-binding factor 1 (GATA1) to recruit more Histone deacetylases 3/4 (HDA3/4) to E-cadherin promoter region, resulting in suppression of E-cadherin expression, which leads to epithelial-mesenchymal transition in breast cancer cells 23. PAK7 can promote the phosphorylation and the nuclear translocation of p65 subunit of nuclear factor-kappaB (NF-κB), therefore promoting the proliferation and cell-cycle progression in breast cancer by increasing the expression of cyclin D1 in vitro and in vivo 25. In addition, another study also reveals the PAK5-Egr1-MMP2 signaling pathway to be a critical regulator of cell migration and invasion in breast cancer cell 24. However, the underlying mechanisms of PAK5 on breast cancer cell proliferation and migration still remains to be fully elucidated.
The Wnt signaling pathway was originally discovered in the study of oncogenic retroviruses. In 1982, Roel Nusse and Harold Varmus used mouse mammary adenocarcinoma virus to infect mice in order to study the viral-induced breast cancer caused by the virus. And murine proto-oncogene int1 was found for the first time 27. The Int1 gene is highly conserved among many species, including humans and Drosophila. In Drosophila, the homolog gene of int1 is Wingless (Wg). Further studies found a lot of int1-related genes, so researchers collectively called them Wg and int family, referred to as the Wnt family 28. In normal physiological state, activation of the Wnt signaling pathway is essential in embryonic development and can maintain cell division and migration 29. However, in the pathological state, the over activation of Wnt/β-catenin signaling pathway plays a role in promoting the development of breast cancer and other tumors 3. In the process of breast cancer, Wnt/β-catenin signaling pathway can promote proliferation and migration of tumor cells via activating target genes, such as c-myc, cyclin D1 and so on30. Inhibition of Wnt/β-catenin signaling pathway can inhibit cell proliferation and migration, ultimately inhibiting lung metastasis of breast cancer 31. The activation of Wnt/β-catenin signaling pathway is closely related to the development of breast cancer.
In our present study, we sought to further explore the role of PAK7 in breast cancer. In addition, we further explored the mechanism of PAK7 in promoting breast cancer by activating the Wnt/β-catenin signaling pathway, which will provide a new idea for better understanding of breast cancer pathogenesis and breast cancer targeted therapy.
Materials and Methods
Human breast cancer samples and tissue microarray (TMA)
Twenty cases of breast cancer tissues and paired paracancerous tissues were collected from Affiliated Zhongnan Hospital of Wuhan University and diagnosed by the Department of Pathology. All patients were informed and agreed. Our study was approved by the Ethics Board of Zhongnan Hospital and was performed in accordance with all relevant principles of the Declaration of Helsinki. The human breast cancer tissue microarray (the TMA ID: BC081120a) was purchased from Alenabio company (Xi'an, China) which contains 110 cases of breast cancer tissue and cancer adjacent normal breast tissue, including the clinicopathologic characteristics such as TNM stage, Pathological grade, et al.
Plasmids, siRNA and antibodies
The PAK7 overexpression plasmid was generated by cloning the genomic PAK7 gene (NM_020341.3), with a 2160-bp sequence on each flanking side, into retroviral transfer plasmid pflag-CMV and pEGFP-C1 (Clontech Laboratories Inc.) to generate plasmid pflag-PAK7 and pEGFP-PAK7. The β-catenin expression plasmid was generated by cloning the genomic CTNNB1 gene (NM_001330729.1), with a 2325-bp sequence on each flanking side, into retroviral transfer plasmid pEGFP-C1 and pCherry-N1 (Clontech Laboratories Inc.) to generate plasmid pEGFP-β-catenin and pCherry-β-catenin. The TOP/FOP flash reporter plasmids containing wild-type (CCTTTGATC; TOP flash) or mutated (CCTTTGGCC; FOP flash) TCF/LEF DNA binding sites were purchased from Upstate Biotechnology.
The RNA oligos containing 21 nucleotides were synthesized in sense and anti-sense directions corresponding to human PAK7 (Genbank accession number NM_020341) siRNA1 at nucleotides 2519 bp (sense: 5'- CCUCGGACAUCCAUUCUUATT -3' and anti-sense: 5'- UAAGAAUGGAUGUCCGAGGTT -3'), siRNA2 at nucleotides 1119 bp (sense: 5'- CUGGAUUAUUCAUUCCAAUTT -3' and anti-sense: 5'- AUUGGAAUGAAUAAUCCAGTT -3'), and siRNA3 at nucleotides 669 bp (sense: 5'- GCAAGGAAACCUCCAUCAATT -3' and anti-sense: 5'- UUGAUGGAGGUUUCCUUGCTT-3') with dTdT overhangs at each 3' terminus (Shanghai GenePharma Co., Ltd.). One negative siRNA control (sense 5'-UUCUCCGAACGUGUCACGU-3' and anti-sense 5'-ACGUGACACGUUCGGAGAA-3') was supplied.
The following primary antibodies were used for western blotting and analyses: PAK7 (Proteintech, USA), flag (Proteintech, USA), GFP (Proteintech, USA), GSK3β (Proteintech, USA), p-GSK3β (Cell Signaling Technology, USA), β-catenin (Proteintech, USA), p-β-catenin (Cell Signaling Technology, USA), c-myc (Proteintech, USA), cyclin D1 (Proteintech, USA) and GAPDH (Proteintech, USA),and the goat anti-rabbit IgG (Proteintech, USA) and goat anti-mouse IgG (Proteintech, USA).
Cell culture and Transfection
Human breast cancer cell lines MDA-MB-231, MCF-7 cell lines, normal breast cell line MCF-10A and human embryonic kidney cells HEK293T were obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in RPMI-1640 medium or Dulbecco's modified Eagle's medium (DMEM, HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, Milano, Italy), 100 U per ml penicillin and 100 mg per ml streptomycin at 37 ℃ and 5% CO2.
Transfection was performed using Lipofectamine 2000 reagent (Invitrogen Co., Ltd.). The day prior to transfection, cells were trypsinized, diluted with fresh medium and then transferred to 6-well plates. Transfection of siRNAs (or plasmids) was carried out using Lipofectamine 2000 reagent. Lipids and siRNAs (or plasmids) were diluted into the DMEM, respectively. Diluted lipids were mixed with diluted siRNAs (or plasmids) and the mixture was incubated for 25 min at room temperature for complex formation. After the addition of DMEM to each well containing cells to a level of 2 ml, the entire mixture was added to the cells in one well resulting in a final concentration of 100 pmol for the siRNAs (4ug for plasmids). After transfection 48 h, the cells were used for biological effect detection or RT-PCR and Western blotting analysis.
Cell proliferation assay
For cell growth assays, cells were seeded in 96-well plates at a density of 1 × 103 per well, and the cell growth rate was assessed by a CCK-8 kit (Dojindo, Japan) according to the manufacturer's instructions. For the colon formation assay, 200 cells were seeded in 6-well plates. After one week in culture, cell colonies were counted by crystal violet staining. The results are expressed as the mean ± SD of three independent experiments.
Migration assays
Wound healing and transwell assays were performed to measure the ability of cell migration. For the wound healing assay, cells were seeded in 6-well plates and cultured for 24 and 48 hours, respectively. A pipette tip (200 μl) was used to make a straight scratch. Cell wound images were taken by a microscope at 0, 24 and 48 hours for examining wound healing.
For the transwell migration assay, 2×104 cells were seeded in the upper transwell chamber insert (Corning, USA). The lower chamber was filled with the complete culture medium containing 10% serum. Cells were allowed to migrate towards the serum gradient for 24 hours. Migrated cells were stained with 1% crystal violet and counted using a phase-contrast microscope. Five random fields were counted per experiment.
TOP/FOP Flash analysis
Human embryonic kidney cells HEK 293T were plated in 24-well plates and were allowed to settle for 12 hours. After 24 h, cells were transfected with pflag-PAK7 plasmid or negative control plasmid (400 ng/well), along with TOP Flash or FOP Flash palsmid (400 ng/well) and pRL-TK (10 ng/well) using Lipofectamine 2000 reagent according to the manufacturer's instruction. Luciferase and renilla signals were measured 48 hours after transfection by using the Dual Luciferase Reporter Assay Kit (Promega) according to a protocol provided by the manufacturer. Each experiment was performed in triplicate and repeated at least three times.
Apoptosis assay and cell cycle assay by flow cytometry
Cell cycle was quantified by flow cytometry using PI staining, and cells were trypsinized and then fixed by incubating in 70% ethanol at 4°C overnight. After fixation, cells were centrifuged for 4 min at 1500 rpm, and the pellet resuspended and incubated for 40 min at 4°C in 1 ml of PBS. Then, the cells were centrifuged again and resuspended in PBS containing 5 mg/ml propidium iodide and 50 μg/mL RNase A. After incubation for 20 min at room temperature, fluorescence intensity was analyzed using a flow cytometry.
Cell apoptosis was quantified by flow cytometry using Annexin V-FITC/PI Apoptosis Detection Kit (BD, New York, USA). After washing with PBS twice, cells were stained with Annexin V-FITC/PI according to the manufacturer's instructions. Then cells were analyzed by flow cytometry (FACScan, BD Biosciences) and apoptotic fractions were acquired.
Reverse transcription and quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells and tissues using Trizol reagent (Invitrogen, Carlsbad, CA). Total RNA (2 μg) was used for first-strand cDNA synthesis with GeneAmp™ RNA PCR Core Kit (Thermo Scientific, USA). Then 2μL cDNA was used to analyze the expression of target genes. qPCR (Applied Biosystem Inc.) was used for mRNA quantification analysis. The primers for mRNA analysis for qPCR are PAK7: forward primer 5'-GGAAGCAACAGAGACGAGACT-3' and reverse primer 5'-TTGTCAGGAGGATGGAGTCAC-3', GAPDH: forward primer 5'-GGTGAAGGTCGGAGTCAACG-3', reverse primer 5'-CCATGTAGT TGAGGTCAATGA AG-3'.
Western Blotting
Proteins were extracted from cells using RIPA lysis buffer and using a BCA Protein Assay Kit (Beyotime Biotechnology Co., Jiangsu, China) to measure proteins' concentration. Equal amounts of total protein (10 mg) were loaded, run on 10% SDS-polyacrylamide gel and transferred to PVDF membranes (Millipore, Billerica, MA). The membranes were blocked with Tris-buffered saline containing 5% nonfat milk for 2 h, then probed with primary antibodies of target protein, including PAK7, flag, GSK3β, p-GSK3β, β-catenin, p-β-catenin, c-myc, cyclin D1 and GAPDH at 4℃ overnight, followed by incubation with horseradish peroxidase (HRP)-linked secondary antibodies (1:5000, Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at room temperature, and detected by ECL reagents (Pierce).
Co-Immunoprecipitation and co-localization analysis
For co-Immunoprecipitation (co-IP) analysis, the protocol was referred to our previous study 32. HEK 293T cells were plated in 10-cm dishes. The cells in Dish 1 and 2 were co-transfected with pflag-PAK7 and pEGFP-β-catenin plasmids. The cells in Dish 3 and 4 were co-transfected with pflag-NC negative control and pEGFP-β-catenin plasmids. After 48 h, the cells were washed twice with ice-cold phosphate-buffered saline (PBS) and then incubated on ice for 15 min in RIPA lysis buffer supplemented with protease inhibitor cocktail. Total cell lysate was centrifuged at 12, 000 rpm for 15 min at 4℃. 300 µg of lysate were incubated with 1µg the anti-flag antibody (Proteintech) for Dish 1, anti-GFP antibody (Proteintech) for Dish 2, anti-flag antibody for Dish 3 and anti-GFP antibody for Dish 4, for 1 h at 4℃, followed by addition of 20 μl agarose beads (Santa Cruz Biotechnology) for overnight at 4 ℃. Agarose beads were washed five times in RIPA lysis buffer supplemented with protease inhibitor cocktail. Complexes were released from the agarose beads by boiling for 5 min in 2× gel electrophoresis loading buffer. The immunocomplex was separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and immunoblotting was used to detect PAK7 with anti-flag and to detect β-catenin with anti-GFP antibody, respectively.
For co-localization analysis, the cells grown on a chamber slide (BD Biosciences, San Jose, CA) were co-transfected with pEGFP-PAK7 and pCherry-β-catenin plasmids. Forty-eight hours after transfection, the cells were fixed with 4% paraformaldehyde in PBS for 30 min and permeabilized by further treatment with 0.2% Triton X-100 for 10 min, followed by DAPI (4',6-Diamidine-2'-phenylindole dihydrochloride) staining for 15min. The images were taken using confocal microscopy (Olympus, Japan).
Immunohistochemistry
Slides were deparaffinized, dehydrated through graded alcohols and rinsed with distilled water. Antigen retrieval was carried out by bringing slides to a boil in 1 mM Sodium Citrate Buffer pH 6.0 followed by 20 min at a sub-boiling temperature. Endogenous peroxidase activity was blocked by incubating the slides with 3% H2O2 for 10 min. Slides were washed in PBS, blocked with serum for 30 min and incubated with a rabbit antibody for PAK7 (1:100 dilution, Proteintech, USA) at 4°C overnight in a humidified container. After washing 3 times with PBS, biotin-conjugated anti-rabbit secondary antibody was incubated for 2 h at 37°C at a dilution of 1:300. 3, 3΄-diaminobenzidine (DAB) (Vector laboratories) was used to stain slides for 2 min and coloring reaction stopped using distilled water. Slides were counterstained with Hematoxylin and Eosin Staining Kit (Beyotime Biotechnology Co., Jiangsu, China) to visualize cell nuclei, followed by bluing in running tap water. Finally, the slides were dehydrated, dipped in xylene and mounted. The Pannoramic MIDI automatic digital slide scanner (3DHISTECH Ltd., Budapest, HUNGARY) was used for image processing and quantifications. Quantification of PAK7 staining was done using IHC profiler in ImageJ33.
Statistical analysis
The data were presented as the mean ± standard deviation (S.D.). The variance analysis between groups was performed using a one-way ANOVA. Pearson Correlation Coefficient was used to determine significance of the correlation between PAK7 and β-catenin, c-myc expression. χ2-tests were performed to determine significance of the relationship between expression of PAK7 and clinicopathologic features in breast cancer tissue microarray. P<0.05 was considered statistically significant.
Results
PAK7 mRNA and protein levels were increased in breast cancer, which was associated with clinicopatholgical characteristics
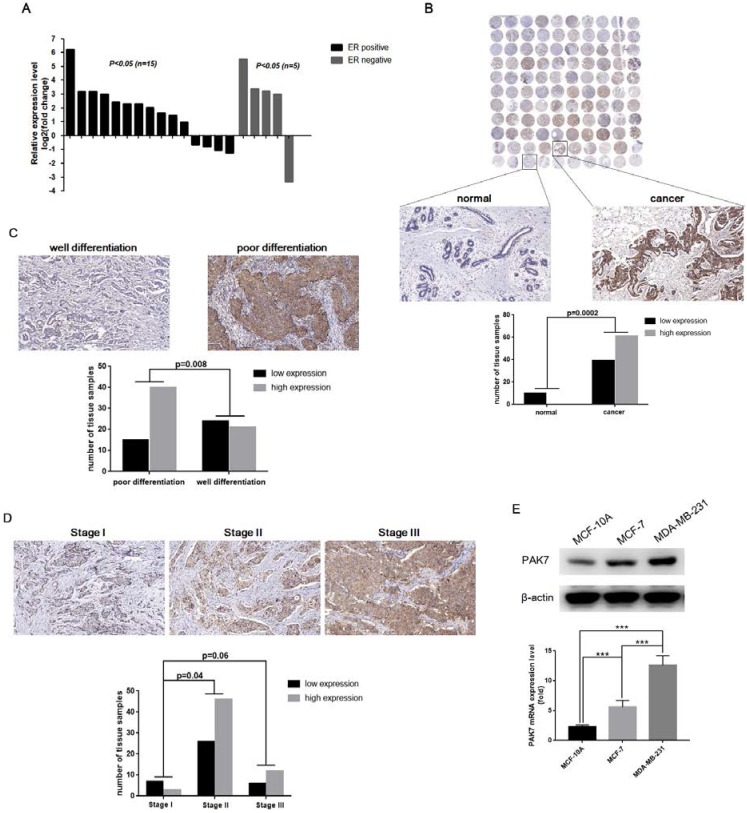
To determine the difference of PAK7 expression in breast tissues and cells, we used RT-qPCR to measure PAK7 mRNA levels in 20 pairs of breast cancer tissues, including 15 pairs of ER positive breast cancer tissues and 5 pairs of ER negative breast cancer tissues, and their adjacent normal tissues. As is shown in Figure 1A, compared to adjacent tissues, the mRNA level of PAK7 was significantly increased in 11 cases of ER positive breast cancer tissues and 4 cases of ER negative breast cancer tissues (P<0.05). To detect the protein expression of PAK7 during tumorigenesis, we examined tissue microarray including 100 cases of breast cancer by immunohistochemistry. Compared to normal breast tissue, PAK7 protein expression (the brown staining areas) was significantly increased in breast cancer tissues (P=0.0002), which was higher in poorly differentiated breast cancer tissues than well-differentiated breast cancer tissues (P=0.008) (Figure 1B-C). Furthermore, as is shown in Table 1, PAK7 protein expression was higher in the stage II than that in the stage I (P=0.04), according to the TNM staging of breast cancer (Figure 1D). In addition, compared to normal breast cell MCF-10A, PAK7 mRNA and protein expression were significantly increased in breast cancer cells MCF-7 and MDA-MB-231. Moreover, the expression level of PAK7 in the more malignant MDA-MB-231 cells was even higher than that in the less malignant MCF-7 cells (P<0.001) (Figure 1E). These data suggest that PAK7 is closely related to the progression of breast cancer.
Figure 1.
PAK7 is significantly upregulated in human breast cancer tissues and cells. A, PAK7 mRNA expression was upregulated in breast cancer tissues (15/20) than paired normal breast tissues which was analyzed by RT-qPCR (x-axis represent 20 pairs of tissue samples). B, PAK7 protein expression was increased in breast cancer, which was detected in 110 cases of human breast cancer tissue microarray by immunohistochemistry. C, The protein levels of PAK7 in poor differentiation of breast cancer tissues were higher than that in well differentiation of breast cancer tissues. D, The protein levels of PAK7 were higher in stage II than that in stage I, according to TNM staging of breast cancer. E, PAK7 mRNA and protein expression were detected by RT-qPCR and western blotting in normal breast cell line MCF-10A and breast cancer cell lines MCF-7 and MDA-MB-231. Values represent the mean ± SD from three independent measurements. The brown staining indicates expression levels of PAK7 protein by immunohistochemistry. *P < 0.05, ***P < 0.001.
Table 1.
PAK7 expression and clinicopathologic characteristics of TMA
| Cases (n=100) |
PAK7 protein expression in breast cancer tissue (n) | P value | ||
|---|---|---|---|---|
| Low expression | High expression | |||
| Age | ||||
| <60 | 86 | 35 | 51 | >0.05 |
| ≥60 | 14 | 5 | 9 | |
| T stage | ||||
| T1 | 11 | 7 | 4 | >0.05 |
| T2 | 66 | 24 | 42 | |
| T3~4 | 23 | 8 | 15 | |
| N stage | ||||
| N0 | 78 | 28 | 50 | >0.05 |
| N1~2 | 22 | 9 | 13 | |
| TNM stage | ||||
| I | 10 | 7 | 3 | 0.04 |
| II | 72 | 26 | 46 | |
| III~ IV | 18 | 6 | 12 | |
| Differentiation | ||||
| well/moderate | 45 | 24 | 21 | 0.008 |
| poor | 55 | 15 | 40 | |
*P values were based on χ2-test.
PAK7 could promote proliferation of breast cancer cells, and inhibit cell apoptosis
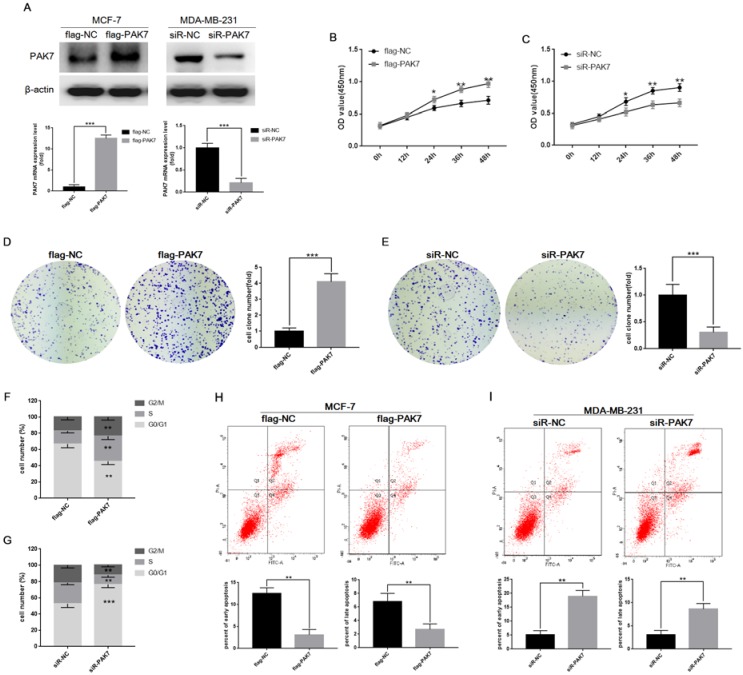
Due to the strong correlation between PAK7 expression levels and breast cancer, we then used gain- and loss-of-expression approaches to determine the biological functions of PAK7 in breast cancer. We transfected PAK7 overexpression plasmids into MCF-7 cells and small interfering RNA (siRNA) of PAK7 into MDA-MB-231 cells, respectively (Figure 2A). CCK-8 assay and clone formation assay were used to detect cell proliferation. The results showed that overexpression of PAK7 can significantly promote the proliferation of MCF-7 cells (Figure 2B and 2D), while knockdown of PAK7 significantly inhibit the proliferation of MDA-MB-231 cells (Figure 2C and 2E). In addition, we further found that overexpression of PAK7 can significantly increase the proportion of MCF-7 cells in S/G2 phase (Figure 2F), while knockdown of PAK7 increased the proportion of MDA-MB-231 cells in G1 phase by flow cytometry (Figure 2G). Apoptosis is also an important factor affecting tumor growth. We found that overexpression of PAK7 can significantly inhibit the early apoptosis and late apoptosis levels of MCF-7 cells (Figure 2H), while knockdown of PAK7 significantly promote the early apoptosis and late apoptosis of MDA-MB-231 cells by flow cytometry (Figure 2I).
Figure 2.
PAK7 promotes proliferation of breast cancer cells, and inhibits cell apoptosis. A, The cell were transfected with flag-PAK7 overexpression plasmid or flag-NC negative control vector, and small interfering RNA of PAK7 (siR-PAK7) or negative control (siR-NC) were detected by RT-qPCR and western blotting in MCF-7 and MDA-MB-231 cells. B, PAK7 overexpression enhances the proliferation of MCF-7 cells which was detected by the CCK-8 assay. C, PAK7 knockdown inhibits the proliferation of MDA-MB-231 cells which was detected by the CCK-8 assay. D, PAK7 overexpression enhances the proliferation of MCF-7 cells which was detected by the colon formation assay. E, PAK7 knockdown inhibits the proliferation of MDA-MB-231 cells which was detected by the colon formation assay. F, PAK7 overexpression increases the portion of MCF-7 cells in S and G2/M phase which was detected by flow cytometry. G, PAK7 knockdown increases the portion of MDA-MB-231 cells in G1 phase, and decreases the portion of cells in S and G2/M phase which was detected by flow cytometry. H, PAK7 overexpression inhibits early apoptosis and late apoptosis of MCF-7 cells which was detected by flow cytometry. I, PAK7 knockdown promotes early apoptosis and late apoptosis of MDA-MB-231 cells which was detected by flow cytometry. Values represent the mean ± SD from three independent measurements. *P < 0.05, **P < 0.01, ***P < 0.001.
PAK7 could significantly promote the migration of breast cancer cells
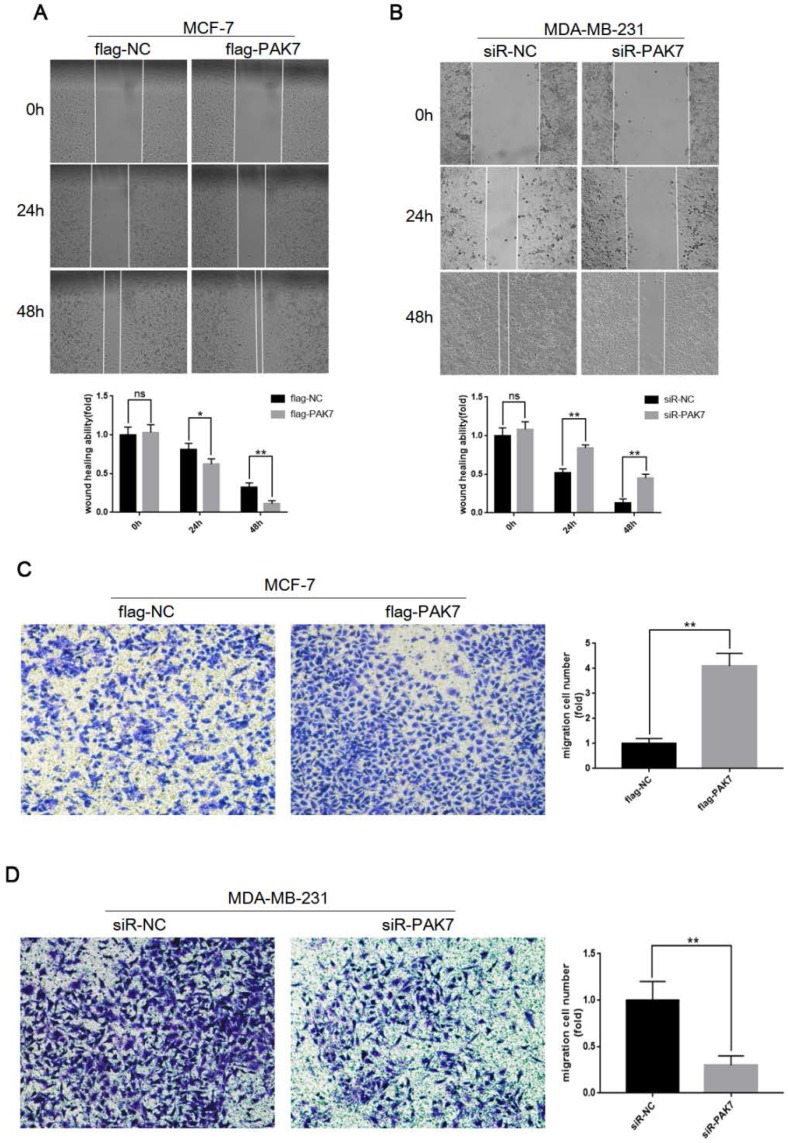
In order to detect the effect of PAK7 expression on cell migration, we transfected the PAK7 overexpression plasmid into MCF-7 cells and siRNA of PAK7 into MDA-MB-231 cells, respectively. Then, wound healing assay and Transwell assay were used to detect cell migration ability. The results of wound healing assay showed that overexpression of PAK7 can significantly promote the migration of MCF-7 cells (Figure 3A), while knockdown of PAK7 significantly inhibited the migration of MDA-MB-231 cells (Figure 3B). In addition, we also detected the effect of PAK7 on the migration ability of breast cancer cells by Transwell assay, and the results were consistent with the wound healing assay (Figure 3C-D). These data showed that PAK7 could promote the migration of breast cancer cells.
Figure 3.
PAK7 significantly promote the migration of breast cancer cells. A, PAK7 overexpression enhances the migration ability of MCF-7 cells which was detected by Wound healing assay. B, PAK7 knockdown suppresses the migration ability of MDA-MB-231 which was detected by Wound healing assay. C, PAK7 overexpression enhances the migration ability of MCF-7 cells which was detected by Transwell assay. D, PAK7 knockdown suppresses the migration ability of MDA-MB-231 which was detected by Transwell assay. Values represent the mean ± SD from three independent measurements. *P < 0.05, **P < 0.01, ***P < 0.001.
PAK7 activates Wnt/β-catenin signaling pathway in breast cancer cells
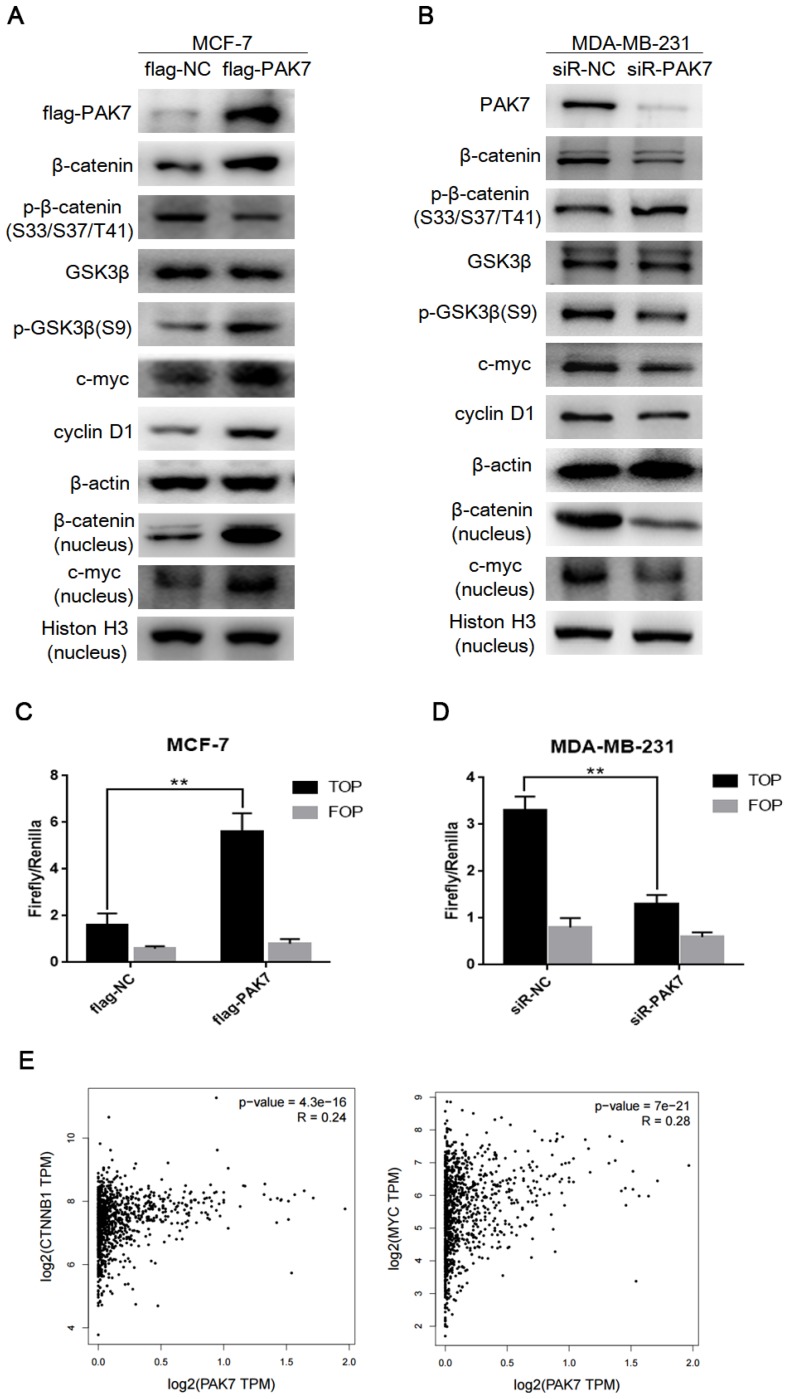
It is known that the activation of Wnt/β-catenin signaling promotes cell proliferation and migration, which is closely related to the occurrence and development of breast cancer 34. Thus, we determined whether PAK7-mediated tumor progression is through Wnt/β-catenin signaling in breast cancer. We found that PAK7 overexpression dramatically up-regulated the expression of β-catenin, p-GSK3β (S9) and downstream target genes cyclin D1 and c-myc, but didn't affect protein expression level of GSK-3β in MCF-7 cells, besides that, the expression of β-catenin and c-myc in nucleus also increased (Figure 4A). In contrast, knockdown of PAK7 in MDA-MB-231 cells led to down-regulation of β-catenin, p-GSK3β (S9), c-myc, cyclin D1, but didn't affect GSK3β, moreover, the expression of β-catenin and c-myc in nucleus also decreased (Figure 4B). In addition, TOP/FOP flash assay is often used to detect activity of Wnt/β-catenin signaling pathway. Thus, we co-transfected PAK7 overexpression plasmids together with TOP flash or the control FOP flash, and PAK7 siRNA together with TOP flash or the control FOP flash in MCF-7 and MDA-MB-231 cells, respectively. The results showed that overexpression of PAK7 can significantly enhance the TOP flash reporter gene activity, but there was no effect on the control FOP flash reporter while the TOP flash reporter gene activity was significantly inhibited by knockdown of PAK7, but there was also no effect on the control FOP flash reporter (Figure 4C-D). Furthermore, we also detect the effect of PAK7 overexpression in MDA-MB-231 cells and PAK7 knockdown in MCF-7 cells, whose results were consistent with that of PAK7 overexpression in MCF-7 cells and PAK7 knockdown in MDA-MB-231 cells (Figure S1). According to these data, we detected the correlation between PAK7 and Wnt/β-catenin signaling pathway in GEPIA database by bioinformatics methods 35, which showed the expression of PAK7 was positively related to β-catenin (CTNNB1) and c-myc (MYC), eventhough the correlation among them was somewhat weak (Figure 4E).The above results suggest that the increase of PAK7 expression in breast cancer can activate Wnt/β-catenin signaling pathway.
Figure 4.
PAK7 activates Wnt/β-catenin signaling pathway in breast cancer cells. A, The effect of transfecting with flag-PAK7 overexpression plasmid or flag-NC negative control vector on the protein levels of flag-PAK7, β-catenin, p-β-catenin (S33/S37/T41), GSK3β, p-GSK3β(S9), c-myc, cyclin D1 and β-catenin (nucleus), c-myc (nucleus) in MCF-7 cells. B, The effect of transfecting with small interfering RNA of PAK7 (siR-PAK7) or negative control (siR-NC) vector on the protein levels of flag-PAK7, β-catenin, p-β-catenin (S33/S37/T41), GSK3β, p-GSK3β(S9), c-myc, cyclin D1 and β-catenin (nucleus), c-myc (nucleus) in MDA-MB-231 cells. C, PAK7 overexpression increases the activity of Wnt/β-catenin signaling pathway which was detected by TOP/FOP flash assay in MCF-7 cells. D, PAK7 knockdown inhibits the activity of Wnt/β-catenin signaling pathway which was detected by TOP/FOP flash assay in MDA-MB-231 cells. E, The expression of PAK7 was positively correlated with β-catenin (CTNNB1), c-myc (MYC) expression which was analyzed in GEPIA database by bioinformatics methods. Values represent the mean ± SD from three independent measurements. *P < 0.05, **P < 0.01, ***P < 0.001.
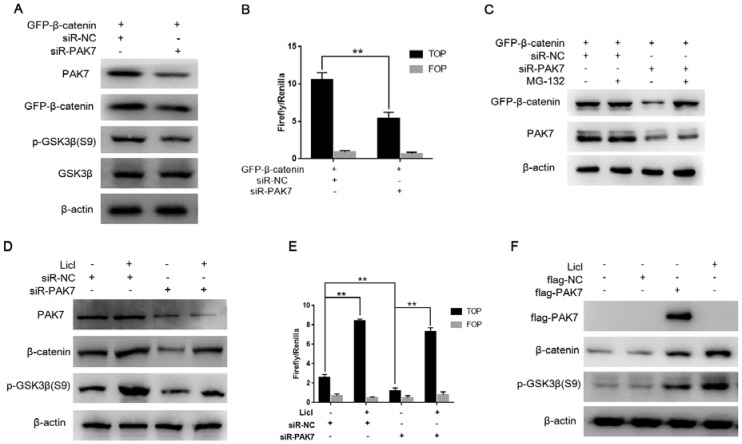
PAK7 inhibits the degradation of β-catenin via phosphorylating GSK3β
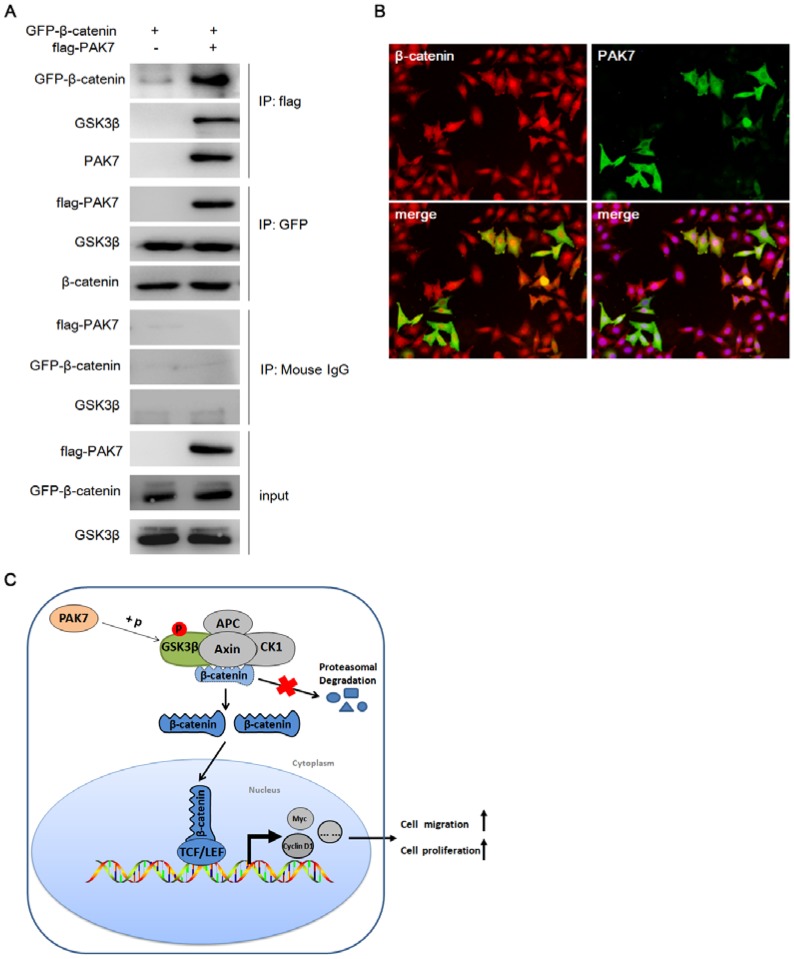
GSK3β can phosphorylate β-catenin at Ser 33 and Ser 37, promoting degradation of β-catenin by intracellular proteasome, and thus inhibit Wnt/β-catenin signaling pathway. In HEK 293T cells, we co-transfected GFP-β-catenin and siR-PAK7, the Western blotting result showed that knockdown of PAK7 could significantly inhibit the expression of β-catenin and p-GSK3β (S9), but didn't affect GSK3β (Figure 5A). And then we conducted TOP/FOP flash assay which also showed that knockdown of PAK7 significantly suppressed the activity of TOP flash reporter via inhibiting β-catenin (Figure 5B). When β-catenin is phosphorylated by GSK3β at Ser 33 and Ser 37, the intracellular proteasome will degrade the phosphorylated β-catenin. Therefore, when we treated with proteasome inhibitor MG132, the effect of PAK7 knockdown on inhibiting β-catenin was offset (Figure 5C). There results showed that PAK7 inhibited the degradation of β-catenin, and the mechanism seemed to be associated with phosphorylation of GSK3β. The activity of GSK3β depends on whether phosphorylation occurs at Ser 9. When phosphorylated at Ser 9, the kinase activity of GSK3β is reduced, which will inhibit phosphorylation of β-catenin at Ser 33 and Ser 37. Licl is often used as an inhibitor of GSK3β, which promotes phosphorylation of GSK3β at Ser 9. When PAK7 was knocked down, the level of phosphorylated GSK3β (S9) was significantly decreased and the level of β-catenin was significantly increased. However, when treated with 20mM Licl at the same time for 12 hours, Licl eliminated the effect of PAK7 knockdown on phosphorylated GSK3β (S9) and β-catenin expression (Figure 5D). In addition, TOP/FOP flash assay also showed that Licl can significantly reverse the impact of PAK7 knockdown on activity of Wnt/β-catenin signaling pathway (Figure 5E). Furthermore, we found that PAK7 overexpression had similar effects with Licl treatment, which both significantly upregulated expression level of β-catenin and p-GSK3β (S9) (Figure 5F). These results suggest that PAK7 indeed play a role in inhibiting the degradation of β-catenin through phosphorylating GSK3β. In order to further explore the regulation mechanism of PAK7 on GSK3β and β-catenin, we detected whether there were direct interaction between them by co-immunoprecipitation and immunofluorescence co-localization. GFP-tagged β-catenin and flag-tagged PAK7 were cotransfected into HEK293T cells, and then the cell lysate was immunoprecipitated with GFP and flag antibodies respectively. The results showed that there was indeed direct interaction between PAK7, GSK3β and β-catenin (Figure 6A). After then, pEGFP-PAK7 (GFP tagged) and pCherry-β-catenin (Cherry tagged) were cotransfected into HEK293T cells. Co-localization (yellow fluorescence) of PAK7 (green fluorescence) and β-catenin (red fluorescence) was observed in the cytoplasm and nucleus (Figure 6B). It was known that β-catenin, as a phosphorylation substrate for GSK3β, had been found to have a direct interaction with GSK3β. In our study, we identified GSK3β acts as a phosphorylation substrate for PAK7, so that PAK7 interacts with GSK3β and β-catenin as a multi-protein complex in cells.
Figure 5.
PAK7 inhibits the degradation of β-catenin via phosphorylating GSK3β. A, PAK7 knockdown suppressed p-GSK3β (S9) expression. pEGFP-β-catenin was transfected into HEK293T cells along with siR-PAK7 or negative control. Forty-eight hours after transfection, cells were harvested for western blotting analysis to detect the expression of PAK7, GFP-β-catenin, GSK3β, p-GSK3β. B, PAK7 knockdown inhibited β-catenin mediated transcriptional activity of TCF. HEK293T cells were cotransfected with siR-PAK7 or siR-NC, pEGFP-β-catenin, TOP flash (TOP) or FOP flash (FOP), and Renilla. 48 h after transfection, cells were harvested for luciferase activity assay. C, Inhibiting proteasomes degradation by MG132 reverses the effect of PAK7 knockdown on β-catenin degradation. HEK293T cells were cotransfected with pEGFP-β-catenin and siR-PAK7 or siR-NC. Forty-four hours after transfection, the cells were treated with 30μM MG132. Cells were harvested 4 hours later and subjected to western blotting analysis to detect the expression of GFP-β-catenin and PAK7. D, Inhibiting GSK3β activity by Licl reduced β-catenin expression inhibition by PAK7 knockdown. siR-PAK7 or siR-NC was transfected into MDA-MB-231 cells. 36 hours after transfection, the cells were treated with 30 mM Licl for 12 hours and then harvested for western blotting analysis to detect the expression of GFP-β-catenin, p-GSK3β (S9) and PAK7. E, siR-PAK7 or siR-NC was transfected into MDA-MB-231 cells. 36 hours after transfection, the cells were treated with 30 mM Licl for 12 hours and harvested for luciferase activity assay. F,MDA-MB-231 cells were transfected with flag-NC and flag-PAK7, treated with Licl for 48 hours and harvested for western blot assay.
Figure 6.
PAK7 interacts with β-catenin and GSK3β. A, PAK7 was binding to β-catenin and GSK3β by immunoprecipitation. β-catenin (GFP tagged) was cotransfected with PAK7 (flag tagged) or empty vector into HEK293T cells. Immunoprecipitation was performed with flag antibody and GFP antibody, respectively. β-catenin, GSK3β and PAK7 were analyzed with a GFP antibody, GSK3β antibody, and flag antibody, respectively. B, PAK7 and β-catenin were co-localized in the cell cytoplasm and nucleus. PAK7 (GFP tagged) and pCherry-β-catenin (Cherry tagged) were cotransfected into HEK293T cells. Co-localization (yellow fluorescence) of PAK7 (green fluorescence) and β-catenin (red fluorescence) was detected in the cytoplasm and nucleus. C, Working model for the regulation of β-catenin degradation by PAK7 via phosphorylating GSK3β in Wnt/β-catenin signaling pathway.
Discussion
PAK7 is a member of the PAK family of serine/threonine protein kinase 13, and exerts a variety of biological effects when it is activated. An increasing body of evidence suggests that increased expression of PAK7 was observed in a variety of human malignancies. However, the role of PAK7 in breast cancer was poorly investigated. In our present study, the result of breast cancer tissue microarray showed that the protein expression of PAK7 in breast cancer tissues was significantly increased, which was positively correlated with the pathological differentiation and TNM stage of breast cancer, suggesting that PAK7 is closely related to the malignant progression and prognosis of breast cancer. Then we found higher expression level of PAK7 in more malignant breast cancer cell MDA-MB-231 than that in MCF-7 and normal breast cell MCF-10A, which may not be related to the methylation of PAK7 (data not shown)36. In vitro study, we found that overexpression of PAK7 can significantly promote cell proliferation and migration, while knockdown of PAK7 significantly inhibited cell proliferation and migration. In addition, PAK7 can also promote escape of apoptosis in breast cancer cell, and knockdown of PAK7 significantly increased the level of apoptosis in breast cancer cells.
A number of studies have shown that PAK7 promotes cancer cell proliferation and migration in colorectal cancer, epithelial ovarian cancer, gastric cancer, glioma, liver cancer, pancreatic cancer and breast cancer 37. In addition, the role of PAK7 in inhibiting apoptosis is also one of the important mechanisms involved in tumor progression. There were studies confirmed that PAK7 could significantly inhibit camptothecin-induced apoptosis in colorectal cancer cells. Moreover, overexpression of PAK7 could inhibit camptothecin-induced activation of caspase-8 and poly ADP ribose polymerase in colorectal cancer cells SW480, which are two important factors to promote apoptosis 38. In addition to inhibit camptothecin-induced apoptosis in colorectal cancer cells, PAK7 was also found to protect pancreatic cancer cells from apoptosis, and inhibition of PAK7 expression significantly promoted apoptosis of MiaPaCa-2 cells 20, 21. PAK7 inhibits the mitochondrial localization of Bcl-2-associated death promoter (BAD) by inhibiting phosphorylation of BAD on Ser-112 in a protein-independent manner and thus inhibits the apoptotic cascade 39. In this study, we also found that overexpression of PAK7 could inhibit the apoptosis of MCF-7 cells in breast cancer cells, while knockdown of PAK7 significantly promoted the apoptosis of MDA-MB-231 cells, suggesting that the role of PAK7 in inhibiting apoptosis is one of the important mechanisms involving breast cancer progression.
Cyclin-dependent kinases (CDKs) are a family of protein kinases that regulate the cell cycle 40, 41. Cyclin D1 forms protein complex with CDK4 or CDK6, the activity of which is required for cell cycle G1/S transition 42. The upregulation of cyclin D1 expression could accelerate the cell cycle progression and eventually lead to tumor cell proliferation 43, 44. PAK7 can upregulate the expression of CDK2, CDC25A and cyclin D1, accelerating cell cycle G1/S transition to promote tumor cell proliferation. Recent studies also found that PAK7 can promote the phosphorylation and the nuclear translocation of p65 subunit of nuclear factor-kappaB (NF-κB). Furthermore, p65 can directly bind to the promoter of cyclin D1 and promote its protein expression 25. In this study, we also observed that overexpression of PAK7 in MCF-7 cells could increase the expression of cyclin D1 and c-myc, while PAK7 knockdown could significantly inhibit the expression of cyclin D1, c-myc in MDA-MB-231 cells. In addition, it was found by flow cytometry that PAK7 knockdown induced G1/S phase arrest in MDA-MB-231 cells, while PAK7 overexpression increased proportion of MCF-7 cells in S/G2 phase, suggesting that PAK7 could promote breast cancer cells proliferation by accelerating G1/S transition. It is well known that cyclin D1 and c-myc are the target genes of Wnt/β-catenin signaling pathway 45, which suggests that whether PAK7 interacts with the Wnt/β-catenin signaling pathway in breast cancer. Firstly, we found that there was a significant correlation between PAK7 and Wnt/β-catenin signaling pathway by bioinformatics analysis. Then, we found that the overexpression of PAK7 significantly increased the expression of β-catenin, c-myc and cyclin D1 in MCF-7 cells breast cancer cells, while PAK7 also significantly enhanced the activity of TOP/FOP reporter. The expression of β-catenin, c-myc and cyclin D1 was significantly suppressed by knocking down PAK7 in MDA-MB-231 cells, meanwhile, the activity of TOP/FOP reporter was also significantly inhibited, suggesting that PAK7 played a role in promoting breast cancer cell proliferation and migration via activating Wnt/β-catenin signaling pathway. However, there was no research on the mechanism of PAK7 regulating Wnt/β-catenin signaling in breast cancer.
In the Wnt/β-catenin pathway, the Wnt protein binds to the Frizzled family receptor and its co-receptor on the cell membrane, and the disheveled protein in the cytoplasm receives biological signal and continues to transmit it down, resulting in the accumulation of β-catenin in the cytoplasm, eventually leading β-catenin to translocating into the nucleus where interact with the TCF/LEF family proteins to form a transcriptional regulation complex, and then activate a series of cell proliferation and migration associated genes, such as c-myc and cyclin D145. In this study, we found that overexpression of PAK7 could significantly upregulate the expression of β-catenin, while knockdown of PAK7 significantly decreased the expression of β-catenin, thus regulating the activity of Wnt/β-catenin signaling pathway. Our further study found that PAK7 could inhibit degradation of β-catenin. It was known that Axin, APC, GSK3β and CK1α form a destruction complex in the cytoplasm 46, 47. When the destruction complex binds to β-catenin, β-catenin is ubiquitinated and subsequently degraded by cellular proteasome 48, 49. With the help of casein kinase 1 (CK1), which carries out an initial phosphorylation of β-catenin at Thr 41, GSK3β follows on phosphorylating β-catenin at Ser 33 and Ser 37 a second time. This targets β-catenin for ubiquitination and degradation by proteasomes, which prevents it from translocating into the nucleus, where it acts as a transcription factor for activating proliferation and metastasis associated genes. Therefore, the decreased expression level or activity of GSK3β will inhibit the degradation of β-catenin, which results in accumulating of β-catenin in the cytoplasm and then enter the nucleus to activate the target genes expression 50-52. GSK3β could be phosphorylated at Ser 9 by serine/threonine kinases, such as Akt, protein kinase A (PKA), and protein kinase C (PKC), which leads to decreased kinase activities 53-55. Thus, GSK3β phosphorylation at Ser 9 has been used as a marker of decreased GSK3β activity. In the present study, we found that overexpression of PAK7 could significantly upregulate the expression of p-GSK3β (S9), while knockdown of PAK7 significantly decreased the expression of p-GSK3β (S9), but had no effect on the expression level of GSK3β, which suggested that PAK7 may regulate GSK3β kinase activity, thereby affecting the Wnt/β-catenin signaling pathway. To further validate our hypothesis, we used Licl, the GSK3β inhibitor, to inhibit GSK3β activity and simultaneously examined the effect of PAK7 knockdown on β-catenin expression. We found that Licl could eliminate the inhibitory effect of PAK7 knockdown on the expression of β-catenin. In addition, TOP/FOP flash assay also showed that Licl could significantly reverse the effect of PAK7 knockdown on the activity of Wnt/β-catenin signaling pathway. These results suggest that the mechanism of PAK7 regulating Wnt/β-catenin signaling pathway is indeed mediated by GSK3β. According to these results, we further found that PAK7 could directly interact with GSK3β and β-catenin by co-Immunoprecipitation and immunofluorescence co-localization assay, which also confirmed our previous assumption that PAK7 inhibited the degradation of β-catenin by binding and promoting phosphorylation of GSK3β at Ser-9, promoting the accumulation of β-catenin in the cytoplasm and then entering nuclear to activate target genes expression (Figure 6C).
In conclusion, we found that the expression of PAK7 was significantly increased in breast cancer tissues and positively correlated with the malignancy of breast cancer. PAK7 could activate the Wnt/β-catenin signaling pathway by inhibiting the kinase activity of GSK3β, exerting the effect of promoting cell proliferation and migration in breast cancer. In this study, we also found that PAK7 and β-catenin also co-localized in the nucleus, so we couldn't exclude whether PAK7 directly interact with β-catenin and regulate Wnt/β-catenin through other mechanisms, which is also an important scientific question we need to explore in the future.
Supplementary Material
Supplementary figure S1.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81472765) and the Key Project of Science and Technology Department of Hubei Province (2015CFA070).
Abbreviations
- PAK7
P21-activated kinase 7
- GSK3β
Glycogen synthase kinase 3β
- TMA
tissue microarray
- APC
Adenomatous polyposis coli
- TCF
Transcription Factor 4.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Zeng H, Zheng R, Zhang S, Zou X, Chen W. Female breast cancer statistics of 2010 in China: estimates based on data from 145 population-based cancer registries. 2014;6(5):466–470. doi: 10.3978/j.issn.2072-1439.2014.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. 2013;13(1):11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 4.Radu M, Semenova G, Kosoff R, Chernoff J. PAK signalling during the development and progression of cancer. 2014;14(1):13–25. doi: 10.1038/nrc3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye DZ, Field J. PAK signaling in cancer. 2012;2(2):105–116. doi: 10.4161/cl.21882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaffer ZM, Chernoff J. p21-activated kinases: three more join the Pak. 2002;34(7):713–717. doi: 10.1016/s1357-2725(01)00158-3. [DOI] [PubMed] [Google Scholar]
- 7.Abo A, Qu J, Cammarano MS, Dan C, Fritsch A, Baud V, Belisle B, Minden A. PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. 1998;17(22):6527–6540. doi: 10.1093/emboj/17.22.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang F, Li X, Sharma M, Zarnegar M, Lim B, Sun Z. Androgen receptor specifically interacts with a novel p21-activated kinase, PAK6. 2001;276(18):15345–15353. doi: 10.1074/jbc.M010311200. [DOI] [PubMed] [Google Scholar]
- 9.Dan C, Nath N, Liberto M, Minden A. PAK5, a new brain-specific kinase, promotes neurite outgrowth in N1E-115 cells. 2002;22(2):567–577. doi: 10.1128/MCB.22.2.567-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong W, An Z, Wang Y, Pan X, Fang W, Jiang B, Zhang H. P21-activated kinase 5 is overexpressed during colorectal cancer progression and regulates colorectal carcinoma cell adhesion and migration. 2009;125(3):548–555. doi: 10.1002/ijc.24428. [DOI] [PubMed] [Google Scholar]
- 11.Zhu G, Li X, Guo B, Ke Q, Dong M, Li F. PAK5-mediated E47 phosphorylation promotes epithelial-mesenchymal transition and metastasis of colon cancer. 2016;35(15):1943–1954. doi: 10.1038/onc.2015.259. [DOI] [PubMed] [Google Scholar]
- 12.Li D, Yao X, Zhang P. The overexpression of P21-activated kinase 5 (PAK5) promotes paclitaxel-chemoresistance of epithelial ovarian cancer. 2013;383(1-2):191–199. doi: 10.1007/s11010-013-1767-7. [DOI] [PubMed] [Google Scholar]
- 13.Gu J, Li K, Li M, Wu X, Zhang L, Ding Q, Wu W, Yang J, Mu J, Wen H. et al. A role for p21-activated kinase 7 in the development of gastric cancer. 2013;280(1):46–55. doi: 10.1111/febs.12048. [DOI] [PubMed] [Google Scholar]
- 14.Zhai J, Qu S, Li X, Zhong J, Chen X, Qu Z, Wu D. miR-129 suppresses tumor cell growth and invasion by targeting PAK5 in hepatocellular carcinoma. 2015;464(1):161–167. doi: 10.1016/j.bbrc.2015.06.108. [DOI] [PubMed] [Google Scholar]
- 15.Aburatani T, Inokuchi M, Takagi Y, Ishikawa T, Okuno K, Gokita K, Tomii C, Tanioka T, Murase H, Otsuki S. et al. High expression of P21-activated kinase 5 protein is associated with poor survival in gastric cancer. 2017;14(1):404–410. doi: 10.3892/ol.2017.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu X, Wang C, Wang X, Ma G, Li Y, Cui L, Chen Y, Zhao B, Li K. Efficient inhibition of human glioma development by RNA interference-mediated silencing of PAK5. 2015;11(2):230–237. doi: 10.7150/ijbs.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han ZX, Wang XX, Zhang SN, Wu JX, Qian HY, Wen YY, Tian H, Pei DS, Zheng JN. Downregulation of PAK5 inhibits glioma cell migration and invasion potentially through the PAK5-Egr1-MMP2 signaling pathway. 2014;31(4):234–241. doi: 10.1007/s10014-013-0161-1. [DOI] [PubMed] [Google Scholar]
- 18.Chen H, Miao J, Li H, Wang C, Li J, Zhu Y, Wang J, Wu X, Qiao H. Expression and prognostic significance of p21-activated kinase 6 in hepatocellular carcinoma. 2014;189(1):81–88. doi: 10.1016/j.jss.2014.01.049. [DOI] [PubMed] [Google Scholar]
- 19.Fang ZP, Jiang BG, Gu XF, Zhao B, Ge RL, Zhang FB. P21-activated kinase 5 plays essential roles in the proliferation and tumorigenicity of human hepatocellular carcinoma. 2014;35(1):82–88. doi: 10.1038/aps.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giroux V, Iovanna JL, Garcia S, Dagorn JC. Combined inhibition of PAK7, MAP3K7 and CK2alpha kinases inhibits the growth of MiaPaCa2 pancreatic cancer cell xenografts. 2009;16(9):731–740. doi: 10.1038/cgt.2009.22. [DOI] [PubMed] [Google Scholar]
- 21.Giroux V, Iovanna J, Dagorn JC. Probing the human kinome for kinases involved in pancreatic cancer cell survival and gemcitabine resistance. 2006;20(12):1982–1991. doi: 10.1096/fj.06-6239com. [DOI] [PubMed] [Google Scholar]
- 22.Han K, Zhou Y, Gan ZH, Qi WX, Zhang JJ, Fen T, Meng W, Jiang L, Shen Z, Min DL. p21-activated kinase 7 is an oncogene in human osteosarcoma. 2014;38(12):1394–1402. doi: 10.1002/cbin.10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Ke Q, Shao Y, Zhu G, Li Y, Geng N, Jin F, Li F. GATA1 induces epithelial-mesenchymal transition in breast cancer cells through PAK5 oncogenic signaling. 2015;6(6):4345–4356. doi: 10.18632/oncotarget.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang XX, Cheng Q, Zhang SN, Qian HY, Wu JX, Tian H, Pei DS, Zheng JN. PAK5-Egr1-MMP2 signaling controls the migration and invasion in breast cancer cell. 2013;34(5):2721–2729. doi: 10.1007/s13277-013-0824-x. [DOI] [PubMed] [Google Scholar]
- 25.Zhang YC, Huo FC, Wei LL, Gong CC, Pan YJ, Mou J, Pei DS. PAK5-mediated phosphorylation and nuclear translocation of NF-kappaB-p65 promotes breast cancer cell proliferation in vitro and in vivo. 2017;36(1):146. doi: 10.1186/s13046-017-0610-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dummler B, Ohshiro K, Kumar R, Field J. Pak protein kinases and their role in cancer. 2009;28(1-2):51–63. doi: 10.1007/s10555-008-9168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nusse R, van Ooyen A, Cox D, Fung YK, Varmus H. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. 1984;307(5947):131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- 28.Clevers H. Wnt/beta-catenin signaling in development and disease. 2006;127(3):469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 29.Nusse R. Wnt signaling in disease and in development. 2005;15(1):28–32. doi: 10.1038/sj.cr.7290260. [DOI] [PubMed] [Google Scholar]
- 30.Micalizzi DS, Farabaugh SM, Ford HL. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. 2010;15(2):117–134. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DiMeo TA, Anderson K, Phadke P, Fan C, Perou CM, Naber S, Kuperwasser C. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. 2009;69(13):5364–5373. doi: 10.1158/0008-5472.CAN-08-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bao Y, Li K, Guo Y, Wang Q, Li Z, Yang Y, Chen Z, Wang J, Zhao W, Zhang H. et al. Tumor suppressor PRSS8 targets Sphk1/S1P/Stat3/Akt signaling in colorectal cancer. 2016;7(18):26780–26792. doi: 10.18632/oncotarget.8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varghese F, Bukhari AB, Malhotra R, De A. IHC Profiler: an open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. 2014;9(5):e96801. doi: 10.1371/journal.pone.0096801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan J, Wu J, Li M. MicroRNA-374a activates Wnt/beta-catenin signaling to promote breast cancer metastasis. 2013;123(2):566–579. doi: 10.1172/JCI65871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian Q, Shi X, Lei Z, Zhan L, Liu RY, Zhao J, Yang B, Liu Z, Zhang HT. Methylated +58CpG site decreases DCN mRNA expression and enhances TGF-beta/Smad signaling in NSCLC cells with high metastatic potential. 2014;44(3):874–882. doi: 10.3892/ijo.2014.2255. [DOI] [PubMed] [Google Scholar]
- 37.Wen YY, Zheng JN, Pei DS. An oncogenic kinase: putting PAK5 forward. 2014;18(7):807–815. doi: 10.1517/14728222.2014.918103. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Gong W, Qing H, Geng Y, Wang X, Zhang Y, Peng L, Zhang H, Jiang B. p21-activated kinase 5 inhibits camptothecin-induced apoptosis in colorectal carcinoma cells. 2010;31(6):575–582. doi: 10.1007/s13277-010-0071-3. [DOI] [PubMed] [Google Scholar]
- 39.Cotteret S, Jaffer ZM, Beeser A, Chernoff J. p21-Activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. 2003;23(16):5526–5539. doi: 10.1128/MCB.23.16.5526-5539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malumbres M. Cyclin-dependent kinases. 2014;15(6):122. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ingham M, Schwartz GK. Cell-Cycle Therapeutics Come of Age. 2017;35(25):2949–2959. doi: 10.1200/JCO.2016.69.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Wang X, Xie Y, Ying Z, Liu W, Ping L, Zhang C, Pan Z, Ding N, Song Y. et al. The mTOR kinase inhibitor everolimus synergistically enhances the anti-tumor effect of the Bruton's tyrosine kinase (BTK) inhibitor PLS-123 on Mantle cell lymphoma. 2018;142(1):202–213. doi: 10.1002/ijc.31044. [DOI] [PubMed] [Google Scholar]
- 43.Lee MH, Yang HY. Regulators of G1 cyclin-dependent kinases and cancers. 2003;22(4):435–449. doi: 10.1023/a:1023785332315. [DOI] [PubMed] [Google Scholar]
- 44.Ferraz C, Lorenz S, Wojtas B, Bornstein SR, Paschke R, Eszlinger M. Inverse correlation of miRNA and cell cycle-associated genes suggests influence of miRNA on benign thyroid nodule tumorigenesis. 2013;98(1):E8–16. doi: 10.1210/jc.2012-2564. [DOI] [PubMed] [Google Scholar]
- 45.Bienz M, Clevers H. Armadillo/beta-catenin signals in the nucleus-proof beyond a reasonable doubt? 2003;5(3):179–182. doi: 10.1038/ncb0303-179. [DOI] [PubMed] [Google Scholar]
- 46.Minde DP, Anvarian Z, Rudiger SG, Maurice MM. Messing up disorder: how do missense mutations in the tumor suppressor protein APC lead to cancer? 2011;10:101. doi: 10.1186/1476-4598-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minde DP, Radli M, Forneris F, Maurice MM, Rudiger SG. Large extent of disorder in Adenomatous Polyposis Coli offers a strategy to guard Wnt signalling against point mutations. 2013;8(10):e77257. doi: 10.1371/journal.pone.0077257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao TP, Kuhl M. An updated overview on Wnt signaling pathways: a prelude for more. 2010;106(12):1798–1806. doi: 10.1161/CIRCRESAHA.110.219840. [DOI] [PubMed] [Google Scholar]
- 49.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conde-Perez A, Gros G, Longvert C, Pedersen M, Petit V, Aktary Z, Viros A, Gesbert F, Delmas V, Rambow F. et al. A caveolin-dependent and PI3K/AKT-independent role of PTEN in beta-catenin transcriptional activity. 2015;6:8093. doi: 10.1038/ncomms9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chua HH, Tsuei DJ, Lee PH, Jeng YM, Lu J, Wu JF, Su DS, Chen YH, Chien CS, Kao PC. et al. RBMY, a novel inhibitor of glycogen synthase kinase 3beta, increases tumor stemness and predicts poor prognosis of hepatocellular carcinoma. 2015;62(5):1480–1496. doi: 10.1002/hep.27996. [DOI] [PubMed] [Google Scholar]
- 52.Gassen NC, Hartmann J, Zannas AS, Kretzschmar A, Zschocke J, Maccarrone G, Hafner K, Zellner A, Kollmannsberger LK, Wagner KV. et al. FKBP51 inhibits GSK3beta and augments the effects of distinct psychotropic medications. 2016;21(2):277–289. doi: 10.1038/mp.2015.38. [DOI] [PubMed] [Google Scholar]
- 53.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. 1995;378(6559):785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 54.Fang X, Yu S, Tanyi JL, Lu Y, Woodgett JR, Mills GB. Convergence of multiple signaling cascades at glycogen synthase kinase 3: Edg receptor-mediated phosphorylation and inactivation by lysophosphatidic acid through a protein kinase C-dependent intracellular pathway. 2002;22(7):2099–2110. doi: 10.1128/MCB.22.7.2099-2110.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fang X, Yu SX, Lu Y, Bast RC Jr, Woodgett JR, Mills GB. Phosphorylation and inactivation of glycogen synthase kinase 3 by protein kinase A. 2000;97(22):11960–11965. doi: 10.1073/pnas.220413597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure S1.