Abstract
The alterations of MET have been detected in non-small-cell lung cancer (NSCLC). However, the prognostic impact of MET gene copy number gain (CNG) has not been consistent among studies. We performed this meta-analysis to evaluate the prognostic value of high MET CNG in patients with NSCLC. A systematic computerized search of the electronic databases including PubMed, EMBASE, Google scholar, and Cochrane Library (up to November 2017) was carried out. From twenty-one studies, 7,647 patients were included in the pooled analysis of hazard ratios (HRs) with 95% confidence intervals (CIs) for disease-free survival or overall survival. Compared with patients with NSCLC showing low MET CNG, those with tumors harboring high MET CNG showed significantly worse survival (HR = 1.45, 95% CI: 1.16-1.80, p = 0.001). Subgroup analyses showed that high MET CNG significantly correlated with a poor prognosis especially in patients with adenocarcinoma (HR = 1.41, 95% CI: 1.11-1.79, p = 0.005) and Asian populations (HR = 1.58, 95% CI: 1.32-1.88, p < 0.00001).
In conclusion, this meta-analysis indicates that high MET CNG is an adverse prognostic factor in patients with NSCLC. Subgroup analyses suggest that high MET CNG is associated with a worse prognosis, especially in patients with adenocarcinoma and Asian populations. However, large prospective studies using standardized methods based on the homogeneous populations are warranted to validate the prognostic value of MET amplification in patients with NSCLC.
Keywords: MET copy number, MET amplification, non-small-cell lung cancer, meta-analysis
Introduction
Lung cancer is the leading cause of cancer-related death worldwide 1. Despite recent advances in molecular targeted therapies, non-small-cell lung cancer (NSCLC) remains a lethal disease 1,2. Therefore, there is a critical need to identify novel therapeutic targets promoting NSCLC pathogenesis and develop more efficacious targeted agents. The mesenchymal-epidermal transition (MET)/hepatocyte growth factor (HGF) pathway has recently emerged as a potential therapeutic target in several types of human cancers including NSCLC 3,4.
MET, located at 7q21-q31, is a proto-oncogene that encodes the tyrosine kinase receptor for HGF.5 The MET/HGF signaling pathway regulates multiple cellular functions, including differentiation, cell cycle progression, proliferation, and angiogenesis 3,6. Therefore, dysregulation of the MET signaling pathway has been implicated in the pathogenesis of cancer, such as tumor cell proliferation and survival, invasion, and metastasis 7,8. The MET/HGF pathway has also cross-talks with the epidermal growth factor receptor (EGFR) network at both PI3K/Akt and MAPK nodes, suggesting mutual compensation 9. Different molecular alterations in the MET gene have been found to determine the MET/HGF pathway activation: point mutations, amplifications, genetic polymorphisms, enhanced transcription, and autocrine activation 10-12.
Alterations in the MET gene, including overexpression, amplification, and mutations, have been also observed in NSCLC 13-17. MET protein overexpression is variable in NSCLC, ranging from 5% to 74.6% 13,14. Some studies have reported that high MET expression is associated with a worse prognosis in patients with NSCLC 14,15. The rate of MET amplification (true amplification or copy gain) in NSCLC has been reported to range from 1% to 39% 18-39, depending on the histology and detection methods. Recently, MET amplification has been proposed as a potential mode of resistance to EGFR tyrosine kinase inhibitors in NSCLC 40-42. In addition, several studies reported a negative prognostic impact of high MET gene copy number gain (CNG) in patients with NSCLC 20, 21,29,30,32,37,39,43,44. However, the data are limited and other studies have failed to confirm this finding 17,19,22-28,31,33-36,38.
Because the prognostic impact of MET amplification has been inconsistent among studies, we performed this meta-analysis to gain a better insight into the prognostic role of high MET CNG in patients with NSCLC.
Materials and Methods
Publication searching strategy
The current study was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines 45. A systematic computerized search of the electronic databases including PubMed, EMBASE, Google scholar, and Cochrane Library (up to November 2017) was performed. The search used the following keywords variably combined: “MET”, “MET copy number”, “MET amplification”, “non-small-cell lung cancer or NSCLC”, and “lung cancer”. The related articles function in PubMed was used to identify all relevant articles.
Inclusion criteria
Eligible studies should meet the following inclusion criteria: (i) clinical trials and prospective or retrospective cohort studies investigating the correlation of MET amplification or CNG with disease-free survival (DFS) or overall survival (OS) in patients with NSCLC; (ii) the use of adequate detection methods including fluorescence in situ hybridization (FISH), silver in situ hybridization (SISH), bright-field in situ hybridization (BISH), or quantitative, real-time polymerase chain reaction (qPCR); (iii) results providing sufficient data for hazard ratios (HRs) with 95% confidence intervals (CIs) for DFS or OS; (iv) Studies published in peer-reviewed journals; and (v) articles written in English.
Data extraction
Two investigators (BJK and HSK) independently screened relevant studies and extracted the data from each eligible study. If these two authors did not agree, the principle investigator (JHK) was consulted to settle the dispute through discussion.
The following data were extracted from the included studies: the first author, year of publication, country, inclusion period, number of patients, stage, histology, detection method of MET amplification, cut-off criteria of high MET CNG, and HRs with their 95% CIs for DFS or OS. When there were both univariate and multivariate analysis for survival, the data were extracted preferentially from multivariate analysis.
Statistical analysis
The survival outcomes were stratified according to MET CNG (low vs. high). Statistical values were obtained directly from the original articles. When papers had no HR and/or its 95% CI, the Engauge Digitizer (version 9.1) was used to estimate them from Kaplan-Meier curves. The effect size of DFS and OS was pooled through HR and its 95% CI. Subgroup analyses were performed according to the histological subtypes (adenocarcinoma or squamous cell carcinoma) and ethnic source (Asian or non-Asian). The heterogeneity across studies was estimated by the Q statistics and I2 inconsistency test. The fixed-effect model (Mantel-Haenszel method) was used for pooling homogeneous outcomes (p ≥ 0.1 and I2 ≤ 50%), and the random-effects model (DerSimonian-Laird method) was selected when significant heterogeneity was observed (p < 0.01 and I2 > 50%).
The RevMan version 5.2 was used to combine the data. The plots show a summary estimate of the results from all the studies combined. The size of the squares represents the estimate from each study, reflecting the statistical 'weight' of the study. Outcomes are provided as forest plots with diamonds representing the estimate of the pooled effect and the width of diamond implying its precision. The line of no effect is number one for binary outcomes, which depicts statistical significance if not crossed by the diamond 46. HR > 1.0 implies worse survival for patients with high MET CNG.
The possibility of publication bias was assessed with the Egger's test and visual inspection of the funnel plot 47,48. Statistical significance was considered for a p-value of less than 0.05 for summary HR and publication biases.
Results
Results of search
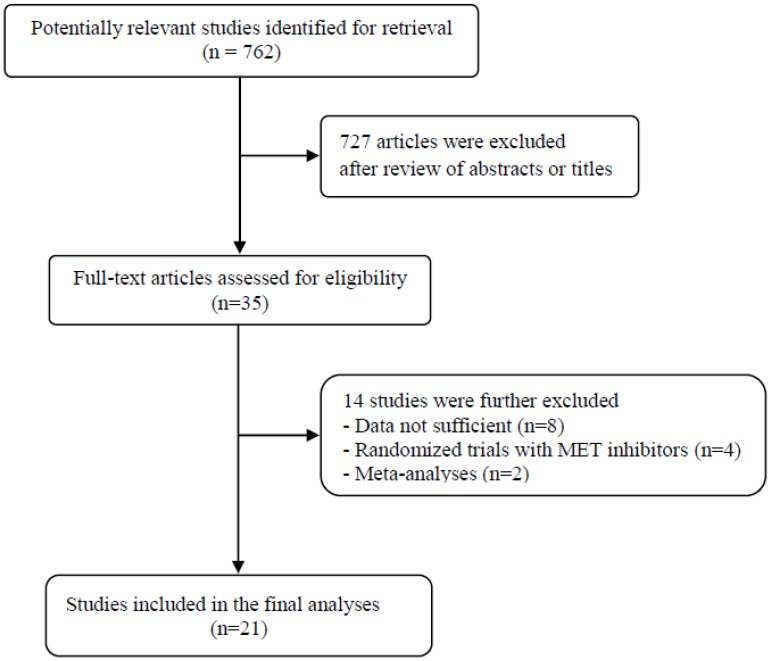
A total of 762 potentially relevant articles were initially found, but 727 of them were excluded after careful screening of the titles and abstracts. Of the remaining 35 potentially eligible studies, 14 were further excluded by the inclusion criteria. Finally, 21 studies were included in the meta-analysis 19-39. Figure 1 shows the search flow diagram of this meta-analysis.
Figure 1.
Flow diagram of search process
Characteristics of the included studies
Table 1 summarizes the main characteristics and clinical outcomes of the eighteen studies. Except for two with a prospective cohort 26,32, most were retrospective studies. The detection method and cut-off criteria for high MET CNG were variable across studies: FISH were most commonly used 20,21,23,24,26,27,32-37, but various cut-off criteria were adopted 19-39. Frequencies of high MET CNG in the included studies ranged from 1% to 38.9% depending on the technique used and the cut-point for positivity. Twelve studies were conducted in Asian populations and six were performed in non-Asian countries. Thirteen studies contained NSCLC of all histological subtypes, while 7 studies involved either adenocarcinoma (ADC) 22,24,26,30,32,33 or squamous cell carcinoma (SQCC) 21. One study involved patients with stage I ADC 30, and another one involved patients with advanced (stage III-IV) ADC harboring EGFR mutations 32. While patients with MET CNG had a better OS in one study 34, seven studies reported high MET CNG as an adverse prognostic factor 20,21,29,30,32,37,39. The remaining 13 studies failed to observe statistically significant impact of high MET CNG on survival, irrespective of whether these studies used univariate or multivariate analyses.
Table 1.
Summary of the 21 included studies
| First author (year) [ref] |
Country | No. of pts | Inclusion period | Stage | Histology | Method | Cut-off | No. of pts with high MET CNG (%) |
HR for DFS (95% CI) |
HR for OS (95% CI) |
Adjusted variables |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Okuda (2008) 19 |
Japan | 213 | 2003-2007 | I-IV | NSCLC | qPCR | > 3 copies | 12 (5.6%) | NA | 1.88 (0.93-3.81) p = 0.081 |
Age, sex, smoking, histology, stage, differentiation, EGFR mutations |
| Cappuzzo (2009) 20 |
Italy | 447 | 2000-2004 | I-IV | NSCLC | FISH | Cappuzzo system | 48 (11.1%) | NA | 1.52 (1.01-2.28) p = 0.04 |
Sex, smoking, histology, stage, grade, EGFR-FISH |
| Go (2010) 21 |
Korea | 97 | 1995-2000 | I-IV | SQCC | FISH | Cappuzzo system or UCCC criteria |
8 (8.2%) or 30 (30.9%) |
NA | 3.37 (1.38-8.23) P = 0.008 |
Age, sex, smoking, stage, EGFR-FISH |
| Onitsuka (2010) 22 |
Japan | 183 | 2003-2007 | IA-IIIB | ADC | qPCR | ≥1.31 copies | 8 (4.4%) | NA | 2.0 (0.85-4.72) p = 0.112 |
Age, sex, stage (T, N), EGFR or KRAS mutations, p-Met or HGF expression |
| Chen (2011) 23 |
Taiwan | 208 | 1996-2004 | I-IV | NSCLC | FISH | ≥3 copies | 22 (10.9%) | NA | 1.84 (0.81-4.19) p = 0.147 |
Age, sex, smoking, histology, stage, EGFR copy, EGFR or KRAS mutations |
| Tachibana (2012) 24 |
Japan | 106 | 2001-2008 | I-III | ADC | FISH | > 3 copies | 11 (10.4%) | NA | 1.79 (0.35-9.0) p = 0.483 |
Vascular or pleural invasion, lymphatic permeation, nuclear grade, LN status, MET or HGF expression |
| Tsuta (2012) 25 |
Japan | 844 |
1997-2007 | I-IV | NSCLC | BISH | UCCC criteria | 92 (10.9%) | NA | 1.295 (0.92-1.82) p = 0.135 |
Univariate |
| Tanaka (2012) 26 |
Japan | 138 | 2004-2009 | I-IV | ADC | FISH | Cappuzzo system or MET/CEP7≥2.0 |
21 (15.2%) or 6 (4.3%) |
3.28 (1.02-10.53) p = 0.05 |
1.60 (0.21-12.16) | Univariate |
| Park (2012) 27 |
Korea | 380 | 1994-2001 | I-IV | NSCLC | FISH | Cappuzzo system or UCCC criteria |
27 (7.1%) or 42 (11.1%) |
NA | 1.11 (0.7-1.76) p = 0.675 * ADC: 1.56 (0.66-3.69) * SQCC: 1.0 (0.45-2.24) |
Age, sex, smoking, histology, stage, MET or EGFR IHC, EGFR- FISH |
| Dziadziuszko (2012) 28 |
USA | 140 | NA | I-IV | NSCLC | SISH | Cappuzzo system | 14 (10%) | 0.88 (0.72-1.07) p = 0.19 |
0.90 (0.73-1.10) p = 0.29 |
Demographic and clinical features (not specified) |
| Sun (2013) 29 |
China | 61 | 2004-2008 | I-IV | NSCLC | qPCR | > 3 copies | 11 (18%) | NA | 9.49 (2.5-35.85) p = 0.001 |
Stage, MET expression |
| Jin (2014) 30 |
Korea | 141 | 2003-2009 | I | ADC | SISH | ≥3.4 copies | 34 (24.1%) | 5.474 (2.16-13.85) p < 0.001 |
NA | NA |
| Kowalczuk (2014) 31 |
Poland | 151 | 2003-1020 | I-IIIA | NSCLC | qPCR | > 3 copies | 28 (18.5%) | 1.21 (0.92-1.59) p = 0.175 |
1.03 (0.76-1.40) p = 0.849 * ADC: 1.17 (0.79-1.74) * SQCC: 0.65 (0.35-1.20) |
Age, sex, smoking, stage, LN status, MET mRNA in tumor, Log2 (MET mRNA RQ), MET mRNA RQ |
| Noro (2015) 32 |
Japan | 35 | 2008-2010 | III-IV | ADC with EGFR mutations | FISH | Colorado criteria | 11 (31.4%) | NA | 2.25 (1.07-4.74) p = 0.03 |
Univariate |
| Park (2015) 33 |
Korea | 316 | 2004-2011 | I-IV | ADC | FISH | UCCC criteria | 123 (38.9%) | NA | 1.32 (0.88-1.98) p = 0.183 |
Age, differentiation, LN status, stage, EGFR-TKI, MET IHC, MET amplification |
| Inoue (2015) 34 |
Japan | 240 | 1990-2011 | I-III | NSCLC | FISH | MET/CEP7 >2.0 | 28 (15.8%) | NA | 1.32 (0.55-3.17) | Univariate |
| Tran (2016) 35 |
Australia | 300 | 1994-2002 2010-2012 |
I-III | NSCLC | FISH | UCCC criteria | 22 (8.1%) | NA | 0.35 (0.15-0.8) p < 0.05 |
Stage, vessel or lymphatic invasion, perineural invasion, histology, grade, MET IHC |
| Song (2016) 36 |
China | 791 | 2011-2014 | I-IV | NSCLC without EGFR mutations | FISH | MET/CEP7 >2.2 | 8 (1%) | NA | 1.97 (0.62-6.3) p = 0.251 |
Age, sex, stage, smoking, histology, MET expression |
| Tong (2016) 37 |
China (Hong Kong) |
687 | 1995-2011 | I-IV | NSCLC | FISH | MET/CEP7 ≥5 | 8 (1.2%) | NA | 3.44 (1.4-8.48) p = 0.007 |
Age,sex, smoking, stage, LN status, tumor size, MET mutation, MET IHC, EGFR mutations |
| Bubendorf (2017) 38 |
Europe | 1572 | NA | I-III | NSCLC | SISH | Cappuzzo system | 65 (4.1%) | 0.86 (0.53-1.40) | 0.84 (0.47-1.49) | Univariate |
| Al-Saad (2017) 39 |
Norway | 298 | 1990-2004 | I-III | NSCLC | SISH | > 3 copies | 18 (6%) | NA | 2.29 (1.21-4.35) p = 0.011 |
Differentiation, performance status, stage, |
NSCLC, non-small-cell lung cancer; ADC, adenocarcinoma; SQCC, quamous cell carcinoma; EGFR, epidermal growth factor receptor; FISH, fluorescence in situ hybridization; SISH, silver in situ hybridization; BISH, bright-field in situ hybridization; qPCR, real-time, quantitative polymerase chain reaction; pts, patients; HR, hazard ratio; CI, confidence interval; DFS, disease-free survival; OS, overall survival; IHC, immunohistochemistry; LN, lymph node; NA, not available; Cappuzzo system: MET FISH-positive group was defined by mean MET gene copy number ≥5 copies per cell; UCCC(University of Colorado Cancer Center) criteria: MET status classified into two groups according to the frequency of tumor cells with specific copy numbers of the MET gene and chromosome 7 centromere (FISH-positive: MET/CEP ratio ≥ 2; > 15 copies of MET signals in > 10% of tumor cells; small gene cluster [4-10 copies]; or innumerable tight gene clusters in > 10% of tumor cells); Colorado criteria: MET/CEP ratio ≥ 2 and mean MET per cell ≥ 5 copies.
Prognostic impacts of MET CNG
Impact of high MET CNG on DFS
From five studies 26,28,30,31,38, a total of 2,141 patients were included in the meta-analysis of HRs for DFS. The random-effects model was selected because there was significant heterogeneity among studies (X2 = 20.46, p = 0.0004, I2= 80%). There was no significant correlation between high MET CNG and DFS (HR = 1.37, 95% CI: 0.88-2.12, p = 0.16) (Data not shown).
Impact of high MET GCN on OS
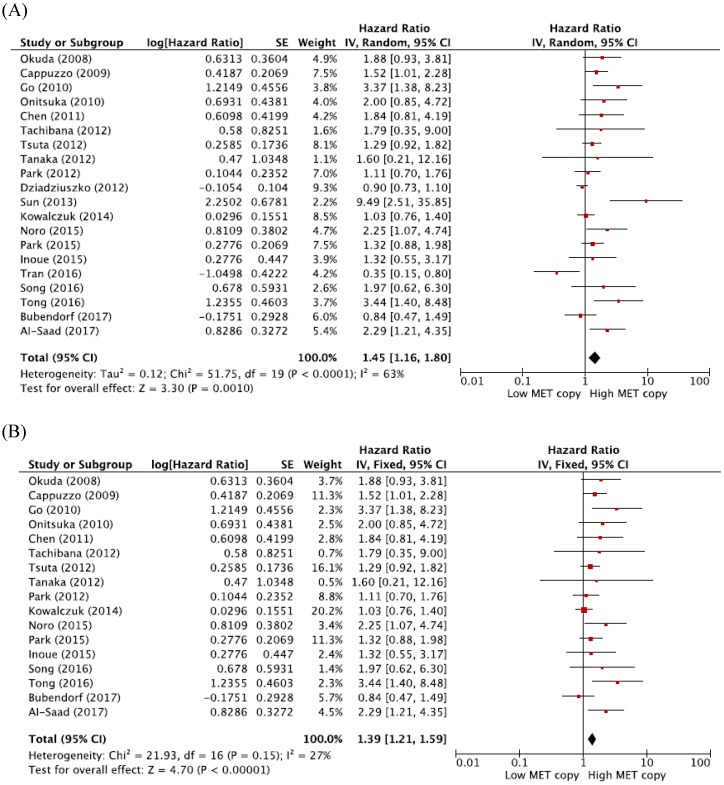
From 20 studies 19-29, 31-39, a total of 7,506 patients were included in the meta-analysis of HRs for OS. Compared with patients with NSCLC showing low MET CNG, patients with tumors harboring high MET CNG showed significantly worse survival (HR = 1.45, 95% CI: 1.16-1.80, p = 0.001) (Figure 2A). There was a significant heterogeneity among studies (X2 = 51.75, p < 0.0001, I2= 63%). Further analyses indicated that the observed heterogeneity was associated with the inclusion of the three studies by Dziadziuszko et al. 28, Sun et al. 29, and Tran et al. 35. When excluding these studies from the meta-analysis for OS, less heterogeneity was observed (X2 = 21.93, p = 0.15, I2= 27%) and the pooled results remained significant (HR = 1.39, 95% CI: 1.21-1.59, p < 0.0001) (Figure 2B)
Figure 2.
Meta-analysis of hazard ratios for overall survival. (A) Forest plot of 20 studies. (B) Forest plot of 17 studies excluding 3 with a significant heterogeneity.
Subgroup-analyses according to the histology and ethnicity
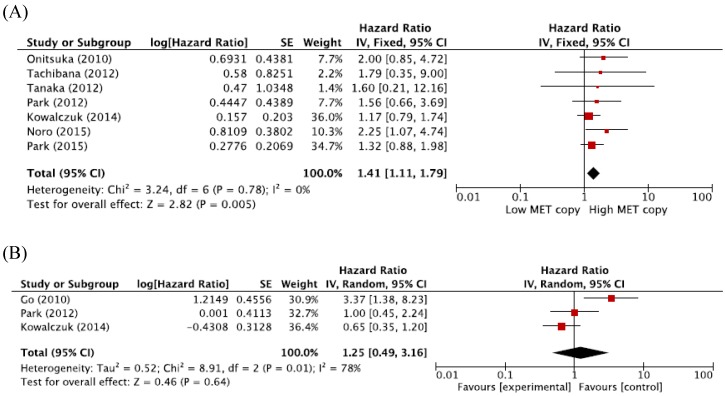
When we pooled HRs for OS according to histological subtypes (ADC or SQCC), the combined HRs were 1.41 (95% CI: 1.11-1.79, p = 0.005, fixed-effect model) for ADC and 1.25 (95% CI: 0.49-3.16, p = 0.64, random-effects model) for SQCC (Figure 3A and 3B).
Figure 3.
Subgroup meta-analysis of hazard ratios for overall survival according to the histology. (A) Forest plot of patients with adenocarcinoma. (B) Forest plot of patients with squamous cell carcinoma.
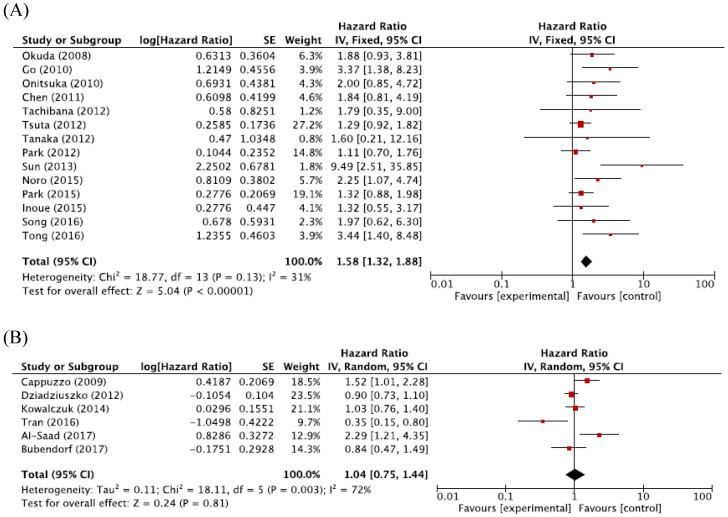
For Asian populations, high MET CNG was significantly associated with a worse prognosis (HR = 1.58, 95% CI: 1.32-1.88, p < 0.00001, fixed-effect model), but this significant correlation was not observed for non-Asian populations (HR = 1.04, 95% CI: 0.75-1.44, p = 0.81, random-effects model) (Figure 4A and 4B).
Figure 4.
Figure 5: Subgroup meta-analysis of hazard ratios for overall survival according to the ethnicity. (A) Forest plot of Asian patients. (B) Forest plot of non-Asian patients.
Publication bias
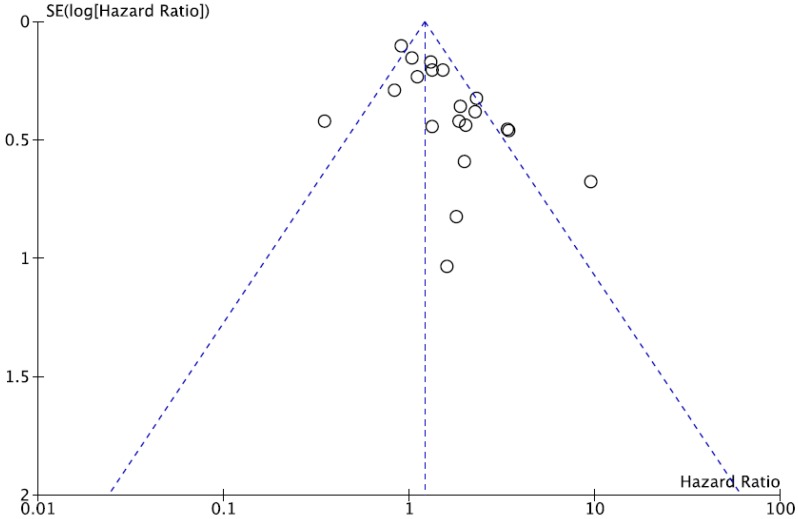
We did not perform publication bias test for DFS because there was a small number of studies included. Begg's funnel plot and Egger's test indicated no evidence of substantial publication bias for OS (Begg's p = 0.113, Egger's p = 0.148) (Figure 5).
Figure 5.
Funnel plot for publication bias regarding overall survival.
Discussion
The MET signaling pathway has recently emerged as a potential molecular target for the treatment of NSCLC. However, the survival impact of MET amplification is still controversial. In the current meta-analysis, we evaluated the prognostic value of MET CNG in patients with NSCLC.
Out of the included 21 studies, only five reported the data regarding DFS. The pooled analysis failed to show a significant correlation between high MET CNG and DFS (HR = 1.37, p = 0.16). Because a limited number of studies were included in the meta-analysis and there was a significant heterogeneity among them (I2 = 80%), our finding seemed not sufficient to determine the correlation between high MET CNG and DFS in patients with resected NSCLC. In terms of OS, however, the meta-analysis revealed that high MET CNG was associated with a worse prognosis (HR = 1.45, p = 0.001).
Subgroup analysis in the current study showed that high MET CNG significantly correlated with shorter survival especially in patients with ADC (HR = 1.41, p = 0.005). Because the subgroup analysis included a limited number of studies (only 3) in the SQCC group and there was a significant heterogeneity across them (I2 = 78%), however, the prognostic significance of MET CNG in NSCLC according to the histology is still unclear. Go et al. reported that tumors with high MET CNG showed a significantly worse prognosis than those without MET amplification (HR = 3.37, p = 0.008) in NSCLC patients with SQCC 21. These results suggest that MET amplification may be involved in the carcinogenesis of both ADC and SQCC in NSCLC. Subgroup analysis also showed a different prognostic impact of MET CNG between Asian (HR = 1.58, p < 0.00001) and non-Asian populations (HR = 1.04, p = 0.81). The effect of dual positivity for MET amplification and EGFR mutations might account for the different impacts of high MET CNG on the prognosis between Asian and non-Asian populations. However, the subgroup analysis included only 6 studies in non-Asian subset and there was a significant heterogeneity (I2 = 72%) among them. Therefore, these data seemed not sufficient to determine the prognostic value of high MET CNG in non-Asian populations. Further studies are still needed to verify the prognostic role of MET amplification according to the histology or ethnicity in patients with NSCLC.
There has been a strong rationale for the development of MET inhibitors in NSCLC 49. Increased MET signaling is linked to a worse prognosis for NSCLC 14-17. Moreover, MET amplification has been described as a mechanism of acquired resistance to EGFR-TKIs in EGFR-mutant NSCLC 40-42. The survival efficacy of MET inhibitors (onartuzumab and tivantinib) has been controversial, even in patients with NSCLC showing MET overexpression 50-57. A couple of randomized trials reported that the addition of a MET inhibitor to erlotinib, an EGFR-TKI, was associated with improved survival in NSCLC patients with MET overexpression 50,56.
However, other randomized studies failed to show survival benefit of adding a MET inhibitor to first-line platinum-based chemotherapy or erlotinib in patients with high MET expression 51-53,55. In the METLung study of 499 patients with NSCLC showing high MET expression (MET immunohistochemistry 2+ or 3+), especially, the addition of onartuzumab to erlotinib failed to prolong OS over placebo (HR = 1.27, p = 0.067) 51. These results suggest that MET protein status may not be the optimal predictor for MET inhibitors in NSCLC. MET overexpression is a late event consecutive to the transformed phenotype, deriving from transcriptional up-regulation of MET in absence of gene amplification or mutations or ligand-dependent autocrine or paracrine mechanism 10-12. Thus, targeting MET pathway in the tumors with MET overexpression as a late event would probably not draw survival benefit. Therefore, there is a need to identify more specific biomarkers for defining the subset of patients benefiting from MET targeted agents. Emerging data have suggested that splice-site mutations (MET exon 14) or MET amplification lead to significant responses to MET inhibitors 54,58-61. In a randomized phase II study of erlotinib plus tivantinib or placebo in patients with previously treated NSCLC, the combination of erlotinib and tivantinib showed a consistent trend toward benefit in the small cohort with increased MET CNG 54. In EGFR mutant NSCLC with showing acquired resistance to EGFR TKIs, the observed response rate of combining an EFGR inhibitor and a MET inhibitor (INC280) was 0% among patients with a mean MET/cell of < 5 and 40% among those with mean MET/cell ≥ 5 60. In addition, a recent phase I study of crizotinib (a MET/ALK/ROS1 inhibitor) in 14 patients with MET amplification reported a 0%, 17%, and 67% response rate in the low- (1.8 ≤ MET/CEP ratio ≤ 2.2), intermediate- (2.2 < MET/CEP ratio < 5), and high-MET amplification (MET/CEP ratio ≥ 5) groups, respectively 61. Thus, MET amplification may represent a better predictive biomarker for MET targeted therapies, especially MET TKIs. However, determining the appropriate measure and threshold for MET amplification is one of the major challenges for clinical development of MET targeted therapies 62.
A variety of methods, such as qPCR, FISH, SISH, and BISH, are currently used for assessing MET amplification 43,44,62. There are no standardized methods for detecting high MET CNG. In this meta-analysis, FISH was the most commonly used technique for determining MET CNG. FISH can be applied to formalin-fixed paraffin-embedded tumor tissues, thus facilitating the exclusive estimation of tumor cells 62. However, the cut-off criteria for high MET CNG are various across studies 20,43,44,62,63. The discrepancies in the prognostic value of MET CNG among studies might be attributable to the different cut-off points for defining MET amplification. Therefore, the definition of a reliable guideline for high MET CNG is an essential prerequisite for assessing the prognostic role of MET amplification and developing MET inhibitors in NSCLC.
Our study has several inherent limitations that needed to be discussed. First, the included studies showed considerable differences in the detection methods and cut-off criteria, types and numbers of target genes, tumor histology and stage, treatment, and other demographic or clinicopathological data. Second, there was a significant heterogeneity among the included studies (X2 = 51.75, p < 0.0001, I2 = 63%), which was primarily associated with the three studies by Dziadziuszko et al. 28, Sun et al. 29, and Tran et al. 35. Sun et al. 29 employed RT-PCR that used the same primers and similar PCR conditions as others 19,22,31. Tran et al. 36 employed FISH with the UCCC criteria 63, and Dziadziuszko et al. 28 used SISH that is a newly approved technology for gene copy assessment. Therefore, technical issue did not seem to address the heterogeneity. However, this variation might correlate with no standardized criteria for defining high MET CNG. Third, most studies were retrospectively performed and therefore might carry the biases of the retrospective design. Fourth, there already were two meta-analyses regarding the prognostic impact of MET CNG in NSCLC in the literature 43,44. However, they included a relatively small number of articles reported before 2014. Our meta-analysis included additional 10 studies published between 2014 and 2017 30-39, and most of them used more solid criteria for MET amplification. Finally, articles published only in English were included, which might bias the results.
In conclusion, our meta-analysis indicates that high MET CNG is an adverse prognostic factor in patients with NSCLC. Subgroup analysis suggests that high MET CNG is associated with a worse prognosis, especially in patients with ADC and Asian populations. However, large prospective studies using standardized methods based on the homogeneous populations are warranted to validate the prognostic value of MET amplification in patients with NSCLC.
References
- 1.Jung KW, Won YJ, Oh CM, Kong HJ, Lee DH, Lee KH. Community of Population-Based Regional Cancer Registries. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2014. Cancer Res Treat. 2017;49:292–305. doi: 10.4143/crt.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Blumenschein GR Jr, Mills GB, Gonzalez-Angulo AM. Targeting the hepatocyte growth factor-cMET axis in cancer therapy. J Clin Oncol. 2012;30:3287–96. doi: 10.1200/JCO.2011.40.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peters S, Adjei AA. MET: a promising anticancer therapeutic target. Nat Rev Clin Oncol. 2012;9:314–26. doi: 10.1038/nrclinonc.2012.71. [DOI] [PubMed] [Google Scholar]
- 5.Furge KA, Zhang YW, Vande Woude GF. Met receptor tyrosine kinase: enhanced signaling through adapter proteins. Oncogene. 2000;19(49):5582–9. doi: 10.1038/sj.onc.1203859. [DOI] [PubMed] [Google Scholar]
- 6.Rosário M, Birchmeier W. How to make tubes: signaling by the Met receptor tyrosine kinase. Trends Cell Biol. 2003;13:328–35. doi: 10.1016/s0962-8924(03)00104-1. [DOI] [PubMed] [Google Scholar]
- 7.Gentile A, Trusolino L, Comoglio PM. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 2008;27:85–94. doi: 10.1007/s10555-007-9107-6. [DOI] [PubMed] [Google Scholar]
- 8.Zhang YW, Su Y, Volpert OV, Vande Woude GF. Hepatocyte growth factor/scatter factor mediates angiogenesis through positive VEGF and negative thrombospondin 1 regulation. Proc Natl Acad Sci USA. 2003;100:12718–23. doi: 10.1073/pnas.2135113100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai AZ, Abella JV, Park M. Crosstalk in Met receptor oncogenesis. Trends Cell Biol. 2009;19:542–51. doi: 10.1016/j.tcb.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Kentsis A, Reed C, Rice KL, Sanda T, Rodig SJ, Tholouli E. et al. Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia. Nat Med. 2012;18:1118–22. doi: 10.1038/nm.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danilkovitch-Miagkova A, Zbar B. Dysregulation of Met receptor tyrosine kinase activity in invasive tumors. J Clin Invest. 2002;109:863–7. doi: 10.1172/JCI15418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pérol M. Negative results of METLung study: an opportunity to better understand the role of MET pathway in advanced NSCLC. Transl Lung Cancer Res. 2014;3:392–4. doi: 10.3978/j.issn.2218-6751.2014.09.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura Y, Niki T, Goto A, Morikawa T, Miyazawa K, Nakajima J. et al. c-Met activation in lung adenocarcinoma tissues: an immunohistochemical analysis. Cancer Sci. 2007;8:1006–13. doi: 10.1111/j.1349-7006.2007.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pyo JS, Kang G, Cho WJ, Choi SB. Clinicopathological significance and concordance analysis of c-MET immunohistochemistry in non-small cell lung cancers: a meta-analysis. Pathol Res Pract. 2016;212:710–6. doi: 10.1016/j.prp.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Masuya D, Huang C, Liu D, Nakashima T, Kameyama K, Haba R. et al. The tumour-stromal interaction between intratumoral c-Met and stromal hepatocyte growth factor associated with tumour growth and prognosis in non-small-cell lung cancer patients. Br J Cancer. 2004;90:1555–62. doi: 10.1038/sj.bjc.6601718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon D, Koh J, Kim S, Go H, Kim YA, Keam B. et al. MET exon 14 skipping mutation in triple-negative pulmonary adenocarcinomas and pleomorphic carcinomas: An analysis of intratumoral MET status heterogeneity and clinicopathological characteristics. Lung Cancer. 2017;106:131–7. doi: 10.1016/j.lungcan.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Casadevall D, Gimeno J, Clavé S, Taus Á, Pijuan L, Arumí M. et al. MET expression and copy number heterogeneity in nonsquamous non-small cell lung cancer (nsNSCLC) Oncotarget. 2015;6:16215–26. doi: 10.18632/oncotarget.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duan J, Yang X, Zhao J, Zhuo M, Wang Z, An T, Bai H, Wang J. Correlation among genetic variations of c-MET in Chinese patients with non-small cell lung cancer. Oncotarget. 2017;9:2660–7. doi: 10.18632/oncotarget.23474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuda K, Sasaki H, Yukiue H, Yano M, Fujii Y. Met gene copy number predicts the prognosis for completely resected non-small cell lung cancer. Cancer Sci. 2008;99:2280–5. doi: 10.1111/j.1349-7006.2008.00916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cappuzzo F, Marchetti A, Skokan M, Rossi E, Gajapathy S, Felicioni L. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol. 2009;27:1667–74. doi: 10.1200/JCO.2008.19.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Go H, Jeon YK, Park HJ, Sung SW, Seo JW, Chung DH. High MET gene copy number leads to shorter survival in patients with non-small cell lung cancer. J Thorac Oncol. 2010;5:305–13. doi: 10.1097/JTO.0b013e3181ce3d1d. [DOI] [PubMed] [Google Scholar]
- 22.Onitsuka T, Uramoto H, Ono K, Takenoyama M, Hanagiri T, Oyama T. et al. Comprehensive molecular analyses of lung adenocarcinoma with regard to the epidermal growth factor receptor, K-ras, MET, and hepatocyte growth factor status. J Thorac Oncol. 2010;5:591–6. doi: 10.1097/JTO.0b013e3181d0a4db. [DOI] [PubMed] [Google Scholar]
- 23.Chen YT, Chang JW, Liu HP, Yu TF, Chiu YT, Hsieh JJ. et al. Clinical implications of high MET gene dosage in non-small cell lung cancer patients without previous tyrosine kinase inhibitor treatment. J Thorac Oncol. 2011;6:2027–35. doi: 10.1097/JTO.0b013e3182307e92. [DOI] [PubMed] [Google Scholar]
- 24.Tachibana K, Minami Y, Shiba-Ishii A, Kano J, Nakazato Y, Sato Y. et al. Abnormality of the hepatocyte growth factor/MET pathway in pulmonary adenocarcinogenesis. Lung Cancer. 2012;75:181–8. doi: 10.1016/j.lungcan.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 25.Tsuta K, Kozu Y, Mimae T, Yoshida A, Kohno T, Sekine I. et al. c-MET/phospho-MET protein expression and MET gene copy number in non-small cell lung carcinomas. J Thorac Oncol. 2012;7:331–9. doi: 10.1097/JTO.0b013e318241655f. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka A, Sueoka-Aragane N, Nakamura T, Takeda Y, Mitsuoka M, Yamasaki F. et al. Co-existence of positive MET FISH status with EGFR mutations signifies poor prognosis in lung adenocarcinoma patients. Lung Cancer. 2012;75:89–94. doi: 10.1016/j.lungcan.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Park S, Choi YL, Sung CO, An J, Seo J, Ahn MJ. et al. High MET copy number and MET overexpression: poor outcome in non-small cell lung cancer patients. Histol Histopathol. 2012;27:197–207. doi: 10.14670/HH-27.197. [DOI] [PubMed] [Google Scholar]
- 28.Dziadziuszko R, Wynes MW, Singh S, Asuncion BR, Ranger-Moore J, Konopa K. et al. Correlation between MET gene copy number by silver in situ hybridization and protein expression by immunohistochemistry in non-small cell lung cancer. J Thorac Oncol. 2012;7:340–7. doi: 10.1097/JTO.0b013e318240ca0d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun W, Song L, Ai T, Zhang Y, Gao Y, Cui J. Prognostic value of MET, cyclin D1 and MET gene copy number in non-small cell lung cancer. J Biomed Res. 2013;27:220–30. doi: 10.7555/JBR.27.20130004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin Y, Sun PL, Kim H, Seo AN, Jheon S, Lee CT. et al. MET gene copy number gain is an independent poor prognostic marker in Korean stage I lung adenocarcinomas. Ann Surg Oncol. 2014;21:621–8. doi: 10.1245/s10434-013-3355-1. [DOI] [PubMed] [Google Scholar]
- 31.Kowalczuk O, Kozlowski M, Niklinska W, Kisluk J, Niklinska BJ, Niklinski J. Increased MET gene copy number but not mRNA level predicts postoperative recurrence in patients with non-small cell lung cancer. Transl Oncol. 2014;7:605–12. doi: 10.1016/j.tranon.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noro R, Seike M, Zou F, Soeno C, Matsuda K, Sugano T. et al. MET FISH-positive status predicts short progression-free survival and overall survival after gefitinib treatment in lung adenocarcinoma with EGFR mutation. BMC Cancer. 2015;15:31. doi: 10.1186/s12885-015-1019-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park S, Koh J, Kim DW, Kim M, Keam B, Kim TM. et al. MET amplification, protein expression, and mutations in pulmonary adenocarcinoma. Lung Cancer. 2015;90:381–7. doi: 10.1016/j.lungcan.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 34.Inoue Y, Matsuura S, Kurabe N, Kahyo T, Mori H, Kawase A. et al. Clinicopathological and Survival Analysis of Japanese Patients with Resected Non-Small-Cell Lung Cancer Harboring NKX2-1, SETDB1, MET, HER2, SOX2, FGFR1, or PIK3CA Gene Amplification. J Thorac Oncol. 2015;10:1590–600. doi: 10.1097/JTO.0000000000000685. [DOI] [PubMed] [Google Scholar]
- 35.Tran TN, Selinger CI, Kohonen-Corish MR, McCaughan B, Kennedy C, O'Toole SA. et al. Alterations of MET gene copy number and protein expression in primary non-small-cell lung cancer and corresponding nodal metastases. Clin Lung Cancer. 2016;17:30–8.e1. doi: 10.1016/j.cllc.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Song Z, Wang X, Zheng Y, Su H, Zhang Y. MET Gene Amplification and Overexpression in Chinese Non-Small-Cell Lung Cancer Patients Without EGFR Mutations. Clin Lung Cancer. 2017;18:213–9.e2. doi: 10.1016/j.cllc.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 37.Tong JH, Yeung SF, Chan AW, Chung LY, Chau SL, Lung RW. et al. MET Amplification and Exon 14 Splice Site Mutation Define Unique Molecular Subgroups of Non-Small Cell Lung Carcinoma with Poor Prognosis. Clin Cancer Res. 2016;22:3048–56. doi: 10.1158/1078-0432.CCR-15-2061. [DOI] [PubMed] [Google Scholar]
- 38.Bubendorf L, Dafni U, Schöbel M, Finn SP, Tischler V, Sejda A. et al. Prevalence and clinical association of MET gene overexpression and amplification in patients with NSCLC: Results from the European Thoracic Oncology Platform (ETOP) Lungscape project. Lung Cancer. 2017;111:143–9. doi: 10.1016/j.lungcan.2017.07.021. [DOI] [PubMed] [Google Scholar]
- 39.Al-Saad S, Richardsen E, Kilvaer TK, Donnem T, Andersen S, Khanehkenari M. et al. The impact of MET, IGF-1, IGF1R expression and EGFR mutations on survival of patients with non-small-cell lung cancer. PLoS One. 2017;12(e):0181527. doi: 10.1371/journal.pone.0181527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E. et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO. et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 42.Ninomiya K, Ohashi K, Makimoto G, Tomida S, Higo H, Kayatani H. et al. MET or RAS amplification is an acquired resistance mechanism to the third-generation EGFR inhibitor aquotinib. Sci Rep. 2018;8:1955. doi: 10.1038/s41598-018-20326-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo B, Cen H, Tan X, Liu W, Ke Q. Prognostic value of MET gene copy number and protein expression in patients with surgically resected non-small cell lung cancer: a meta-analysis of published literatures. PLoS One. 2014;9:e99399. doi: 10.1371/journal.pone.0099399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dimou A, Non L, Chae YK, Tester WJ, Syrigos KN. MET gene copy number predicts worse overall survival in patients with non-small cell lung cancer (NSCLC); a systematic review and meta-analysis. PLoS One. 2014;9:e107677. doi: 10.1371/journal.pone.0107677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panic N, Leoncini E, de Belvis G, Ricciardi W, Boccia S. Evaluation of the endorsement of the preferred reporting items for systematic reviews and meta-analysis (PRISMA) statement on the quality of published systematic review and meta-analyses. PLoS One. 2013;8:e83138. doi: 10.1371/journal.pone.0083138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wald NJ, Bestwick JP. Presentation of meta-analysis plots. J Med Screen. 2015;22:49–51. doi: 10.1177/0969141314556490. [DOI] [PubMed] [Google Scholar]
- 47.Eagger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sterne JA, Sutton AJ, Ioannidis JP, Terrin N, Jones DR, Lau J. et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ. 2011;343:d4002. doi: 10.1136/bmj.d4002. [DOI] [PubMed] [Google Scholar]
- 49.Landi L, Minuti G, D'Incecco A, Salvini J, Cappuzzo F. MET overexpression and gene amplification in NSCLC: a clinical perspective. Lung Cancer (Auckl) 2013;4:15–25. doi: 10.2147/LCTT.S35168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spigel DR, Ervin TJ, Ramlau RA, Daniel DB, Goldschmidt JH Jr, Blumenschein GR Jr. et al. Randomized phase II trial of onartuzumab in combination with erlotinib in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2013;31:4105–14. doi: 10.1200/JCO.2012.47.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spigel DR, Edelman MJ, O'Byrne K, Paz-Ares L, Mocci S, Phan S. et al. Results from the phase III randomized trial of onartuzumab plus erlotinib versus erlotinib in previously treated stage IIIB or IV non-small-cell lung cancer: METLung. J Clin Oncol. 2017;35:412–20. doi: 10.1200/JCO.2016.69.2160. [DOI] [PubMed] [Google Scholar]
- 52.Hirsch FR, Govindan R, Zvirbule Z, Braiteh F, Rittmeyer A, Belda-Iniesta C. et al. Efficacy and safety results from a phase II, placebo-controlled study of onartuzumab plus first-line platinum-doublet chemotherapy for advanced squamous cell non-small-cell lung cancer. Clin Lung Cancer. 2017;18:43–9. doi: 10.1016/j.cllc.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 53.Wakelee H, Zvirbule Z, De Braud F, Kingsley CD, Mekhail T, Lowe T. et al. Efficacy and safety of onartuzumab in combination with first-line bevacizumab- or pemetrexed-based chemotherapy regimens in advanced non-squamous non-small-cell lung cancer. Clin Lung Cancer. 2017;18:50–9. doi: 10.1016/j.cllc.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 54.Sequist LV, von Pawel J, Garmey EG, Akerley WL, Brugger W, Ferrari D. et al. Randomized phase II study of erlotinib plus tivantinib versus erlotinib plus placebo in previously treated non-small-cell lung cancer. J Clin Oncol. 2011;29:3307–15. doi: 10.1200/JCO.2010.34.0570. [DOI] [PubMed] [Google Scholar]
- 55.Yoshioka H, Azuma K, Yamamoto N, Takahashi T, Nishio M, Katakami N. et al. A randomized, double-blind, placebo-controlled, phase III trial of erlotinib with or without a c-Met inhibitor tivantinib (ARQ 197) in Asian patients with previously treated stage IIIB/IV nonsquamous nonsmall-cell lung cancer harboring wild-type epidermal growth factor receptor (ATTENTION study) Ann Oncol. 2015;26:2066–72. doi: 10.1093/annonc/mdv288. [DOI] [PubMed] [Google Scholar]
- 56.Scagliotti G, von Pawel J, Novello S, Ramlau R, Favaretto A, Barlesi F. et al. Phase III multinational, randomized, double-blind, placebo-controlled study of tivantinib (ARQ 197) plus erlotinib versus erlotinib alone in previously treated patients with locally advanced or metastatic nonsquamous non-small-cell lung cancer. J Clin Oncol. 2015;33:2667–74. doi: 10.1200/JCO.2014.60.7317. [DOI] [PubMed] [Google Scholar]
- 57.Kim JH, Kim HS, Kim BJ. MET inhibitors in advanced non-small-cell lung cancer: a meta-analysis and review. Oncotarget. 2017;8:75500–8. doi: 10.18632/oncotarget.20824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paik PK, Drilon A, Fan PD, Yu H, Rekhtman N, Ginsberg MS. et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 2015;5:842–9. doi: 10.1158/2159-8290.CD-14-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smolen GA, Sordella R, Muir B, Mohapatra G, Barmettler A, Archibald H. et al. Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752. Proc Natl Acad Sci USA. 2006;103:2316–21. doi: 10.1073/pnas.0508776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bang YJ, Su WC, Nam DH, Lim WTx, Wan-Teck Lim. et al. Phase I study of the safety and efficacy of INC280 in patients with advanced MET-dependent solid tumors. J Clin Oncol. 2014;32(Suppl 15):2520. [Google Scholar]
- 61.Camidge DR, Ou S-HI, Shapiro G, Otterson GAx. et al. Efficacy and safety of crizotinib in patients with advanced c-MET-amplified non-small cell lung cancer. J Clin Oncol. 2014;32(Suppl 15):8001. [Google Scholar]
- 62.Noonan SA, Berry L, Lu X, Gao D, Barón AE, Chesnut P. et al. Identifying the appropriate FISH criteria for defining MET copy number-driven lung adenocarcinoma through oncogene overlap analysis. J Thorac Oncol. 2016;11:1293–304. doi: 10.1016/j.jtho.2016.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Varella-Garcia M. Stratification of non-small cell lung cancer patients for therapy with epidermal growth factor receptor inhibitors: the EGFR fluorescence in situ hybridization assay. Diagn Pathol. 2006;1:19. doi: 10.1186/1746-1596-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]