Abstract
Circular RNA (circRNA) is associated with human cancers, however, few studies have reported its value in the diagnosis and prognosis prediction of osteosarcoma (OS). In this study, we investigated the expression level of eight selected cancer-related circRNAs including circ-Cdr1as, circ_HIPK3 and circ-ITCH in OS cell lines, tissues and plasmas by quantitative real-time polymerase chain reaction (qRT-PCR) and found that only circ_HIPK3 could stably down-regulate in the OS cell lines, tissues and plasmas than the corresponding controlled. One-way analysis of variance was further conducted to analyze the relationship between circ_HIPK3 expression level and clinic pathological factors of OS patients. Receiver operating characteristic (ROC) curve was built to evaluate the diagnostic values of circ_HIPK3. Circ_HIPK3 expression was significantly correlated with Enneking stage (P=0.042) and lung metastasis (P=0.036). The area under the ROC curve was 0.783 and the sensitivity and specificity were 0.56 and 0.84, respectively. Kaplan-Maier analysis also showed that lower expression of circ_HIPK3 correlated with shorter overall survival time and poor prognosis of OS patients. Besides, function analysis demonstrated that circHIPK3 overexpression significantly suppressed OS cell proliferation, migration and invasion in vitro. Overall, our data suggest that circ_HIPK3 may become a novel potential biomarker for diagnosis and treatment target of OS.
Keywords: circRNA, circ_HIPK3, osteosarcoma, biomarker, progression
Introduction
Osteosarcoma (OS) is the most common type of primary bone cancer in childhood and adolescence1. Although wide tumor excision combined with the adjuvant chemotherapy and radiotherapy is effective, the cure rate of osteosarcoma patients remain dismal because of the metastatic lesions to lung and multi-drug resistance2. The poor prognosis of osteosarcoma is partially due to the lack of a better molecular biomarker to detect it at early stage3. Therefore, identification of clinically relevant biomarkers for diagnosis and prognosis is therefore urgently needed for OS patients.
Circular RNA (circRNA) is a new class of endogenously expressed non-coding RNA, which is characterized by covalently closed loop structures with neither 5' to 3' polarity nor polyadenylated tail4. They are common in mammalian cells and regulate gene expression at the transcriptional or post-transcriptional level by interacting with microRNAs (miRNAs) or other molecules 5-7. Recently, circRNAs have been reported to mediate cancer progression and used to be biomarkers for the diagnosis and prognosis of cancer8. Xuan et al9 found that hsa_circRNA_100855 and hsa_circRNA_104912 could serve as novel and stable biomarkers for the diagnosis and progress of laryngeal squamous cell cancer tissues (LSCC). Wang et al10 reported that hsa_circ_001988 was down-regulated in colorectal cancer tissues and may become a novel potential biomarker in the diagnosis and treatment of colorectal cancer. Qin et al11 found that hsa_circ_0001649 was significantly down-regulated in hepatocellular carcinoma (HCC) and might serve as a novel potential biomarker for HCC.
However, there is seldom report about circRNA as biomarker to predict diagnosis and prognosis in OS. In the current study, we examined the expression level of eight reported cancer-related circRNAs such as circ-Cdr1as12-15, circHIPK316-18, circ-sirt719 and so on, in OS cell lines, tissues and plasmas by real-time quantitative reverse transcription-polymerase chain reactions (qRT-PCRs). Then tests were performed between clinical information and circRNA expression level by analysis of variance (ANOVA) or t-test, Kaplan-Maier analysis and a receiver operating characteristics (ROC) curve was established to estimate the value of circRNA expression as a biomarker in OS. Besides, the molecular function of circHIPK3 in OS progression in vitro was also demonstrated.
Material and method
Clinical specimens
A total of 82 primary osteosarcoma patients who received the same chemotherapy regimen before surgery and underwent complete resection surgery at Shanghai Tenth People's Hospital between 2006 and 2016 were included in this study. The study was approved by the Ethics Committee of Shanghai Tenth People's Hospital, and written informed consent was obtained from all the patients. All patients' slides were reviewed to confirm the diagnosis and to classify the tumor according to Enneking Stage. Patients with evidence of other malignancies or diseases were excluded from this study. Besides, we collected blood samples from 50 patients with osteosarcoma before surgery, other 10 patients with benign bone tumor (four cases of osteoclastoma, six cases of fibrous dysplasia) and 20 age- and sex-matched healthy individuals as the control group. All the resected specimens were placed immediately into liquid nitrogen and stored at - 80 ℃. The plasma extracted from the blood samples were collected using standard procedures. The clinical parameters of osteosarcoma patients in this study are shown in Table 1.
Table 1.
Clinical parameters of osteosarcoma patients enrolled in this study
| Pathological characteristics | Cases (n) | circHIPK3 expression | P value | |
|---|---|---|---|---|
| High (37) | Low (45) | |||
| Gender | ||||
| Male | 49 | 22 | 27 | 0.84 |
| Female | 33 | 15 | 18 | |
| Age | ||||
| ≥25 | 24 | 11 | 13 | 0.63 |
| <25 | 58 | 26 | 32 | |
| Location | 0.45 | |||
| Distal of Femur | 38 | 16 | 22 | |
| Proximal of Tibia | 28 | 12 | 16 | |
| Other | 16 | 9 | 7 | |
| Lung Metastasis | 0.036 | |||
| Yes | 26 | 5 | 21 | |
| No | 56 | 32 | 24 | |
| Enneking stage | 0.042 | |||
| I+IIA | 26 | 16 | 10 | |
| IIB/III | 56 | 21 | 35 | |
Cell culture
Six human osteosarcoma cell lines (SaoS2, HOS, KH-OS, MG63, 143B and U2-OS) were purchased from American Type Culture Collection and cultured in DMEM supplemented with 10% FBS (Gibco, Gran Island, NY, USA), 100 U/mL of penicillin and 100 mg/mL of streptomycin (Invitrogen). Cultures were maintained at 37°C in a humidified CO2 (5%) atmosphere. Normal osteoblast cells (hFOB1.19) obtained from the Chinese Cell Bank of the Chinese Academy of Sciences (Shanghai, China) were cultured in Ham's F12/ DMEM supplemented with 10% FBS, 100 U/mL penicillin and 100 mg/mL streptomycin. Cultures were maintained at 33.5°C in a humidified CO2 (5%) atmosphere.
RNA isolation
Total RNA from each specimen was quantified using a NanoDrop ND-1000 spectrophotometer (OD 260 nm, NanoDrop, Wilmington, DE, USA). RNA integrity was assessed using standard denaturing agarose gel electrophoresis, and the purity was estimated by the ratio of absorbance at 260 to 280 nm (A260/A280).
Quantitative real-time reverse transcription polymerase chain reaction
qRT-PCR, the gold standard for data verification, was used to verify the differential expression of circRNAs that were detected on the microarray. Total RNA was extracted and 1 mg of total RNA was converted to cDNA with Primer Script RT Mix (Takara, Dalian, China). PCR was performed with the SYBR Green PCR kit (Takara) on an ABI PRISM 7500 fast Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Reaction parameters were: 95°C for 20 s, then 40 cycles of 95°C for 10 s and 60°C for 45 s. Relative gene expression was calculated with the 2/(DDCT) method, using GAPDH as an internal control. Primers used for amplifying specific genes in this study are shown in Table 2.
Table 2.
Primers used for PCR validation
| Gene | Forward and Reverse primer |
|---|---|
| hsa_circ_001569 | F:5'TCCCCTGAACATTCTCCCCAT3' |
| R:5'GAAAGCACTTGGTGAAGTCGG3' | |
| circ-Cdr1as | F:5'GTGTCTCCAGTGTATCGGCG3' |
| R:5'TACTGGCACCACTGGAAACC3' | |
| circHIPK3 | F:5'TGGAGACTGGGGGAAGATGA3' |
| R:5'CACACTAACTGGCTGAGGGG3' | |
| cir-ITCH | F:5'ACAGAGACAACCGAGAAACAGTG3' |
| R:5'GCCTTGATACTTGTTACCGTCGA3' | |
| circ-Foxo3 | F:5'GTGGGGAACTTCACTGGTGCTAAG3' |
| R:5'GGGTTGATGATCCACCAAGAGCTCTT3' | |
| circSMARCA5 | F:5'CTCCAAGATGGGCGAAAG3' |
| R:5'TGTGTTGCTCCATGTCTAATCA3' | |
| circ-sirt7 | F:5'GGAGCCATCGGCGGGGCTCA3' |
| R:5'CTGGTCCCTGGCAAGTGTA3' | |
| circ-mcm5 | F:5'CTCAGGCTGTTCTTCCTTGG3' |
| R:5'CCAGGGAACCCAGATCAAT3' | |
| GAPDH | F:5' CATGAGAAGTATGACAACAGCCT 3' |
| R:5' AGTCCTTCCACGATACCAAAGT 3' |
Cell transfection
The synthetic circHIPK3 sequence was subcloned into the pcDNA3.1 vector (Invitrogen). CircHIPK3 ectopic expression was achieved through pcDNA3.1- circHIPK3 transfection using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions, and an empty pcDNA3.1 vector was used as a control. Cells were collected 48h after transfection. CircHIPK3 expression levels were determined by qRT-PCR.
CCK-8 assay
Cells were incubated in 10% CCK-8 diluted in normal culture medium at 37℃ until visual color conversion occurred. Proliferation rates were determined at 24, 48, and 72 h after transfection. The absorbance of each well was measured with a microplate reader set at 570 nM.
Colony Formation Assay
Cells were seeded in six-well plates and were incubated for 24 h. The colonies were stained with crystal violet solution 14 days later. The colony number in each well was counted and calculated.
Wound Healing Assay and Cell Invasion Assay
Stable transfected cells were seeded onto six-well plates and cultured overnight. Wounds were created by scratching the cell layer with a sterile plastic pipette tip and were washed with phosphate buffer saline. Cells were further cultured with medium containing 1% FBS in 24 h.
For the invasion assays, a 24-well transwell chamber with the upper chamber coated with Matrigel (BD Biosciences) was used. 1.0×105 cells in 100 mL serum-free DMEM were seeded in the top chamber, and 500 mL medium containing 10% FBS was placed into the lower chamber. After incubation for 24 h, cells on the upper membrane surface were wiped off using a cotton swab, and the cells that had traversed the membrane were stained by crystal violet and counted.
Statistical analysis
All statistical analyses were performed using SPSS 22.0 software (IBM) and Graphpad Prism 5.0. Differences between OS tissues and paired adjacent nontumorous tissues and between paired preoperative and postoperative OS patient plasma samples were analyzed using the Student's t test. The correlations between circRNA levels and clinicopathological factors were further analyzed by one-way analysis of variance (ANOVA). A receiver operating characteristic (ROC) curve was established to evaluate its diagnostic value. The cutoff value of circRNA was analyzed by SigmaPlot 12.3. Overall survival was calculated by Kaplan-Meier survival analysis and compared using the log-rank test. p values<0.05 were considered statistically significant.
Results
Hsa_circ_001569, circ-Cdr1as, circHIPK3 and circ-Foxo3 were down-regulated in osteosarcoma cell lines
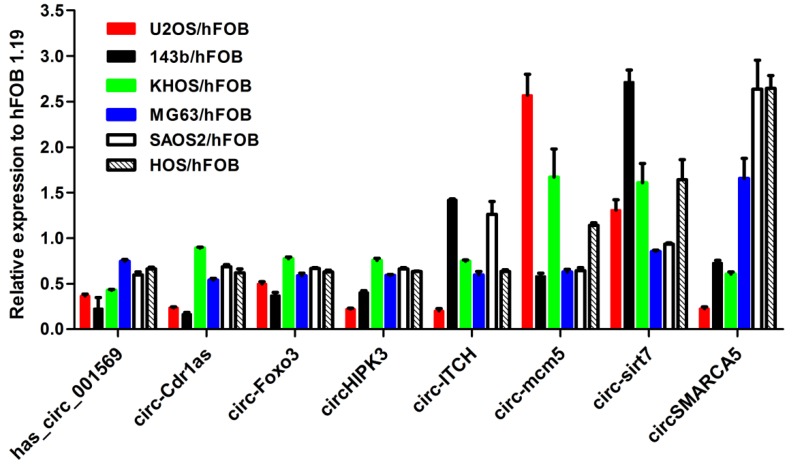
We first examined the expression of the eight-selected cancer-related circRNAs in the six osteosarcoma cell lines (SaoS2, HOS, KH-OS, MG63, 143B and U2-OS) and the normal osteoblast cell hFOB 1.19. As shown in Figure 1, hsa_circ_001569, circ-Cdr1as, circHIPK3 and circ-Foxo3 other than circ-ITCH, circSMARCA5, circ-sirt7, circ-mcm5 were consistently down-regulated in the six osteosarcoma cell lines compared with hFOB 1.19(Fig.1). Then the four circRNAs were chosen for further detection in tissues and plasmas.
Figure 1.
Expression level of the selected eight circRNAs in six osteosarcoma cell lines compared to normal osteoblast cell line hFOB1.19
CircHIPK3 expression was down-regulated in osteosarcoma tissues and plasmas, related to poor prognosis
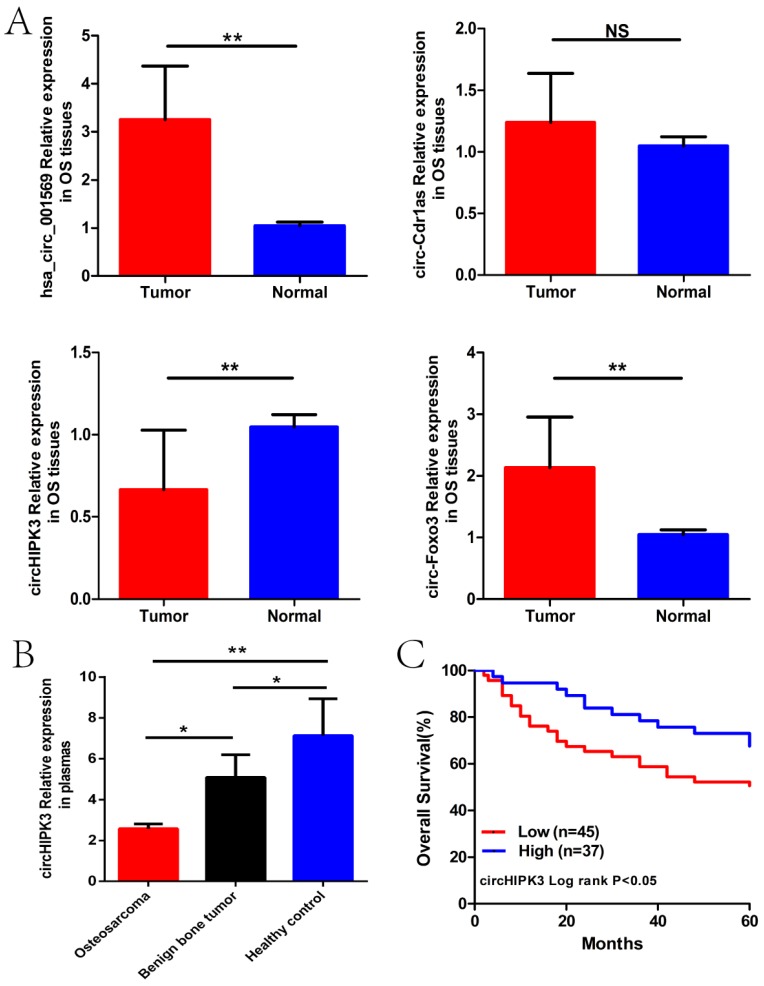
We next detected the expression levels of hsa_circ_001569, circ-Cdr1as, circHIPK3 and circ-Foxo3 in the osteosarcoma tissues and plasmas previously described. As was shown in Figure 2A and B, only circHIPK3 was stably significantly lower in osteosarcoma tissues and plasmas than those in corresponding controlled (P<0.01). Subsequently, we classified the 82 osteosarcoma patients into high expression and low expression groups according to the median expression level used as the cut-off. Kaplan-Maier analysis showed that patients with lower expression of circHIPK3 had shorter overall survival time than those with higher circHIPK3 expression (P<0.05; Fig. 2C).
Figure 2.
CircHIPK3 expression was down-regulated in osteosarcoma tissues and plasmas, related to poor prognosis. (A) Expression level of four circRNAs in 82 pairs of OS and paracancerous tissues. (B)Expression level of circHIPK3 in plasmas from 50 patients with osteosarcoma before surgery, 10 patients with benign bone tumor and 20 matched healthy individuals. (C) Patients with lower circHIPK3 expression had a shorter overall survival time than those with higher circHIPK3 expression. *P < 0.05, **P < 0.01.
Potential diagnostic values of circHIPK3 in osteosarcoma
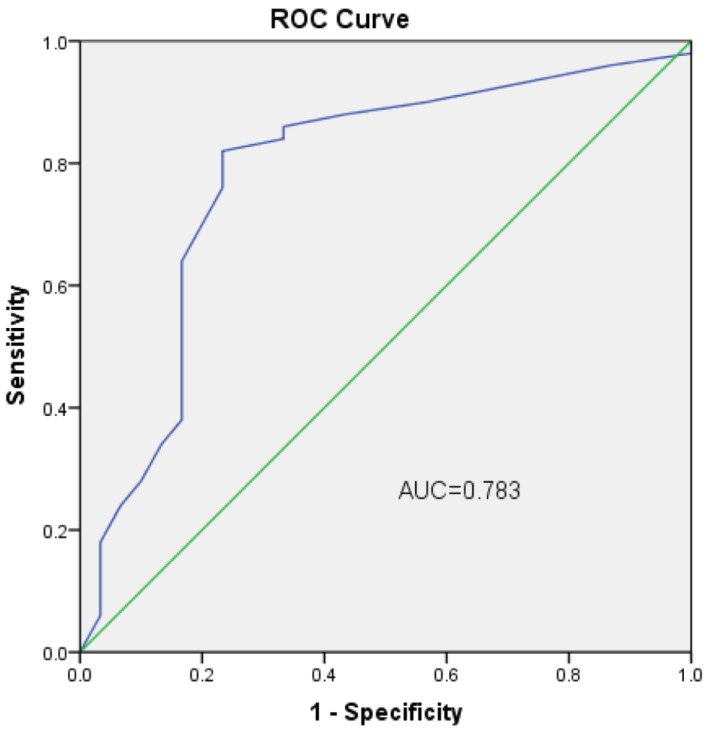
We next performed an analysis to evaluate the potential diagnostic value of circHIPK3. The results showed that circHIPK3 expression was associated with several clinicopathological features of patients with OS. As shown in Table 1, circHIPK3 levels were related to Enneking stage (P=0.042) and lung metastasis (P=0.036) other than age, gender and tumor location (P>0.05) according to statistical analysis. These results clearly showed that lung metastasis and advanced cancer were associated with lower expression levels of circHIPK3. Furthermore, to estimate whether circHIPK3 could be used as an indicator for diagnosis of osteosarcoma, a ROC curve was built and the area under the ROC curve was 0.783 (Fig. 3). The sensitivity and specificity were 0.56 and 0.84, respectively. In addition, the cutoff value was 29.3. These results suggested that circHIPK3 could be used as a biomarker for diagnosis and prognosis prediction of osteosarcoma.
Figure 3.
ROC curve of the plasma circHIPK3 in the 50 osteosarcoma patients and 20 healthy donors.
CircHIPK3 overexpression suppressed OS cell proliferation and clone formation
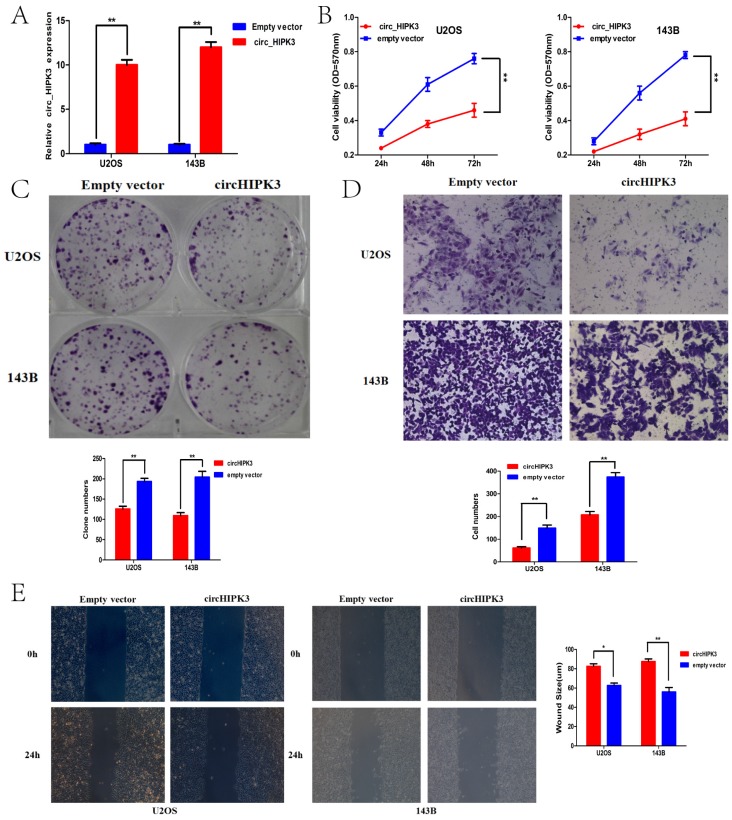
Considering the expression level of circHIPK3 in the previously described six OS cell lines, U2OS and 143B with the most down-regulated of circHIPK3 expression were chosen as the further experimental candidates. To further identify the role of circHIPK3 in osteosarcoma, CCK-8 assay and colony formation analysis were performed to examine the effect of circHIPK3 on cell proliferation and vitality in the U2OS and 143B cells. U2OS (or 143B) cells were transfected with the circHIPK3 expression vector or empty vector. Satisfactory transfection efficiency was obtained at 48 h post-transfection and confirmed by qPCR (Fig.4A). CCK-8 assay showed that the cell proliferation rates of U2OS (or 143B) cells transfected with circHIPK3 were lower than the cells transfected with empty vector in the 24h, 48h or 72h (Fig.4B). Besides, colony formation assays also revealed that cell proliferation was significantly suppressed in circHIPK3-overexpressing U2OS (or 143B) cells compared with empty vector transfected cells (Fig. 4C). These results demonstrated that overexpression of circHIPK3 suppressed proliferation in OS cells.
Figure 4.
CircHIPK3 overexpression suppressed cell proliferation, migration and invasion of OS cells in vitro. (A) qRT-PCR analysis of the effect on overexpression of circHIPK3 by vector transfection in the U2OS and 143B cell lines. (B) CCK-8 assays were performed to examine the cell proliferation rate of U2OS and 143B cells after circHIPK3 overexpression. (C) Clone formation assays were performed to examine cell vitality after transfection. (D) Transwell assays were performed to identify the capacity of cell invasion after transfection. (E) Wound healing assays were performed to examine the capacity of cell migration after transfection.*P < 0.05; **P < 0.01.
CircHIPK3 overexpression inhibited OS cell migration and invasion
Transwell and wound healing assays were conducted to further demonstrate the role of circHIPK3 in OS progression. Our data showed that circHIPK3 overexpression significantly reduced cell migration as assayed by the number of cells migrating through the Transwell membrane in both U2OS and 143B cells (Fig.4D). Wound healing assay also showed the similar results (Fig.4E). Taken together, these data suggested that circHIPK3 might serve as a tumor-suppressor in the OS progression.
Discussion
In recent years, with the rapid development and widespread application of RNA sequencing, researchers have found that many exonic transcripts can form circRNAs through non-linear reverse splicing or gene rearrangement20. Moreover, they account for a large proportion of all spliced transcripts. CircRNAs are widely expressed in human cells, and their expression levels could be 10-fold or higher compared to their linear isomers21, 22. Compared with other noncoding RNA, such as miRNAs and long noncoding RNAs (lncRNAs), these properties provide circRNAs with the potential to become ideal biomarkers in the diagnosis of cancers 8. For example, hsa_circ_0000190 23, hsa_circ_00198810 and hsa_circ_000164911 have been found to be biomarkers in the diagnosis of gastric cancer, colorectal cancer and hepatocellular carcinoma respectively.
In the current study, we selected eight cancer-related circRNAs, including circ-Cdr1as and circ-ITCH, previously reported to be involved in the progression of colorectal cancer, hepatocellular carcinoma, ESCC, prostate cancer, gastric cancer and so on. Of them, circ-Cdr1as, also named ciRS-7, has been found to occur as a super sponge or competing endogenous RNA (ceRNA) of miR-7 to competitively sequester and quench the activity of miR-7, which has tremendous effects on the development of a variety of cancer, such as breast cancer, hepatocellular carcinoma, and cervical cancer13, 24, 25. Besides, circRNA_001569 targeting miR-145 promotes cell proliferation and invasion of colorectal cancer26. Circ-ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway 27. Circ-KLHDC10 is enriched and stable in exosomes and could serve as a promising biomarker for liver cancer diagnosis 28.
However, the expression, function and possible clinical significance of these circRNAs were still unknown in OS. Here, we first detected the eight circRNAs in 6 kinds of OS cell lines and the normal osteoblast cell (hFOB1.19), 82 paired OS tissues and corresponding nontumorous tissues, 50 plasma specimens from patients with OS, 10 plasma specimens from patients with benign bone tumor and 20 plasma specimens from healthy controlled individuals by qRT-PCR. We found that only circHIPK3 was consistently down-regulated in the OS cell lines, tissues and plasmas compared with the control. Then we further analyzed and found that down-regulation of circHIPK3 was associated with Enneking Stage, lung metastasis, poor prognosis of OS patients and ROC analyses show that circHIPK3 may be used as a novel biomarker for OS with high degrees of accuracy, specificity, and sensitivity.
As is known to us, circHIPK3, derived from Exon2 of the HIPK3 gene, is an important circRNA in disease progression. CircHIPK3 consists solely of a large second exon (1,099 bp) from the HIPK3 gene flanked on either side by long introns, which include many complementary Alu repeats to further promote its circularization. These features indicate that circHIPK3 is formed by “direct splicing” and stably expressed in different cell lines and tissues16-18. There have been several reports about its function and regulatory mechanism in many diseases, including cancers. For example, Shan K et al17 found that circHIPK3 could mediate retinal vascular dysfunction in diabetes mellitus. Li Y et al18 reported that circHIPK3 sponge miR-558 to suppress heparanase expression in bladder cancer cells. Zheng Q et al16 found that circHIPK3 promotes proliferation of human liver cancer HuH-7 cells, human colon cancer HCT-116 cells, and human cervical cancer HeLa cells via sponging multiple miRNAs.
To our knowledge, this is the first report about circHIPK3 and OS. We found the decreased expression of circHIPK3 could serve as a biomarker for diagnosis and prognosis of OS. Besides, functional analysis revealed that overexpression of circHIPK3 significantly suppressed cell proliferation, migration and invasion of OS cells in vitro. But more samples are needed in the future work to further identify the results. In addition, it's necessary to explore the further function and regulatory mechanism of circHIPK3 in OS.
In summary, the expression of circHIPK3 was down regulated in OS cell lines, tissues and plasmas. Furthermore, its expression was associated with Enneking stage, lung metastasis and poor prognosis of OS patients. Overexpressing circHIPK3 significantly suppressed OS cell proliferation, migration and invasion in vitro. These results suggest that circHIPK3 may be a potential novel biomarker for diagnosis and treatment target of OS.
Acknowledgments
This project was supported by a Grant from the National Natural Science Foundation of China (No.81572630), the Fundamental Research Funds for the Central Universities (NO.22120170216), Shanghai Pujiang Program of Shanghai Science and Technology Commission (NO.13PJD023) and Shanghai Jiaotong University Medical-Engineering Cross Research Fund (NO.YG2012MS49).
References
- 1.Wang Y, Zhang Y, Yang T. et al. Long non-coding RNA MALAT1 for promoting metastasis and proliferation by acting as a ceRNA of miR-144-3p in osteosarcoma cells. Oncotarget. 2017;8:59417–34. doi: 10.18632/oncotarget.19727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao J, Han X, Qi X. et al. TUG1 promotes osteosarcoma tumorigenesis by upregulating EZH2 expression via miR-144-3p. Int J Oncol. 2017;51:1115–23. doi: 10.3892/ijo.2017.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai L, Lv J, Zhang Y. et al. The lncRNA HNF1A-AS1 is a negative prognostic factor and promotes tumorigenesis in osteosarcoma. J Cell Mol Med. 2017;21:2654–2662. doi: 10.1111/jcmm.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qu S, Yang X, Li X. et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–8. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Zhong Z, Huang M, Lv M. et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–17. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, Liu H, Hou L. et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang YY, Zhao P, Zou TN. et al. Circular RNA hsa_circ_0001982 Promotes Breast Cancer Cell Carcinogenesis Through Decreasing miR-143. DNA Cell Biol. 2017;36:901–908. doi: 10.1089/dna.2017.3862. [DOI] [PubMed] [Google Scholar]
- 8.Zhang HD, Jiang LH, Sun DW. et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 9.Xuan L, Qu L, Zhou H. et al. Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res. 2016;8:932–9. [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Zhang Y, Huang L. et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8:16020–5. [PMC free article] [PubMed] [Google Scholar]
- 11.Qin M, Liu G, Huo X. et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–9. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 12.Xu L, Zhang M, Zheng X. et al. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17–27. doi: 10.1007/s00432-016-2256-7. [DOI] [PubMed] [Google Scholar]
- 13.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan H, Li T, Jiang Y. et al. Overexpression of Circular RNA ciRS-7 Abrogates the Tumor Suppressive Effect of miR-7 on Gastric Cancer via PTEN/PI3K/AKT Signaling Pathway. J Cell Biochem. 2018;119:440–446. doi: 10.1002/jcb.26201. [DOI] [PubMed] [Google Scholar]
- 15.Geng HH, Li R, Su YM. et al. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PloS One. 2016;11:e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Q, Bao C, Guo W. et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. EMBO Rep. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shan K, Liu C, Liu BH. et al. Circular Non-Coding RNA HIPK3 Mediates Retinal Vascular Dysfunction in Diabetes Mellitus. Circulation. 2017;136:1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Zheng F, Xiao X. et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–59. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Zhang XO, Chen T. et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 20.Yang Z, Xie L, Han L. et al. Circular RNAs: Regulators of Cancer-Related Signaling Pathways and Potential Diagnostic Biomarkers for Human Cancers. Theranostics. 2017;7:3106–17. doi: 10.7150/thno.19016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Ding W, Sun T. et al. Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. FEBS J. 2018;285:220–232. doi: 10.1111/febs.14191. [DOI] [PubMed] [Google Scholar]
- 22.Greene J, Baird AM, Brady L. et al. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front Mol Biosci. 2017;4:38. doi: 10.3389/fmolb.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167–71. doi: 10.1016/j.cca.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 24.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–12. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 25.Peng L, Yuan XQ, Li GC. The emerging landscape of circular RNA ciRS-7 in cancer (Review) Oncol Rep. 2015;33:2669–74. doi: 10.3892/or.2015.3904. [DOI] [PubMed] [Google Scholar]
- 26.Xie H, Ren X, Xin S. et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–91. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li F, Zhang L, Li W. et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–13. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Zheng Q, Bao C. et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–4. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]