Abstract
CRISPR/Cas9-based therapeutics, especially those that can correct gene mutations via homology directed repair (HDR), have the potential to revolutionize the treatment of genetic diseases. However, HDR-based therapeutics are challenging to develop because they require simultaneous in vivo delivery of Cas9 protein, guide RNA and donor DNA. Here, we demonstrate that a delivery vehicle composed of gold nanoparticles conjugated to DNA and complexed with cationic endosomal disruptive polymers can deliver Cas9 ribonucleoprotein and donor DNA into a wide variety of cell types, and efficiently correct the DNA mutation that causes Duchenne muscular dystrophy in mice via local injection, with minimal off-target DNA damage.
Gene editing therapeutics based on the CRISPR/Cas9 system have tremendous potential for treating genetic diseases1–4. Primarily two types of gene editing therapies are being considered for the CRISPR/Cas9 system: (1) therapies based on non-homologous end joining (NHEJ), which permanently silence disease-causing genes by inducing indel mutations, and (2) therapies based on homology directed repair (HDR), which correct disease-causing gene mutations to their wild type sequence. HDR based therapies have the potential to cure the vast majority of genetic diseases because this class of therapeutics can correct mutated genes back to their wild type sequence. There is therefore great interest in developing HDR based therapeutics. However, gene editing via HDR in vivo is challenging because HDR requires the delivery of Cas9, gRNA, and donor DNA.
Gene therapy with adeno-associated viruses (AAVs) is currently the most advanced methodology for delivering Cas9 in vivo5,6. However, developing Cas9 therapeutics based on AAV delivery is challenging because a large fraction of the human population has a pre-existing immunity towards AAV, making them ineligible for AAV based therapies. In addition, AAV based Cas9 delivery has the potential to cause significant off-target genomic damage, due to the sustained expression of Cas97,8. AAV also has a small packing size and multiple viruses are needed to deliver Cas9 RNP and donor DNA in vivo, and this decreases the HDR efficiency of AAV based Cas9 delivery methods. Finally, although AAV based Cas9 delivery has generated several exciting pre-clinical demonstrations in vivo9–11, the viral titers needed to generate therapeutic levels of editing have been orders of magnitude higher than the clinically accepted levels.
There is therefore great interest in developing non-viral Cas9 based therapeutics that can induce HDR12. However, developing delivery vehicles that can induce HDR in vivo has been challenging because of the multiple components involved. The only non-viral demonstration of HDR in vivo has been via the hydrodynamic delivery of plasmid DNA that expresses Cas9, gRNA, and donor DNA13. The translational potential of hydrodynamic based delivery of plasmids is unclear because of the dramatic changes in blood pressure it causes.
Direct delivery of the Cas9 RNP is also being considered as a therapeutic strategy for generating HDR and has tremendous promise for clinical translation14, because of the established protocols for producing proteins on a large scale and for clinical use, and the well characterized clinical track record of protein therapeutics. Delivery strategies have been developed for delivering the Cas9 RNP in vitro and in vivo15–18. Lipofectamine and polyethylenimine (PEI) have been the most successful Cas9 RNP delivery vehicles, and have been able to deliver Cas9 RNP into the ear and in tumors to knock-out genes via NHEJ. However, the application of lipid products or PEI to induce HDR in vivo has not been successfully demonstrated, and will be potentially problematic due to the challenges associated with delivering multiple macromolecules in vivo. Therefore, the development of vehicles that can simultaneously deliver Cas9 RNP and donor DNA and induce HDR in vivo remains a central problem in the field of therapeutic gene editing.
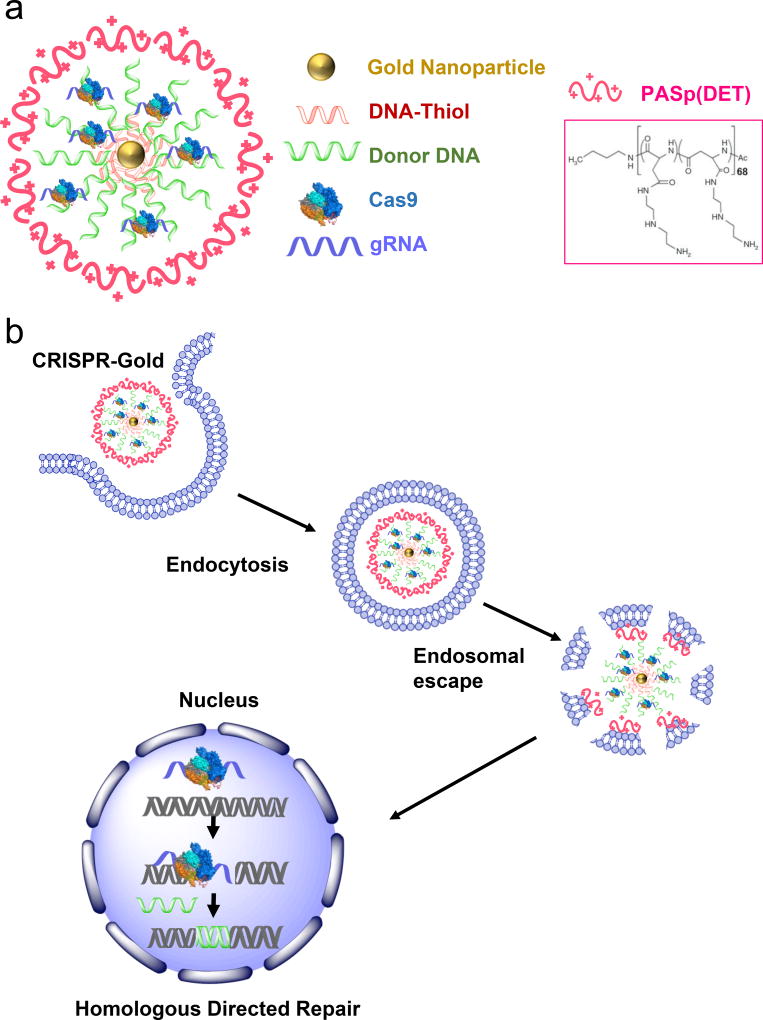
In this report, we present such a vehicle, CRISPR-Gold, which can directly deliver Cas9 RNP and donor DNA in vivo via local administration and induce HDR. CRISPR-Gold is composed of gold nanoparticles conjugated with DNA, which are complexed with donor DNA, Cas9 RNP, and the endosomal disruptive polymer PAsp(DET) (Fig. 1). CRISPR-Gold is designed to be internalized by cells via endocytosis due to the cationic PAsp(DET) that complexes the components of CRISPR-Gold19,20. After endocytosis, the PAsp(DET) polymer on CRISPR-Gold triggers endosomal disruption, and causes the release of CRISPR-Gold into the cytoplasm (Fig. 1). Importantly, once in the cytoplasm, glutathione releases the DNA from the gold core of CRISPR-Gold, which causes the rapid release of Cas9 RNP and donor DNA21.
Figure 1. CRISPR-Gold can deliver Cas9 ribonucleoprotein and donor DNA in vivo and induce homology directed DNA (HDR) repair.
a) CRISPR-Gold is composed of 15 nm gold nanoparticles conjugated to thiol modified oligonucleotides, which are hybridized with donor ssODN and subsequently complexed with Cas9 RNP, and the endosomal disruptive polymer PAsp(DET).
b) CRISPR-Gold is internalized by cells in vitro and in vivo via endocytosis, triggers endosomal disruption, and releases Cas9 RNP and donor DNA into the cytoplasm. Nuclear delivery results in HDR.
Results and discussion
Design and synthesis of CRISPR-Gold
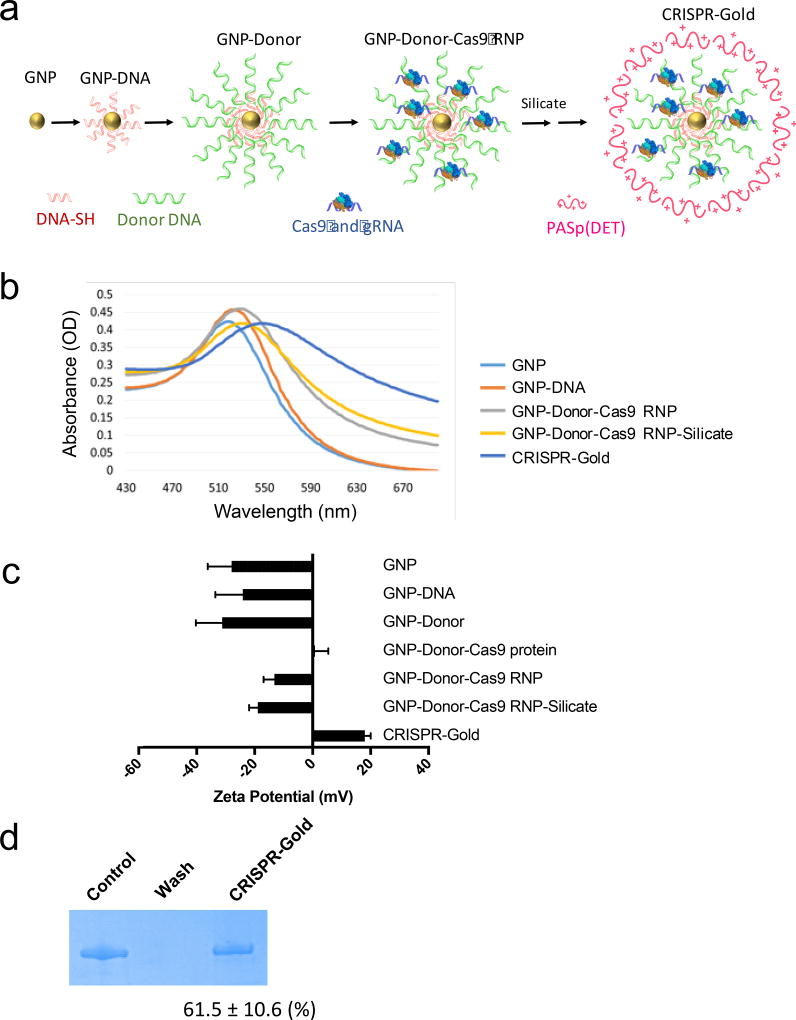
Non-viral gene editing via HDR requires developing materials that can simultaneously deliver Cas9 RNP and donor DNA into cells. A key challenge in delivering both proteins and nucleic acids into cells is developing materials that can simultaneously complex both classes of macromolecules. CRISPR-Gold addresses this problem by taking advantage of the ability of Cas9 to bind gRNA and its affinity to the donor DNA coating the gold nanoparticles22,23. In addition, gold nanoparticles bind a large variety of proteins, via non-specific electrostatic forces, and could also have affinity for Cas9 RNP24,25. Gold nanoparticles were selected as the core of CRISPR-Gold because they can be coated with a densely packed layer of DNA and because gold nanoparticles are taken up by a variety of different cell types21,26–28. The synthesis of CRISPR-Gold is shown in Figure 2a and in Supplementary Fig. 1. The first step in the synthesis is the facile reaction of thiol-terminated DNA with gold nanoparticles, followed by hybridization with the donor DNA. Cas9 RNP is then adsorbed onto the particles, via the binding affinity of Cas9 RNP to the DNA loaded onto the gold nanoparticles, and its potential non-specific affinity for gold nanoparticles. A layer of silica was then deposited onto the nanoparticle to increase the negative charge density, and then finally complexed with the cationic endosomal disruptive polymer PAsp(DET)29. The synthesis of CRISPR-Gold was monitored with absorbance analysis, transmission electron microscopy (TEM), and dynamic light scattering (Fig. 2b and Supplementary Fig. 2 and 3). The adsorption of the silica and the complexation of PAsp(DET) were monitored by zeta potential analysis, which demonstrated that large changes in zeta potential occurred in both of these steps, which shows the coating of each material onto CRISPR-Gold (Fig. 2c). TEM analysis of CRISPR-Gold indicated that 5 minutes after formulation, it is composed of single gold nanoparticles. The structure CRISPR-Gold is dynamic and aggregation was observed after 30 min, presumably due to inter-particle electrostatic interactions.
Figure 2. Synthesis and characterization of CRISPR-Gold.
a) Synthesis of CRISPR-Gold. Gold nanoparticles 15 nm in diameter were conjugated with a 5′ thiol modified single stranded DNA (DNA-SH), and hybridized with single stranded donor DNA. Cas9 and gRNA are loaded and then a silicate and PAsp(DET) polymer coating are added.
b) Absorbance spectra analysis. The absorption maxima of CRISPR-Gold intermediates are red shifted as CRISPR-Gold is sequentially constructed. Unmodified GNPs had an absorbance peak at 518 nm and the peak shifts to 522 nm for GNP-DNA, 528 nm for GNP-DNA-Cas9 RNP, and 546 nm for CRISPR-Gold.
c) Zeta potential analysis. Zeta potential measurements were performed on CRISPR-Gold and all of the synthetic intermediates generated during the construction of CRISPR-Gold. Zeta potential changes demonstrate the sequential synthesis of CRISPR-Gold.
d) Cas9 loading analysis. CRISPR-Gold was formulated and the unbound Cas9 RNP was removed via spin filtration. Gel electrophoresis was performed on CRISPR-Gold and the SDS gel was stained with Coomassie blue to quantify the amount of Cas9 RNP bound to CRISPR-Gold. The control represents the original amount of Cas9 RNP added to the CRISPR-Gold formulation. The wash represents the unbound Cas9 RNP, removed by filtration.
Analysis of CRISPR-Gold complexes with Cas9 RNP
The ability of CRISPR-Gold to complex Cas9 RNP was investigated. CRISPR-Gold was synthesized following the procedures described above and the particles were purified via spin filtration through a 300kDa molecular weight cut-off filter, washed, and analyzed via gel electrophoresis. The percent of Cas9 RNP bound to the GNPs was determined by comparing the recovered Cas9 with the original amount of Cas9 mixed with the particles. Figure 2d demonstrates that CRISPR-Gold has a 61.5% encapsulation efficiency for complexing Cas9 RNP, and thus has the complexation efficiency needed for developing Cas9 delivery vehicles. In addition, the activity of Cas9 RNP complexed to CRISPR-Gold was investigated. The enzymatic activity of Cas9 RNP released from CRISPR-Gold was examined by assaying its ability to cleave target DNA. Cas9 mediated DNA cleavage was analyzed via gel electrophoresis30. Cas9 RNP released from CRISPR-Gold was still active, and cleaved target template DNA (Supplementary Fig. 4). We also performed binding experiments with Cas9 RNP and unmodified gold nanoparticles, and observed that Cas9 RNP bound unmodified gold nanoparticles, but the binding was unstable and did not survive multiple wash cycles (Supplementary Fig. 5).
Investigation of CRISPR-Gold’s ability to induce HDR in HEK cells
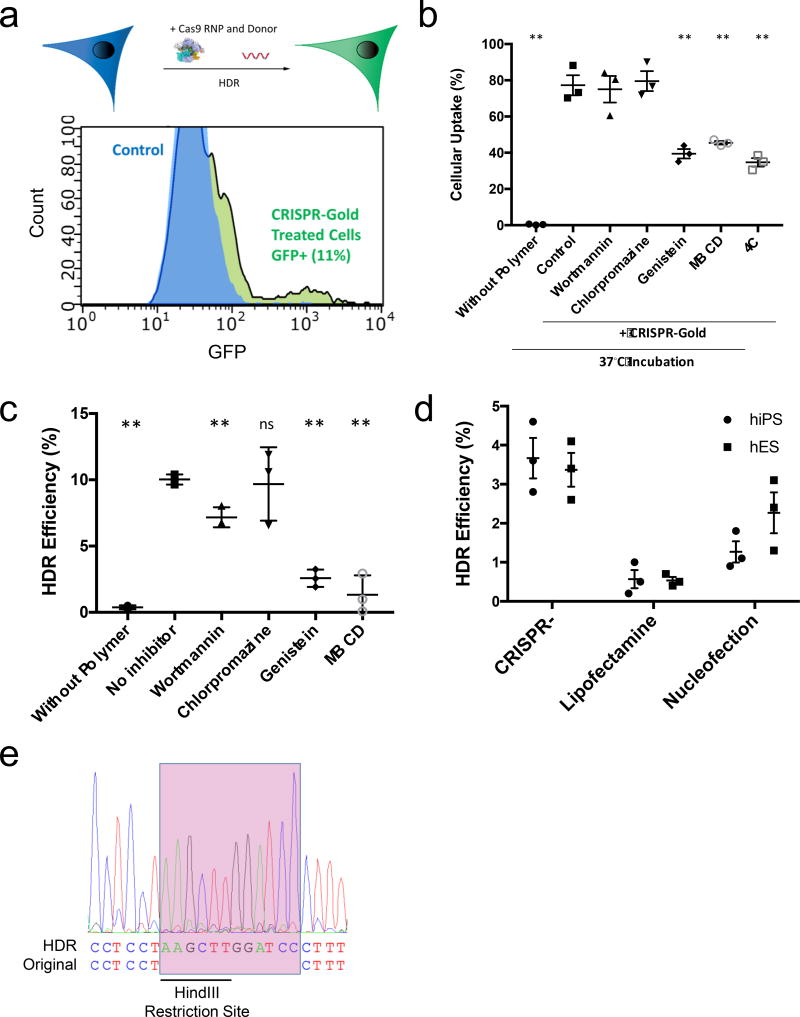
We performed HDR experiments on HEK293 cells expressing the blue fluorescent protein (BFP) to investigate the ability of CRISPR-Gold to induce HDR in cells31. CRISPR-Gold containing a single-stranded donor oligonucleotide (ssODN) that converts the BFP gene into the GFP gene and a gRNA that cuts the BFP gene were synthesized (described in Supplementary Fig. 6). CRISPR-Gold was incubated with BFP-HEK cells (8 µg/mL Cas9 protein) and the level of HDR experienced by the BFP-HEK cells was determined by flow cytometry. Figure 3a shows that CRISPR-Gold induced 11.3% of the BFP-HEK cells to express GFP via HDR. This result was further confirmed by sequencing, which demonstrated that the GFP sequence in the edited cells exactly matched the donor DNA sequence (Supplementary Fig. 7). In addition, we performed experiments to determine the ratio of donor DNA to gRNA in CRISPR-Gold that generates the highest level of HDR in cells. CRISPR-Gold was made with various ratios of donor DNA to gRNA and was then incubated with BFP-HEK cells to measure the level of HDR induced. A 1:1 ratio of gRNA to donor DNA was determined to be optimal for inducing HDR (Supplementary Fig. 8 and see Supplementary Fig. 9 for fluorescent microscopy image).
Figure 3. CRISPR-Gold induces HDR in vitro.
a) CRISPR-Gold can induce HDR in BFP-HEK cells by delivering Cas9 RNP and a donor DNA. 11.3% of BFP-HEK cells were converted to GFP expressing cells, after treatment with CRISPR-Gold. GFP expression due to BFP sequence editing was determined by flow cytometry.
b) The uptake of CRISPR-Gold in HEK293 cells is dependent on non-clathrin mediated endocytosis. CRISPR-Gold containing Alexa647 labeled sgRNA was added to HEK293 cells and the uptake efficiency was measured using flow cytometry. Various inhibitors and low temperature were used to study the cellular uptake mechanism. Also, the uptake of CRISPR-Gold without the PAsp(DET) polymer coating was investigated. Genistein, MBCD, and 4°C all significantly inhibited the uptake of CRISPR-Gold, whereas clathrin based endocytosis inhibitors had no effect. Mean ± S.E, n=3. **, p < 0.01.
c) Non-clathrin mediated endocytosis plays a critical role in CRISPR-Gold mediated HDR. The inhibitor of clathrin mediated endocytosis, chlorpromazine, has minimal effects on CRISPR-Gold mediated HDR, whereas the caveolae/raft mediated endocytosis inhibitor MBCD and genistein reduced CRISPR-Gold mediated HDR by 80%. Mean ± S.E, n=3. **, p < 0.01, ns= not significant.
d) CRISPR-Gold induces HDR in hES and hiPS cells with higher efficiency than lipofectamine or nucleofection. Cells were treated with CRISPR-Gold that contained a donor sequence with a HindIII cleavage site, and a guide RNA that targeted the CXCR4 gene. The CXCR4 gene was amplified by PCR and the HDR efficiency was determined by quantifying HindIII cleavage of the CXCR4 PCR amplicon. Mean ± S.E, n=3. A one-way ANOVA test had a p= 0.0066 for hES samples and p=0.0023 for hiPS samples.
e) Sanger sequencing demonstrates that CRISPR-Gold induces HDR in hES cells. PCR of the CXCR4 gene was performed on CRISPR-Gold treated hES cells, and sequencing confirms the presence of the 12 bp insertion (pink box).
In addition, we investigated the dose response of CRISPR-Gold, its cell culture toxicity, and the mechanism by which CRISPR-Gold delivers Cas9 RNP and donor DNA. We observed that CRISPR-Gold had a maximum HDR efficiency at 8 µg/mL of Cas9, and that at doses above this level the HDR efficiency was lowered due to CRISPR-Gold-mediated cellular toxicity (see Supplementary Fig. 10 and 11). We also performed flow cytometry uptake experiments with fluorescently labeled CRISPR-Gold (gRNA labeled)32, in the presence of a panel of inhibitors to determine the mechanism by which cells internalize CRISPR-Gold. Figure 3b demonstrates that the caveolae/raft-dependent endocytosis inhibitors, genistein and methyl-β-cyclodextrin (MBCD), dramatically reduced the cellular uptake of CRISPR-Gold, whereas the inhibitor of clathrin-mediated endocytosis, chlorpromazine, had no effect on uptake. Incubation of CRISPR-Gold with cells at 4°C also inhibited uptake. Collectively these experiments suggest that CRISPR-Gold is dependent on caveolae/raft-dependent endocytosis. Cellular uptake experiments with CRISPR-Gold were also performed with formulations that did not contain the polymer PAsp(DET). Figure 3b demonstrates that PAsp(DET) is essential for stimulating cellular uptake, and suggests that it is playing a key role in triggering endocytosis.
Finally, we performed HDR experiments with CRISPR-Gold, using the same panel of inhibitors described above, to investigate the relationship between cellular uptake and HDR efficiency. Figure 3c demonstrates that genistein and MBCD caused a significant reduction in HDR efficiency, whereas the clathrin-mediated endocytosis inhibitor, chlorpromazine, had no effect on HDR efficiency, suggesting that CRISPR-Gold’s ability to induce HDR in cells is largely dependent upon caveolae/raft-dependent endocytosis.
CRISPR-Gold’s HDR efficiency in primary cells and stem cells
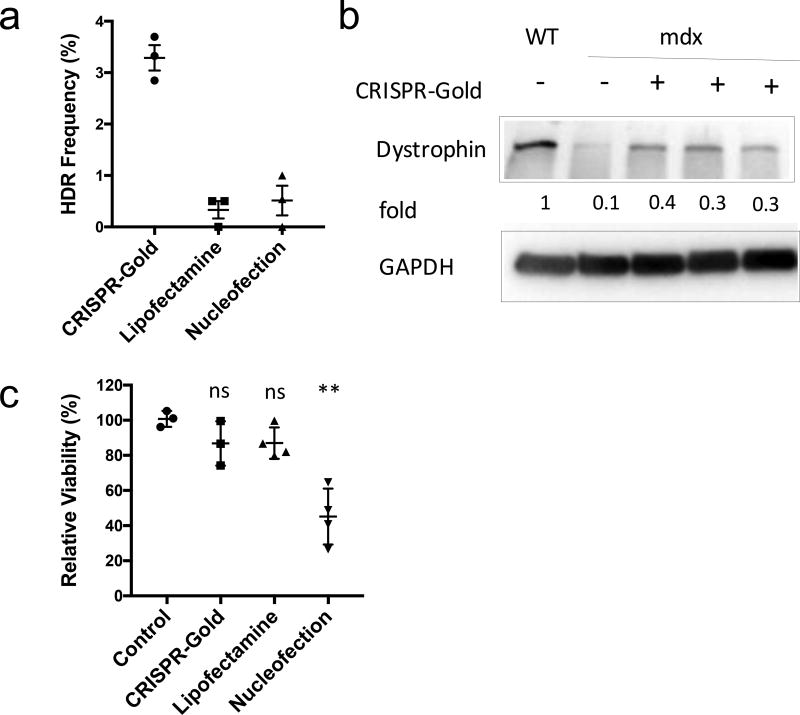
The ability of CRISPR-Gold to induce HDR in a panel of therapeutically relevant cell types was tested to identify potential therapeutic applications of CRISPR-Gold. CRISPR-Gold’s delivery efficiency was investigated in embryonic stem (hES) cells, human induced pluripotent stem (hiPS) cells, primary bone marrow derived dendritic cells (BMDCs), and primary myoblasts from mdx mice. CRISPR-Gold formulations were synthesized that were designed to edit the CXCR4 gene or the dystrophin gene, and their ability to perform gene editing in cell culture was analyzed. Figures 3d, 3e and 4 demonstrate that CRISPR-Gold was able to target the CXCR4 in hES cells, hiPS cells, BMDCs, and the dystrophin gene in myoblasts with an HDR efficiency between 3–4% (Supplementary Fig. 12–14)8,30,33,34. Importantly, CRISPR-Gold was significantly more effective at inducing HDR in these cell types, and less toxic than either lipofectamine or nucleofection methods (Fig. 3 and 4)35. These results demonstrate that CRISPR-Gold can simultaneously deliver Cas9 protein, gRNA, and donor DNA into a wide range of cells.
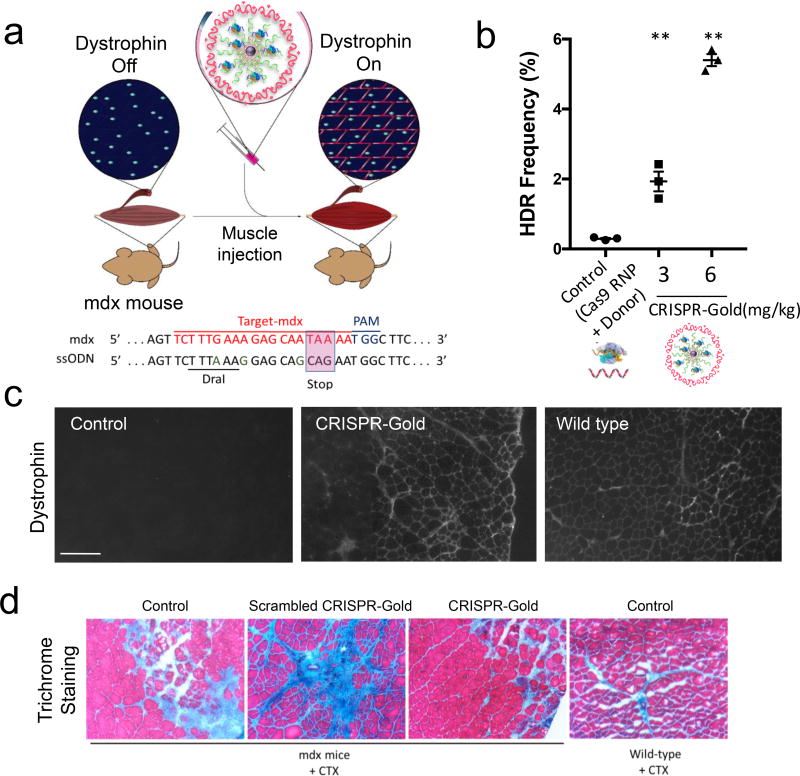
Figure 4. CRISPR-Gold induces HDR and promotes the expression of dystrophin protein in primary myoblasts.
a) The dystrophin gene was edited with CRISPR-Gold in primary myoblasts from the mdx mouse. CRISPR-Gold corrected the nonsense mutation in the dystrophin gene of mdx myoblasts with an HDR efficiency of 3.3%, which is significantly higher than either nucleofection or lipofectamine. No correction was observed in the negative control, composed of CRISPR-Gold without gRNA (data not shown). Mean ± S.E, n=3. A one-way ANOVA test had a p= 0.0002.
b) Dystrophin protein is expressed in myotubes that were differentiated from CRISPR-Gold edited primary mdx myoblasts. Western blot analysis was conducted to quantify the levels of dystrophin protein in muscle cells. The fold expression was determined by dividing the band pixel density of each group with the dystrophin band intensity from WT (C57.B6) myotubes (n=1). GAPDH was used as a loading control.
c) CRISPR-Gold causes minimal toxicity to primary myoblasts, whereas nucleofection caused significant toxicity. Primary mdx myoblasts were transfected with the indicated methods and cell viability was measured 2 days after the transfections with the cell counting kit-8. Relative viability to control, mean ± S.E, n=6. *, p < 0.05, ns = statistically not significant to control.
CRISPR-Gold mediated gene editing in a reporter mouse model
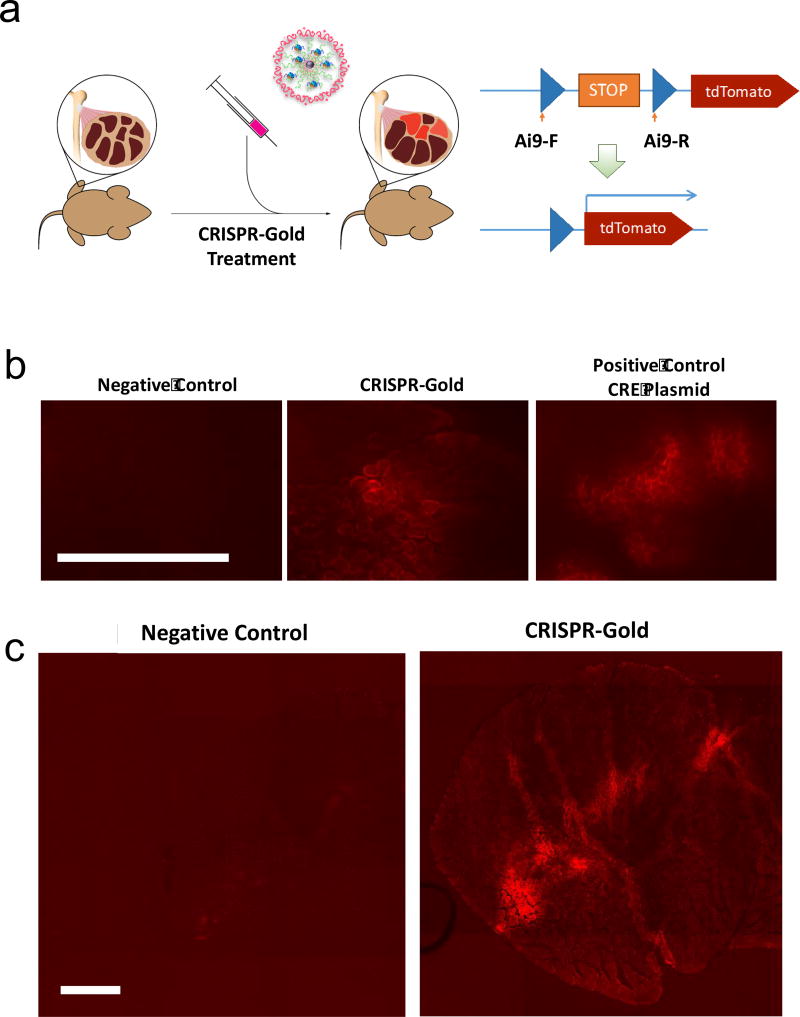
We performed experiments on Ai9 mice to determine if CRISPR-Gold could deliver the Cas9 RNP in vivo and generate double-stranded breaks. We used Ai9 mice for these experiments because gene deletions in the Ai9 DNA sequence results in the expression of the fluorescent tdTomato protein, which can be easily monitored. Ai9 mice were given one intramuscular injection of CRISPR-Gold, designed to induce tdTomato fluorescence via gene deletion, and after 2 weeks, the expression of tdTomato was determined in 10 µm Tibialis Anterior (TA) muscle sections (see Supplementary Fig. 15 for more details). Figure 5 and Supplementary Fig. 16 demonstrate that CRISPR-Gold can deliver Cas9 RNP and generate gene deletions in Ai9 mice, and that the gene editing effect of CRISPR-Gold was observable millimeters away from the injection site. CRISPR-Gold can therefore efficiently deliver Cas9 RNP in vivo and edit genomic DNA.
Figure 5. CRISPR-Gold induces gene editing in the muscle tissue of Ai9 mice.
a) CRISPR-Gold was injected into Ai9 mice and gene deletion of the stop sequence in the Ai9 gene was determined by tdTomato expression.
b) Representative images of tdTomato fluorescence in the gastrocnemius muscle after a single CRISPR-Gold injection. A plasmid expressing Cre recombinase was injected with lipofectamine as a positive control. Scale bar is 500 µm.
c) tdTomato expression is observed in a broad area of the muscle after injection with CRISPR-Gold. The entire TA muscle cross-section image shows tdTomato expression after CRISPR-Gold injection. Scale bar is 500 µm.
Gene editing in mdx mice with CRISPR-Gold
CRISPR-Gold has numerous potential applications because of its ability to complex Cas9 RNP and donor DNA, and deliver Cas9 RNP in vivo. We selected Duchenne muscular dystrophy (DMD) as an initial medical application for CRISPR-Gold. DMD is an early onset lethal disease, caused by mutations in the dystrophin gene; it is the most common congenital myopathy, and approximately 30% of DMD patients have single base mutations or small deletions that could be potentially treated with HDR-based therapeutics36. Several therapeutic strategies have been developed to regenerate functional dystrophin in patients, ranging from exon skipping with antisense oligonucleotides to gene therapy with dystrophin minigenes37–39. However, despite significant efforts, the development of effective DMD therapies remains a major challenge. The disease is currently without a cure and patients have mostly palliative care, such as steroids to diminish muscle inflammation. CRISPR/Cas9 based therapeutics have great potential for treating DMD because they can correct dystrophin gene mutations, after a single injection, and cure DMD. However, at present, the only gene editing strategy available for treating DMD is based on NHEJ induced exon skipping, which generates a truncated form of dystrophin, with suboptimal functionality and used viral delivery of Cas99–11,40.
CRISPR-Gold was able to correct a point mutation in the dystrophin gene, in primary myoblasts from the mdx mouse, and induce the expression of dystrophin protein in myotubes that were differentiated from mdx myoblasts (Fig. 4a and 4b). Encouraged by these results, we investigated if CRISPR-Gold could correct the dystrophin mutation in mdx mice, after an intramuscular injection (Fig. 6a). In the first studies, CRISPR-Gold (two different doses: 3 and 6 mg/kg) was injected into the gastrocnemius and tibialis anterior muscles of 4-week-old mdx mice simultaneous with cardiotoxin (CTX), which activates the proliferation of muscle stem cells by muscle damage. After two weeks, the muscles were harvested and analyzed for HDR in the dystrophin gene, the expression of dystrophin protein and muscle fibrosis. Remarkably, CRISPR-Gold was able to correct the mutated dystrophin gene in mdx mice to the wild type sequence after a single injection, and restore the expression of dystrophin protein in muscle tissue (Fig. 6b and 6c). Figure 6b demonstrates that CRISPR-Gold can induce HDR in the dystrophin gene. Specifically, 5.4% of the dystrophin gene in mdx mice was corrected back to the wild type gene after CRISPR-Gold treatment, at 6 mg/kg, and this correction rate was approximately 18 times higher than treatment with Cas9 RNP and donor DNA by themselves, which had only a 0.3% correction rate (see Supplementary Fig. 17 for PCR analysis). Robust dystrophin protein expression was also observed in 10 µm cryo-sections of the injected muscle tissue (Fig. 6c and Supplementary Fig. 18 and Supplementary Fig. 19). In contrast, minimal levels of dystrophin expression were observed in the negative control, composed of Cas9 RNP and donor DNA injected without particles. In addition, Figure 6d demonstrates that CRISPR-Gold treated mdx mice had reduced levels of muscle fibrosis indicative of better tissue health.
Figure 6. CRISPR-Gold promotes HDR in the dystrophin gene and dystrophin protein expression, and reduces muscle fibrosis in mdx mice, with CTX stimulation.
a) CRISPR-Gold was injected into the hind leg muscle of 8-week-old mdx mice (n=3), simultaneously with CTX. Two weeks later, the mice were sacrificed and analyzed for dystrophin expression by immunofluorescence, for HDR by deep sequencing and for their degree of fibrosis. Bottom panel: dystrophin mutation sequence and donor DNA design. Donor DNA sequence designed to repair the non-sense mutation are marked in the pink box, nucleotides marked in green (A, G, G) are silent mutations that prevent Cas9 activity on the edited sequence.
b) CRISPR-Gold induced genome editing in the dystrophin gene was confirmed by deep sequencing. Deep sequencing of genomic DNA from muscle tissues injected with CRISPR-Gold revealed a 5.4% HDR efficiency. The negative control composed of Cas9 RNP and donor DNA had an HDR efficiency of only 0.3%. Mean ± S.E, n=3. **, p < 0.01.
c) CRISPR-Gold causes dystrophin expression in muscle tissue of mdx mice. The CRISPR-Gold injected muscle shows dystrophin expression (immuno-fluorescence), whereas control mdx mice did not express dystrophin protein. The scale bar is 200 µm.
d) CRISPR-Gold reduces muscle fibrosis in mdx mice. Trichrome staining was performed on the TA muscle cryosectioned to 10 µm, 2 weeks after the injection of CRISPR-Gold. CTX was co-injected in all three groups of mdx mice: No treatment control, scrambled CRISPR-Gold, and CRISPR-Gold. Images were acquired at the areas of muscle injury and regeneration. Fibrotic tissue appears blue, while muscle fibers appear red. Wild-type mice treated with CTX were analyzed 5 days after injection. The width of each image is 0.7 mm.
Muscle function in mdx mice treated with CRISPR-Gold
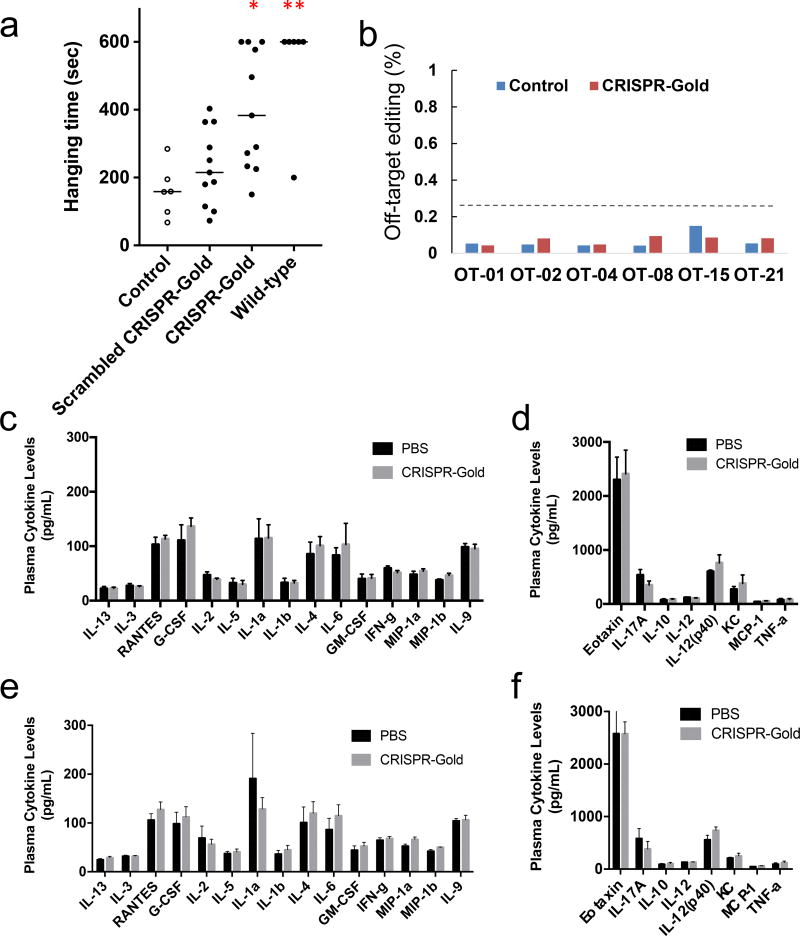
Encouraged by the above results, we performed additional experiments to determine the translational potential of CRISPR-Gold as a therapeutic for DMD under clinically relevant conditions (no CTX). Mdx mice were injected with CRISPR-Gold or the appropriate controls (without CTX), and a four-limb hanging test was performed on the mice to evaluate the therapeutic benefits of CRISPR-Gold. Promisingly, CRISPR-Gold was able to enhance animal strength and agility in mdx mice under these clinically relevant conditions. Specifically, CRISPR-Gold treated mdx mice showed a two-fold increase in hanging time in the four-limb hanging test in comparison to mdx mice injected with scrambled CRISPR-Gold (Figure 7a). CRISPR-Gold showed a 0.8% rate of HDR in the dystrophin gene without CTX (Supplementary Fig. 20). Additionally, deep sequencing analysis was performed to quantify the degree of off-target DNA damage it caused, and CRISPR-Gold’s degree of off-target genomic damage was minimal and similar to the levels of sequencing error (0.005 – 0.2%), Figure 7b. These results demonstrate that CRISPR-Gold can induce HDR in muscle tissue with minimal off-target genomic damage, effectively edit the dystrophin mutation in mdx mice to the wild type sequence, and improve animal strength under clinically relevant conditions
Figure 7. CRISPR-Gold induced dystrophin editing enhances muscle function with minimal off-target effects and without elevation of systemic inflammatory cytokines.
a) CRISPR-Gold improves strength and agility in mdx mice without CTX. A four-limb hanging test was conducted in mdx mice treated with CRISPR-Gold, two weeks after the initial CRISPR-Gold injection. CRISPR-Gold significantly enhanced muscle strength in comparison to mice treated with scrambled CRISPR-Gold. Mean ± S.E, n=11 for injected mdx mice, n=6 for control mdx mice and wild-type mice, *, p < 0.05, **, p < 0.01, statistically significant to scrambled CRISPR-Gold group.
b) In vivo off-target effects are insignificant in CRISPR-Gold injected mouse muscle tissue. The major predicted and reported off-target sites for the mdx gRNA were analyzed with deep sequencing. Control: scrambled CRISPR-Gold injected mouse and CRISPR-Gold: CRISPR-Gold injected mouse without CTX. Each read-out was more than 30,000 reads and all off-target mutations were within the range of deep sequencing error.
c–f) CRISPR-Gold does not elevate plasma cytokine levels as compared to PBS injections. c–d) Plasma cytokine levels after a single injection of CRISPR-Gold. e–f) Plasma cytokine levels after two injections of CRISPR-Gold. c, e) group of high plasma level cytokines. d, f) group of low plasma level cytokines.
Analysis of CRISPR-Gold immunogenicity
Cas9 is a bacterial protein, with potential immunogenicity, and the immune response generated from CRISPR-Gold could therefore be problematic and limit its translational potential. To examine the possibility of an immune response to CRISPR-Gold we injected CRISPR-Gold into the gastrocnemius muscle of mdx mice, at 6 mg/kg of Cas9 protein. 24 hours and 2 weeks after the CRISPR-Gold injection, the systemic cytokine profile was analyzed. Figures 7c–d demonstrates that CRISPR-Gold does not cause an acute up-regulation of inflammatory cytokines in plasma, thus suggesting the absence of a broad immune response towards CRISPR-Gold. Also, plasma cytokine levels were stable 2 weeks after the injection (Supplementary Fig. 21). Additionally, we stained the CRISPR-Gold treated muscle tissue for CD45 and CD11b positive cells, which are frequently found in muscle tissue undergoing inflammation and muscle regeneration41. We observed higher numbers of CD45+ and CD11+ leukocytes in tissue injected with CRISPR-Gold, as compared to the controls: untreated mdx and wild-type muscles (Supplementary Fig. 22–24). Macrophage-promoted clearance of nanoparticles/microparticles from muscle is expected, and intramuscular leukocytes are frequently found in the vicinity of FDA approved biomaterials, such as PLGA microparticles42.
In addition, we performed experiments where CRISPR-Gold was injected into mice, twice, three days apart, and analyzed for plasma cytokines and weight loss, to determine if CRISPR-Gold could be administered multiple times without toxicity. Figures 7e–f and Supplementary Fig. 25 demonstrate that CRISPR-Gold does not cause an up-regulation of inflammatory cytokines in plasma, after multiple injections, or weight loss, suggesting that CRISPR-Gold can be used multiple times safely, and that it has a high therapeutic window for gene editing in muscle tissue.
Outlook
In this report, we present a delivery vehicle termed CRISPR-Gold that can deliver Cas9 protein, gRNA, and donor DNA both in vitro and in vivo and edit genes via HDR. CRISPR-Gold offers a new therapeutic strategy for treating DMD patients that suffer from point mutations and small deletions. For this class of patients, CRISPR-Gold has the potential to correct their mutation back to the wild type sequence, and regenerate fully functional wild-type dystrophin, without the use of viruses. More broadly, the results presented here, demonstrate that non-viral delivery vehicles can generate HDR in vivo and have great potential for treating genetic diseases.
Methods
S1. Materials
Oligonucleotides were purchased from Integrated DNA Technologies (IDT, Coralville, IA). Gold nanoparticles (15 nm) were purchased from BBI solutions. Sodium citrate and 4-(2-hydroxyethyl) piperazine-1-ethanesulfonate (HEPES) were purchased from Mandel Scientific (Guelph, ON). Sodium silicate and cardiotoxin (C9759) were purchased from Sigma Aldrich (St. Louis, MO). Phusion High-fidelity DNA Polymerase was purchased from NEB (Ipswich, MA). The MEGAscript T7 kit, the MEGAclear kit, the PageBlue solution, the propidium iodide, and the PureLink genomic DNA kit were purchased from Thermo Fischer (Waltham, MA). Mini-PROTEAN TGX Gels (4–20%) were purchased from Bio-Rad (Hercules, CA). MTeSR-1 media gentle cell dissociation reagent was purchased from StemCell Technologies (Vancouver, Canada). Matrigel was purchased from BD Biosciences (San Jose, CA). DMEM media, non-essential amino acids, penicillin-streptomycin, DPBS and 0.05% trypsin were purchased from Life Technologies (Carlsbad, CA). EMD Millipore Amicon Ultra-4 100kDa was purchased from Millipore (Germany).
S2. Expression and purification of Cas9
The full-length catalytically active Streptococcus pyogenes Cas9 was expressed from an expression vector that was previously published in the manuscript by Jinek et al1,2. It was composed of a custom pET-based expression vector encoding an N-terminal 6×His-tag followed by the maltose-binding protein (MBP) and a TEV protease cleavage site, as well as two SV40 nuclear localization signal (NLS) peptides at its C-terminus. Recombinant Cas9 protein was expressed in the Escherichia coli strain BL21 (DE3) (Novagen) and further purified to homogeneity as previously described1,2. Purified Cas9 protein was stored in 50 mM HEPES at pH 7.5 with 300 mM NaCl, 10% glycerol, and 100 µM TCEP at −80°C. S. pyogenes Cas9 D10A nickase was expressed and purified following the same procedure3. Cas9 protein concentration was determined by a NanoDrop 2000 (Thermo Scientific) from the absorbance at 280 nm.
S3. In Vitro T7 Transcription of sgRNA
Oligonucleotide primers for sgRNA synthesis were purchased from IDT, with the forward primer containing a T7 promoter sequence. The DNA template for in vitro sgRNA transcription was prepared by overlapping PCR4. Briefly, the T7 forward template (20 nM), T7Rev-Long template (20 nM), T7 forward primer (1 µM), and T7 reverse primer (1 µM), were mixed with Phusion Polymerase (NEB) and PCR amplification was performed according to the manufacturer’s protocol. RNA in vitro transcription was performed with the MEGAscript T7 kit (Thermo Fisher) and purification of the resulting RNA was conducted using the MEGAclear kit, following the manufacturer’s protocol. The transcribed sgRNA was eluted into 50 mM HEPES at pH 7.5 with 300 mM NaCl, 10% (vol/vol) glycerol, and 100 µM TCEP. The concentration of sgRNA was determined with a Nanodrop 2000 (Thermo Scientific) and the final sgRNA products were stored at −80 °C for subsequent experiments.
S4. Synthesis of PAsp(DET)
Poly{N-[N-(2-aminoethyl)-2-aminoethyl] aspartamide} (PAsp(DET)) was synthesized as previously reported5,6. Briefly, poly(β-benzyl L-aspartate) (PBLA) was synthesized by the ring-opening polymerization of the β–benzyl-L-aspartate N-carboxy-anhydride (BLA-NCA) with initiation by n-butylamine. The polymerization proceeded for 48 hours at 37 °C under an argon atmosphere. The degree of polymerization of the benzyl-L-aspartate (BLA) unit was calculated to be 55 from the 1H NMR spectrum (DMSO-d6, 80°C). The resulting PBLA was reacted with diethylenetriamine (DET) to obtain PAsp(DET). After one hour of reaction, the reaction mixture was added dropwise into cold HCl. The polymer product was purified by dialysis against 0.01 M HCl and then against deionized water overnight at 4 °C. The dialyzed solution was lyophilized to obtain the final product.
S5. Synthesis of CRISPR-Gold
A representative synthesis of CRISPR-Gold is described in this section. Gold nanoparticles (GNP) (15 nm in diameter, 450 nM) were reacted with a 5’ thiol modified single stranded oligonucleotide (DNA-SH), 25 bases in length (200 µM), which had a region complementary to the donor DNA sequence, the reaction was performed in an Eppendorf tube in 20 mM HEPES buffer, in a 100 µL volume. The NaCl concentration of the reaction was increased a 100 mM per hour up to 400 mM (final volume 150 µL) by adding 1M NaCl solution, and the reaction was allowed to proceed overnight. Unconjugated DNA-SH was removed by centrifugation at 17,000 g for 15 minutes, and washed two times with 20 mM HEPES buffer. The resulting GNP-DNA conjugate was hybridized with the donor oligonucleotide, generating GNP-Donor. The donor DNA (100 µM concentration, 10 µL) was added to the GNP-DNA solution in 20 mM HEPES with 50mM NaCl (100 µL), and incubated at 65 °C for 10, then gradually brought to room temperature (−2 °C/min). The GNP-Donor solution was stored at 4 °C until further use. CRISPR-Gold was synthesized using a layer-by-layer method. Cas9 (8 µg in 10 µL) and gRNA (2 µg in 10 µL) were mixed in 80 µL of Cas9 buffer (50 mM HEPES (pH 7.5), 300 mM NaCl, and 10% (vol/vol) glycerol) for 5 min at RT, and this solution was then added to the GNP-Donor solution (0.45 pmole of GNP), generating GNP-Donor-Cas9 RNP. Freshly diluted sodium silicate (6 mM, 2 µL) was added to the GNP-Donor-Cas9 RNP solution and incubated for 5 min at RT. The mixture was centrifuged using an EMD Millipore Amicon Ultra-4 100 kDa at 3,000 rpm for 5 min to remove the unbound Cas9 RNP. The recovered GNP-Donor-Cas9 RNP-silicate was resuspended in 20mM HEPES buffer (100 µL) and PAsp(DET) was added to generate a final concentration of 100 µg/mL and incubated for up to 15 min at RT to form the last layer of CRISPR-Gold.
S6. Absorbance spectra, particle size (DLS and TEM), and zeta potential analysis
The synthetic intermediates in the synthesis of CRISPR-Gold, GNPs, GNP-DNA, GNP-DNA-donor DNA, GNP-Cas9 RNP and GNP-Cas9 RNP-Silicate were synthesized following the protocols described in S5 and characterized by UV-vis spectroscopy. The absorbance spectra of each sample were measured with a UV-vis spectrophotometer (NanoDrop 2000, Thermo Scientific). Dynamic light scattering (DLS) and zeta potential measurements were also made on each intermediate at 25 °C. Zeta potential measurements were made with a Zetasizer Nano ZS instrument (Malvern Instruments Ltd., Worcestershire, UK), and electrophoretic mobility was measured in a folded capillary cell (DTS 1060, Malvern Instruments). The zeta potential was calculated using the Smoluchowski equation. The size of the particles measured with DLS is reported in a number-based measurement mode. Each sample was prepared and incubated for a few minutes, to form particles, and then transferred to the capillary cell. An equilibration time ranging from and 2–5 minutes was needed to optimize the DLS measurements and collect accurate DLS data. Transmission electron microscopy (TEM) was conducted using a FEI Tecnai 12 microscope in the electron microscope lab at UC Berkeley. The samples were prepared on copper TEM grids (3.05 mm, 400 mesh).
S7. Gel electrophoresis analysis of CRISPR-Gold to determine Cas9 protein content
The ability of CRISPR-Gold to complex Cas9 was determined via gel electrophoresis, after separation of the unbound Cas9 RNP from CRISPR-Gold with a spin-filter (300 K MWCO). GNP-DNA (0.45 pmole) was incubated with Cas9 (8 µg) and gRNA (2 µg) for 5 min at RT in 100 µL of PBS. A 300kDa concentrator (Vivaspin 500, 300K MWCO) was used to remove the unbound components of CRISPR-Gold, and in particular the Cas9 and the gRNA. We performed preliminary experiments and determined that both components, the Cas9 and the gRNA, flowed through the 300kDa molecular cut-off membrane after spinning at 2,000 g for 3 min, and could be separated from CRISPR-Gold. After concentrating the GNP-Cas9 RNP through a 300kDa molecular cut-off membrane, the flow through (filtrate) was collected for analysis. Similar analysis was performed on CRISPR-Gold. CRISPR-Gold without purification (control), the flow through solution from the wash step, and CRISPR-Gold after purification were analyzed via gel electrophoresis. Gel electrophoresis was performed using a 4–20% Mini-PROTEAN TGX Gel (Bio-rad) in Tris/SDS buffer. Heparin (100 µg) was added to the CRISPR-Gold sample to dissociate the PAsp(DET) polymer from the particle to facilitate gel electrophoresis. Nucleic acid staining was conducted with Sybr Safe and then the gel was imaged with ChemiDoc MP using ImageLab software (Bio-rad); subsequently, PageBlue solution (Thermo Fischer) staining was conducted and the gel was imaged again with the same software. The protein content in the particles was quantified via densitometry analysis on the respective gel bands. The percent binding of CRISPR-Gold was determined by comparing the amount of Cas9 present in CRISPR-Gold after purification (retentate) versus the amount of Cas9 present in the unpurified CRISPR-Gold sample. Experiments were also performed to determine if gold nanoparticles by themselves (without DNA modification) bound Cas9 RNP, following the identical procedure as described above. The percent of Cas9 RNP bound to the GNPs was determined by comparing the recovered Cas9 RNP with the amount of Cas9 RNP added to the GNPs before washing. All the Cas9 RNP binding experiments were performed in triplicate.
S8. Enzymatic activity of Cas9 released from CRISPR-Gold
The enzymatic activity of Cas9 released from CRISPR-Gold was analyzed via gel electrophoresis. Purified samples of GNP-Cas9 RNP and CRISPR-Gold from S7 were prepared and incubated in 40 µL PBS containing 5 mM beta-mercaptoethanol at 37°C for 1 hour, to release Cas9 from the GNPs. The particles were centrifuged at 17,000 g for 10 min, and 10 uL of the supernatants were collected and incubated with a PCR amplicon (200 ng) that contained a Cas9 cleavage site. After incubation at 37°C for 2 hr, the samples were analyzed by gel electrophoresis using a 4–20% Mini-PROTEAN TGX Gel (Bio-rad), stained with SYBR safe (Thermo Fischer) and imaged with a ChemiDoc MP using ImageLab software (Bio-rad).
S9. Cell culture
S9.1 BFP expressing HEK cell culture
BFP-HEK cells were generated by infection of HEK293T cells with a BFP-containing lentivirus, followed by FACS-based enrichment using the protocol published by Richardson et al7. The lenti-virus was generated by transfection of HEK293FT cells with a custom lentiviral vector containing a BFP gene driven by the pEF1 promoter, cloned into a Lenti X1 DEST Blast backbone by Gateway cloning (Life Technologies, Inc.). Reporter cell lines were generated by infection of HEK293T cells with lentivirus, at low MOI (as estimated by FACS 3 days post-infection). BFP-positive cells were enriched by FACS, grown out, and sorted into clones by FACS. A clone with high constitutive BFP fluorescence (>99% BFP-positive) after expansion was selected as a reporter for BFP-GFP conversion by CRISPR-Gold-mediated HDR. To edit BFP-HEK to GFP, cells were plated at a density of 5×104 cells per well in a 24 well plate, a day before CRISPR-Gold experiments, and cultured in DMEM with 10% FBS, 1× MEM non-essential amino acids, and 100 µg/mL Pen Strep.
S9.2 Stem cell culture
Human H9 embryonic stem (hES) cells and human induced pluripotent stem (hiPS) cells were cultured according to the procedure described by Downing et al8. Cell culture plates were coated with Matrigel diluted to 12.5 µL/mL in DMEM and incubated for an hour at 37 °C1. MTeSR-1 medium (StemCell Technologies) was added to the cells every day and the cells were passaged passaged into 24 well plates, at a density of 2×104 cells per well, 3 days prior to Cas9 transfection. Gentle cell dissociation reagent (StemCell Technologies) was used for cell detachment, according to the manufacturer’s protocol.
S9.3 Mouse primary bone marrow derived dendritic cell culture
Bone marrow cells were obtained from the tibias and femurs of mice following the procedure of Matheu et al9. Bone marrow cells were plated in complete medium containing granulocyte-macrophage colony-stimulating factor (GM-CSF) (10 ng/mL; Peprotech) for 6 days to allow for differentiation into DCs. Cas9 transfection was conducted on Day 6.
S9.4 Isolation and culture of primary myoblasts from mdx mice
Primary myoblasts were obtained from C57BL/10ScSn-Dmdmdx/J (mdx) mice following the previously reported protocol from Conboy et al. and Rando et al.10,11. Briefly, the gastrocnemius and tibialis anterior muscles were harvested and incubated in collagenase for tissue dissociation. Isolated myoblasts were maintained on Matrigel coated culture plates for a few weeks with fresh medium replacement every 24hr. Differentiation of the primary myoblasts to myotubes was conducted after the CRISPR-Gold treatment. After overnight incubation with CRISPR-Gold, the medium was switched to a differentiation medium (DMEM, 2% bovine growth serum, pen/strep), and cultured for an additional 5 days to allow for dystrophin expression. The myotubes were then lysed, and protein was collected for Western Blotting.
S10. Cell transfection
S10.1 CRISPR-Gold treatment
For all of the in vitro cell experiments, 105 cells were seeded in a 24 well plate one day before the transfection. Unless otherwise indicated, the cells in a 1 mL volume, were treated with 0.45 pmole GNP-Donor (determined by absorbance), Cas9 (8 µg), gRNA (2 µg), 2 µL sodium silicate (6 mM), and 10 µg PAsp(DET). Cas9 and gRNA solution were mixed in Cas9 buffer (50 mM Hepes (pH 7.5), 300 mM NaCl, and 10% (vol/vol) glycerol) for 5 min at RT, and added to the GNP-Donor solution. Freshly diluted sodium silicate (6 mM, 2 µL) was added to the GNP solution and incubated for 5 min at RT. The reaction mixture was centrifuged using an EMD Millipore Amicon Ultra-4 100 kDa at 3,000 rpm for 5 min to remove unbound sodium silicate. The recovered gold nanoparticles were resuspended in 20mM HEPES buffer (100 µL) and PAsp(DET) polymer was added to a final concentration of 100 µg/mL and incubated up to 15 min at RT to form the last layer of CRISPR-Gold. For experiments in which less or more than 8 µg/mL of Cas9 were used, the other components of CRISPR-Gold were changed accordingly, following the same ratios as described above. The CRISPR-Gold solution was added to cells to generate a final Cas9 concentration of 8 µg/mL of Cas9 protein. The cells were incubated with CRISPR-Gold in serum-free OptiMEM for 4 hours, and then the medium was changed to fresh culture media (DMEM).
S10.2 Nucleofection
Cells were detached by 0.05 % trypsin or a gentle dissociation reagent and spun down at 600 g for 3 min, then washed with PBS. Nucleofection was then conducted using an Amaxa 96-well Shuttle system following the manufacturer’s protocol, using 10 µL of Cas9 RNP and DNA donor (Cas9 : 100 pmole, gRNA : 120 pmole, DNA donor : 100 pmole) (1.6 mg/mL of Cas9 protein and later diluted to 16 µg/mL in cell culture). The nucleofection program was chosen to match the cell type used for the experiment. After the nucleofection, 500 µL of growth media was added, and the cells were incubated at 37°C in tissue culture plates. The cell culture media was changed 16 hr after the nucleofection, and the cells were incubated for a total of 3 days before genomic DNA extraction and analysis was conducted.
S10.3 Lipofection
Lipofectamine transfection with Cas9 was performed following the protocol described in Zuris et al. using 4.4 µg of Cas9, 1.2 µg of gRNA, and 1.2 µL of Lipofectamine 2000 in a 100 µL total volume12. Additionally, donor DNA (250 ng) was mixed with lipofectamine (500 nL) and added to the transfection media, which contained the Cas9 RNP lipofectamine solution. The lipofection was conducted in OptiMEM media without serum, and an equal volume of 2× growth media was added to the cells after 1hr of lipofection to minimize cytotoxicity. The medium was changed 16 hr after the lipofection and the cells were incubated for a total of 3 days before genomic DNA extraction and analysis.
S11. Flow cytometry analysis & fluorescence microscopy
Flow cytometry was used to quantify the expression levels of BFP and GFP in BFP-HEK cells treated with CRISPR-Gold. The BFP-HEK cells were analyzed 7 days after Cas9 treatment. The cells were washed with PBS and detached by 0.05% trypsin. BFP and GFP expression was quantified using BD LSR Fortessa X-20 and Guava easyCyte™.
S12. Sanger sequencing of the BFP/GFP gene
The GFP+ population was sorted from BFP-HEK cells that had been treated with CRISPR-Gold (7 days after treatment). Cells were detached by 0.05% trypsin treatment, and the GFP+ edited cells were sorted using a BD influx cell sorter (BD Biosciences) in the Berkeley flow cytometry facility. Genomic DNA was extracted from the GFP+ sorted cells and PCR amplification of the BFP/GFP gene was conducted, following the procedure described in S13. Sanger sequencing was conducted by Quintara Inc (CA, USA) and the sequence was analyzed with apE software.
S13. PCR amplification of genomic DNA from transfected cells
Genomic DNA of 2 × 104 to 2 × 105 cells was extracted after transfection using the Purelink genomic DNA kit (Thermo Fisher). The concentration of genomic DNA was measured with a Nanodrop 2000 (Thermo Scientific). The target genomic DNA sequences (BFP, CXCR4, and dystrophin) were amplified using primer sets and Phusion polymerase with high efficiency (HE) or GC buffer, according to the manufacturer’s protocol. All primer sets were designed to anneal outside of the homology arms of the donor DNA in order to avoid amplifying the donor DNA. The PCR products were analyzed on a 1.5% (wt/vol) agarose gel casted with SYBR Safe (Thermo Fischer).
S14. Analysis of genome editing efficiency with restriction enzyme digestion and Surveyor assay
HDR was determined by the restriction enzyme digestion method, and indel mutations were determined by the surveyor assay. The HDR efficiency in cells was determined with restriction enzyme digestion of PCR amplified target genes. Donor DNAs were designed to insert restriction enzyme sites, cleavable by either HindIII or DraI, into the target gene locus. The PCR amplicon of the CXCR4 locus and the PCR amplicon of the DMD locus were incubated with 10 units of HindIII and DraI, respectively. After 2 hr to 16 hr of incubation at 37 °C, the products were analyzed by gel electrophoresis using a 4–20% Mini-PROTEAN TGX Gel (Bio-rad) and stained with SYBR green (Thermo Fischer). Individual band intensity was quantified using ImageJ and the HDR efficiency was calculated using the following equation: (b + c) / (a + b + c) × 100 (a = uncleaved PCR amplicon, b and c = the cleavage products). The surveyor assay was conducted to estimate the total DNA editing (NHEJ +HDR) by cutting mismatched heteroduplex DNA from mutant or HDR modified DNA. Hybridization and SURVEYOR incubation were performed as described in Schumann et al.13
S15. Cell viability assays
The relative cell viability of cells transfected with CRISPR-Gold, nucleofection, and lipofection was determined with a cell counting kit (Dojindo) using regular culture media supplemented with 10% (v/v) CCK solution. Cells were plated in a 24 well plate at a seeding density of 105 cell/well and CRISPR-Gold was treated to the cells as described in S10.1. The CCK assay was conducted 2 days after the transfection. Relative cell viability was defined as percent viability compared to untreated controls.
S16. Sanger sequencing of hES cells edited with CRISPR-Gold
The CXCR4 PCR amplicon of CRISPR-Gold treated hES cells were cloned into plasmids using a Zero Blunt TOPO PCR cloning kit (Life Technologies), following the manufacturer’s instruction. Briefly, TOP10 E.coli were transformed with plasmids containing the PCR amplicons and cultured on LB plates containing kanamycin. Sanger sequencing of the CXCR4 gene cloned into the E.coli colonies was conducted by Quintara Bioscience (CA, USA).
S17. Inhibitor studies with CRISPR-Gold
Cell culture experiments with various cellular uptake inhibitors and under conditions of low temperature were performed to investigate the mechanism of CRISPR-Gold uptake. L2 sgRNA was labeled with Alexa647-NHS-ester following the method from Lee et al.4. Briefly, 5’ amine modified L2 sgRNA was incubated with Alexa647-NHS-ester (100 fold molar excess) in pH9 sodium bicarbonate buffer overnight at RT and purified with desalting column. CRISPR-Gold was formulated with 647-L2 sgRNA as described in S10.1. Wortmannin (150 ng/mL), chlorpromazine (1.5 µg/mL), Genistein (5 µg/mL), and methyl-β-cyclodextrin (7.5 mg/mL) were added to HEK-293 cells for 1 h under regular culture conditions (serum). The cells were washed with PBS twice and then treated with CRISPR-Gold at a concentration of 8 µg/mL Cas9 protein concentration. All samples were incubated at 37°C except for the 4°C sample, which was incubated at 4°C for 1 hour without any inhibitors. The cells were analyzed 16hr after the CRISPR-Gold treatment to quantify the Alexa 647+ cell population using flow cytometry.
In a separate set of experiments, BFP-HEK cells were treated with CRISPR-Gold designed to convert the BFP gene into the GFP gene via HDR. CRISPR-Gold was formulated with L2 sgRNA as described in S10.1. Wortmannin (150 ng/mL), chlorpromazine (1.5 µg/mL), Genistein (5 µg/mL), and methyl-β-cyclodextrin (7.5 mg/mL) were added to BFP-HEK-293 cells for 1 h under regular culture conditions (serum). The cells were washed with PBS twice and then treated with CRISPR-Gold at concentration of 8 µg/mL Cas9 protein concentration. All samples were incubated at 37°C. HDR efficiency was analyzed 3 days after the treatment following the procedures described in S10.1 and S11.
S18. Western blot analysis for dystrophin protein production
Myoblasts were treated with CRISPR-Gold following the procedure described in S10. After differentiation, cells were harvested and protein extracts for Western blot analysis were made following the procedure of Lu QL, et al., 200514. Briefly, cells were lysed in RIPA buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.5% deoxycholate, and 1% Nonidet P-40) containing proteinase inhibitor, and the total protein concentration was determined using the BCA protein assay kit (Thermo Scientific, USA). 150 µg of protein per sample was loaded onto a 4–20% gradient polyacrylamide gel (Bio-Rad, CA, Cat # 4561094), and run 6 hours at 35 volts, the protein content of the gel was then transferred onto nitrocellulose membranes. The membranes were blocked with 5% milk in Tris Buffered Saline (TBS) plus 0.1% Tween20 followed by an overnight incubation with dystrophin antibody (dilution 1 : 200, Abcam, UK) in TBS plus 0.1% Tween20 to detect dystrophin. GAPDH (dilution 1 : 2000, Thermo Scientific, USA) was used as a sample loading control. Blots were washed with 0.2% Tween-20 in TBS 3 times, and then incubated with a horseradish peroxidase (HRP)-coupled secondary antibody (Azures Biosystems, USA). Antibody binding was detected using an enhanced chemiluminescent detection system (Amersham).
S19. In vivo delivery of CRISPR-Gold in Ai9 mice
Ai9 (Jackson Laboratory, #007909) mice were purchased from Jackson Laboratory. All animal studies were performed following authorized protocols, and animals were treated in accordance with the policies of the animal ethics committee of the University of California Berkeley and ACUC. Three groups of Ai9 mice were used for this experiment. The experimental groups were: Control (no injection, n=3), CRISPR-Gold (mice injected with CRISPR-Gold, n=3), and positive control (Cre plasmid and lipofectamine injection, n=1). Each group of mice were selected randomly. Injections were performed in 4-week-old Ai9 mice in the tibialis anterior (TA) muscle (10 µL per muscle) and gastrocnemius muscles (10 µL per muscle), using a Hamilton syringe. Two weeks after the injection, the muscles were harvested and analyzed. CRISPR-Gold particles were formed as described in S5. For all Ai9 mice in vivo experiments, 6.75 pmole GNP-DNA, 120 µg Cas9 (6 mg/kg dose), 30 µg sgRNAs (15 µg of Ai9-F and 15 µg of Ai9-R), 30 µL sodium silicate (6 mM), and 150 µg PAsp(DET) were mixed and incubated for 5 min to formulate CRISPR-Gold, and then were injected into the mice. For the positive control, Cre plasmid (20 µg) and Lipofectamine 2000 (40 µL) were mixed and incubated for 10 min before the injection.
S20. High throughput automated imaging of a whole muscle section
Glass slides with whole muscle sections were imaged with Molecular Devices ImageXpress Micro. On average, about 288 images were taken per slide with a 10×objective lens. The images were analyzed using MetaXpress software to merge the images and create a montage of the whole muscle section.
S21. In vivo delivery of CRISPR-Gold in mdx mice treated with cardiotoxin
Male C57BL/10ScSn (wild-type) mice and C57BL/10ScSn-Dmdmdx/J (mdx) mice that contain a nonsense mutation in exon 23 of the dystrophin gene were purchased from Jackson Laboratory. All animal studies were performed following authorized protocols, and animals were treated in accordance with the policies of the animal ethics committee of the University of California at Berkeley and ACUC. Three groups of mdx mice were used for this experiment. The experimental groups were:
Control 1 no GNP group, consisting of mice injected with Cas9 RNP and donor DNA without gold nanoparticles (n=3).
Control 2 no gRNA group, consisting of mice injected with CRISPR-Gold without gRNA (n=1),
CRISPR-Gold group, consisting of mice injected with CRISPR-Gold containing RNP and donor DNA (n=3).
**Each group of mice were selected randomly. The investigators were not blinded to the group allocation during the experiment. CRISPR-Gold treatments were performed in 2-month-old mdx mice in the tibialis anterior (TA) muscle (10 µL per muscle) and gastrocnemius muscles (10 µL per muscle), using a Hamilton syringe. CRISPR-Gold particles were formed as described in S5. For all mdx in vivo experiments, 6.75 pmole GNP, 120 µg Cas9 (6 mg/kg dose), 30 µg gRNA, 30 µL sodium silicate (6 mM), and 150 µg PAsp(DET) were mixed and incubated for 5 min to formulate CRISPR-Gold, and then were injected into the mice. The injection mix contained cardiotoxin (0.1mg/ml) mixed with lidocaine hydrochloride. For the no GNP control, 120 µg Cas9 (6 mg/kg dose) + 30 µg gRNA were formulated in a 10 µL volume containing cardiotoxin (0.1mg/ml) mixed with lidocaine hydrochloride, and injected into mdx mice. The experiments were conducted in a non-blinded and non-randomized way. For the experiments with a Cas9 dose of 3 mg/kg, all of the other CRISPR-Gold reagents were scaled back accordingly. Two weeks after the injection, the mice were sacrificed and the muscles were analyzed by deep sequencing and histology for dystrophin expression and fibrosis.
S22. In vivo delivery of CRISPR-Gold in mdx mice without cardiotoxin treatment
Male C57BL/10ScSn (wild-type) mice and C57BL/10ScSn-Dmdmdx/J (mdx) mice that contain a nonsense mutation in exon 23 of the dystrophin gene were purchased from Jackson Laboratory. All animal studies were performed following authorized protocols, and animals were treated in accordance with the policies of the animal ethics committee of the University of California at Berkeley. Four groups of mice were used for this experiment. The experimental groups were:
Negative Control, consisting of mdx mice without injection (n=6).
Control scrambled CRISPR-Gold, consisting of mice injected with CRISPR-Gold containing scrambled gRNA (n=11).
CRISPR-Gold, consisting of mice injected with CRISPR-Gold containing RNP and donor DNA (n=11).
wild-type C57BL/10ScSn mice n=6.
**The injections were conducted in 4-week-old mdx mice in the tibialis anterior (TA) muscle (10 µL per muscle), gastrocnemius muscle (10 µL per muscle), and forelimb muscle (10 µL per muscle) using a Hamilton syringe. Two weeks after the injection, the mice received a second round of injections with exactly the same composition. Two weeks after the second injection, mice were sacrificed and the muscles were analyzed by deep sequencing and for dystrophin expression. A four-limb hanging test was conducted on CRISPR-Gold treated mdx mice, at the age of 6 weeks, which was two weeks after the initial injection. Mice were placed on a hand made square apparatus with a grid structure. The apparatus was inverted and positioned 25 cm up from the cage to discourage intentional dropping. Soft bedding was prepared to prevent the mice from harming themselves if they fell. The maximum hanging time out of three trials was recorded for a duration of 600 sec, the chosen fixed hanging time limit was determined following the procedure of Aartsma-Rus et al.15 The wild type mice were also tested at the age of 6 weeks. An unpaired student t test was conducted using Prism 7 software. The experiments were conducted in a blinded manner.
S23. Deep sequencing analysis of CRISPR-Gold treated muscle tissue
The genomic region of the Cas9 target sequence was amplified by PCR using Phusion high-fidelity polymerase according to the manufacturer’s protocol. Target genes were amplified first with primer sets used for HDR detection and amplified again with deep sequencing primers to eliminate the potential of donor sequence amplification. The amplicons were purified using the ChargeSwitch PCR clean-up kit (Thermo Fischer). The Nextflex rapid illumine DNA-seq library prep kit was used to attach illumine adapters and PCR amplify the product for five cycles. PCR clean-up was performed one additional time. The Berkeley Sequencing facility performed DNA quantification using a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA). A bioAnalyzer was then used for size analysis and qPCR quantification. The library was sequenced with the Illumina HiSeq2500 in the Vincent Coates Genomic Sequencing Laboratory at UC Berkeley. The analysis was conducted using the CRISPR Genome Analyzer16.
S24. Immunofluorescence of dystrophin, CD45, and CD11b
Gastrocnemius and tibialis anterior muscles were frozen, sectioned to 10 micron, and fixed with 70% ETOH at 4C overnight. After blocking for 1 hr with 0.1% TritonX100, slides were incubated with a primary antibody against dystrophin (Santa Cruz Biotechnology-47760 or 358922) or alternatively, with anti-CD11b antibody or anti-CD45 antibody (F10-89-4, EMD Millipore 05-1410) antibody in PBS, 1% FBS overnight. After 3 × 5 minute washes with PBS, and 1%FBS, slides were incubated with the respective secondary antibodies (Santa Cruz Biotechnology-2005, 362282, donkey anti-rabbit 546 Life Technologies A11010, donkey anti-rat 488 Life Technologies A21206) for 2 hours at RT in PBS, 1% FBS that also contained Hoechst nuclear dye. Slides were imaged with a Zeiss Axioscope fluorescent microscope. All images were taken at 40× magnification.
C57BL/6 mice were treated with cardiotoxin and then stained for CD45 and CD11b. The procedure for treating C57BL/6 mice with cardiotoxin is described below. Mice were anesthetized with isoflurane, and the hind leg muscles (Tibialis Anterior, and/or Gastrocnemius) were injured by percutaneous injections of 5–10 microliters of cardiotoxin-1 (1mg/mL, Sigma). Typically, such small focal injuries completely heal in 5 days. Animals were monitored for general signs of health (activity, inquisitiveness), and specifically the regeneration site (e.g., leg) were examined for signs of tissue damage or necrosis (which did not occur).
S25. Trichrome staining
Muscle sections were stained using a Gomori Trichrome Stain Kit (Polysciences #24205-1) according to the manufacturer’s instructions. Briefly, ethanol fixed sections were fixed again, overnight at room temperature in Bouin’s fixative, then stained with Wiegert’s Iron Hematoxylin, then with Gomori Trichrome Stain, then with 1% acetic acid, and then finally dehydration, clearing and mounting steps were performed.
S26. PCR amplification of genomic DNA from CRISPR-Gold edited muscle tissue
Muscle genomic DNA from either control mice (Cas9 RNP + donor DNA without GNP) or CRISPR-Gold treated mice was amplified with primers designed to only amplify the HDR edited sequence. PCR was conducted using the forward primer (AAAGGAGCAGCAGAATGGCT), the reverse primer (CCACCAACTGGGAGGAAAG), and Phusion polymerase with GC buffer according to the manufacturer’s protocol. The PCR products were analyzed on a 1.5% (wt/vol) agarose gel casted with SYBR Safe (Thermo Fischer).
S27. Off-target deep sequencing analysis
Deep sequencing was performed on CRISPR-Gold treated mdx mice (without CTX) to investigate the frequency of off-target genomic damage. Potential off-target loci were determined using CRISPR off-target prediction programs, which were identical to off-target loci identified by Long et al.17. PCR was conducted using primers listed in the Supplementary Tables. The amplicons were purified using the ChargeSwitch PCR clean-up kit (Thermo Fischer). The Nextflex rapid illumine DNA-seq library prep kit was used to attach illumine adapters and PCR amplify the product for five cycles. PCR clean-up was then performed a second time. The Berkeley Sequencing facility performed DNA quantification using a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA). BioAnalyzer for size analysis and qPCR quantification followed. The library was sequenced with the Illumina HiSeq2500 in the Vincent Coates Genomic Sequencing Laboratory at UC Berkeley using the 150PE read. The analysis was conducted using the CRISPR Genome Analyzer (54.80.152.219). Figure 7b in the main manuscript presents the off-target mutation frequency of control and the CRISPR-Gold injected mouse samples (without CTX).
S28. Bio-Plex cytokine assay and weight loss
Systemic inflammation and toxicity induced by CRISPR-Gold was assessed by measuring the concentrations of 22 murine cytokines in mice that had been injected with CRISPR-Gold, and their weight loss. The plasma cytokine concentrations were measured with Bio-Plex kits (Bio-Rad, USA, Cat#: M60009RDPD). CRISPR-Gold mediated inflammation was assayed under three different conditions, described below:
Condition 1: A single CRISPR-Gold injection in mice, mouse sacrifice after 24 hr – measurement of plasma cytokines and weight loss.
Condition 2: A single CRISPR-Gold injection in mice, mouse sacrifice after 2 weeks – measurement of plasma cytokines.
Condition 3: Two CRISPR-Gold injections in mice, 3 days apart, mouse sacrifice 24 hr after the 2nd injection and measurement of plasma cytokines and weight loss.
Two groups of mdx mice were used for each condition, the control PBS group and the CRISPR-Gold group (n = 3 for each group). The mice were injected with CRISPR-Gold at a dose of 6 mg/kg of Cas9 protein, per injection. The mice were injected following the method described in S22, in the gastrocnemius and TA muscles, using a 10 µL volume. Mice were sacrificed either 24 hours or 2 weeks after the last CRISPR-Gold injection and their plasma was collected and analyzed. All samples for the cytokine assay were prepared using a Bio-Plex Cell Lysis Kit (BioRad, USA, Cat#: 171304011). The assays were run according to manufacturer’s recommended procedures. The plates were read in the Bio-Plex 200 System and the data was analyzed using the BioPlex Manager 4.1 software. The assays were performed in duplicate and all data points were analyzed except for one cytokine that was not detected in the assay.
S29. Statistics
Statistical analysis was conducted using Prism7 software. A Student-t-test was conducted for two sample analysis and a One-way ANOVA with post-hoc Tukey HSD was conducted for multiple sample analysis.
Supplementary Material
Table 1.
Primers for HDR analysis
| mdx | |
| Forward | GAGAAACTTCTGTGATGTGAGGACATATAAAG |
| Reverse | CAATATCTTTGAAGGACTCTGGGTAAAATATC |
| CXCR4 | |
| Forward | TTAATTCTCTTGTGCCCTTAGCCCACTACTTCAG |
| Reverse | GGACAGGATGACAATACCAGGCAGGATAAGGCC |
Table 2.
Sequencing Primers
| mdx | |
| Forward | GCGTGTTAGTGTAAATGAACTTCTA |
| Reverse | CCACCAACTGGGAGGAAAG |
| CXCR4 | |
| Forward | GAGAAGCATGACGGACAAGTACAGGCTG |
| Reverse | TTGACTGTGTAGATGACATGGACTGCCT |
| BFP | |
| Forward | TGTCCGGCGAGGGCGAGGGCGAT |
| Reverse | CGTCCTTGAAGAAGATGGTGCGC |
Table 3.
Thiol oligonucleotides for conjugation to GNP
| GFP-SH | /5ThioMC6-D/GCGCTCCTGGACGTAGCCTTCGGGCAT |
| mdx-SH | /5ThioMC6-D/AAATTCTGACAGATATTTCTGGCATATTTC |
| CXCR4-SH | /5ThioMC6-D/AATAGATGACATGGACTGCC |
Table 4.
Donor DNA
| MDX | TGATATGAATGAAACTCATCAAATATGCGTGTTAGTGTAAATGAACTTCTATTTAATTTTGAGGCTCTGCAAAGTTCTTTAAAGGAGCAGCAGAATGGCTTCAACTATCTGAGTGACACTGTGAAGGAGATGGCCAAGAAAGCACCTTCAGAAATATGCCAGAAATATCTGTCAGAATTT |
| CXCR4 | CCTGGTCATGGGTTACCAGAAGAAACTGAGAAGCATGACGGACAAGTACAGGCTGCACCTGTCAGTGGCCGACCTCCTAAGCTTGGATCCCTTTGTCATCACGCTTCCCTTCTGGGCAGTTGATGCCGTGGCAAACTGGTACTTTGGGAACTTCCTATGCAAGGCAGTCCATGTCATCTA |
| GFP | GATGCCACCTACGGCAAGCTGACCCTGAAGTTCATCTGCACCACCGGCAAGCTGCCCGTGCCCTGGCCCACCCTCGTGACCACCGTGACGTACGGCGTGCAGTGCTTCAGTCGCTACCCCGACCACATGAAGCAGCACGACTTCTTCAAGTCCGCCATGCCCGAAGGCTACGTCCAGGAGCGC |
Table 5.
sgRNA T7 Template Forward Primer
| CXCR4 | GATCCTAATACGACTCACTATAGAAGCGTGATGACAAAGAGGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC |
| Mdx | GATCCTAATACGACTCACTATAGGTCTTTGAAAGAGCAATAAAAGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC |
| GFP-L2 | GATCCTAATACGACTCACTATAGCTGAAGCACTGCACGCCATGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC |
| CXCR4-F1 (Cas9-nickase) | GATCCTAATACGACTCACTATAGTACAGGCTGCACCTGTCAGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC |
| DMD | GATCCTAATACGACTCACTATAGTCTTTGAAAGAGCAACAAAAGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC |
| Ai9-F | GGATCCTAATACGACTCACTATAgAAAGAATTGATTTGATACCGGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC |
| Ai9-R | GGATCCTAATACGACTCACTATAgGTATGCTATACGAAGTTATTGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC |
| gRNA primers | |
| T7Rev-Long | AAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC |
| T7 Forward Primer : GGATCCTAATACGACTCACTATAG | |
| T7 Reverse Primer : AAAAAAGCACCGACTCGG | |
Ai9 sequence was designed based on Tabebordbar et al.18
Table 6.
Off-target primers
| OT-01_F | TATGCCACTTCTTCAAAGAGATGAT |
| OT-01_R | AACAAGCAAACAATTCAAAGGATAG |
| OT-02_F | AAGAAGATATGGCATTGCTGGTA |
| OT-02_R | TCTGGAAACAAAAAGGCAATG |
| OT-04_F | CTATGAGTTTACCACCCTAATGTGC |
| OT-04_R | CTTATGCTTGTTCAGGCAAATACC |
| OT-08_F | TTTCTTGGAGCTGTAGTGTGTACTG |
| OT-08_R | GGAATAGAGTGAGCATTGTTCTGAT |
| OT-15_F | TTAAGCGGAAAGATAAGCTGAAGTA |
| OT-15_R | GGACCAATGTTACTGGAACACATAC |
| OT-21_F | CATGAAGATACAGAAACATCCCAGT |
| OT-21_R | GGAGTGGCACCCTCCTTAC |
Acknowledgments
This work was supported by the grants from NIH, U01 268201000043C-0-0-1, R56 AI107116-01 to IC, and RO1 AI088023-03, Grants from Calico, Roger’s and SENS to IC. In addition, this work was supported by the WM Keck Foundation, the Moore Foundation, Li Ka Shing Foundation, and the Center of Innovation (COI) Program of the Japan Science and Technology Agency (JST). K.L. is a Siebel Fellow of the Siebel Scholars Foundation. F.J. is a Merck Fellow of the Damon Runyon Cancer Research Foundation (DRG-2201-14). M.D. is a CIRM post-doctoral fellow, and is supported by CIRM training grant TG2-01164. J.A.D is Howard Hughes Medical Institute Investigator. We thank M. West in the CIRM/QB3 Shared Stem Cell Facility and H. Nolla T. Shovha in the Berkeley FACS facility for technical assistance; D. Schaffer and L. S. Qi, B. Staahl, S. Lin, and S. Yang for advice and technical support. This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303.
Footnotes
Author Contributions
K.L. planned and performed experiments shown in all figures, analyzed and interpreted the data and wrote the manuscript, M.C. designed, planned and performed the in vivo studies shown in Figure 5 and 6, and wrote the manuscript. H.M.P. and H.L performed in vivo experiments, analyzed and interpreted the data. F.J. purified Cas9 protein and analyzed gel data, H.J.K. and K.K. synthesized the PAsp(DET) polymer and performed the experiment shown in Supplementary Table 1 and contributed to the data interpretation, W.H. and S.L. cultured stem cells and performed the experiment shown in Supplementary Fig. 7, M.A.D. and J.E.C. generated the BFP-HEK cells and supported deep sequencing analysis, V.A.M., K.C., H.M.P. and A.R. performed in vitro and in vivo DNA analysis work, C.S, M.M. and T.S performed muscle histology studies, F.L. and N.L.B performed deep sequencing analysis, J.E.C. and J.A.D. contributed to the design of the studies and data interpretation. I.C. and N.M. planned and integrated the work, interpreted the data, and wrote the manuscript.
Competing interests:
K.L., H. P, and N.M. are co-founders of GenEdit Inc. J.A.D. is a co-founder of Caribou Biosciences, Editas Medicine, and Intellia Therapeutics.
List of DNA sequences
Please see Tables 1–6 in the Supplementary Information for a list of the DNA sequences used in this manuscript.
Data Availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information
References
- 1.Jinek M, et al. A Programmable Dual-RNA – Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–822. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong L, et al. Multiplex Genome Engineering Using CRISPR/Cas System. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho SW, Kim S, Kim JM, Kim J-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–2. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 5.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–190. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin H, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–33. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schumann K, et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. U. S. A. 2015;112:201512503. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin S, Staahl B, Alla RK, Doudna Ja. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:1–13. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabebordbar M, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson CE, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long C, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dever DP, et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin H, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32:551–3. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeWitt MA, et al. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci. Transl. Med. 2016;8:360ra134. doi: 10.1126/scitranslmed.aaf9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun W, et al. Self-Assembled DNA Nanoclews for the Efficient Delivery of CRISPR-Cas9 for Genome Editing. Angew. Chemie Int. Ed. 2015;54:12029–12033. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramakrishna S, et al. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24:1020–7. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X, et al. Improved delivery of Cas9 protein/gRNA complexes using lipofectamine CRISPRMAX. Biotechnol. Lett. 2016;38:919–929. doi: 10.1007/s10529-016-2064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyata K, et al. Polyplexes from poly(aspartamide) bearing 1,2-diaminoethane side chains induce pH-selective, endosomal membrane destabilization with amplified transfection and negligible cytotoxicity. J. Am. Chem. Soc. 2008;130:16287–16294. doi: 10.1021/ja804561g. [DOI] [PubMed] [Google Scholar]
- 20.Kim HJ, et al. Introduction of stearoyl moieties into a biocompatible cationic polyaspartamide derivative, PAsp(DET), with endosomal escaping function for enhanced siRNA-mediated gene knockdown. J. Control. Release. 2010;145:141–148. doi: 10.1016/j.jconrel.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 21.Ding Y, et al. Gold nanoparticles for nucleic acid delivery. Mol. Ther. 2014;22:1075–83. doi: 10.1038/mt.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh D, Sternberg SH, Fei J, Ha T, Doudna JA. Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nat. Commun. 2016;7:1–8. doi: 10.1038/ncomms12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walkey CD, et al. Protein corona fingerprinting predicts the cellular interaction of gold and silver nanoparticles. ACS Nano. 2014;8:2439–2455. doi: 10.1021/nn406018q. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Peng Q. Protein-gold nanoparticle interactions and their possible impact on biomedical applications. Acta Biomater. doi: 10.1016/j.actbio.2017.03.055. doi: https://doi.org/10.1016/j.actbio.2017.03.055. [DOI] [PubMed]
- 26.Chithrani BD, Ghazani AA, Chan WCW. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 27.Chou LYT, Zagorovsky K, Chan WCW. DNA assembly of nanoparticle superstructures for controlled biological delivery and elimination. Nat. Nanotechnol. 2014;9:148–55. doi: 10.1038/nnano.2013.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rouge JL, Hao L, Wu XA, Briley WE, Mirkin CA. Spherical Nucleic Acids as a Divergent Platform for Synthesizing RNA-Nanoparticle Conjugates through Enzymatic Ligation. ACS Nano. 2014;8:8837–8843. doi: 10.1021/nn503601s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith RC, Riollano M, Leung A, Hammond PT. Layer-by-layer platform technology for small-molecule delivery. Angew. Chemie - Int. Ed. 2009;48:8974–8977. doi: 10.1002/anie.200902782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–84. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016;34:339–44. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 32.Lee K, et al. Synthetically modified guide RNA and donor DNA are a versatile platform for CRISPR-Cas9 engineering. Elife. 2017;6:e25312. doi: 10.7554/eLife.25312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L, et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41:9049–9061. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Long C, et al. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuris JA, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2014;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hegde MR, et al. Microarray-based mutation detection in the dystrophin gene. Hum. Mutat. 2008;29:1091–1099. doi: 10.1002/humu.20831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakamura A, Takeda S. Exon-skipping therapy for Duchenne muscular dystrophy. The Lancet. 2011;378:546–547. doi: 10.1016/S0140-6736(11)61028-3. [DOI] [PubMed] [Google Scholar]
- 38.Mendell JR, et al. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann. Neurol. 2013;74:637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- 39.Kornegay JN, et al. Widespread Muscle Expression of an AAV9 Human Mini-dystrophin Vector After Intravenous Injection in Neonatal Dystrophin-deficient Dogs. Mol. Ther. 2010;18:1501–1508. doi: 10.1038/mt.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu L, et al. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol. Ther. 2015 doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pavlath GK, Horsley V. Cell fusion in skeletal muscle--central role of NFATC2 in regulating muscle cell size. Cell cycle (Georgetown Tex.) 2003;2:420–423. [PubMed] [Google Scholar]
- 42.Sy JC, et al. Sustained release of a p38 inhibitor from non-inflammatorymicrospheres inhibits cardiac dysfunction. Nature Materials. 2008:863–869. doi: 10.1038/nmat2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.