Abstract
Background
Bladder cancer (BC) is the most common urological malignant tumor. In BC, aberrant DNA methylation is believed to be associated with carcinogenesis. Therefore, the identification of key genes and pathways could help determine the potential molecular mechanisms of BC development.
Material/Methods
Microarray data on gene expression and gene methylation were downloaded from the Gene Expression Omnibus (GEO) database. Abnormal methylated/expressed genes were analyzed by GEO2R and statistical software R. Gene Ontology term enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed using the DAVID database and KOBAS 3.0. STRING and Cytoscape software were used to construct protein–protein interaction (PPI) networks and analyze modules of the PPI network.
Results
A total of 71 hypomethylated/upregulated genes were significantly enriched in cell–cell adhesion and blood vessel development. KEGG pathway analysis highlighted p53 signaling and metabolic pathways. Five core genes in the PPI network were determined: CDH1, DDOST, CASP8, DHX15, and PTPRF. Additionally, 89 hypermethylated/downregulated genes were found. These genes were enriched mostly in cell adhesion and signal transduction. KEGG pathway analysis revealed enrichment in focal adhesion. The top 5 core genes in the PPI network were GNG4, ADCY9, NPY, ADRA2B, and PENK. We found most of the core genes were also significantly altered in the Cancer Genome Atlas database.
Conclusions
Abnormal methylated/expressed genes and key signaling pathways involved in BC were identified through integrated bioinformatics analysis. In the future, these genes may serve as biomarkers for diagnosis and therapeutic targets in BC.
MeSH Keywords: Computational Biology, Signal Transduction, Urinary Bladder Neoplasms
Background
Bladder cancer (BC) is a common malignant urologic cancer. There are 2 major subsets of BC: non-muscle-invasive and muscle-invasive BC. BC is the fifth most common malignancy in the United States, with an estimated 76 960 new cases and 16 390 deaths in 2016 [1]. Chronic infection, smoking, and occupation are the main contributory factors of BC [2]. Aberrant DNA methylation, an epigenetic change, is believed to be associated with carcinogenesis [3–5]. The functions of key genes, including tumor suppressor genes and cell transformation genes, can be affected by changes in methylation, explaining how aberrant DNA methylation is involved in various processes of cancer development [3]. Aberrant DNA methylation has also been found in BC. Unlike genetic alterations, DNA methylation is reversible. Therefore, regulation of DNA methylation may represent a new strategy for cancer treatment [6–8]. Although some clinical results exist, such as anemia being a prognostic factor of BC [9], molecular and genetic biomarkers are considered as promising diagnostic options [10]. A few studies on have shown altered DNA methylation in cancer, but the roles of key differentially methylated genes (DMGs) and differentially expressed genes (DEGs) in BC remain unclear.
Recent technological advances in microarray analysis have provided an ideal and reliable method to identify important genetic or epigenetic modifications in cancer development. Thus, there have been many studies on DEGs in BC identified by gene expression profiling using microarrays [11,12]. Because of limited numbers of samples, individual studies are often not sufficient to demonstrate the underlying mechanism in caner progression. At present, we can obtain more reliable and accurate results by advanced bioinformatics analysis. To obtain novel abnormal methylated/expressed genes and key signaling pathways in BC, we extracted and analyzed information from both microarray-based gene expression and methylation profiling conducted in BC.
Material and Methods
Microarray data
We extracted gene expression (GSE3167 and GSE65635) and methylation (GSE37816 and GSE33510) profiling data from the Gene Expression Omnibus (GEO) database at the National Center for Biotechnology Information.
The BC-associated dataset GSE3167 submitted by Lars Dyrskjot based on the GPL96 platform (Affymetrix Human Genome U133A Array) was obtained from the GEO database and includes 51 BC samples and 9 normal samples. The BC-associated dataset GSE65635 submitted by Maria Vladimirovna Suntsova based on the GPL14951 platform (Illumina HumanHT-12 WG-DASL V4.0 R2 Expression BeadChip) was obtained from the GEO database and includes 8 BC samples and 4 normal samples. Regarding microarray-based gene methylation profiling, GSE37816 includes 18 BC samples and 6 normal samples, while GSE33510 includes 149 BC samples and 7 normal samples. Both of these methylation microarray analyses used the GPL8490 platform (Illumina HumanMethylation27 BeadChip).
Data processing
GEO provides users with a useful tool called GEO2R that can be used to analyze microarray data. We used GEO2R to identify DEGs and statistical software R (version 3.3.2, http://www.r-project.org/) with the minfi R package to identify DMGs. We set P<0.05 and |t| >2 as the thresholds to identify DEGs and DMGs. First, we identified up- and downregulated genes from the 2 gene expression profiling datasets (GSE3167 and GSE65635). Second, we identified hypo- and hypermethylated genes from GSE37816 and GSE33510. Finally, we identified hypomethylated/upregulated genes via overlapping the hypomethylated and overexpression gene lists and identified hypermethylated/downregulated genes via overlapping the hypermethylated and low-expression gene lists. Statistical software R and the pheatmap R package were used for bidirectional hierarchical clustering.
Functional and pathway enrichment analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) provides a set of data-mining tools and thus was used to perform Gene Ontology (GO) term enrichment analysis. GO term enrichment analysis includes biological process, cellular component, and molecular function. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed using KOBAS 3.0. P<0.05 was set as the screening criterion.
Protein–protein interaction (PPI) network construction and module analysis
PPI analysis is used to search core genes and gene modules related to carcinogenesis. In this study, PPI network analysis of the hypomethylated/upregulated genes and hypermethylated/downregulated genes were performed using the Search Tool for the Retrieval of Interacting Genes (STRING) database. The interaction score was set at 0.4. Subsequently, modules within the PPI network were screened by Molecular Complex Detection (MCODE) in Cytoscape software. An MCODE score >3 and number of nodes ≥3 were set as the screening criteria.
Validation of core genes in The Cancer Genome Atlas (TCGA) database
The TCGA database, which contains a tremendous amount of data on various types of cancers, was used to confirm our results. Most of the core genes selected by PPI analysis were verified in TCGA database.
Results
Identification of DEGs and DMGs in BC
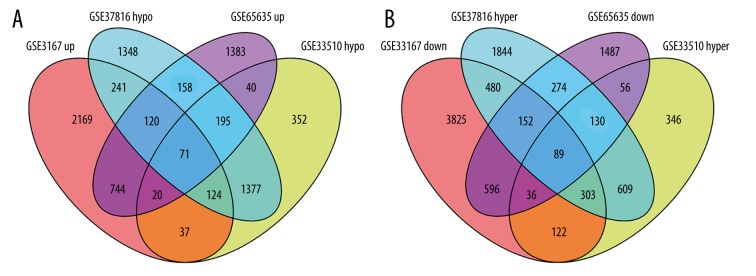
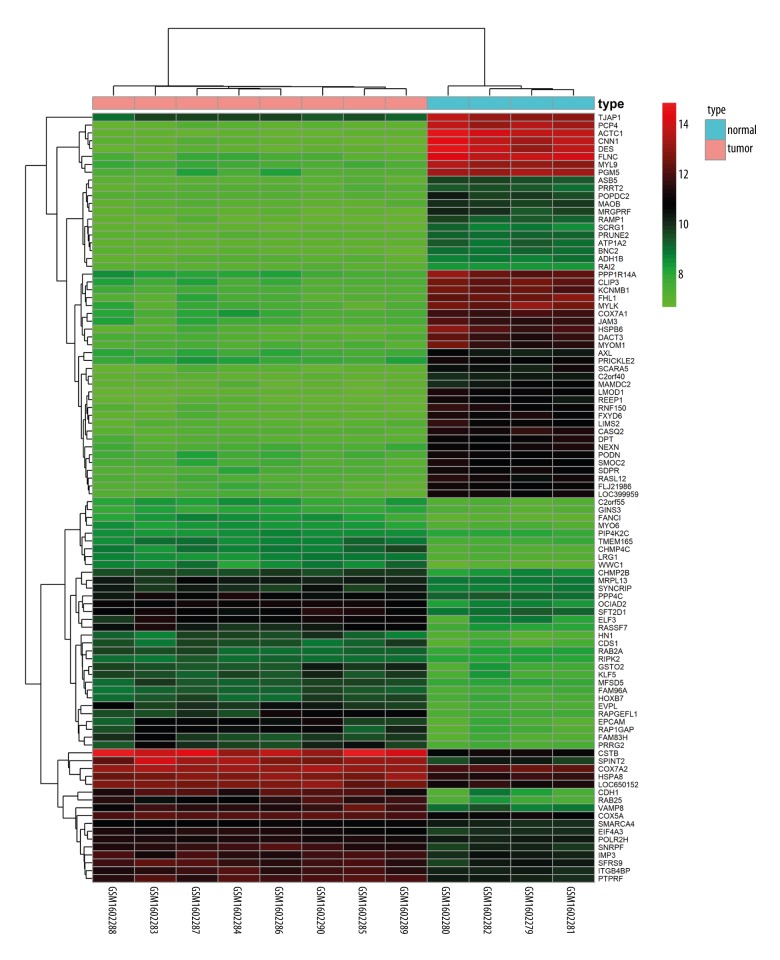
After obtaining DEGs and DMGs, 955 overlapping upregulated genes (3526 in GSE3167 and 2731 in GSE65635) and 873 overlapping low-expression genes (5603 in GSE3167 and 2820 in GSE65635) were found from the gene expression microarray analysis. There were 1131 overlapping hypermethylated genes (3881 in GSE37816 and 1691 in GSE33510) and 1767 overlapping hypomethylated genes (3634 in GSE37816 and 2216 in GSE33510) identified by the gene methylation microarray analysis. Further analysis of overlapping genes revealed 71 hypomethylated/upregulated genes and 89 hypermethylated/downregulated genes (Figure 1A, 1B; Supplementary Tables 1, 2). Visualization of part of aberrantly expressed genes (GSE65635) was illustrated by heat map (Figure 2).
Figure 1.
Identification of aberrantly methylated differentially expressed genes in gene expression datasets (GSE3167 and GSE65635) and gene methylation datasets (GSE37816 and GSE33510). (A) hypomethylation and upregulated genes. (B) hypermethylation and downregulated genes.
Figure 2.
Representative heat map of the top 100 differentially expressed genes in dataset GSE65635 (50 upregulated genes and 50 downregulated genes). Red indicates upregulation and green indicates downregulation.
GO term analysis
GO enrichment analysis results are presented in Table 1. Regarding the biological processes (BP), the hypomethylated/upregulated genes were significantly enriched in cell–cell adhesion, positive regulation of cell growth, and blood vessel development. In the molecular function (MF), the hypomethylated/high-expression genes were enriched in response to poly (A) RNA binding, KDEL sequence binding, and cadherin binding involved in cell–cell adhesion. In the cellular component (CC), the analysis revealed that enrichment mainly occurred at endoplasmic reticulum membrane and cytosol.
Table 1.
Gene ontology analysis of aberrantly methylated-differentially expressed genes in bladder cancer.
| Category | Term | Count | % | P value |
|---|---|---|---|---|
| Hypomethylation and high expression | ||||
| GOTERM_BP_DIRECT | GO: 0098609~cell-cell adhesion | 5 | 7.0423 | 0.027 |
| GOTERM_BP_DIRECT | GO: 0008284~positive regulation of cell proliferation | 7 | 9.8592 | 0.013 |
| GOTERM_BP_DIRECT | GO: 0001568~blood vessel development | 3 | 4.2254 | 0.011 |
| GOTERM_BP_DIRECT | GO: 0009636~response to toxic substance | 5 | 7.0423 | 0.0004 |
| GOTERM_BP_DIRECT | GO: 0051056~regulation of small GTPase mediated signal transduction | 4 | 5.6338 | 0.018 |
| GOTERM_MF_DIRECT | GO: 0044822~poly(A) RNA binding | 10 | 14.085 | 0.043 |
| GOTERM_MF_DIRECT | GO: 0005046~KDEL sequence binding | 2 | 2.8169 | 0.008 |
| GOTERM_MF_DIRECT | GO: 0098641~cadherin binding involved in cell-cell adhesion | 6 | 8.4507 | 0.007 |
| GOTERM_MF_DIRECT | GO: 0016853~isomerase activity | 3 | 4.2254 | 0.003 |
| GOTERM_MF_DIRECT | GO: 0005515~protein binding | 49 | 69.014 | 0.003 |
| GOTERM_CC_DIRECT | GO: 0005789~endoplasmic reticulum membrane | 11 | 15.493 | 0.0016 |
| GOTERM_CC_DIRECT | GO: 0043005~neuron projection | 6 | 8.4507 | 0.0021 |
| GOTERM_CC_DIRECT | GO: 0005829~cytosol | 22 | 30.986 | 0.011 |
| GOTERM_CC_DIRECT | GO: 0005739~mitochondrion | 12 | 16.901 | 0.012 |
| GOTERM_CC_DIRECT | GO: 0031234~extrinsic component of cytoplasmic side of plasma membrane | 3 | 4.2254 | 0.028 |
| Hypermethylation and low expression | ||||
| GOTERM_BP_DIRECT | GO: 0007155~cell adhesion | 10 | 11.236 | 0.00035 |
| GOTERM_BP_DIRECT | GO: 0008284~positive regulation of cell proliferation | 8 | 8.9888 | 0.007 |
| GOTERM_BP_DIRECT | GO: 0007165~signal transduction | 12 | 13.483 | 0.023 |
| GOTERM_BP_DIRECT | GO: 0030198~extracellular matrix organization | 5 | 5.618 | 0.015 |
| GOTERM_BP_DIRECT | GO: 0006936~muscle contraction | 4 | 4.4944 | 0.015 |
| GOTERM_MF_DIRECT | GO: 0031697~beta-1 adrenergic receptor binding | 2 | 2.2472 | 0.014 |
| GOTERM_MF_DIRECT | GO: 0097110~scaffold protein binding | 3 | 3.3708 | 0.023 |
| GOTERM_MF_DIRECT | GO: 0003700~transcription factor activity, sequence-specific DNA binding | 10 | 11.236 | 0.046 |
| GOTERM_CC_DIRECT | GO: 0008076~voltage-gated potassium channel complex | 6 | 6.7416 | 0.00005 |
| GOTERM_CC_DIRECT | GO: 0005886~plasma membrane | 31 | 34.831 | 0.005 |
| GOTERM_CC_DIRECT | GO: 0005887~integral component of plasma membrane | 13 | 14.607 | 0.031 |
| GOTERM_CC_DIRECT | GO: 0005578~proteinaceous extracellular matrix | 5 | 5.618 | 0.036 |
| GOTERM_CC_DIRECT | GO: 0008328~ionotropic glutamate receptor complex | 2 | 2.2472 | 0.036 |
The biological processes enriched by the hypermethylated/low-expression genes included positive regulation of cell proliferation, cell adhesion, and signal transduction, and the molecular function included scaffold protein binding, transcription factor activity, and sequence-specific DNA binding. In the cellular component, the hypermethylated/low-expression genes were enriched in integral component of plasma membrane and proteinaceous extracellular matrix.
KEGG pathway analysis
According to KEGG analysis (Table 2), the hypomethylated/high-expression genes were significantly enriched in the fructose and mannose metabolism, p53 signaling, and metabolic pathways. Hypermethylated/low-expression genes were enriched in calcium signaling, insulin secretion, cAMP signaling pathway, and focal adhesion pathways.
Table 2.
KEGG pathway analysis of aberrantly methylated-differentially expressed genes in bladder cancer.
| Pathway name | ID | Gene num | P-value | Genes |
|---|---|---|---|---|
| Hypomethylation and high expression | ||||
| p53 signaling pathway | hsa04115 | 3 | 0.00029 | SFN, ATP1B1, SERPINB5 |
| Metabolic pathways | hsa01100 | 8 | 0.00171 | TSTA3, DHCR7, B4GALT4, AHCY, IDH1, TUSC3, HSD17B4, DDOST |
| Fructose and mannose metabolism | hsa00051 | 2 | 0.00179 | TSTA3, ENOSF1 |
| Hypermethylation and low expression | ||||
| Insulin secretion | hsa04911 | 5 | 1.88E-06 | ABCC8, ADCY9, KCNMA1, RYR2, VAMP2 |
| Calcium signaling pathway | hsa04020 | 6 | 4.02E-06 | RYR2, P2RX1, ADCY9, PTGFR, PDE1C, MYLK |
| cAMP signaling pathway | hsa04024 | 5 | 9.80E-05 | ADCY9, GHSR, VIPR2, RYR2, NPY |
| Focal adhesion | hsa04510 | 5 | 0.00011 | RELN, ITGA8, MYLK, CCND2, THBS4 |
PPI network analysis
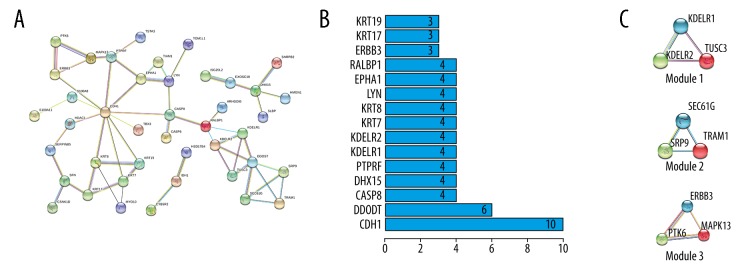
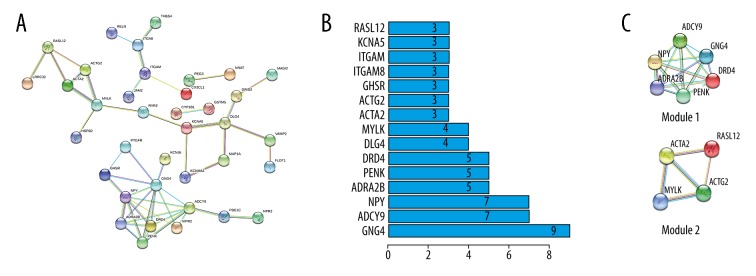
PPI network analysis was performed by STRING database. The PPI network of hypomethylated/upregulated genes is shown in Figure 3A. We screened the data and found 3 modules via MCODE in Cytoscape software (Figure 3C). KOBAS 3.0 was used for module analysis and demonstrated functions related to Vibrio cholerae infection in Module 1, protein export in Module 2, and proteoglycans in cancer Module 3 (Table 3). The top core genes were chosen: CDH1, DDOST, CASP8, DHX15, and PTPRF (Figure 3B). The PPI network of hypermethylated/downregulated genes was constructed (Figure 4A) and 2 key modules were found (Figure 4C). Module 1 demonstrated functions of GABAergic synapse and Module 2 functions of vascular smooth muscle contraction (Table 3). The top core genes were revealed: GNG4, ADCY9, NPY, ADRA2B, and PENK (Figure 4B).
Figure 3.
PPI network and the top 3 modules of hypomethylation/high-expression genes. (A) PPI network. (B) Core genes and the number of gene cords. (C) Top modules 1–3).
Table 3.
Module analysis of the protein–protein interaction network.
| Category | Module | Function description | FDR | Nodes | Genes |
|---|---|---|---|---|---|
| Hypomethylation and high expression | |||||
| 1. Vibrio cholerae infection | 2.09E-05 | 3 | KDELR2, KDELR1, TUSC3 | ||
| 2. Protein export | 4.55E-06 | 3 | SEC61G, SRP9, TRAM1 | ||
| 3. Proteoglycans in cancer | 0.00346724 | 3 | PTK6, ERBB3, MAPK13 | ||
| Hypermethylation and low expression | |||||
| 1. GABAergic synapse | 0.000879043 | 6 | ADRA2B, PENK, ADCY9, DRD4, NPY, GNG4 | ||
| 2. Vascular smooth muscle contraction | 9.22E-07 | 4 | RASL12, ACTA2, MYLK, ACTG2 | ||
Figure 4.
PPI network and the top 2 modules of hypermethylation/low-expression genes. (A) PPI network. (B) Core genes and the number of gene cords. (C) Top modules 1–2).
Validate the identified genes in TCGA database
To confirm our results, the identified core genes were validated in the TCGA database and results are shown in Table 4. In TCGA, the methylation and expression status of the core genes were also significantly altered in BC samples. These findings suggest that our results were reliable.
Table 4.
Validation of the hub genes in TCGA database.
| Category | Hub gene | Methylation status | P value | Expression status | P value |
|---|---|---|---|---|---|
| Hypomethylation and high expression | |||||
| CDH1 | Hypomethylation | 9.26E-08 | Up-regulated | 0.007185916 | |
| DDOST | Up-regulated | 2.45E-11 | |||
| CASP8 | Hypomethylation | 9.13E-12 | Up-regulated | 0.001398258 | |
| DHX15 | |||||
| PTPRF | Up-regulated | 0.005353039 | |||
| Hypermethylation and low expression | |||||
| GNG4 | Hypermethylation | 9.09E-09 | |||
| ADCY9 | Hypermethylation | 3.21E-06 | Down-regulated | 8.54E-13 | |
| NPY | Hypermethylation | 9.48E-11 | |||
| ADRA2B | Hypermethylation | 6.15E-11 | Down-regulated | 0.00036077 | |
| PENK | Hypermethylation | 1.40E-07 | Down-regulated | 2.98E-05 | |
Discussion
Aberrant methylation is an important molecular mechanism in BC initiation and development. In the present study, 71 hypomethylated/upregulated genes and 89 hypermethylated/downregulated genes were identified by a variety of bioinformatics analysis. These genes enriched certain pathways and affected by abnormal methylation. Knowledge of the aberrantly methylated/expressed key genes in BC would provide novel insights into its diagnosis, treatment, and prognosis.
In the GO analysis, we found hypomethylated/upregulated genes were significantly enriched in biological processes including cell–cell adhesion, positive regulation of cell growth, and blood vessel development. Regarding molecular function, enrichment of poly (A) RNA binding, KDEL sequence binding, and cadherin binding involved in cell–cell adhesion was found. Cell adhesion is an important factor in cancer dissemination and development of multiple types of cancers, including BC [13–16]. Cell growth and blood vessel growth are indispensable for the development of BC. RNA binding-related proteins, such as IGF2BP3 and MSI2, were shown to contribute to cancer cell malignancy and cancer progression [17,18]. The regulation of hypomethylation for poly (A) RNA binding genes might be associated with the occurrence of BC. KEGG pathway analysis demonstrated that the hypomethylated/upregulated genes significantly enriched in pathways included fructose and mannose metabolism, p53 signaling, and metabolic pathways. Acute myeloid leukemia cells can efficiently utilize fructose, and this promotes the malignant progression of leukemia [19]. D-Mannose was suggested as a novel biomarker for early detection of colorectal cancer [20]. The p53 signaling pathway has been shown to play an important role in BC [21,22]. Aberrant metabolism of BC is correlated with bladder tumorigenesis [23]. In brief, the dysregulation of fructose and mannose metabolism, p53 signaling, and metabolic pathways might affect BC progression from multiple aspects.
For the hypomethylated/upregulated genes, PPI network analysis was performed. The top 5 core genes were: CDH1, DDOST, CASP8, DHX15, and PTPRF. The CDH1 protein is a subunit of the APC/C complex, which is related to the regulation of cell division. The inactivation of CDH1 in vivo results in tumor cell death [24]. Maruyama et al. [25] first reported that the methylation status of CDH1 was significantly correlated with poor survival in BC. Some drugs induce apoptosis in BC by regulating CASP8 [26,27]. DHX15 plays multiple roles in tumor progression. In prostate cancer, DHX15 enhances transcriptional activity of the androgen receptor and promotes prostate cancer progression [28]. Additionally, DHX15 suppresses tumor proliferation by downregulating the expression of NF-κB in glioma [29]. PTPRF is considered a potential biomarker candidate for prostate cancer and non-small cell lung cancer [30,31]. These results suggest that these core genes play an important role in BC development. Thus, the functions and methylation status of these top core genes in BC warrant additional research.
In the PPI network, module analysis of hypomethylated/high-expression genes suggested that Vibrio cholerae infection, protein export, and proteoglycans in cancer are related to BC development. Various proteoglycans, which are key molecular effectors located at the cell surface, perform multiple functions in tumor cell growth, the tumor microenvironment, metastasis, and angiogenesis [32]. In addition, important alterations of small leucine-rich proteoglycans in BC have been reported [33]. In our study, PTK6, ERBB3, and MAPK13 were correlated with function of proteoglycans in cancers (Table 3). PTK6 has been demonstrated to be a potential predictive protein in bladder cancer patients [34]. ERBB3 was reported to be involved in treatment resistance [35]. In high-grade serous ovarian cancer, MAPK13 can serve as a potential predictive protein [36]. These results all suggest that PTK6, ERBB3, and MAPK13 genes can serve as potential biomarkers in BC.
In the GO analysis, we found that the hypermethylated/low-expression genes enriched in BP included positive regulation of cell proliferation, cell adhesion, and signal transduction. Cancer is normally considered a genetic disease. Aberrant expression of genes related to positive regulation of cell proliferation, cell adhesion, and signal transduction can affect tumor development and metastasis. Our findings suggest that these hypermethylated/low-expression genes represent promising targets for BC treatment or diagnosis. Regarding molecular functions, hypermethylated/low-expression genes were enriched in scaffold protein binding, transcription factor activity, and sequence-specific DNA binding. Scaffold proteins are an important regulatory factor in many key signaling pathways [37,38]. There are various scaffold proteins exerting different functions in carcinogenesis. Grp1-associated scaffold protein promotes the function of p53 tumor suppressor genes and inhibits cancer development [39]. In our study, we found that many hypermethylated/low-expression genes were enriched in scaffold protein binding. Additional research is needed to determine the function of these hypermethylated/low-expression genes enriched in scaffold protein binding. KEGG analysis revealed enrichment of the genes in the calcium signaling pathway, insulin secretion, cAMP signaling pathway, and focal adhesion, many of which are associated with BC. Calcium signaling is associated with carcinoma in situ in the bladder [40]. Activation of the cAMP/PKA signaling pathway may inhibit BC cell invasion [41]. Focal adhesion kinase (FAK) is a non-receptor protein-tyrosine kinase that is triggered by some growth factors and integrin. Some findings suggest that FAK is involved in oncogenic signaling of invasion and migration in BC [42]. In addition, inhibition of FAK induces apoptosis in bladder cancer cells. FAK may function as an important regulator of extracellular signaling-mediated apoptosis in bladder cancer and could be used as a novel therapeutic target in the treatment of BC [43]. Our results suggest that these signaling pathways are candidate research targets in BC.
We performed PPI network analysis on hypermethylated/downregulated genes. The top 5 core genes were: GNG4, ADCY9, NPY, ADRA2B, and PENK. ADCY9 is an enzyme that modulates signal transduction, and its expression is correlated with colon cancer TNM staging [44]. MiR-181b promotes cell proliferation and reduces apoptosis by downregulating ADCY9 expression in cervical cancer cells [45]. GNG4 is a hypermethylated/low-expression gene identified in glioblastoma, and overexpression of GNG4 inhibited the proliferation of glioblastoma cells [46]. NPY is a 36-amino acid peptide that acts via G-protein-coupled receptors (Y1–Y6). Peripherally, NPY affects smooth muscle contraction of blood vessels, blood pressure, and atherogenic processes [47]. Activation of the Y2 receptor stimulated angiogenesis in various animal models [48]. The hypermethylation status of PENK has been validated in BC [49]. In multiple types of cancers, PENK serves as a potential diagnostic marker [50,51]. We found that all top core genes were deeply involve in cancer proliferation or progression, suggesting that these core genes may serve as prognostic biomarkers or therapeutic targets in BC.
According to core modules analysis in the PPI network, hypermethylated/downregulated genes were closely related to GABAergic synapse and vascular smooth muscle contraction. We found that all top 5 core genes with hypermethylated/low-expression were involved in the function of GABAergic synapse. This suggests hypermethylated/low-expression genes related to GABAergic synapse might play an important role in BC development. Until now, the effects of GABAergic synapse in BC and its regulation by aberrant methylation have been unclear.
Several limitations to this investigation should be acknowledged. The clinical application value of those core genes in BCs need further evaluation. Moreover, we only chose related data from the TCGA database as a validated cohort. Our study results need to be validated in more databases.
Conclusions
In conclusion, our research identified some aberrantly expressed genes and pathways potentially regulated by abnormal methylated in BC by integrated analysis of gene expression and methylation microarrays. An analysis of multiple microarray data makes our results more reliable and precise than individual microarray data analysis. This provided a better understanding of the molecular mechanisms underlying the occurrence and progression of BC. Core genes such as CDH1, DDOST, CASP8, DHX15, PTPRF, GNG4, ADCY9, NPY, ADRA2B, and PENK may became new markers in accurate diagnosis and therapy of BC. Further work is needed to determine the role of these identified genes in BC progression.
Supplementary Tables
Supplementary Table 1.
The complete list of the 89 hypermethylated/downregulated genes.
| Hypermethylated/low-expression genes | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| NPR2 | HLF | RYR2 | ACTA2 | LY6H | CCND2 | TBX4 | ZCCHC14 | PEG3 | ZNF219 |
| CHST3 | RNF122 | STXBP6 | SEMA3B | PHYHIP | SSBP3 | CYP1B1 | MYLK | EIF5A2 | EMILIN1 |
| POU6F1 | RUSC2 | NPY | PDLIM4 | ITGAM | FLOT1 | RAX | LCAT | THBS4 | SPARCL1 |
| DLG4 | P2RX1 | ARMC4 | DRD4 | AOX1 | PLEKHA4 | SPON1 | NOVA1 | LRRC32 | ST6GALNAC5 |
| RARB | SYDE1 | ADCY9 | NNAT | GNG4 | PTGFR | VAMP2 | HS1BP3 | PHF1 | RASL12 |
| KCNMA1 | ADRA2B | MAP3K14 | CUL3 | ZNF154 | HSF4 | KCNA5 | STC2 | GYPC | BHMT2 |
| GSTM5 | VIPR2 | ROBO4 | CKMT2 | HAND2 | NCAM2 | NEFH | ABCC8 | ITGA8 | GHSR |
| PDE1C | HPSE2 | HSPB2 | BLZF1 | ARHGEF4 | FEZ1 | CX3CL1 | GRID2 | LTC4S | MAGI2 |
| HAAO | MAP1A | KCNJ6 | FAM50B | ACTG2 | PENK | CDO1 | RELN | JAM2 | |
Supplementary Table 2.
The complete list of the 71 hypomethylated/upregulated genes.
| Hypomethylated/high-expression genes | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| HMGN1 | KRT7 | SRP9 | VAMP8 | ARHGEF18 | LYPD3 | ENOSF1 | TOM1L1 | CASP6 | TBX3 |
| SLBP | CCDC59 | SNRPB2 | KRT17 | MRPS15 | MARCKSL1 | SPINT1 | KRT20 | ERBB3 | SERPINB5 |
| KDELR2 | EXOSC10 | CDH1 | KRT8 | B4GALT4 | MAPK13 | CASP8 | LYN | ID3 | HSD17B4 |
| TRAM1 | MKNK2 | CCDC47 | RALBP1 | ARHGDIB | TSTA3 | TRAK1 | KDELR1 | FXYD3 | S100A11 |
| DDOST | HDAC1 | ISG20L2 | SLC25A5 | KTN1 | SFN | KRT19 | CYB5R2 | IDH1 | BPHL |
| CSNK1D | AHR | AHCY | SEC61G | FAM3C | SPINT2 | TIAM1 | PTK6 | EPHA1 | GNA15 |
| GRSF1 | DHX15 | TUSC3 | GDF15 | PTPRF | S100A8 | MAP7 | DHCR7 | SH2D3A | MYO10 |
| SCAP | |||||||||
Acknowledgements
We thank Dr. Lei Peng and Dr. Chungang Zhai for their technical assistance.
Footnotes
Conflict of interest
None.
Source of support: This study was supported by grants from the Natural Science Foundation of China (81372335)
References
- 1.Li HT, Duymich CE, Weisenberger DJ, Liang G. Genetic and epigenetic alterations in bladder cancer. Int Neurourol J. 2016;20(Suppl 2):S84–94. doi: 10.5213/inj.1632752.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, et al. Global cancer statistics. Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 4.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11(3):191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17(3):330–39. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 6.Kim WJ, Kim YJ. Epigenetic biomarkers in urothelial bladder cancer. Expert Rev Mol Diagn. 2009;9(3):259–69. doi: 10.1586/erm.09.5. [DOI] [PubMed] [Google Scholar]
- 7.Besaratinia A, Cockburn M, Tommasi S. Alterations of DNA methylome in human bladder cancer. Epigenetics. 2013;8(10):1013–22. doi: 10.4161/epi.25927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kandimalla R, van Tilborg AA, Zwarthoff EC. DNA methylation-based biomarkers in bladder cancer. Nat Rev Urol. 2013;10(6):327–35. doi: 10.1038/nrurol.2013.89. [DOI] [PubMed] [Google Scholar]
- 9.Chen C, Hu L, Li X, Hou J. Preoperative anemia as a simple prognostic factor in patients with urinary bladder cancer. Med Sci Monit. 2017;23:3528–35. doi: 10.12659/MSM.902855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ilijazi D, Abufaraj M, Hassler MR, et al. Waiting in the wings: The emerging role of molecular biomarkers in bladder cancer. Expert Rev Mol Diagn. 2018;18(4):347–56. doi: 10.1080/14737159.2018.1453808. [DOI] [PubMed] [Google Scholar]
- 11.Dhawan D, Paoloni M, Shukradas S, et al. Comparative gene expression analyses identify luminal and basal subtypes of canine invasive urothelial carcinoma that mimic patterns in human invasive bladder cancer. PLoS One. 2015;10(9):e0136688. doi: 10.1371/journal.pone.0136688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez A, Loizaga A, Arceo R, et al. A pilot study on the potential of rna-associated to urinary vesicles as a suitable non-invasive source for diagnostic purposes in bladder cancer. Cancers (Basel) 2014;6(1):179–92. doi: 10.3390/cancers6010179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juengel E, Meyer dos Santos S, Schneider T, et al. HDAC inhibition suppresses bladder cancer cell adhesion to collagen under flow conditions. Exp Biol Med (Maywood) 2013;238(11):1297–304. doi: 10.1177/1535370213498975. [DOI] [PubMed] [Google Scholar]
- 14.Zhao L, Zhao Y, He Y, Mao Y, et al. miR-19b promotes breast cancer metastasis through targeting MYLIP and its related cell adhesion molecules. Oncotarget. 2017;8(38):64330–43. doi: 10.18632/oncotarget.19278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frantzi M, Klimou Z, Makridakis M, et al. Silencing of Profilin-1 suppresses cell adhesion and tumor growth via predicted alterations in integrin and Ca2+ signaling in T24M-based bladder cancer models. Oncotarget. 2016;7(43):70750–68. doi: 10.18632/oncotarget.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryan RT. Cell adhesion and urothelial bladder cancer: the role of cadherin switching and related phenomena. Philos Trans R Soc Lond B Biol Sci. 2015;370(1661):20140042. doi: 10.1098/rstb.2014.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao W, Lu D, Liu L, et al. Insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) promotes lung tumorigenesis via attenuating p53 stability. Oncotarget. 2017;8(55):93672–87. doi: 10.18632/oncotarget.21280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Jin H, Mao G, et al. Msi2 plays a carcinogenic role in esophageal squamous cell carcinoma via regulation of the Wnt/beta-catenin and Hedgehog signaling pathways. Exp Cell Res. 2017;361(1):170–77. doi: 10.1016/j.yexcr.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Chen WL, Wang YY, Zhao A, et al. Enhanced fructose utilization mediated by SLC2A5 is a unique metabolic feature of acute myeloid leukemia with therapeutic potential. Cancer Cell. 2016;30(5):779–91. doi: 10.1016/j.ccell.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long Y, Sanchez-Espiridion B, Lin M, et al. Global and targeted serum metabolic profiling of colorectal cancer progression. Cancer. 2017;123(20):4066–74. doi: 10.1002/cncr.30829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C, Ge Q, Zhang Q, et al. Targeted p53 activation by saRNA suppresses human bladder cancer cells growth and metastasis. J Exp Clin Cancer Res. 2016;35:53. doi: 10.1186/s13046-016-0329-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madka V, Mohammed A, Li Q, et al. Targeting mTOR and p53 signaling inhibits muscle invasive bladder cancer in vivo. Cancer Prev Res (Phila) 2016;9(1):53–62. doi: 10.1158/1940-6207.CAPR-15-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massari F, Ciccarese C, Santoni M, et al. Metabolic phenotype of bladder cancer. Cancer Treat Rev. 2016;45:46–57. doi: 10.1016/j.ctrv.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 24.Eguren M, Porlan E, Manchado E, et al. The APC/C cofactor Cdh1 prevents replicative stress and p53-dependent cell death in neural progenitors. Nat Commun. 2013;4:2880. doi: 10.1038/ncomms3880. [DOI] [PubMed] [Google Scholar]
- 25.Maruyama R, Toyooka S, Toyooka KO, et al. Aberrant promoter methylation profile of bladder cancer and its relationship to clinicopathological features. Cancer Res. 2001;61(24):8659–63. [PubMed] [Google Scholar]
- 26.Choi EO, Park C, Hwang HJ, et al. Baicalein induces apoptosis via ROS-dependent activation of caspases in human bladder cancer 5637 cells. Int J Oncol. 2016;49(3):1009–18. doi: 10.3892/ijo.2016.3606. [DOI] [PubMed] [Google Scholar]
- 27.Hsu FT, Sun CC, Wu CH, et al. Regorafenib induces apoptosis and inhibits metastatic potential of human bladder carcinoma cells. Anticancer Res. 2017;37(9):4919–26. doi: 10.21873/anticanres.11901. [DOI] [PubMed] [Google Scholar]
- 28.Jing Y, Nguyen MM, Wang D, et al. DHX15 promotes prostate cancer progression by stimulating Siah2-mediated ubiquitination of androgen receptor. Oncogene. 2017;37(5):638–50. doi: 10.1038/onc.2017.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito S, Koso H, Sakamoto K, Watanabe S, et al. RNA helicase DHX15 acts as a tumour suppressor in glioma. Br J Cancer. 2017;117(9):1349–59. doi: 10.1038/bjc.2017.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitmore TE, Peterson A, Holzman T, et al. Integrative analysis of N-linked human glycoproteomic data sets reveals PTPRF ectodomain as a novel plasma biomarker candidate for prostate cancer. J Proteome Res. 2012;11(5):2653–65. doi: 10.1021/pr201200n. [DOI] [PubMed] [Google Scholar]
- 31.Soulières D, Hirsch FR, Shepherd FA, et al. PTPRF expression as a potential prognostic/predictive marker for treatment with erlotinib in non-small-cell lung cancer. J Thorac Oncol. 2015;10(9):1364–69. doi: 10.1097/JTO.0000000000000624. [DOI] [PubMed] [Google Scholar]
- 32.Iozzo RV, Sanderson RD. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J Cell Mol Med. 2011;15(5):1013–31. doi: 10.1111/j.1582-4934.2010.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Appunni S, Anand V, Khandelwal M, et al. Altered expression of small leucine-rich proteoglycans (Decorin, Biglycan and Lumican): Plausible diagnostic marker in urothelial carcinoma of bladder. Tumour Biol. 2017;39(5):1010428317699112. doi: 10.1177/1010428317699112. [DOI] [PubMed] [Google Scholar]
- 34.Xu XL, Ye YL, Wu ZM, et al. Overexpression of PTK6 predicts poor prognosis in bladder cancer patients. J Cancer. 2017;8(17):3464–73. doi: 10.7150/jca.21318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redlich N, Robinson AM, Nickel KP, et al. Anti-Trop2 blockade enhances the therapeutic efficacy of ErbB3 inhibition in head and neck squamous cell carcinoma. Cell Death Dis. 2018;9(1):5. doi: 10.1038/s41419-017-0029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie H, Wang W, Sun F, et al. Proteomics analysis to reveal biological pathways and predictive proteins in the survival of high-grade serous ovarian cancer. Sci Rep. 2017;7(1):9896. doi: 10.1038/s41598-017-10559-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw AS, Filbert EL. Scaffold proteins and immune-cell signalling. Nat Rev Immunol. 2009;9(1):47–56. doi: 10.1038/nri2473. [DOI] [PubMed] [Google Scholar]
- 38.Zheng Y, Zhang C, Croucher DR, et al. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature. 2013;499(7457):166–71. doi: 10.1038/nature12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venkataraman A, Coleman D, Nevrivy DJ, et al. Grp1-associated scaffold protein regulates skin homeostasis after ultraviolet irradiation. Photochem Photobiol Sci. 2014;13(3):531–40. doi: 10.1039/c3pp50351h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herbsleb M, Christensen OF, Thykjaer T, et al. Bioinformatic identification of FGF, p38-MAPK, and calcium signalling pathways associated with carcinoma in situ in the urinary bladder. BMC Cancer. 2008;8:37. doi: 10.1186/1471-2407-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ou Y, Zheng X, Gao Y, et al. Activation of cyclic AMP/PKA pathway inhibits bladder cancer cell invasion by targeting MAP4-dependent microtubule dynamics. Urol Oncol. 2014;32(1):47e21–8. doi: 10.1016/j.urolonc.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 42.Kong DB, Chen F, Sima N. Focal adhesion kinases crucially regulate TGFbeta-induced migration and invasion of bladder cancer cells via Src kinase and E-cadherin. Onco Targets Ther. 2017;10:1783–92. doi: 10.2147/OTT.S122463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong D, Chen F, Sima NI. Inhibition of focal adhesion kinase induces apoptosis in bladder cancer cells via Src and the phosphatidylinositol 3-kinase/Akt pathway. Exp Ther Med. 2015;10(5):1725–31. doi: 10.3892/etm.2015.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi H, Wang K, Jin JF, et al. Elevated adenylyl cyclase 9 expression is a potential prognostic biomarker for patients with colon cancer. Med Sci Monit. 2018;24:19–25. doi: 10.12659/MSM.906002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L, Wang YL, Liu S, et al. miR-181b promotes cell proliferation and reduces apoptosis by repressing the expression of adenylyl cyclase 9 (AC9) in cervical cancer cells. FEBS Lett. 2014;588(1):124–30. doi: 10.1016/j.febslet.2013.11.019. [DOI] [PubMed] [Google Scholar]
- 46.Pal J, Patil V, Mondal B, et al. Epigenetically silenced GNG4 inhibits SDF1alpha/CXCR4 signaling in mesenchymal glioblastoma. Genes Cancer. 2016;7(3–4):136–47. doi: 10.18632/genesandcancer.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sliwinska-Mosson M, Borowiecka K, Milnerowicz H. [Neuropeptides Y, YY, PP and their clinical significance]. Postepy Hig Med Dosw (Online) 2013;67:631–36. doi: 10.5604/17322693.1058890. [in Polish] [DOI] [PubMed] [Google Scholar]
- 48.Abe K, Tilan JU, Zukowska Z. NPY and NPY receptors in vascular remodeling. Curr Top Med Chem. 2007;7(17):1704–9. doi: 10.2174/156802607782340948. [DOI] [PubMed] [Google Scholar]
- 49.Wolff EM, Chihara Y, Pan F, et al. Unique DNA methylation patterns distinguish noninvasive and invasive urothelial cancers and establish an epigenetic field defect in premalignant tissue. Cancer Res. 2010;70(20):8169–78. doi: 10.1158/0008-5472.CAN-10-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roperch JP, Incitti R, Forbin S, et al. Aberrant methylation of NPY, PENK, and WIF1 as a promising marker for blood-based diagnosis of colorectal cancer. BMC Cancer. 2013;13:566. doi: 10.1186/1471-2407-13-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salhia B, Kiefer J, Ross JT, et al. Integrated genomic and epigenomic analysis of breast cancer brain metastasis. PLoS One. 2014;9(1):e85448. doi: 10.1371/journal.pone.0085448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
The complete list of the 89 hypermethylated/downregulated genes.
| Hypermethylated/low-expression genes | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| NPR2 | HLF | RYR2 | ACTA2 | LY6H | CCND2 | TBX4 | ZCCHC14 | PEG3 | ZNF219 |
| CHST3 | RNF122 | STXBP6 | SEMA3B | PHYHIP | SSBP3 | CYP1B1 | MYLK | EIF5A2 | EMILIN1 |
| POU6F1 | RUSC2 | NPY | PDLIM4 | ITGAM | FLOT1 | RAX | LCAT | THBS4 | SPARCL1 |
| DLG4 | P2RX1 | ARMC4 | DRD4 | AOX1 | PLEKHA4 | SPON1 | NOVA1 | LRRC32 | ST6GALNAC5 |
| RARB | SYDE1 | ADCY9 | NNAT | GNG4 | PTGFR | VAMP2 | HS1BP3 | PHF1 | RASL12 |
| KCNMA1 | ADRA2B | MAP3K14 | CUL3 | ZNF154 | HSF4 | KCNA5 | STC2 | GYPC | BHMT2 |
| GSTM5 | VIPR2 | ROBO4 | CKMT2 | HAND2 | NCAM2 | NEFH | ABCC8 | ITGA8 | GHSR |
| PDE1C | HPSE2 | HSPB2 | BLZF1 | ARHGEF4 | FEZ1 | CX3CL1 | GRID2 | LTC4S | MAGI2 |
| HAAO | MAP1A | KCNJ6 | FAM50B | ACTG2 | PENK | CDO1 | RELN | JAM2 | |
Supplementary Table 2.
The complete list of the 71 hypomethylated/upregulated genes.
| Hypomethylated/high-expression genes | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| HMGN1 | KRT7 | SRP9 | VAMP8 | ARHGEF18 | LYPD3 | ENOSF1 | TOM1L1 | CASP6 | TBX3 |
| SLBP | CCDC59 | SNRPB2 | KRT17 | MRPS15 | MARCKSL1 | SPINT1 | KRT20 | ERBB3 | SERPINB5 |
| KDELR2 | EXOSC10 | CDH1 | KRT8 | B4GALT4 | MAPK13 | CASP8 | LYN | ID3 | HSD17B4 |
| TRAM1 | MKNK2 | CCDC47 | RALBP1 | ARHGDIB | TSTA3 | TRAK1 | KDELR1 | FXYD3 | S100A11 |
| DDOST | HDAC1 | ISG20L2 | SLC25A5 | KTN1 | SFN | KRT19 | CYB5R2 | IDH1 | BPHL |
| CSNK1D | AHR | AHCY | SEC61G | FAM3C | SPINT2 | TIAM1 | PTK6 | EPHA1 | GNA15 |
| GRSF1 | DHX15 | TUSC3 | GDF15 | PTPRF | S100A8 | MAP7 | DHCR7 | SH2D3A | MYO10 |
| SCAP | |||||||||