Abstract
Epstein-Barr virus (EBV) is an oncogenic virus that infects over 90% of the world's adult population. EBV can establish life-long latent infection in host due to the balance between EBV and host immune system. EBV latency is associated with various malignancies such as nasopharyngeal carcinoma, gastric carcinoma and Burkitt's lymphoma. EBV is the first human virus that has the capability to encode microRNAs (miRNAs). Remarkably, EBV-encoded miRNAs are abundantly expressed in latently-infected cells and serve important function in viral infection and pathogenesis. Increasing evidence indicates that EBV miRNAs target the host mRNAs involved in cell proliferation, apoptosis and transformation. EBV miRNAs also inhibit the expression of viral antigens, thereby enabling infected cells to escape immune recognition. Intriguingly, EBV miRNAs directly suppress host antiviral immunity by interfering with antigen presentation and immune cell activation. This review will update the current knowledge about EBV miRNAs implicated in host immune responses. An in-depth understanding of the functions of EBV miRNAs in host antiviral immunity will shed light on the EBV-host interactions and provide potential therapeutic targets for the treatment of EBV-associated malignancies.
Keywords: Epstein-Barr virus, antiviral immunity, microRNA, therapeutic target, EBV-associated malignancies
Introduction
Epstein-Barr virus (EBV), a member of the human herpesvirus family, was initially identified in Burkitt's lymphoma (BL) in 1964 1, 2. EBV infects more than 90% of the human population 3. EBV can not be completely cleared once entry into the host cells and ultimately establishes life-long latent infection in host, indicating that EBV has developed elaborate strategies to evade host immune surveillance 4. During latent infection, EBV genome exists as a circular episome and replicates by recruiting the cellular replication machinery 5. Four latency types (0, I, II, III) are defined depending on the expression pattern of EBV latent genes, and at the same time are linked to a particular tumor type 6, 7. In some cases, primary EBV infection causes infectious mononucleosis (IM) in adolescents 8. Accumulating evidence indicates that latent EBV infection is associated with a variety of human malignancies such as Burkitt's lymphoma (BL), Hodgkin's lymphoma (HL), nasopharyngeal carcinoma (NPC) and gastric carcinoma (GC) 9, 10.
microRNAs (miRNAs) are a large class of small non-coding RNAs with a length of 21-23 nucleotides (nt) 11. They are usually generated from either exons or introns of both protein-coding and non-coding transcripts that are processed by RNA polymerase II (Pol II) 12-14. Exceptionally, a minority of miRNAs are derived from RNA Pol III transcripts. For example, murine gammaherpesvirus 68 (MHV-68)-encoded miRNAs are located downstream of viral tRNAs and transcribed by RNA Pol III 2. miRNAs are able to post-transcriptionally control gene expression by binding to target mRNAs 15. The canonical pattern for miRNA target recognition involves conserved Watson-Crick paring to the 5' region of the miRNA 16. It has been reported that only 2-7 nucleotides (nt) within a miRNA (the seed region) perfectly match a region on its target mRNA 17. However, some seed-match regions that contain single mismatches, bulges, G:U wobbles, insertion or deletion are functional sites 18. Imperfect miRNA-mRNA hybrids with bulges or G:U wobbles can also cause mRNA degradation or translational suppression 19. Surprisingly, the miRNA-mRNA interaction often violates the seed rule. There is now substantial evidence that the 'non-seed' sequence of a miRNA is required for the miRNA targeting of its target mRNA. For instance, the non-seed sites of a white spot syndrome virus (WSSV) miRNA (WSSV-miR-N32) are essential for its binding to the target mRNAs 20. A single miRNA can target multiple genes, which constitutes complicated miRNA regulatory networks 21. miRNAs are proven to serve important functions in a number of cellular processes, including cell proliferation, metabolism and apoptosis 22, 23.
miRNAs have been found in various organisms including algae, worms, plants, and mammals 24. Intriguingly, viruses also encode miRNAs. In 2004, Tuschl et al. 25 identified the first virus-encoded miRNAs in EBV. To date, EBV has been found to encode about 44 miRNAs that directly target cellular or viral mRNAs 26. These miRNAs are distributed by three clusters: the BamH I fragment A rightward transcript (BART)-cluster 1, BART-cluster 2 and the BamH I fragment H rightward open reading frame 1 (BHRF1)-cluster 27. EBV infection is commonly asymptomatic and can persist in the host for life, which is ascribed to the delicate balance between EBV and host immune system 28. Notably, EBV expresses high levels of miRNAs during all phases of its life cycle, implying that these miRNAs may be involved in the interplay between EBV and host immune system 29. The function of EBV miRNAs has been intensively investigated. Previous studies verified that EBV miRNAs were implicated in viral infection by directly targeting viral mRNAs 30, 31. Moreover, EBV miRNAs directly subvert the host defense system by regulating immune-related genes 32, 33. In this review, we provide an overview of the biogenesis and function of EBV miRNAs with emphasis on the modulatory roles of EBV miRNAs in host immune responses. An in-depth understanding of the functions of EBV miRNAs in host antiviral immunity will shed light on the complex EBV-host interactions and provide potential therapeutic targets for the treatment of EBV-associated malignancies.
Biosynthesis of EBV miRNAs
To date, the biosynthetic mechanism of cellular miRNAs has been intensively studied. The biogenesis of miRNAs mainly depends on several processing steps. In general, the miRNA gene is first transcribed to generate a long primary miRNA transcript (pri-miRNA) by RNA Pol II 13. Pri-mRNA is a capped and polyadenylated transcript that incorporates one or more stem-loop structures. Pri-mRNA is processed by the Drosha/DiGeorge syndrome chromosomal region 8 (DGCR8) complex into a hairpin shaped precursor (pre-miRNA) of ~70 nt within the nucleus 34. Pre-miRNA is then exported to the cytoplasm by exportin-5 and further cleaved by the endonuclease Dicer to generate a double-stranded miRNA (miRNA duplex) of ~22 nt 35. One strand of the miRNA duplex is incorporated into the RNA-induced silencing complex (RISC) which contains the endonuclease Argonaute 2 (Ago2) 36. miRNA-guided RISC binds to its target mRNA and inhibits target gene expression by facilitating mRNA cleavage and translation repression 37.
It is intriguing whether EBV miRNAs are generated in the way that is similar to cellular miRNAs. Previously, Lo et al. 38 amplified viral DNA fragments harboring the template for BART miRNAs from EBV-positive NPC cells. The amplified DNA fragments were cloned into the pCMV4 vectors. These recombinant vectors almost express all BART miRNAs despite the absence of intact virus. Similarly, Marquitz et al. 39 generated recombinant pcDNA3 vectors covering the BART miRNA coding regions. The recombinant pcDNA3 vectors were transfected into EBV-negative GC cells. Consequently, the BART miRNAs were expressed exogenously in the GC cells. Moreover, Skalsky et al. 40, 41 constructed EBV miRNA expression vectors by cloning ~200 nt regions encompassing each EBV pre-miRNA into expression vectors. The successful expression of these EBV miRNAs was confirmed by indicator assays. These studies demonstrate that the viral gene products are not required for the biogenesis of EBV miRNAs.
EBV BART miRNAs are located within introns of the latent BART transcripts that are expressed in all types of EBV latency 42-44. The BART miRNAs can be detected in all forms of latency 25, 45 and display similar expression patterns to that of BART transcripts 43. The BART miRNAs are thought to be originated from introns prior to splicing of the BART primary transcripts 46. BART miRNA synthesis is correlated with the accumulation of a spliced mRNA in which exon 1 located within the BART primary transcript is directly joined to exon 3. Blocking formation of this spliced transcript may inhibit generation of mature BART miRNAs. Inhibition of RNA Pol II activity reduces the expression levels of EBV BART miRNAs 47. Remarkably, sequences essential for Pol III activity are not present in the promoter region of BART miRNAs. Moreover, knockdown of Drosha or Dicer also suppresses the expression of BART miRNAs. It has been confirmed that both strands of a miRNA duplex can form mature miRNAs 48-50. Likewise, both strands of the BART miRNA original hairpin can be processed into functional mature miRNAs 47.
EBV miR-BHRF1-1 is located within the 5' UTR region of the BHRF1 gene and overlaps the TATA box of the EBV replication-activated BHRF1 promoter, while miR-BHRF1-2 and miR-BHRF1-3 are in the 3' UTR region of the BHRF1 gene 51. The BHRF1 miRNAs are generally expressed in type III latency, in which all the EBNAs are transcribed from Cp or Wp 52, 53. Xing et al. 54 found that the BHRF1 miRNAs were generated by processing of an intron present within the Cp or Wp-initiated long primary EBNA transcript. However, during latency I, the only latency associated gene expressed is EBNA1, transcribed from a separate promoter (Qp), and BHRF1 miRNAs are detected at low levels. Because the Qp promoter differs from the Cp and Wp promoters in being located between the BHRF1 and EBNA1 open reading frames (ORFs), viral pre-miRNAs initiating at Qp could not be processed to generate the BHRF1 miRNAs 42. Additionally, the BHRF1 miRNAs may be produced by processing of the 5' and 3' UTRs within latent BHRF1 transcripts 43. During EBV replication, lytic BHRF1 transcripts initiated from the alternative lytic promoter BHRF1p generate only the two downstream BHRF1 miRNAs, miR-BHRF1-2 and -3 55, 56. More importantly, depletion of Drosha represses the processing of the BHRF1 pre-miRNAs in cells transfected with constructs encoding BHRF1 miRNAs 57.
Collectively, these studies confirm the crucial role of Drosha and Dicer in the processing of EBV miRNA precursors, demonstrating that EBV miRNA processing is reliant on the host machinery. So far, there is no proof that EBV encodes additional miRNA-processing enzymes or RISC components, hinting that EBV miRNAs may be transcribed and generated in the same way as cellular miRNAs.
Expression of EBV miRNAs in virus-positive tumor cells
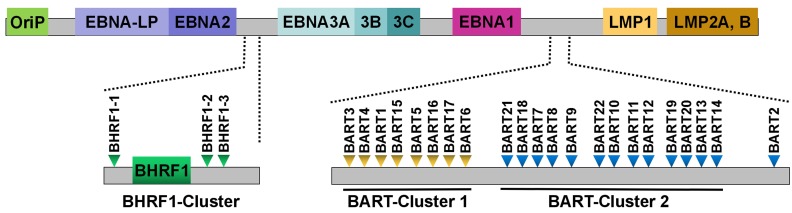
EBV was the first virus in which viral miRNAs were found 25. It encodes 25 EBV miRNA precursors and 44 mature miRNAs 58. EBV miRNAs are derived from two regions of the viral genome: BHRF1 and BART (Figure 1). Pfeffer et al. 25 were the first to find that EBV is capable of encoding miRNAs. They identified five viral miRNAs, miR-BHRF1-1 to -3 and miR-BART1 to 2, in EBV-associated Burkitt's lymphoma cells. These EBV miRNAs are derived from five different dsRNA precursors that are located in two distinct clusters (BHRF1 and BART). One cluster is located around the BHRF1 gene, which encodes miR-BHRF1-1 to -3. Other EBV miRNAs are positioned in intronic regions of the BART transcript. Subsequently, fourteen additional viral miRNAs are identified in EBV-positive effusion lymphoma cell lines, and all of these miRNAs are derived from a miRNA cluster located within introns of the BART gene 42. A total of 22 novel miRNAs are identified in EBV-positive Burkitt's lymphoma cells by using a combined computational and microarray-based approach, which significantly increases the number and complexity of EBV miRNAs during latent infection 59. Zhu et al. 52 characterized miRNA expression profiles of EBV-positive nasopharyngeal carcinoma (NPC) tumor samples by using small RNA deep sequencing. Consequently, they identified two new and highly abundant EBV miRNAs, miR-BART21 and miR-BART22. At the same time, Cosmopoulos et al. 60 also identified miR-BART22 as a novel EBV miRNA by comprehensively analyzing the expression profile of EBV miRNAs in primary NPC tumors. Ultimately, four novel mature BART miRNAs, including miR-BART12-5p, miR-BART15-5p, miR-BART16-3p and miR-BART22-5p, are discovered in clinical NPC tissues 58.
Figure 1.
Schematic representation of the genomic locations of EBV-encoded miRNAs. The relative positions of EBV latent genes on the genome are indicated. EBV encodes 25 pre-miRNAs located within two regions of the genome: BHRF1 and BART. The regions of the BHRF1-cluster and BART-clusters are expanded to show individual miRNAs.
EBV miRNAs are differentially expressed in EBV-positive tumor cells. In particular, expression of EBV miRNAs differs depending on the type of viral latency. BART miRNAs are present in EBV-associated NPC while BHRF1 miRNAs are undetectable in NPC biopsy samples 60. Likewise, miR-BART3, miR-BART5, miR-BART7 and miR-BART10 are detectable in EBV-associated gastric carcinoma (EBVaGC) cells 61. These findings indicate that the BART miRNAs are abundantly expressed in epithelial tumor cells harboring the virus in type II latency (NPC and GC). Deep sequencing was previously applied to determine EBV miRNA expression in EBV-positive NK/T-cell lymphomas 62. Of note, miR-BART7, miR-BART5, miR-BART11-5p, miR-BART1-5p and miR-BART19-3p are highly expressed in NK/T-cell lymphoma (latency type III). Nearly all BART miRNAs are present in EBV-associated B-cell lymphoma (latency type I) 63. The viral BHRF1 miRNAs are found in cells undergoing type III EBV latency, such as a range of EBV-infected B-cell tumors and lymphoblastoid cell lines (LCLs) 32. This association with the specific latency stage (latency III) mainly attributes to the fact that BHRF1 miRNAs are derived from the Cp- and/or Wp-initiated EBNA transcript that is expressed only in latency III 43. Collectively, BART miRNAs are widely expressed in all types of EBV-associated latency, while the expression of BHRF1 miRNAs is restricted to a specific latency type in tumor cells.
The BART and BHRF1 miRNAs are also expressed during the viral lytic cycle in EBV-positive tumor cells 42. Several miRNAs, such as miR-BART1-3p, miR-BART3-3p, miR-BART7, miR-BART10-3p and miR-BHRF1-2, are upregulated during lytic replication. It has been known that miR-BHRF1-2 and miR-BHRF1-3 are positioned within the 3' UTR of the early lytic transcript encoding BHRF1 54. In addition, BART mRNA expression is induced upon lytic reactivation. Therefore, the expression of viral miRNAs derived from BHRF1 and BART regions is increased during EBV lytic replication. A quantitative analysis of EBV-encoded miRNA expression in lytically induced EBV-positive Burkitt's lymphoma cells further reveals the expression patterns of viral miRNAs 43. Due to the onset of lytic BHRF1 transcription, miR-BHRF1-2 and miR-BHRF1-3 are rapidly induced at 24 h post lytic induction. The viral Wp and Cp promoters are activated at 48 h. Compared with miR-BHRF1-2 and miR-BHRF1-3, the upregulation of miR-BHRF1-1 is delayed and is not observed until the onset of Wp/Cp-initiated EBNA transcription at 48 h or later. Although robust BART transcription is observed during EBV replication, BART miRNA levels maintain relatively steady. This study demonstrates that changes in transcriptional levels of BHRF1 and BART do not necessarily correlate with alteration in EBV miRNA abundance.
The mRNA targets of EBV miRNAs
Viral mRNA targets of EBV miRNAs
EBV miRNAs can alter the expression of viral genes to regulate EBV infection and pathogenesis. miR-BART2 specially binds to the viral mRNA encoding EBV DNA polymerase (also called BALF5) and leads to degradation of the BALF5 mRNA 30. miR-BART2 upregulation can inhibit the viral lytic infection, while its expression level reduces during EBV lytic cycle as BALF5 expression is increased 64. Thus, miR-BART2 supports viral latency via suppressing viral DNA replication by targeting BALF5. miR-BART20-5p inhibits the expression of EBV immediate-early genes Zta and Rta 31. Zta and Rta cooperatively induce the expression of EBV lytic genes as well as viral replication 65-67. These findings indicate that EBV utilizes its own miRNAs to maintain latent infection in host cells by inhibiting viral lytic cycle.
EBV miRNAs can inhibit the expression of the latency-associated membrane proteins LMP1 and LMP2A. LMP1, one of the virus-encoded proteins, is critical for cell proliferation and transformation 68. LMP1 also regulates a variety of molecules involved in immune responses by acting as an activator of multiple cellular pathways such as phosphatidylinositol 3-kinase (PI3-K), mitogen-activated protein kinase (MAPK), interferon regulatory factor 7 (IRF7), nuclear factor-ĸB (NF-ĸB), Janus kinase (JAK)/signal transducer and activator of transcription (STAT) and activator protein (AP-1) signaling pathways 69, 70. However, high levels of LMP1 leads to suppression of cell proliferation and increased cellular sensitivity to pro-apoptotic stresses 71, 72. EBV miRNAs (miR-BART5-5p and miR-BART19-5p) from BART-clusters have been reported to suppress LMP1 expression, thereby maintaining the balance between the growth-promoting effect of LMP1 and its pro-apoptotic function 73, 74. LMP2A is a highly immunogenic antigen that is able to stimulate specific T-cell responses 75. In latently infected cells, miR-BART22 may serve as an antisense regulator of LMP2A 76, 77. Upregulation of miR-BART22 leads to a decrease in the protein level of LMP2A but not affecting its mRNA abundance, indicating that miR-BART22 regulates LMP2A expression at the post-transcriptional level. Additionally, EBV miRNAs suppress the expression of other latent genes. The EBV nuclear antigen 2 (EBNA2) is important for type III latency by elevating the expression level of EBV latent genes, and it plays a significant role in EBV-induced cellular transformation 78-80. miR-BART6-5p directly targets Dicer to regulate the host miRNA production, and it also suppresses EBNA2 expression, suggesting the modulatory role of miR-BART6-5p in EBV latency and carcinogenesis 81.
Cellular targets of EBV miRNAs
EBV miRNAs can downregulate cellular transcripts. Wnt inhibitory factor 1 (WIF1) and adenomatous polyposis coli (APC) are tumor suppressors that play an important role in Wnt signaling pathway 82. miR-BART19-3p inhibits WIF1 expression 83. miR-BART7, miR-BART19-3p and miR-BART17-5p decrease the expression level of APC. miR-BHRF1-1 downregulates expression of another tumor suppressor, p53 84. p53 expression is increased during EBV latency and early lytic phase while reduced during mid and late lytic phases 85, 86. Thus, miR-BHRF1-1 may promote EBV lytic infection by decreasing p53 expression. The basic leucine zipper transcription factor 1 (BACH1), a transcription repressor, is capable of controlling the cell cycle and regulating oxidative stress response 87. miR-BART4 and miR-BHRF1-2 are found to directly target BACH1 40. EBV miRNAs also target other cellular transcripts such as the natural killer (NK) ligand lectin-like transcript 1 (CLEC2D), importin 7 (IPO7), mitochondrial import receptor subunit TOM22 homolog (TOMM22), F-box only protein 9 (FBXO9), caprin family member 2 (CAPRIN2), the CAP-Gly domain-containing linker protein 1 (CLIP1), mitochondrial GTPase GUF1, lysine-specific histone demethylase 4B (KDM4B), transcription regulator zinc finger protein 451 (ZNF451) and deubiquitinating enzyme OTUD1 40, 73, 88-90. In addition, Ye et al. 91 found that EBV miR-BART1 regulates the expression of metabolism-related genes including phosphoserine aminotransferase (PSAT1) and phosphoglycerate dehydrogenase (PHGDH). PSAT1 and PHGDH have been shown to promote proliferation of tumor cells 92, 93. Accordingly, EBV miRNAs may play important roles in the development of EBV-associated tumors.
Biological function of EBV miRNAs
EBV miRNAs suppress tumor cell apoptosis
EBV latently persists in tumor cells and expresses a restricted number of latent proteins to escape host immune surveillance 94. Apoptosis blocks EBV latent infection and prevents viral immune evasion by inducing EBV-associated tumor cell death 95. However, EBV has devised cunning strategies to counteract cell apoptosis and facilitate tumor development. A number of studies have showed that EBV miRNAs are implicated in the initiation of EBV-associated malignancies by regulating cell apoptosis. The BH3-interacting domain death agonist (BID) serves as a caspase substrate that engages the mitochondrial pathway to amplify the apoptotic response 96. miR-BART4-5p is able to restrain cell apoptosis by downregulating BID. MAP3K5, an apoptotic protein, is a crucial early-response gene in p38/MAPK pathway 97. miR-BART22 blocks MAPK pathway and remits cell apoptosis by suppressing MAP3K5 expression 98. miR-BART5, a highly expressed miRNA in EBV-associated epithelial tumor cells, promotes the survival of tumor cells by inhibiting p53-upregulated modulator of apoptosis (PUMA) 99. In addition to PUMA, other pro-apoptotic BH3-only proteins, Bcl-2 interacting mediator of cell death (BIM) and Bcl-2-associated death promoter (BAD), have been shown to be targets of BART miRNAs 39, 100. The translocase of outer mitochondrial membrane 22 homolog (TOMM22) is a mitochondrial receptor for Bcl-2-associated X protein (Bax) 88, 101. miR-BART16 inhibits the pro-apoptotic function of Bax by mediating TOMM22 downregulation 102. Caspase-3 is a known effector caspase that executes apoptosis 103. BHRF1 miRNAs have been shown to prevent apoptosis in EBV-associated Burkitt's lymphoma by downregulating caspase-3 104. Collectively, these findings indicate that EBV miRNAs control the expression of apoptosis-related genes to impair host immune defense against viral infection.
EBV miRNAs alter cell growth by targeting tumor suppressor genes
Tumor suppressor genes play a central role in preventing the initiation and progression of cancers 105. The expression of tumor suppressor genes can be regulated by EBV-encoded miRNAs. In a previous report, EBV miR-BART3 was found to bind to the 3' UTR of tumor suppressor gene DICE1 (deleted in cancer 1) 106. Upregulation of miR-BART3 results in reduced expression of DICE1, thus offsetting its growth-prohibitive function. miR-BART1 and miR-BART7 are involved in the invasion and metastasis of EBV-positive NPC via reverting the PI3K-Akt/EMT pathway by targeting the cellular tumor suppressor PTEN (phosphatase and tensin homolog) 107, 108. miR-BHRF1-2 is a viral miRNA regulator of the tumor suppressor PRDM1 (PR domain zinc finger protein 1) 109. Downregulation of PRDM1 expression by miR-BHRF1-2 promotes the proliferation of EBV-infected cells. Another tumor suppressor forkhead box P1 (FOXP1) is targeted by EBV miR-BART11 110, 111. Therefore, miR-BART11 plays an important role in the progression of EBV-positive epithelial cancers by inhibiting tumor-suppressive effects of FOXP1. As EBV miRNAs target cellular genes involved in tumor suppression, they possess oncogenic functions in EBV-associated tumors.
EBV miRNAs promote cell transformation
Increasing evidence has confirmed the proliferation-promoting and cell-transforming capabilities of EBV miRNAs. Nemo-like kinase (NLK) is a negative regulator of Wnt signaling pathway that inhibits cell proliferation and migration 112, 113. miR-BART14, miR-BART19-3p, miR-BART18-5p and miR-BART10 are able to activate the Wnt pathway by downregulating NLK, thereby promoting cell proliferation and epithelial malignancy 114. Recently, E-cadherin has been found to be a target of miR-BART9 115. Depleting miR-BART9 induces an increased expression of E-cadherin and miR-200 family, suppressing proliferation and invasion of EBVaGC cells. Since downregulation of both E-cadherin and miR-200 family is a vital step in the carcinogenesis of EBVaGC 116, miR-BART9 plays a promotive role in malignant transformation.
Seto et al. 117 previously introduced mutations in viral precursor miRNA transcripts to prevent their processing into mature miRNAs. Phenotypic analysis of B cells infected with the mutant viruses indicated that EBV-encoded BHRF1 miRNAs can induce EBV-associated cellular transformation by strongly facilitating cell proliferation and inhibiting spontaneous apoptosis. Another research group also reveals the important role of EBV miRNAs in cell transformation by constructing an EBV mutant that lacks BHRF1 miRNAs 118. Likewise, they found that knockdown of BHRF1 miRNAs contributes to reduced immortalization efficiency, impeded S phase progression and increased apoptosis in infected B cells. miR-BHRF1-2 and miR-BHRF1-3 suppress the expression of EBNA-LP to facilitate B cell transformation by promoting the cleavage of EBNA-LP mRNAs during pre-miR-BHRF1 excision 119. These studies demonstrate that EBV miRNAs are key players in host cell transformation.
Contrarily, EBV miRNAs can inhibit the development of EBV-positive tumors. For example, miR-BART15-3p induces apoptosis in EBVaGC cells by lowering the expression level of the apoptosis inhibitor BRUCE (BIR repeat containing ubiquitin-conjugating enzyme) 120. miR-BART6-3p downregulates the expression of a long non-coding RNA (lncRNA), LOC553103, and reverses the epithelial-mesenchymal transition (EMT) process, suppressing EBV-associated tumor cell migration and invasion 121. These studies provide novel insights into the functional role of EBV miRNAs in progression of EBV-associated malignancies.
EBV miRNAs control host immune responses
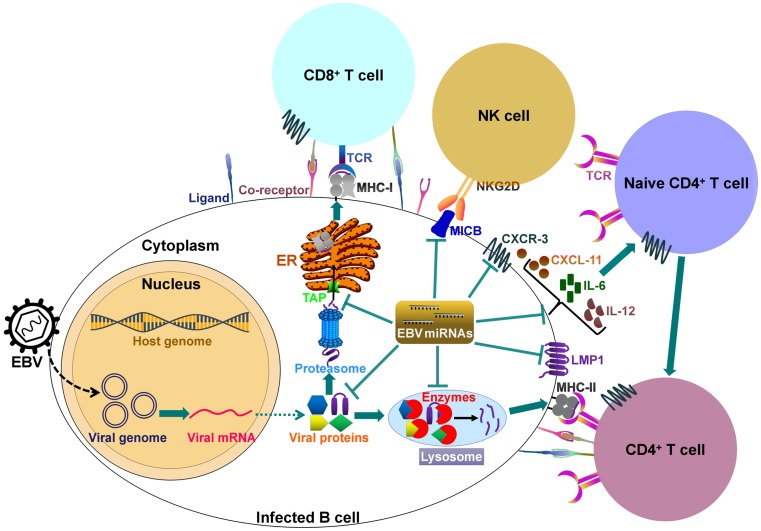
It can be seen that EBV miRNAs facilitate viral infection and pathogenesis by directly targeting host mRNAs involved in cell transformation, apoptosis and tumorigenesis. However, the host immune system plays a pivotal role in preventing viral infection. To successfully establish lifelong infections in host, EBV has to conquer the immune attack from the host. EBV invokes its miRNAs to subvert and elude host immune responses by interfering with cytokine and chemokine signaling networks, and blocking antigen presentation and T cell responses (Figure 2).
Figure 2.
EBV-encoded miRNAs regulate host immune responses. In latently infected cells, the viral DNA genome forms in a circular shape (episome). EBV miRNA biogenesis is dependent on the host machinery. EBV miRNAs reduce the levels of viral antigens (LMP1 and LMP2A) to favor viral persistence in host cells. LMP1 acts as a functional mimic of CD40 and can activate MHCs and costimulatory molecules. Thus, LMP1 downregulation by viral miRNAs may block antigen presentation. More importantly, EBV invokes its miRNAs to subvert host immune responses by downregulating multiple cellular genes or pathways. 1) EBV miRNAs can efficiently interfere with MHC class I-mediated antigen presentation by targeting the antigen transporter TAP2. 2) Viral miRNAs inhibit expression of the lysosomal enzymes (IFI30, LGMN and CTSB), restraining the capability to present antigens to CD4+ T cells through MHC class-II. 3) Viral miRNAs control the expression of inflammatory cytokines (IL-6 and IL-12), thereby suppressing cytokine-mediated cellular immune responses. 4) EBV miRNAs also allow virus-infected cells to escape from NK- and T-cell attack by directly targeting the NK-cell attracting ligand, MICB and the T-cell attracting chemokine, CXCL-11. TCR, T-cell receptor; MHC, major histocompatibility complex; NKG2D, natural killer group 2D; MICB, MHC class I chain-related molecule B; CXCR3, C-X-C chemokine receptor type 3; CXCL-11, C-X-C motif chemokine ligand 11; ER, endoplasmic reticulum.
EBV miRNAs modulate innate immune responses
Importin 7 (IPO7), a receptor for the transport of transcription factors into the nucleus, plays an important role in innate immunity 122. IPO7 is confirmed to be targeted by EBV miR-BART3 88. The IFN-inducible T-cell attracting chemokine CXCL-11, a ligand for the CXCR3 receptor, is produced by various cells including fibroblasts, endothelial cells and leukocytes upon stimulation with IFNs 123. IFN-induced CXCL-11 recruits and activates NK cells 124. Moreover, the interaction between CXCR3 and CXCL-11 is important for the selective homing of Th1 effector cells 125. miR-BHRF1-3 serves as a potential immune regulator by suppressing CXCL-11 32. Retinoic acid inducible gene I (RIG-I) is a pattern recognition receptor (PRR) that triggers the host innate immune response by specifically sensing viral infection 126. The type I IFN induced by RIG-I is a key component of the host innate immune response and exerts antiviral and immuno-regulatory functions 127, 128. Previous study showed that type I IFN plays an important role in controlling EBV infection 129, 130. miR-BART6-3p can inhibit RIG-I-like receptor signaling and type I IFN response by binding to the 3' UTR of RIG-I mRNA 131. The major histocompatibility complex (MHC) class I chain-related molecule B (MICB) is able to enhance NK cell-mediating killing of virus-infected cells by interacting with its receptor natural killer group 2D (NKG2D) 132. miR-BART2-5p inhibits MICB expression on the cell surface and thus protects virus-infected cells from the immune attack by NK cells 33. cAMP response element-binding protein (CREB)-binding protein (CREBBP) functions as a transcriptional coactivator of type I IFN-induced signaling pathway 133. miR-BART16 directly targets CREBBP, blocking the virus-induced type I IFN signaling pathway 134. miR-BART15 is capable of limiting inflammation to promote EBV infection by targeting nucleotide-binding oligomerization domain-like receptor family pyrin domain-containing 3 (NLRP3) inflammasome 135.
EBV miRNAs block cytokine signaling networks
Cell-mediated immunity in response to viral infection is controlled by altered expression of cytokines. EBV miRNAs may control host immune responses through regulation of cytokine signaling networks. IL-1 signaling serves as a key initiator of inflammation and host innate immune responses following viral infection 136, 137. The recent study indicates that miR-BHRF1-2-5p directly targets IL-1 receptor 1 (IL1R1) and blocks IL-1β-triggered NF-κB activation 138. IL-6 is an important mediator of acute inflammatory responses and also exerts stimulatory effects on B cells and T cells 139. miR-BART6-3p can control the expression of IL-6 receptor 140. Inhibition of miR-BART6-3p expression results in upregulation of IL-6 receptor that leads to the activation of NF-κB signaling. The proinflammatory cytokine IL-12 enhances IFN-γ production by CD8+ T cells and is crucial for the differentiation of naive CD4+ T cells to antiviral Th1 cells 141, 142. The release of IL-12 is markedly suppressed by miR-BART1, miR-BART2, miR-BART22 and miR-BHRF1-2 143. EBV miRNA-mediated IL-12 downregulation leads to lowered T-cell activation, facilitating the survival of EBV-infected cells 144. Moreover, these viral miRNAs are derived from different viral transcripts, indicating a tight control of IL-12 in both latent and lytic phases of EBV life cycle. IFN-γ plays an important role in CD8+ T cell immunity via enhancing MHC class-I antigen presentation pathway 145. miR-BART20-5p and miR-BART8 are shown to depress the IFN-γ/STAT1 pathway in nasal NK-cell lymphoma 146. This finding suggests that EBV may exploit its own miRNAs to favor viral replication and tumor growth by inhibiting the IFN-γ signaling pathway.
EBV miRNAs interfere with antigen processing and presentation
EBV miRNAs are found to modulate several genes related to antigen processing and presentation. Lysosomal enzymes are involved in MHC class II epitope processing 147. EBV miRNAs have been reported to control the expression of lysosomal enzymes 143. For example, IFN-γ-inducible lysosomal thiol reductase (IFI30), legumain (LGMN) and cathepsin B (CTSB) are targeted by EBV miR-BART1, miR-BART2 and miR-BHRF1-2, respectively. Costimulatory molecules and MHCs are essential components for efficient antigen presentation and can be indirectly regulated by EBV miRNAs 148. The viral antigen LMP1 can activate the CD40 signaling pathway, increasing expression of costimulatory molecules and MHCs on cell surface 3, 143, 149. Therefore, BART miRNA-mediated LMP1 downregulation may reduce expression of these surface molecules, thereby dampening the antigen presentation pathway 74. EBV miRNAs also limit cell surface levels of HLA class II molecules, providing an alternative way to block MHC class II antigen presentation. Lymphocyte antigen 75 (LY75) delivers antigens from the surface of dendritic cells to lysosomes or late endosomes 150. As a result, LY75 is crucial for antigen presentation on MHC molecules as well as stimulation of CD4+ and CD8+ T cells 151. LY75 can be targeted by miR-BART1-5p 40. Inhibition of LY75 expression by the viral miRNA might render EBV-harboring tumor cells capable to evade immune recognition by CD4+ and CD8+ T cells. Additionally, miR-BHRF1-3 and miR-BART17 directly target the peptide transporter subunit TAP2 which is involved in EBV antigen presentation 144.
EBV miRNAs inhibit antiviral CD4+ and CD8+ T cell responses
The adaptive immune system plays an important role in the clearance of virus infections 152. EBV miRNAs have been shown to control the expression of cellular genes involved in T cell-mediated responses, providing an alternative mechanism of EBV immune evasion. Mucosa-associated lymphoid tissue lymphoma transport protein 1 (MALT1) plays a pivotal role in inducing CD4+ T cell responses 153. EBV miR-BHRF1-2-5p is capable of directly targeting MALT1 154. CLEC2D is expressed on the surface of B cells upon activation of toll-like receptor (TLR) or B-cell receptor (BCR) signaling pathways 155. CLEC2D can activate NK- and T- cells by interacting with CD161, thereby enhancing production of IFN-γ and CD8+ T lymphocytes 156. miR-BART1-3p and miR-BART3-3p are confirmed to target the 3' UTR of CLEC2D 40. T-bet is a key regulator of Th1 development and function 157. T-bet maintains the effector function of CD8+ T cells via multiple mechanisms, such as modulation of CD8+ T cell proliferation and IFN-γ production 158. miR-BART20-5p is able to inhibit T-bet translation and suppress p53 in nasal NK/T-cell lymphoma 159.
EBV miRNAs regulate viral functions
EBV miRNAs regulate viral functions by controlling the expression of viral genes. It has been reported that EBV latent infection is strongly associated with the development of virus-associated malignancies 160. During latent expression, EBV expresses a limited number of antigens to favor its survival and escape from the host immune system 94. EBV miRNAs help the virus to maintain latency in infected cells. miR-BART2 displays complete sequence complementarity to the mRNA encoding viral DNA polymerase BALF5 30. miR-BART2 may facilitate the persistence of EBV latency and promote the development of EBV-associated tumors. Downregulation of Dicer by miR-BART6-5p results in the decreased expression of the viral lytic transactivators Zta and Rta 81. Moreover, miR-BART6-5p limits the expression of EBNA2 which serves an important function in maintaining EBV latent infection 161. These findings suggest that inhibition of Dicer and EBNA2 expression by miR-BART6 controls the latency-lytic switch of EBV and is essential for the maintenance of EBV latency to escape immune surveillance.
During latent infection, EBV miRNAs control the expression of viral antigens to restrict the immunogenicity of infected cells. LMP1 can activate multiple cellular signaling pathways including MAPK, NF-ĸB, and JAK/STAT signaling which robustly elicit host immune responses 162-164. miR-BART5-5p and miR-BART19-5p are able to downregulate the expression of LMP1 73, 74. EBNA1 is required for the maintenance, replication and transcription of EBV episome 165. More importantly, EBNA1 is the only viral antigen that is expressed in all types of EBV-harboring cells 166. Although EBNA1 is proven to be a target of effector T cells, the epitopes of EBNA1 are rarely presented on MHC class I molecules and can not be recognized by T cells 167. Previously, Albanese et al. 144 found that EBV miRNAs directly targeted TAP2 and decreased EBNA1 expression. LMP2A is an immunogenic viral antigen that can be recognized by cytotoxic T cells (CTLs) 168. Viral miR-BART22 is shown to limit the expression of LMP2A 76. These findings herein reviewed collectively imply that EBV miRNAs may facilitate viral escape from host immune surveillance by downregulating viral latent antigens.
EBV miRNAs can be used as diagnostic and prognostic biomarkers for cancers
Early detection and accurate prognosis are two key factors for the treatment of EBV-associated tumors. Recently, the clinical value of EBV miRNAs for diagnosis and prognosis of EBV-associated tumors has been investigated. It has been reported that the expression level of miR-BART1-5p is reflective of NPC progression 169. Detection of miR-BART1-5p is effective for early diagnosis of NPC, even in cases that are mis-diagnosed as negative based on clinical tests. miR-BART10-3p can directly target BTRC gene that encodes β-transducin repeat containing E3 ubiquitin protein ligase (βTrCP) 170. Upregulation of miR-BART10-3p and decreased expression of BTRC are associated with poor prognosis in NPC patients. Moreover, miR-BART10-3p promotes the migration and invasion of NPC cells by exerting its inhibitory role in BTRC expression. These findings indicate that miR-BART10-3p is a potential novel prognostic biomarker and therapeutic target for EBV-associated NPC. miR-BART7 is able to enhance the sensitivity of NPC cells to radiation treatment by targeting GFPT1/TGFβ1 signaling pathway, which may contribute to improved efficacy of NPC radiotherapy 171. miR-BART7 detection may provide a potential indicator for monitoring the progression of EBV-associated NPC. miR-BART7 and miR-BART13 are found to be stably present in the plasma of NPC patients 172. Their expression levels are elevated in NPC patients with disease progression. On the contrary, miR-BART7 and miR-BART13 are undetectable in NPC patients after radiotherapy. Therefore, EBV miR-BART7 and miR-BART13 are regarded as potential serological biomarkers for NPC diagnosis and prognosis.
miR-BART20-5p is highly expressed in patients with EBV-positive gastric carcinoma 173. A multivariate analysis indicates that miR-BART20-5p is associated with worse recurrence-free survival of the patients, which makes miR-BART20-5p a potential biomarker and therapeutic target for EBV-associated gastric carcinoma. Recently, a study reveals miR-BART2-5p upregulation in sera of patients with EBV-associated nasal natural killer/T-cell lymphoma (NNKTL) 174. The high circulating miR-BART2-5p level is strongly linked with disease progression and poor prognosis in patients with NNKTL. Thus, miR-BART2-5p may serve as diagnostic and prognostic biomarkers in patients with NNKTL.
Based on these clues, EBV miRNAs hold particular promise to be utilized as diagnostic and prognostic biomarkers for EBV-associated tumors. To date, there are still no predictive biomarkers available for EBV-associated tumors. Increasing knowledge about the function of EBV miRNAs in immune modulation will help to develop novel diagnostic and therapeutic strategies for EBV-associated tumors. Furthermore, deep investigations into the mechanism underlying regulation of host immune responses by EBV miRNAs would accelerate their clinical application in diagnosis and treatment of EBV-associated tumors.
Concluding remarks and future perspectives
EBV miRNAs are expressed in all phases of viral life cycle and are produced in the way similar to cellular miRNAs. EBV has evolved to exploit its own miRNAs to change the expression of a variety of genes related to viral latency, tumor suppression, cell proliferation, apoptosis and transformation. More importantly, EBV miRNAs play powerful roles in subverting host immune responses. However, more targets of EBV-encoded miRNAs are warranted to be identified to comprehensively uncover the interplay between EBV and host immune system. Moreover, the roles of EBV-encoded miRNAs in immune regulation require to be studied in detail. In particular, the immunological mechanisms underlying the involvement of EBV-encoded miRNAs in viral infection and pathogenesis need more systematic survey. In-depth exploration of EBV miRNAs will be of great importance for comprehensive elucidation of the interaction between EBV and host immune system and also provide novel insights into how EBV succeeds as a viral pathogen. Furthermore, EBV-encoded miRNAs are abundantly expressed in EBV-associated tumors, and they are implicated in tumor development by targeting viral or cellular genes. Controlling the activities of EBV-encoded miRNAs in EBV-positive tumors may represent a promising approach to restoring host anti-tumor immunity, which provides a novel therapeutic strategy for the treatment of EBV-associated tumors.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81701991), Applied Basic Research Programs of Qingdao, China (17-1-1-59-jch) and the China Postdoctoral Science Foundation Funded Project (2016M600520).
References
- 1.Epstein MA, Barr YM, Achong BG. A Second Virus-Carrying Tissue Culture Strain (Eb2) of Lymphoblasts from Burkitt's Lymphoma. Pathol Biol. 1964;12:1233–4. [PubMed] [Google Scholar]
- 2.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA. et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–76. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 3.Albanese M, Tagawa T, Buschle A, Hammerschmidt W. MicroRNAs of Epstein-Barr Virus Control Innate and Adaptive Antiviral Immunity. J Virol. 2017;91:e01667–16. doi: 10.1128/JVI.01667-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price AM, Luftig MA. To be or not IIb: a multi-step process for Epstein-Barr virus latency establishment and consequences for B cell tumorigenesis. PLoS Pathog. 2015;11:e1004656. doi: 10.1371/journal.ppat.1004656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dheekollu J, Chen HS, Kaye KM, Lieberman PM. Timeless-dependent DNA replication-coupled recombination promotes Kaposi's Sarcoma-associated herpesvirus episome maintenance and terminal repeat stability. J Virol. 2013;87:3699–709. doi: 10.1128/JVI.02211-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim H, Iizasa H, Kanehiro Y, Fekadu S, Yoshiyama H. Herpesviral microRNAs in Cellular Metabolism and Immune Responses. Front Microbiol. 2017;8:1318. doi: 10.3389/fmicb.2017.01318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez AF, Esteller M. Viral epigenomes in human tumorigenesis. Oncogene. 2010;29:1405–20. doi: 10.1038/onc.2009.517. [DOI] [PubMed] [Google Scholar]
- 8.Odumade OA, Hogquist KA, Balfour HH Jr. Progress and problems in understanding and managing primary Epstein-Barr virus infections. Clin Microbiol Rev. 2011;24:193–209. doi: 10.1128/CMR.00044-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Liu S, Hu J, Luo X, Li N, A MB. et al. Epstein-Barr virus lytic reactivation regulation and its pathogenic role in carcinogenesis. Int J Biol Sci. 2016;12:1309–18. doi: 10.7150/ijbs.16564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jha HC, Pei Y, Robertson ES. Epstein-Barr Virus: Diseases Linked to Infection and Transformation. Front Microbiol. 2016;7:1602. doi: 10.3389/fmicb.2016.01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 12.Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005;14( Spec No 1 R):121–32. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH. et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia S, Zhai H, Zhao M. MicroRNAs regulate immune system via multiple targets. Discov Med. 2014;18:237–47. [PubMed] [Google Scholar]
- 16.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 18.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009;10:141–8. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 19.Saxena S, Jonsson ZO, Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J Biol Chem. 2003;278:44312–9. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 20.He Y, Ma T, Zhang X. The Mechanism of Synchronous Precise Regulation of Two Shrimp White Spot Syndrome Virus Targets by a Viral MicroRNA. Front Immunol. 2017;8:1546. doi: 10.3389/fimmu.2017.01546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sachdeva M, Mito JK, Lee CL, Zhang M, Li Z, Dodd RD. et al. MicroRNA-182 drives metastasis of primary sarcomas by targeting multiple genes. J Clin Invest. 2016;126:1606. doi: 10.1172/JCI86573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Rooij E. The art of microRNA research. Circ Res. 2011;108:219–34. doi: 10.1161/CIRCRESAHA.110.227496. [DOI] [PubMed] [Google Scholar]
- 23.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Y, Haecker I, Yang Y, Gao SJ, Renne R. gamma-Herpesvirus-encoded miRNAs and their roles in viral biology and pathogenesis. Curr Opin Virol. 2013;3:266–75. doi: 10.1016/j.coviro.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J. et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–6. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 26.Shinozaki-Ushiku A, Kunita A, Isogai M, Hibiya T, Ushiku T, Takada K. et al. Profiling of Virus-Encoded MicroRNAs in Epstein-Barr Virus-Associated Gastric Carcinoma and Their Roles in Gastric Carcinogenesis. J Virol. 2015;89:5581–91. doi: 10.1128/JVI.03639-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barth S, Meister G, Grasser FA. EBV-encoded miRNAs. Biochim Biophys Acta. 2011;1809:631–40. doi: 10.1016/j.bbagrm.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 28.Williams H, Crawford DH. Epstein-Barr virus: the impact of scientific advances on clinical practice. Blood. 2006;107:862–9. doi: 10.1182/blood-2005-07-2702. [DOI] [PubMed] [Google Scholar]
- 29.Qiu J, Cosmopoulos K, Pegtel M, Hopmans E, Murray P, Middeldorp J. et al. A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog. 2011;7:e1002193. doi: 10.1371/journal.ppat.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E. et al. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666–75. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung YJ, Choi H, Kim H, Lee SK. MicroRNA miR-BART20-5p stabilizes Epstein-Barr virus latency by directly targeting BZLF1 and BRLF1. J Virol. 2014;88:9027–37. doi: 10.1128/JVI.00721-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia T, O'Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB. et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68:1436–42. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5:376–85. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 35.Flores-Jasso CF, Arenas-Huertero C, Reyes JL, Contreras-Cubas C, Covarrubias A, Vaca L. First step in pre-miRNAs processing by human Dicer. Acta Pharmacol Sin. 2009;30:1177–85. doi: 10.1038/aps.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–60. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 37.Li Z, Rana TM. Molecular mechanisms of RNA-triggered gene silencing machineries. Acc Chem Res. 2012;45:1122–31. doi: 10.1021/ar200253u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G. et al. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A. 2007;104:16164–9. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marquitz AR, Mathur A, Nam CS, Raab-Traub N. The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim. Virology. 2011;412:392–400. doi: 10.1016/j.virol.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M. et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012;8:e1002484. doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skalsky RL, Kang D, Linnstaedt SD, Cullen BR. Evolutionary conservation of primate lymphocryptovirus microRNA targets. J Virol. 2014;88:1617–35. doi: 10.1128/JVI.02071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R. et al. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amoroso R, Fitzsimmons L, Thomas WA, Kelly GL, Rowe M, Bell AI. Quantitative studies of Epstein-Barr virus-encoded microRNAs provide novel insights into their regulation. J Virol. 2011;85:996–1010. doi: 10.1128/JVI.01528-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hooykaas MJ, Kruse E, Wiertz EJ, Lebbink RJ. Comprehensive profiling of functional Epstein-Barr virus miRNA expression in human cell lines. BMC Genomics. 2016;17:644. doi: 10.1186/s12864-016-2978-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pratt ZL, Kuzembayeva M, Sengupta S, Sugden B. The microRNAs of Epstein-Barr Virus are expressed at dramatically differing levels among cell lines. Virology. 2009;386:387–97. doi: 10.1016/j.virol.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edwards RH, Marquitz AR, Raab-Traub N. Epstein-Barr virus BART microRNAs are produced from a large intron prior to splicing. J Virol. 2008;82:9094–106. doi: 10.1128/JVI.00785-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim DN, Lee SK. Biogenesis of Epstein-Barr virus microRNAs. Mol Cell Biochem. 2012;365:203–10. doi: 10.1007/s11010-012-1261-7. [DOI] [PubMed] [Google Scholar]
- 48.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 49.Zeng Y, Cullen BR. RNA interference in human cells is restricted to the cytoplasm. RNA. 2002;8:855–60. doi: 10.1017/s1355838202020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–90. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cox MA, Leahy J, Hardwick JM. An enhancer within the divergent promoter of Epstein-Barr virus responds synergistically to the R and Z transactivators. J Virol. 1990;64:313–21. doi: 10.1128/jvi.64.1.313-321.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu JY, Pfuhl T, Motsch N, Barth S, Nicholls J, Grasser F. et al. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J Virol. 2009;83:3333–41. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swaminathan S. Noncoding RNAs produced by oncogenic human herpesviruses. J Cell Physiol. 2008;216:321–6. doi: 10.1002/jcp.21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xing L, Kieff E. Epstein-Barr virus BHRF1 micro- and stable RNAs during latency III and after induction of replication. J Virol. 2007;81:9967–75. doi: 10.1128/JVI.02244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pearson GR, Luka J, Petti L, Sample J, Birkenbach M, Braun D. et al. Identification of an Epstein-Barr virus early gene encoding a second component of the restricted early antigen complex. Virology. 1987;160:151–61. doi: 10.1016/0042-6822(87)90055-9. [DOI] [PubMed] [Google Scholar]
- 56.Austin PJ, Flemington E, Yandava CN, Strominger JL, Speck SH. Complex transcription of the Epstein-Barr virus BamHI fragment H rightward open reading frame 1 (BHRF1) in latently and lytically infected B lymphocytes. Proc Natl Acad Sci U S A. 1988;85:3678–82. doi: 10.1073/pnas.85.11.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xing L, Kieff E. cis-Acting effects on RNA processing and Drosha cleavage prevent Epstein-Barr virus latency III BHRF1 expression. J Virol. 2011;85:8929–39. doi: 10.1128/JVI.00336-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen SJ, Chen GH, Chen YH, Liu CY, Chang KP, Chang YS. et al. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS One. 2010;5:e12745. doi: 10.1371/journal.pone.0012745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:733–50. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cosmopoulos K, Pegtel M, Hawkins J, Moffett H, Novina C, Middeldorp J. et al. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. J Virol. 2009;83:2357–67. doi: 10.1128/JVI.02104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim DN, Chae HS, Oh ST, Kang JH, Park CH, Park WS. et al. Expression of viral microRNAs in Epstein-Barr virus-associated gastric carcinoma. J Virol. 2007;81:1033–6. doi: 10.1128/JVI.02271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Motsch N, Alles J, Imig J, Zhu J, Barth S, Reineke T. et al. MicroRNA profiling of Epstein-Barr virus-associated NK/T-cell lymphomas by deep sequencing. PLoS One. 2012;7:e42193. doi: 10.1371/journal.pone.0042193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Imig J, Motsch N, Zhu JY, Barth S, Okoniewski M, Reineke T. et al. microRNA profiling in Epstein-Barr virus-associated B-cell lymphoma. Nucleic Acids Res. 2011;39:1880–93. doi: 10.1093/nar/gkq1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Forte E, Luftig MA. The role of microRNAs in Epstein-Barr virus latency and lytic reactivation. Microbes Infect. 2011;13:1156–67. doi: 10.1016/j.micinf.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petosa C, Morand P, Baudin F, Moulin M, Artero JB, Muller CW. Structural basis of lytic cycle activation by the Epstein-Barr virus ZEBRA protein. Mol Cell. 2006;21:565–72. doi: 10.1016/j.molcel.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 66.Kalla M, Schmeinck A, Bergbauer M, Pich D, Hammerschmidt W. AP-1 homolog BZLF1 of Epstein-Barr virus has two essential functions dependent on the epigenetic state of the viral genome. Proc Natl Acad Sci U S A. 2010;107:850–5. doi: 10.1073/pnas.0911948107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heilmann AM, Calderwood MA, Portal D, Lu Y, Johannsen E. Genome-wide analysis of Epstein-Barr virus Rta DNA binding. J Virol. 2012;86:5151–64. doi: 10.1128/JVI.06760-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morris MA, Dawson CW, Young LS. Role of the Epstein-Barr virus-encoded latent membrane protein-1, LMP1, in the pathogenesis of nasopharyngeal carcinoma. Future Oncol. 2009;5:811–25. doi: 10.2217/fon.09.53. [DOI] [PubMed] [Google Scholar]
- 69.Ahsan N, Kanda T, Nagashima K, Takada K. Epstein-Barr virus transforming protein LMP1 plays a critical role in virus production. J Virol. 2005;79:4415–24. doi: 10.1128/JVI.79.7.4415-4424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kieser A, Sterz KR. The Latent Membrane Protein 1 (LMP1) Curr Top Microbiol Immunol. 2015;391:119–49. doi: 10.1007/978-3-319-22834-1_4. [DOI] [PubMed] [Google Scholar]
- 71.Lu JJ, Chen JY, Hsu TY, Yu WC, Su IJ, Yang CS. Induction of apoptosis in epithelial cells by Epstein-Barr virus latent membrane protein 1. J Gen Virol. 1996;77( Pt 8):1883–92. doi: 10.1099/0022-1317-77-8-1883. [DOI] [PubMed] [Google Scholar]
- 72.Liu Y, Wang X, Lo AK, Wong YC, Cheung AL, Tsao SW. Latent membrane protein-1 of Epstein-Barr virus inhibits cell growth and induces sensitivity to cisplatin in nasopharyngeal carcinoma cells. J Med Virol. 2002;66:63–9. doi: 10.1002/jmv.2112. [DOI] [PubMed] [Google Scholar]
- 73.Riley KJ, Rabinowitz GS, Yario TA, Luna JM, Darnell RB, Steitz JA. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 2012;31:2207–21. doi: 10.1038/emboj.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Verhoeven RJ, Tong S, Zhang G, Zong J, Chen Y, Jin DY. et al. NF-kappaB Signaling Regulates Expression of Epstein-Barr Virus BART MicroRNAs and Long Noncoding RNAs in Nasopharyngeal Carcinoma. J Virol. 2016;90:6475–88. doi: 10.1128/JVI.00613-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Su Z, Peluso MV, Raffegerst SH, Schendel DJ, Roskrow MA. The generation of LMP2a-specific cytotoxic T lymphocytes for the treatment of patients with Epstein-Barr virus-positive Hodgkin disease. Eur J Immunol. 2001;31:947–58. doi: 10.1002/1521-4141(200103)31:3<947::aid-immu947>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 76.Lung RW, Tong JH, Sung YM, Leung PS, Ng DC, Chau SL. et al. Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22. Neoplasia. 2009;11:1174–84. doi: 10.1593/neo.09888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanda T, Miyata M, Kano M, Kondo S, Yoshizaki T, Iizasa H. Clustered microRNAs of the Epstein-Barr virus cooperatively downregulate an epithelial cell-specific metastasis suppressor. J Virol. 2015;89:2684–97. doi: 10.1128/JVI.03189-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bishop GA, Busch LK. Molecular mechanisms of B-lymphocyte transformation by Epstein-Barr virus. Microbes Infect. 2002;4:853–7. doi: 10.1016/s1286-4579(02)01605-2. [DOI] [PubMed] [Google Scholar]
- 79.Hislop AD, Taylor GS, Sauce D, Rickinson AB. Cellular responses to viral infection in humans: lessons from Epstein-Barr virus. Annu Rev Immunol. 2007;25:587–617. doi: 10.1146/annurev.immunol.25.022106.141553. [DOI] [PubMed] [Google Scholar]
- 80.Pagano JS, Blaser M, Buendia MA, Damania B, Khalili K, Raab-Traub N. et al. Infectious agents and cancer: criteria for a causal relation. Semin Cancer Biol. 2004;14:453–71. doi: 10.1016/j.semcancer.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 81.Iizasa H, Wulff BE, Alla NR, Maragkakis M, Megraw M, Hatzigeorgiou A. et al. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J Biol Chem. 2010;285:33358–70. doi: 10.1074/jbc.M110.138362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Najdi R, Holcombe RF, Waterman ML. Wnt signaling and colon carcinogenesis: beyond APC. J Carcinog. 2011;10:5. doi: 10.4103/1477-3163.78111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wong AM, Kong KL, Tsang JW, Kwong DL, Guan XY. Profiling of Epstein-Barr virus-encoded microRNAs in nasopharyngeal carcinoma reveals potential biomarkers and oncomirs. Cancer. 2012;118:698–710. doi: 10.1002/cncr.26309. [DOI] [PubMed] [Google Scholar]
- 84.Li Z, Chen X, Li L, Liu S, Yang L, Ma X. et al. EBV encoded miR-BHRF1-1 potentiates viral lytic replication by downregulating host p53 in nasopharyngeal carcinoma. Int J Biochem Cell Biol. 2012;44:275–9. doi: 10.1016/j.biocel.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 85.Chang SS, Lo YC, Chua HH, Chiu HY, Tsai SC, Chen JY. et al. Critical role of p53 in histone deacetylase inhibitor-induced Epstein-Barr virus Zta expression. J Virol. 2008;82:7745–51. doi: 10.1128/JVI.02717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sato Y, Kamura T, Shirata N, Murata T, Kudoh A, Iwahori S. et al. Degradation of phosphorylated p53 by viral protein-ECS E3 ligase complex. PLoS Pathog. 2009;5:e1000530. doi: 10.1371/journal.ppat.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Warnatz HJ, Schmidt D, Manke T, Piccini I, Sultan M, Borodina T. et al. The BTB and CNC homology 1 (BACH1) target genes are involved in the oxidative stress response and in control of the cell cycle. J Biol Chem. 2011;286:23521–32. doi: 10.1074/jbc.M111.220178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dolken L, Malterer G, Erhard F, Kothe S, Friedel CC, Suffert G. et al. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe. 2010;7:324–34. doi: 10.1016/j.chom.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 89.Gottwein E, Corcoran DL, Mukherjee N, Skalsky RL, Hafner M, Nusbaum JD. et al. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe. 2011;10:515–26. doi: 10.1016/j.chom.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Malterer G, Dolken L, Haas J. The miRNA-targetome of KSHV and EBV in human B-cells. RNA Biol. 2011;8:30–4. doi: 10.4161/rna.8.1.13745. [DOI] [PubMed] [Google Scholar]
- 91.Ye Y, Zhou Y, Zhang L, Chen Y, Lyu X, Cai L. et al. EBV-miR-BART1 is involved in regulating metabolism-associated genes in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2013;436:19–24. doi: 10.1016/j.bbrc.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 92.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ. et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–74. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vie N, Copois V, Bascoul-Mollevi C, Denis V, Bec N, Robert B. et al. Overexpression of phosphoserine aminotransferase PSAT1 stimulates cell growth and increases chemoresistance of colon cancer cells. Mol Cancer. 2008;7:14. doi: 10.1186/1476-4598-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ressing ME, van Gent M, Gram AM, Hooykaas MJ, Piersma SJ, Wiertz EJ. Immune Evasion by Epstein-Barr Virus. Curr Top Microbiol Immunol. 2015;391:355–81. doi: 10.1007/978-3-319-22834-1_12. [DOI] [PubMed] [Google Scholar]
- 95.Zuo L, Yue W, Du S, Xin S, Zhang J, Liu L. et al. An update: Epstein-Barr virus and immune evasion via microRNA regulation. Virol Sin. 2017;32:175–87. doi: 10.1007/s12250-017-3996-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 97.Pressinotti NC, Klocker H, Schafer G, Luu VD, Ruschhaupt M, Kuner R. et al. Differential expression of apoptotic genes PDIA3 and MAP3K5 distinguishes between low- and high-risk prostate cancer. Mol Cancer. 2009;8:130. doi: 10.1186/1476-4598-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen R, Zhang M, Li Q, Xiong H, Liu S, Fang W. et al. The Epstein-Barr Virus-encoded miR-BART22 targets MAP3K5 to promote host cell proliferative and invasive abilities in nasopharyngeal carcinoma. J Cancer. 2017;8:305–13. doi: 10.7150/jca.15753. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 99.Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF. et al. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205:2551–60. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim H, Choi H, Lee SK. Epstein-Barr Virus MicroRNA miR-BART20-5p Suppresses Lytic Induction by Inhibiting BAD-Mediated caspase-3-Dependent Apoptosis. J Virol. 2015;90:1359–68. doi: 10.1128/JVI.02794-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bellot G, Cartron PF, Er E, Oliver L, Juin P, Armstrong LC. et al. TOM22, a core component of the mitochondria outer membrane protein translocation pore, is a mitochondrial receptor for the proapoptotic protein Bax. Cell Death Differ. 2007;14:785–94. doi: 10.1038/sj.cdd.4402055. [DOI] [PubMed] [Google Scholar]
- 102.Kang D, Skalsky RL, Cullen BR. EBV BART MicroRNAs Target Multiple Pro-apoptotic Cellular Genes to Promote Epithelial Cell Survival. PLoS Pathog. 2015;11:e1004979. doi: 10.1371/journal.ppat.1004979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kumar S. Caspase function in programmed cell death. Cell Death Differ. 2007;14:32–43. doi: 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- 104.Zhang J, Huang T, Zhou Y, Cheng ASL, Yu J, To KF. et al. The oncogenic role of Epstein-Barr virus-encoded microRNAs in Epstein-Barr virus-associated gastric carcinoma. J Cell Mol Med. 2018;22:38–45. doi: 10.1111/jcmm.13354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Guo XE, Ngo B, Modrek AS, Lee WH. Targeting tumor suppressor networks for cancer therapeutics. Curr Drug Targets. 2014;15:2–16. doi: 10.2174/1389450114666140106095151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lei T, Yuen KS, Xu R, Tsao SW, Chen H, Li M. et al. Targeting of DICE1 tumor suppressor by Epstein-Barr virus-encoded miR-BART3* microRNA in nasopharyngeal carcinoma. Int J Cancer. 2013;133:79–87. doi: 10.1002/ijc.28007. [DOI] [PubMed] [Google Scholar]
- 107.Cai L, Ye Y, Jiang Q, Chen Y, Lyu X, Li J. et al. Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. Nat Commun. 2015;6:7353. doi: 10.1038/ncomms8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cai LM, Lyu XM, Luo WR, Cui XF, Ye YF, Yuan CC. et al. EBV-miR-BART7-3p promotes the EMT and metastasis of nasopharyngeal carcinoma cells by suppressing the tumor suppressor PTEN. Oncogene. 2015;34:2156–66. doi: 10.1038/onc.2014.341. [DOI] [PubMed] [Google Scholar]
- 109.Ma J, Nie K, Redmond D, Liu Y, Elemento O, Knowles DM. et al. EBV-miR-BHRF1-2 targets PRDM1/Blimp1: potential role in EBV lymphomagenesis. Leukemia. 2016;30:594–604. doi: 10.1038/leu.2015.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shi C, Sakuma M, Mooroka T, Liscoe A, Gao H, Croce KJ. et al. Down-regulation of the forkhead transcription factor Foxp1 is required for monocyte differentiation and macrophage function. Blood. 2008;112:4699–711. doi: 10.1182/blood-2008-01-137018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Song Y, Li X, Zeng Z, Li Q, Gong Z, Liao Q. et al. Epstein-Barr virus encoded miR-BART11 promotes inflammation-induced carcinogenesis by targeting FOXP1. Oncotarget. 2016;7:36783–99. doi: 10.18632/oncotarget.9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ke H, Masoumi KC, Ahlqvist K, Seckl MJ, Rydell-Tormanen K, Massoumi R. Nemo-like kinase regulates the expression of vascular endothelial growth factor (VEGF) in alveolar epithelial cells. Sci Rep. 2016;6:23987. doi: 10.1038/srep23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bilir B, Kucuk O, Moreno CS. Wnt signaling blockage inhibits cell proliferation and migration, and induces apoptosis in triple-negative breast cancer cells. J Transl Med. 2013;11:280. doi: 10.1186/1479-5876-11-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lung RW, Tong JH, To KF. Emerging roles of small Epstein-Barr virus derived non-coding RNAs in epithelial malignancy. Int J Mol Sci. 2013;14:17378–409. doi: 10.3390/ijms140917378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tsai CY, Liu YY, Liu KH, Hsu JT, Chen TC, Chiu CT. et al. Comprehensive profiling of virus microRNAs of Epstein-Barr virus-associated gastric carcinoma: highlighting the interactions of ebv-Bart9 and host tumor cells. J Gastroenterol Hepatol. 2017;32:82–91. doi: 10.1111/jgh.13432. [DOI] [PubMed] [Google Scholar]
- 116.Shinozaki A, Sakatani T, Ushiku T, Hino R, Isogai M, Ishikawa S. et al. Downregulation of microRNA-200 in EBV-associated gastric carcinoma. Cancer Res. 2010;70:4719–27. doi: 10.1158/0008-5472.CAN-09-4620. [DOI] [PubMed] [Google Scholar]
- 117.Seto E, Moosmann A, Gromminger S, Walz N, Grundhoff A, Hammerschmidt W. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010;6:e1001063. doi: 10.1371/journal.ppat.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Feederle R, Linnstaedt SD, Bannert H, Lips H, Bencun M, Cullen BR. et al. A viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog. 2011;7:e1001294. doi: 10.1371/journal.ppat.1001294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Poling BC, Price AM, Luftig MA, Cullen BR. The Epstein-Barr virus miR-BHRF1 microRNAs regulate viral gene expression in cis. Virology. 2017;512:113–23. doi: 10.1016/j.virol.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Choi H, Lee H, Kim SR, Gho YS, Lee SK. Epstein-Barr virus-encoded microRNA BART15-3p promotes cell apoptosis partially by targeting BRUCE. J Virol. 2013;87:8135–44. doi: 10.1128/JVI.03159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.He B, Li W, Wu Y, Wei F, Gong Z, Bo H. et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis. 2016;7:e2353. doi: 10.1038/cddis.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang IV, Wade CM, Kang HM, Alper S, Rutledge H, Lackford B. et al. Identification of novel genes that mediate innate immunity using inbred mice. Genetics. 2009;183:1535–44. doi: 10.1534/genetics.109.107540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Proost P, Mortier A, Loos T, Vandercappellen J, Gouwy M, Ronsse I. et al. Proteolytic processing of CXCL11 by CD13/aminopeptidase N impairs CXCR3 and CXCR7 binding and signaling and reduces lymphocyte and endothelial cell migration. Blood. 2007;110:37–44. doi: 10.1182/blood-2006-10-049072. [DOI] [PubMed] [Google Scholar]
- 124.Egesten A, Eliasson M, Olin AI, Erjefalt JS, Bjartell A, Sangfelt P. et al. The proinflammatory CXC-chemokines GRO-alpha/CXCL1 and MIG/CXCL9 are concomitantly expressed in ulcerative colitis and decrease during treatment with topical corticosteroids. Int J Colorectal Dis. 2007;22:1421–7. doi: 10.1007/s00384-007-0370-3. [DOI] [PubMed] [Google Scholar]
- 125.Bonecchi R, Bianchi G, Bordignon PP, D'Ambrosio D, Lang R, Borsatti A. et al. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J Exp Med. 1998;187:129–34. doi: 10.1084/jem.187.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xu XX, Wan H, Nie L, Shao T, Xiang LX, Shao JZ. RIG-I: a multifunctional protein beyond a pattern recognition receptor. Protein Cell. 2017;9:246–53. doi: 10.1007/s13238-017-0431-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95:15623–8. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM. et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912–20. [PubMed] [Google Scholar]
- 129.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lim WH, Kireta S, Russ GR, Coates PT. Human plasmacytoid dendritic cells regulate immune responses to Epstein-Barr virus (EBV) infection and delay EBV-related mortality in humanized NOD-SCID mice. Blood. 2007;109:1043–50. doi: 10.1182/blood-2005-12-024802. [DOI] [PubMed] [Google Scholar]
- 131.Lu Y, Qin Z, Wang J, Zheng X, Lu J, Zhang X. et al. Epstein-Barr Virus miR-BART6-3p Inhibits the RIG-I Pathway. J Innate Immun. 2017;9:574–86. doi: 10.1159/000479749. [DOI] [PubMed] [Google Scholar]
- 132.Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL. et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–9. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 133.Tang X, Gao JS, Guan YJ, McLane KE, Yuan ZL, Ramratnam B. et al. Acetylation-dependent signal transduction for type I interferon receptor. Cell. 2007;131:93–105. doi: 10.1016/j.cell.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 134.Hooykaas MJG, van Gent M, Soppe JA, Kruse E, Boer IGJ, van Leenen D. et al. EBV MicroRNA BART16 Suppresses Type I IFN Signaling. J Immunol. 2017;198:4062–73. doi: 10.4049/jimmunol.1501605. [DOI] [PubMed] [Google Scholar]
- 135.Haneklaus M, Gerlic M, Kurowska-Stolarska M, Rainey AA, Pich D, McInnes IB. et al. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. J Immunol. 2012;189:3795–9. doi: 10.4049/jimmunol.1200312. [DOI] [PubMed] [Google Scholar]
- 136.Mills KH, Dunne A. Immune modulation: IL-1, master mediator or initiator of inflammation. Nat Med. 2009;15:1363–4. doi: 10.1038/nm1209-1363. [DOI] [PubMed] [Google Scholar]
- 137.Sims JE, Smith DE. The IL-1 family: regulators of immunity. Nat Rev Immunol. 2010;10:89–102. doi: 10.1038/nri2691. [DOI] [PubMed] [Google Scholar]
- 138.Skinner CM, Ivanov NS, Barr SA, Chen Y, Skalsky RL. An Epstein-Barr Virus MicroRNA Blocks Interleukin-1 (IL-1) Signaling by Targeting IL-1 Receptor 1. J Virol. 2017;91:e00530–17. doi: 10.1128/JVI.00530-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gabay C. Interleukin-6 and chronic inflammation. Arthritis Res Ther. 2006;8(Suppl 2):S3. doi: 10.1186/ar1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ambrosio MR, Navari M, Di Lisio L, Leon EA, Onnis A, Gazaneo S. et al. The Epstein Barr-encoded BART-6-3p microRNA affects regulation of cell growth and immuno response in Burkitt lymphoma. Infect Agent Cancer. 2014;9:12. doi: 10.1186/1750-9378-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Henry CJ, Ornelles DA, Mitchell LM, Brzoza-Lewis KL, Hiltbold EM. IL-12 produced by dendritic cells augments CD8+ T cell activation through the production of the chemokines CCL1 and CCL17. J Immunol. 2008;181:8576–84. doi: 10.4049/jimmunol.181.12.8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Annu Rev Immunol. 2003;21:713–58. doi: 10.1146/annurev.immunol.21.120601.140942. [DOI] [PubMed] [Google Scholar]
- 143.Tagawa T, Albanese M, Bouvet M, Moosmann A, Mautner J, Heissmeyer V. et al. Epstein-Barr viral miRNAs inhibit antiviral CD4+ T cell responses targeting IL-12 and peptide processing. J Exp Med. 2016;213:2065–80. doi: 10.1084/jem.20160248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Albanese M, Tagawa T, Bouvet M, Maliqi L, Lutter D, Hoser J. et al. Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific CD8+ T cells. Proc Natl Acad Sci U S A. 2016;113:E6467–E75. doi: 10.1073/pnas.1605884113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.de Araujo-Souza PS, Hanschke SC, Viola JP. Epigenetic control of interferon-gamma expression in CD8 T cells. J Immunol Res. 2015;2015:849573. doi: 10.1155/2015/849573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Huang WT, Lin CW. EBV-encoded miR-BART20-5p and miR-BART8 inhibit the IFN-gamma-STAT1 pathway associated with disease progression in nasal NK-cell lymphoma. Am J Pathol. 2014;184:1185–97. doi: 10.1016/j.ajpath.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 147.Harding CV. Pathways of antigen processing. Curr Opin Immunol. 1991;3:3–9. doi: 10.1016/0952-7915(91)90068-c. [DOI] [PubMed] [Google Scholar]
- 148.Gupta S, Termini JM, Niu L, Kanagavelu SK, Schmidtmayerova H, Snarsky V. et al. EBV LMP1, a viral mimic of CD40, activates dendritic cells and functions as a molecular adjuvant when incorporated into an HIV vaccine. J Leukoc Biol. 2011;90:389–98. doi: 10.1189/jlb.0211068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Busch LK, Bishop GA. The EBV transforming protein, latent membrane protein 1, mimics and cooperates with CD40 signaling in B lymphocytes. J Immunol. 1999;162:2555–61. [PubMed] [Google Scholar]
- 150.Gurer C, Strowig T, Brilot F, Pack M, Trumpfheller C, Arrey F. et al. Targeting the nuclear antigen 1 of Epstein-Barr virus to the human endocytic receptor DEC-205 stimulates protective T-cell responses. Blood. 2008;112:1231–9. doi: 10.1182/blood-2008-03-148072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Roche PA, Furuta K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat Rev Immunol. 2015;15:203–16. doi: 10.1038/nri3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Christiaansen A, Varga SM, Spencer JV. Viral manipulation of the host immune response. Curr Opin Immunol. 2015;36:54–60. doi: 10.1016/j.coi.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Uehata T, Iwasaki H, Vandenbon A, Matsushita K, Hernandez-Cuellar E, Kuniyoshi K. et al. Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell. 2013;153:1036–49. doi: 10.1016/j.cell.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 154.Wang YF, He DD, Liang HW, Yang D, Yue H, Zhang XM. et al. The identification of up-regulated ebv-miR-BHRF1-2-5p targeting MALT1 and ebv-miR-BHRF1-3 in the circulation of patients with multiple sclerosis. Clin Exp Immunol. 2017;189:120–6. doi: 10.1111/cei.12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Germain C, Meier A, Jensen T, Knapnougel P, Poupon G, Lazzari A. et al. Induction of lectin-like transcript 1 (LLT1) protein cell surface expression by pathogens and interferon-gamma contributes to modulate immune responses. J Biol Chem. 2011;286:37964–75. doi: 10.1074/jbc.M111.285312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ramalingam D, Kieffer-Kwon P, Ziegelbauer JM. Emerging themes from EBV and KSHV microRNA targets. Viruses. 2012;4:1687–710. doi: 10.3390/v4091687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Solomou EE, Keyvanfar K, Young NS. T-bet, a Th1 transcription factor, is up-regulated in T cells from patients with aplastic anemia. Blood. 2006;107:3983–91. doi: 10.1182/blood-2005-10-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kao C, Oestreich KJ, Paley MA, Crawford A, Angelosanto JM, Ali MA. et al. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nat Immunol. 2011;12:663–71. doi: 10.1038/ni.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lin TC, Liu TY, Hsu SM, Lin CW. Epstein-Barr virus-encoded miR-BART20-5p inhibits T-bet translation with secondary suppression of p53 in invasive nasal NK/T-cell lymphoma. Am J Pathol. 2013;182:1865–75. doi: 10.1016/j.ajpath.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 160.Thompson MP, Kurzrock R. Epstein-Barr virus and cancer. Clin Cancer Res. 2004;10:803–21. doi: 10.1158/1078-0432.ccr-0670-3. [DOI] [PubMed] [Google Scholar]
- 161.Gordadze AV, Poston D, Ling PD. The EBNA2 polyproline region is dispensable for Epstein-Barr virus-mediated immortalization maintenance. J Virol. 2002;76:7349–55. doi: 10.1128/JVI.76.14.7349-7355.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Dawson CW, Laverick L, Morris MA, Tramoutanis G, Young LS. Epstein-Barr virus-encoded LMP1 regulates epithelial cell motility and invasion via the ERK-MAPK pathway. J Virol. 2008;82:3654–64. doi: 10.1128/JVI.01888-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lavorgna A, Harhaj EW. EBV LMP1: New and shared pathways to NF-kappaB activation. Proc Natl Acad Sci U S A. 2012;109:2188–9. doi: 10.1073/pnas.1121357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Vaysberg M, Lambert SL, Krams SM, Martinez OM. Activation of the JAK/STAT pathway in Epstein Barr virus+-associated posttransplant lymphoproliferative disease: role of interferon-gamma. Am J Transplant. 2009;9:2292–302. doi: 10.1111/j.1600-6143.2009.02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jones K, Nourse JP, Morrison L, Nguyen-Van D, Moss DJ, Burrows SR. et al. Expansion of EBNA1-specific effector T cells in posttransplantation lymphoproliferative disorders. Blood. 2010;116:2245–52. doi: 10.1182/blood-2010-03-274076. [DOI] [PubMed] [Google Scholar]
- 166.Apcher S, Daskalogianni C, Manoury B, Fahraeus R. Epstein Barr virus-encoded EBNA1 interference with MHC class I antigen presentation reveals a close correlation between mRNA translation initiation and antigen presentation. PLoS Pathog. 2010;6:e1001151. doi: 10.1371/journal.ppat.1001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tellam J, Connolly G, Green KJ, Miles JJ, Moss DJ, Burrows SR. et al. Endogenous presentation of CD8+ T cell epitopes from Epstein-Barr virus-encoded nuclear antigen 1. J Exp Med. 2004;199:1421–31. doi: 10.1084/jem.20040191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cho SG, Kim N, Sohn HJ, Lee SK, Oh ST, Lee HJ. et al. Long-term Outcome of Extranodal NK/T Cell Lymphoma Patients Treated With Postremission Therapy Using EBV LMP1 and LMP2a-specific CTLs. Mol Ther. 2015;23:1401–9. doi: 10.1038/mt.2015.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zheng XH, Lu LX, Cui C, Chen MY, Li XZ, Jia WH. Epstein-Barr virus mir-bart1-5p detection via nasopharyngeal brush sampling is effective for diagnosing nasopharyngeal carcinoma. Oncotarget. 2016;7:4972–80. doi: 10.18632/oncotarget.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yan Q, Zeng Z, Gong Z, Zhang W, Li X, He B. et al. EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC. Oncotarget. 2015;6:41766–82. doi: 10.18632/oncotarget.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gao W, Li ZH, Chen S, Chan JY, Yin M, Zhang MJ. et al. Epstein-Barr virus encoded microRNA BART7 regulates radiation sensitivity of nasopharyngeal carcinoma. Oncotarget. 2017;8:20297–308. doi: 10.18632/oncotarget.15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang G, Zong J, Lin S, Verhoeven RJ, Tong S, Chen Y. et al. Circulating Epstein-Barr virus microRNAs miR-BART7 and miR-BART13 as biomarkers for nasopharyngeal carcinoma diagnosis and treatment. Int J Cancer. 2015;136:E301–12. doi: 10.1002/ijc.29206. [DOI] [PubMed] [Google Scholar]
- 173.Kang BW, Choi Y, Kwon OK, Lee SS, Chung HY, Yu W. et al. High level of viral microRNA-BART20-5p expression is associated with worse survival of patients with Epstein-Barr virus-associated gastric cancer. Oncotarget. 2017;8:14988–94. doi: 10.18632/oncotarget.14744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Komabayashi Y, Kishibe K, Nagato T, Ueda S, Takahara M, Harabuchi Y. Circulating Epstein-Barr virus-encoded micro-RNAs as potential biomarkers for nasal natural killer/T-cell lymphoma. Hematol Oncol. 2017;35:655–63. doi: 10.1002/hon.2360. [DOI] [PubMed] [Google Scholar]