Abstract
The ability of a host to curb a viral infection is heavily reliant on the effectiveness of an initial antiviral innate immune response, resulting in the upregulation of interferon (IFN) and, subsequently, IFN-stimulated genes (ISGs). ISGs serve to mount an antiviral state within a host cell, and although the specific antiviral function of a number of ISGs has been characterized, the function of many of these ISGs remains to be determined. Recent research has uncovered a novel role for a handful of ISGs, some of them directly induced by IFN regulatory factor 3 in the absence of IFN itself. These ISGs, most with potent antiviral activity, are also able to augment varying arms of the innate immune response to viral infection, thereby strengthening this response. This new understanding of the role of ISGs may, in turn, help the recent advancement of novel therapeutics aiming to augment innate signaling pathways in an attempt to control viral infection and pathogenesis.
Keywords: Interferon-stimulated genes, Interferon, Innate immunity, Virus
Introduction
Viral infection constitutes a significant portion of human morbidity and mortality, which has led to extensive investigation into the countermeasures employed by the host to combat such infection. The mammalian immune system acts to eradicate viral infection through disrupting pathways and functions imperative to the pathogen's life-cycle, and it is the innate immune system that carries a substantial burden of the defense against viral pathogens. Our innate immune system has evolved a range of receptors to detect viral pathogens, termed pattern recognition receptors (PRRs), which recognize viral proteins and nucleic acid in the process of non-self-recognition. Once activated, PRRs initiate a series of signaling cascades that result in the production of the well-known antiviral cytokine, interferon (IFN). IFN is able to act in both an autocrine and paracrine manner to activate the Janus kinase signal transducer and activator of transcription (JAK-STAT) signaling pathway, resulting in the subsequent downstream expression of hundreds of antiviral host effector proteins, called IFN-stimulated genes (ISGs) which control viral infection in the infected cell and help neighboring cells to resist infection [1]. Much research has focused on the host receptors and proteins involved in the recognition of viral pathogens and the production of IFN, but the research into the ability of the ISGs themselves to control viral infection of host cells is limited.
Work aimed at elucidating the function of some of these ISGs has revealed a broad scope of very specific antiviral mechanisms able to target viruses at varying stages of their life-cycle, with some ISGs having a broader spectrum of viral targets, such as cholesterol-25-hydroxylase and the IFN-induced transmembrane (IFITM) proteins which inhibit the entry of many viruses. MX1 inhibits the early viral replication of multiple viral families and viperin has been shown to inhibit both viral egress and the replication of multiple viruses [2, 3]. However, increasing evidence in recent years has pointed to a role for a small handful of these host ISGs in specific mechanisms of viral restriction as well as in positive augmentation of the early innate pattern-recognition pathways that upregulate the ISGs themselves (Table 1; Fig. 1). Some of these ISGs are present at high basal levels, or can be regulated directly by IFN regulatory factor 3 (IRF3) very early (once a virus has been detected in a host cell), independently of IFN, offering a positive reinforcement of the initial viral detection pathways. This review will briefly discuss the detection of viral pathogens and the production of ISGs, and then focus specifically on this new family of ISGs with its dual roles in combating viral infection.
Table 1.
Interferon-stimulated genes that have specific antiviral targets as well as immune modulatory functions
| ISG | Virus families shown to be susceptible | Ref. | Genome group of susceptible viral family | Component of innate immune system augmented by ISG | Upregulated independently of type I IFN |
|---|---|---|---|---|---|
| PKR | Poxviridae (vaccinia virus) Picornavirdae (encephalomyocarditis virus) Togaviridae (Semliki Forest virus) |
15, 16 | dsDNA ssRNA(+) |
MDA5, TLR3, MAVS, TRAF2, TRAF6 | no, but present at high basal levels |
| ZAP | Togaviridae (Sindbis virus) Filoviridae (Ebolavirus, Marburg virus) Paramyxoviridae (Newcastle disease virus) Orthomyxoviridae (influenza A virus) Retroviridae (murine leukemia virus) |
20, 21 | ssRNA(+) ssRNA(–) |
RIG-I | yes |
| TRIM21 | Picornaviridae (human rhinovirus) Adenovirdae (human adenovirus type 5) |
30, 36 | ssRNA(+) dsDNA |
cGAS, RIG-I, NF-κB, AP-1, IRF3/5/7 | no |
| TRIM56 | Orthomyxoviridae (influenza A and B viruses) Flaviviridae (HCV, bovine viral diarrhea virus, yellow fever, DENV serotype 2) Rhabdoviridae (VSV) Coronaviridae (human coronavirus OC43) |
31–33 | ssRNA(+) ssRNA(–) |
STING, TLR3/TRIF | no |
| Viperin | Flaviviridae (DENV, HCV, West Nile virus, tick-borne encephalitis virus) Herpesviridae (human Cytomegalovirus) Paramyxoviridae (respiratory syncytial virus) Togaviridae (Chikungunya virus, Sindbis virus) Orthomyxoviridae (influenza A virus) Retroviridae (human immunodeficiency virus) Bunyaviridae (Bunyamwera virus) Picornaviridae (rhinovirus) Rhabdoviridae (VSV) |
2 | ssRNA(+) ssRNA(–) dsDNA |
TLR7/9 (IRAK1/TRAF6) NF-κB1/p50, AP-1 | yes |
| DDX60 | Rhabdoviridae (VSV) Flaviviridae (HCV) |
46 | ssRNA(–) ssRNA(+) |
RIG-I, MDA5, LGP2 | no |
ISG, interferon-stimulated gene; Ref., reference; HCV, hepatitis C virus; VSV, vesicular stomatitis virus; DENV, Dengue virus; ssDNA, single-stranded DNA; dsDNA, double-stranded DNA.
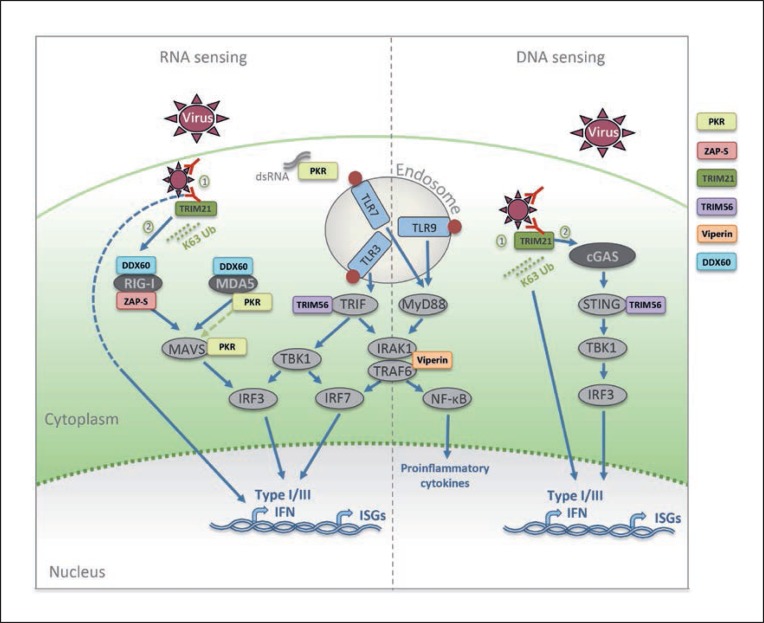
Fig. 1.
When a virus enters a cell, viral PAMPS are recognized by pattern recognition receptors, and activate a number of signaling cascades that result in the production of type I and III interferon (IFN) and inflammatory cytokines to induce an antiviral state. These pathways are augmented and enhanced by a number of IFN-stimulated genes (ISGs). Protein kinase R (PKR) is a double-stranded RNA receptor pivotal in the activation of MAVS (both following MDA5 activation and independently of MDA5 and RIG-I activation). Zinc-finger antiviral protein S (ZAP-S) enhances RIG-I ATPase activity. Tripartite motif 21 (TRIM21) enhances innate immune signaling in 2 ways: (1) it detects the Fc portion of the antibody bound to nonenveloped viruses entering the cytosol and catalyzes the synthesis of K63-linked polyubiquinated (K63 Ub) chains to activate IRFs and NF-κB, and induce a type I IFN response independently of RIG-I and cGAS; (2) it recruits the proteasome instigating premature virion uncoating, exposing PAMPS to RIG-I and cGAS. TRIM56 acts as a scaffold protein promoting IRF3 activation by enhancing the efficient interaction of STING and TBK1 and via an interaction with TRIF. Viperin enhances the K63-linked ubiquitination of IRAK1 by promoting an interaction between IRAK1 and TRAF6. DDX60 can bind dsRNA, and it associates with both RIG-I and MDA5 to enhance their activation.
Innate Immune Recognition of Pathogens and ISG Production
The first PRRs to be described were the Toll-like receptors (TLRs) in Drosophila, and we now know that both cell surface and cytosolic PRRs exists in all metazoan animals. PRRs recognize the pathogen-activated molecular patterns (PAMPS) present in invading pathogenic organisms, such as nucleic acids (double-stranded [ds]RNA, single-stranded [ss]RNA, dsDNA, and 5′triphosphate RNA) and proteins, with the early innate response detection of PAMPS being characterized best in mouse models of infection and also in humans. Once a PRR recognizes a PAMP, a series of signaling pathways is set in motion, involving a number of conserved adaptor molecules, and this results in the production of type I IFN (IFN-β and/or IFN-α), and sometimes type III IFN (IFN-λ), all of which are critical mediators of innate immunity (Fig. 1; reviewed in [1]).
Viral pathogens can trigger the activation of cell-surface or endosomal TLRs, cytoplasmic PRRs such as RIG-I-like receptors (RLRs), including RIG-I and MDA-5 which detect viral RNA, and dsDNA sensors such as cGAS, DAI, and IFI16 which all activate primary signaling adaptors, followed by the activation of IRFs, and culminating in the production of IFN. The ability of these PRRs to detect viral RNA and DNA in comparison to host nucleic acid species is governed mainly by the localization of these receptors, the local concentration of nucleic acid, and the structure of the nucleic acids able to bind to each PRR (reviewed in [4]).
Type I IFNs are produced by most cells, while type III IFNs are produced mainly by epithelial cells, and were recently shown to be produced by dendritic cells (DCs). All the IFNs are essential for the early immune defense against viral infection (reviewed in [5, 6]). Once they have been produced, they are secreted from the virally infected cell, allowing them to bind to their cognate IFN receptor and activate the JAK-STAT pathway [7]. Receptor activation triggers the phosphorylation of both janus kinase I (JAK1) and tyrosine kinase 2 (TYK2), followed by the activation and dimerization of STAT1 and STAT2, which ultimately forms the transcription factor ISGF3 in combination with IRF9. Translocation of ISGF3 to the nucleus allows activation of IFN-stimulated response elements (ISREs) in host antiviral genes, and induces the expression of hundreds of ISGs which act as antiviral effectors to control viral replication and spread (Fig. 1; [3]).
ISGs as Augmenters of Antiviral Innate Signaling
ISGs are the effectors of the initial host antiviral response and are engaged in a wide array of functions in the cell. Although most members of the viral PAMP detection and JAK-STAT signaling pathways are present at baseline levels, many of the PRRs, IRFs, and members of the JAK-STAT pathway are ISGs themselves (reviewed in [3]), allowing for the further amplification of these pathways upon the detection of viral infection in the host cell. Many functional screening efforts have been aimed at determining the specifically antiviral ISGs and characterizing their respective functions, but >25 years on from the discovery of ISGs, the antiviral potential elicited by many of them remains elusive. The research dedicated to this field has focused on demonstrating the mechanisms responsible for the direct antiviral capabilities of ISGs, primarily attributing these properties to direct interactions with particular viral pathogens. However, more recent research efforts have identified a small handful of ISGs that are not only capable of directed antiviral activities, but are also able to act in a synergistic fashion to further augment specific PRR signaling pathways, often by assisting in the posttranslational modification of members of these pathways, and thereby enhancing the innate antiviral response. These ISGs are part of a new group of host proteins that have a multipronged approach to inhibiting viral infection, and include protein kinase R (PKR), zinc-finger antiviral proteins (ZAPs), IFN-regulated members of the tripartite motif (TRIM)-containing family, viperin, and the DExD/H box helicase (DDX60) (Fig. 2, representative diagrams of these genes).
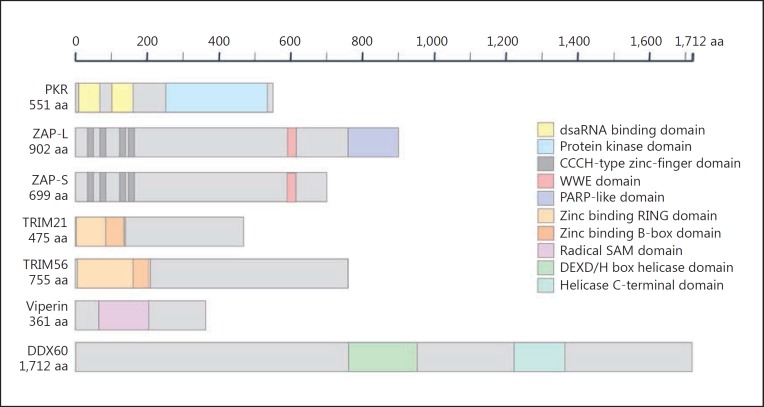
Fig. 2.
Protein domains for interferon (IFN)-stimulated genes (ISGs) able to positively augment the type I IFN response. PKR, protein kinase R; ZAP, zinc-finger antiviral protein; ZAP-L, long ZAP isoform; ZAP-S, short ZAP isoform; TRIM, tripartite motif; DDX60, DExD/H box helicase 60; PARP, poly(ADP-ribose) polymerase; RING, really interesting new gene; SAM, S-adenosyl methionine; WWE, W and E residues; aa, amino acids.
Protein Kinase R
The dsRNA-dependent PKR is upregulated by type I and type III IFNs, but is present in all tissues at basal levels [8]. PKR is an integral antiviral protein that is responsible for terminating protein synthesis in the cell upon the recognition and binding of viral dsRNA in the cytoplasm [9]. This role involves the autophosphorylation and dimerization of PKRs, and the subsequent phosphorylation of the translation-limiting α subunit of eukaryotic initiation factor 2 (eIF2α) [10]. In addition, PKR has since been reported to regulate a variety of signaling pathways, either in a kinase-dependent manner, or acting in a “scaffold-like” function independent of its enzymatic activity, extending its ability to repress viral expression, replication, and propagation [11, 12, 13, 14, 15, 16, 17, 18, 19]. PKR has been found to play a critical role in the posttranscriptional regulation of IFN-β, in response to the activation of the cytosolic RNA sensor, MDA5, but not of RIG-I [17, 18]; interestingly this was in the absence of activation of its primary substrate, eIF2α [17]. PKR is essential for the activation of mitochondrial antiviral signaling protein (MAVS) following the engagement of MDA5 by multiple viruses, and, interestingly, PKR activation can induce IFN in the absence of the MDA5, with MAVS alone being demonstrated to be essential in the PKR-driven upregulation of type I IFN [17].
Numerous other studies have reinforced a positive augmentative role for PKR in innate immune signaling, with reports of an interaction of PKR with the TNF receptor-associated factor (TRAF) family proteins, TRAF2 and TRAF6, to promote MAVS signaling [15]. PKR has also been shown to be an integral component in the TAK1 signaling complex following dsRNA activation of the endosomal TLR3 receptor [16]. The role of PKR in innate immune signaling has been controversial at times, however contradictions to PKR's role in IFN induction, are now thought to be the result of an abolished production of alternate IFN via a positive feedback when PKR inhibits protein synthesis [17, 19]. Clearly PKR plays an important role in the detection of viral dsRNA in the cytoplasm, acting itself as an RNA sensor; however its ability to also augment other innate RNA detection pathways, both cytosolic and endosomal, makes it an integral ISG in the host response to RNA viral infection.
Zinc-Finger Antiviral Protein
ZAP, also known as poly(ADP-ribose) polymerase (PARP)13/ZC3HAV1, has been characterized for its extensive antiviral activity against multiple alphaviruses, filoviruses, and retroviruses, where its antiviral activities involve viral RNA binding, and the degradation or inhibition of translation (reviewed in [20]). ZAP is a member of the PARP family. It was first shown to bind specific viral mRNA [21] and, soon after, to play an active role in the degradation of viral RNA by recruiting p72 DEAD box helicase [22]. This antiviral ability has been accredited to the N-terminal 4 CCCH-type zinc-finger domains that bind to the viral mRNA [21] (Fig. 2).
Recent studies have examined the ability of various PARP family members to augment the type I IFN signaling pathway following nucleic acid stimulation, with ZAP (PARP13) showing the strongest ability to drive a type I IFN response in HEK293T cells following both dsRNA and dsDNA stimulation [23]. However, ZAP has 2 splice-variant isoforms, ZAP-L, the long isoform, and ZAP-S, the shorter isoform lacking the PARP domain [24] (Fig. 2), and it is the shorter isoform that demonstrates the greatest ability to enhance the type I IFN response as well as being preferentially induced following both IFN and nucleic acid stimulation of cells in vitro [23]. This augmentation of the type I IFN pathway has recently been attributed to the ability of ZAP-S to play a critical role in the regulation of the cytosolic RIG-I pathway [23]. As an RNA helicase, RIG-I relies on its ATPase activity to alter its structural conformation, allowing it to activate its downstream adaptor molecule MAVS [25]. The ISG ZAP-S was shown to associate with RIG-I to promote oligomerization and ATPase activity of the receptor, significantly enhancing downstream activation of IRF3 and NF-κB in the presence of the RIG-I ligand, 3′pRNA, in both human HEK293T cells and primary human CD14+ monocytes [23]. ZAP-S binds to both the helicase domain and the carboxyl-terminal region of RIG-I and, interestingly, is bound to the carboxyl-terminal region preceding the RNA ligand binding to RIG-I [23]. This suggests that ZAP-S remains poised, ready to immediately enhance the ATPase activity of RIG-I, once the receptor's conformational change has allowed ZAP-S to interact with both domains. Interestingly, recent work in a ZAP knockout murine model, suggests that ZAP deficiency does not alter the RIG-I-dependent production of type I IFN. However, ZAP deficiency was shown to greatly decrease the antiviral response to the retrovirus, murine leukemia virus in mice, independent of RIG-I [26], with ZAP acting as a cytosolic RNA sensor for the virus, and subsequently mediating its degradation.
The discrepancy as to the role of ZAP-S in human and murine cells may lie in the divergent cell types examined, or may be a factor of species. It is clear that more work needs to be performed to solidify the role of ZAP-S in augmenting the RIG-I pathway in multiple animal species, as well as to delineate why ZAP-L cannot perform similar functions to its shorter counterpart, ZAP-S, which only lacks the PARP domain. It is interesting to note that ZAP-S is also directly IRF3-regulated, resulting in its upregulation prior to the production of type I IFN [27], which enhances the ability of the protein to act swiftly during a viral infection, possibly enhancing RIG-I activation as well as possessing the capacity to specifically inhibit multiple viruses (reviewed in [21]).
TRIM Proteins
The TRIM protein superfamily comprises >80 members in humans, and is a novel class of single-protein RING finger E3 ubiquitin ligases [28], characterized by their N-terminal zinc-binding RING and B-box and coiled-coil (RBCC) domain motif, which mediates homomeric and heteromeric interactions amongst TRIM family members and other proteins (Fig. 2). Around one-third of these family members are also ISGs, with many known to be involved in the foundation of an innate antiviral state [29]. TRIM5α, TRIM19, TRIM79α, TRIM56, TRIM21, and TRIM22 have all been demonstrated to specifically inhibit multiple viruses, including a number of flaviviruses, retroviruses, and hepatitis B virus (reviewed in [30, 31, 32, 33]).
Many TRIM family members are known to play a role in innate signaling pathways, with a recent screening assay suggesting that as many as half of the family members have a positive role in augmenting innate immune signaling events [34]. Of the TRIM family members shown to enhance either the IFN-β, ISRE, or NF-κB pathways using a luciferase-based screening assay, 9 have previously been shown to be ISGs, TRIM5, TRIM6, TRIM9, TRIM14, TRIM21, TRIM25, TRIM38, TRIM56, and TRIM58 [29]. As mentioned above, TRIM21 and TRIM56 were previously described as having specific antiviral activity, and have since been shown to enhance various arms of the innate immune pathway detection of viruses.
TRIM21 can facilitate innate sensing of both DNA and RNA viruses via cGAS- and RIG-I-dependent and independent mechanisms, respectively (reviewed in [35, 36]). TRIM21 was able to detect non-enveloped viruses entering the cytosol with a virally bound antibody, and subsequently induced 2 waves of enhanced PAMP detection, resulting in the increased upregulation of type I IFN and ISGs [36, 37]. Upon initial recognition of the Fc portion of the antibody, TRIM21 catalyzed the synthesis of K63-linked polyubiquinated chains, which was shown to enhance the upregulation of IFN-β and its effector genes in a cGAS- and RIG-I-independent manner, via the enhanced activation of IRFs and NF-κB [37]. Interestingly, TRIM21 can also induce a second wave of ISG upregulation, via recruiting the proteasome to instigate premature viral uncoating and viral protein degradation, thereby exposing viral genomic nucleic acid for subsequent detection by RIG-I and cGAS [36].
TRIM56 is another member of the TRIM family, with extensively studied antiviral properties. It has been shown to be essential in the direct antiviral restriction of multiple viruses as well as in the regulation of the cytosolic dsDNA and TLR3 signaling pathways, either independently or dependently of its E3 ubiquitination activity [31, 32, 38, 39]. It has been shown to specifically inhibit influenza viruses by impeding viral RNA synthesis as well as to limit the replication of the flaviviruses, yellow fever, dengue, and bovine viral diarrhea virus, in a manner that is dependent on its E3 ligase domain in all cases except for in the restriction of influenza [31, 32, 33]. TRIM56 is perhaps one of the widest-acting ISGs, with an ability to not only restrict viruses directly, but also enhance multiple innate signaling pathways, thereby acting indirectly as an antiviral host protein. Its regulation of the cytosolic dsDNA signaling pathway involves a role in the formation of a complex involving the adaptor protein of the dsDNA signaling pathway, STING, and tank-binding kinase-1 (TBK1), required for the phosphorylation of IRF3 and the subsequent production of type I IFN [39]. TRIM56 was shown to interact with STING and target it for K63-linked ubiquitination, thereby inducing its dimerization and subsequent activation to enhance IFN-β production in the presence of dsDNA [39]. This augmentation of the cytosolic dsDNA pathway is dependent on its E3 ubiquitin ligase activity; however, its ability to augment the TLR3 and dsRNA sensing pathways appears to be independent of its ligase activity [38].
Shen et al. [38] were able to demonstrate that TRIM56 overexpression in HeLa cells significantly enhanced the production of ISGs during TLR3 activation by exogenous dsRNA, but not via the activation of the RIG-I pathway, using a Sendai virus infection model. Likewise, siRNA knockdown of TRIM56 not only demonstrated a marked reduction in IFN-β and ISG production via dsRNA stimulation of the TLR3 pathway, it also significantly reduced the production of IFN-stimulated chemokines in response to replicating hepatitis C virus. In studies on the TLR3-mediated activation of IRF3, TRIM56 was found to be essential; this was due to its ability to interact with the TLR3 adaptor protein, TIR-domain-containing adaptor-inducing IFN-β (TRIF). However, as this was also found to be independent of its E3 ubiquitin ligase activity, it is postulated that TRIM56 acts as a scaffold for the assembly of multiple signaling complex factors, as other TRIM proteins do [38, 40]. Interestingly, recent work has suggested that TRIM56 may also be involved in the direct modulation of the JAK-STAT pathway following IFN-α stimulation, but the mechanism involved remains unclear [41].
Clearly, the TRIM family of proteins is an integral part of the early innate immune response to viral pathogens, with there likely being many more of the ISG members of this family being involved than has been described to date. Further research is required to fully understand the role that these proteins play in the development of an antiviral state in the host cell.
Viperin
Viperin was first shown to have antiviral capacities against human Cytomegalovirus, and has since been recognized to inhibit many different viruses, utilizing a variety of mechanisms and protein domains to bind viral proteins and inhibit both viral entry and the replication of specific viruses ([42]; reviewed in [2]). However, the definitive mechanism for viperin's ability to limit many viruses still remains unknown.
Given the diversity of viruses susceptible to viperin (Table 1), it is unlikely that this protein possesses a direct mechanism to inhibit each of them individually. Alternatively, viperin has more recently been viewed as a potential enhancer of immune signaling, acting indirectly to aid the inhibition of multiple viruses. It has been shown to increase the production of type I IFN following TLR7 and TLR9 stimulation in murine plasmacytoid DCs [43]. Mice deficient in viperin displayed a significantly decreased production of IFN-β by plasmacytoid DCs when cells were stimulated with the UV-inactivated Newcastle disease virus and CpG DNA. Additionally, ex vivo experiments in these DCs showed that viperin was localized to lipid bodies, acting as a scaffold for the recruitment of both IRAK1 and TRAF6, and enhancing the K63-linked ubiquitination of IRAK1. Viperin is induced very early following viral infection, and is directly IRF3-regulated. This means it can be induced in the absence of IFN, which potentially underlies its importance as an early enhancer of the innate immune host response to viral infection [2, 44].
DExD/H Box Helicase 60
DDX60 was first shown to be upregulated by the measles virus in human DCs, and then also by direct poly (I:C) and type I IFN stimulation [45]. DDX60 is a cytoplasmic RNA helicase, but lacks a caspase activation and recruitment domain (CARD) present on the cytosolic RNA sensors, RIG-I, and MDA-5. However, it has been shown to be critical for the recruitment and activation of the signaling adaptor MAVS for an efficient type I IFN response [45]. It is also antiviral in its own right, being involved in viral RNA degradation independently of augmentation of the cytosolic RNA sensing pathways [46]. Despite its lack of a CARD domain, DDX60 is an RNA helicase still able to bind dsRNA, and has been shown to associate with the innate RNA receptors RIG-I, MDA5, and LGP2, to augment the signaling pathways associated with these PRRs [45]. Of these receptors, RIG-I has received the most attention while the mechanisms of the interactions with the others remains elusive. The helicase domain of DDX60 enables it to bind viral RNA while the protein's ATP-binding site enables its mediation of downstream RIG-I activation, with DDX60 expression significantly increasing the association of RIG-I to RNA [45]. The DDX60 homolog, DDX60L, has also been demonstrated to play a role in the RIG-I-dependent activation of IFN [47].
The exact role of DDX60 in the detection of dsRNA and activation of the augmentation of the cytosolic RNA PRRs continues to unfold. Two separate models using DDX60 knockout mice presented conflicting results with regard to the functional importance of DDX60 as an augmenter of dsRNA signaling; a significant reduction in (but not an abolishment of) the response of IFN-α and IFN-β to RIG-I ligands was seen in one model but not in the other [45, 48]. Given the variation in primary cell types and viruses utilized in these studies, it is likely that DDX60 may act in both a cell-type and ligand-specific role in its augmentation of RIG-I activation, and further studies will be required to elucidate the extent of its role in the early innate immune response to viral infection.
Concluding Remarks
The process involved in the effective inhibition of a viral pathogen by the innate immune response of a mammalian host is immensely intricate, and the ambiguity that arises as a consequence of the system's functional redundancy presents a challenge in understanding the molecular intricacies. Difficulty comprehending the innate immune response to viral infection may also be due to the differential regulatory mechanisms of the relevant signaling pathways under basal and stimulated conditions. In light of the complexity associated with antiviral innate immunity, the value of immunomodulatory molecules, especially those stimulated by IFN, has become apparent, and the study of ISGs as possible augmenters of innate immunity has greatly increased our understanding of these pathways. Although only a small handful of ISGs able to both directly inhibit the viral life-cycle and augment the early innate immune signaling pathways that direct their upregulation have been discovered to date (Fig. 1), it is likely that many more will be revealed as the field of ISG research expands.
There is no effective treatment for many viral diseases, and vaccines do not exist for every viral pathogen, partly due to high developmental costs and the rapidly evolving nature of some viruses. Immunomodulatory molecules that can activate various arms of the innate immune system have recently gained attention in their potential ability to provide an alternative antiviral treatment, that targets a system capable of inhibiting any known viral pathogen (reviewed in [49, 50]). However, a more detailed analysis of the host proteins controlling these pathways and the antiviral effectors that the pathways upregulate will offer much-needed insight into this field, and help to tailor the advent of novel antiviral treatments in the future.
Disclosure Statement
The authors have no conflicts of interest to declare.
References
- 1.Iwasaki A. A virological view of innate immune recognition. Annu Rev Microbiol. 2012;66:177–196. doi: 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helbig KJ, Beard MR. The role of viperin in the innate antiviral response. J Mol Biol. 2014;426:1210–1219. doi: 10.1016/j.jmb.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 3.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlee M, Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol. 2016;16:566–580. doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wack A, Terczynska-Dyla E, Hartmann R. Guarding the frontiers: the biology of type III interferons. Nat Immunol. 2015;16:802–809. doi: 10.1038/ni.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meurs E, Chong K, Galabru J, Thomas NS, Kerr IM, Williams BR, Hovanessian AG. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 10.Galabru J, Hovanessian A. Autophosphorylation of the protein kinase dependent on double-stranded RNA. J Biol Chem. 1987;262:15538–15544. [PubMed] [Google Scholar]
- 11.Mundschau LJ, Faller DV. Platelet-derived growth factor signal transduction through the interferon-inducible kinase PKR. Immediate early gene induction. J Biol Chem. 1995;270:3100–3106. doi: 10.1074/jbc.270.7.3100. [DOI] [PubMed] [Google Scholar]
- 12.Der SD, Yang YL, Weissmann C, Williams BR. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:3279–3283. doi: 10.1073/pnas.94.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar A, Yang YL, Flati V, Der S, Kadereit S, Deb A, Haque J, et al. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-κB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu WM, Ostertag D, Li ZW, Chang L, Chen Y, Hu Y, Williams B, et al. JNK2 and IKKβ are required for activating the innate response to viral infection. Immunity. 1999;11:721–731. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- 15.Gil J, Garcia MA, Gomez-Puertas P, Guerra S, Rullas J, Nakano H, Alcami J, et al. TRAF family proteins link PKR with NF-κB activation. Mol Cell Biol. 2004;24:4502–4512. doi: 10.1128/MCB.24.10.4502-4512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Z, Zamanian-Daryoush M, Nie H, Silva AM, Williams BR, Li X. Poly(I-C)-induced Toll-like receptor 3 (TLR3)-mediated activation of NFκB and MAP kinase is through an interleukin-1 receptor-associated kinase (IRAK)-independent pathway employing the signaling components TLR3-TRAF6-TAK1-TAB2-PKR. J Biol Chem. 2003;278:16713–16719. doi: 10.1074/jbc.M300562200. [DOI] [PubMed] [Google Scholar]
- 17.Pham AM, Santa Maria FG, Lahiri T, Friedman E, Marie IJ, Levy DE. PKR transduces MDA5-dependent signals for type I IFN induction. PLoS Pathog. 2016;12:e1005489. doi: 10.1371/journal.ppat.1005489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulz O, Pichlmair A, Rehwinkel J, Rogers NC, Scheuner D, Kato H, Takeuchi O, et al. Protein kinase R contributes to immunity against specific viruses by regulating interferon mRNA integrity. Cell Host Microbe. 2010;7:354–361. doi: 10.1016/j.chom.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith EJ, Marie I, Prakash A, Garcia-Sastre A, Levy DE. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or IκB kinase but is blocked by Vaccinia virus E3L protein. J Biol Chem. 2001;276:8951–8957. doi: 10.1074/jbc.M008717200. [DOI] [PubMed] [Google Scholar]
- 20.Kuny CV, Sullivan CS. Virus-host interactions and the ARTD/PARP family of enzymes. PLoS Pathog. 2016;12:e1005453. doi: 10.1371/journal.ppat.1005453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y, Gao G. ZAP-mediated mRNA degradation. RNA Biol. 2008;5:65–67. doi: 10.4161/rna.5.2.6044. [DOI] [PubMed] [Google Scholar]
- 22.Chen G, Guo X, Lv F, Xu Y, Gao G. p72 DEAD box RNA helicase is required for optimal function of the zinc-finger antiviral protein. Proc Natl Acad Sci USA. 2008;105:4352–4357. doi: 10.1073/pnas.0712276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayakawa S, Shiratori S, Yamato H, Kameyama T, Kitatsuji C, Kashigi F, Goto S, et al. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat Immunol. 2011;12:37–44. doi: 10.1038/ni.1963. [DOI] [PubMed] [Google Scholar]
- 24.Kerns JA, Emerman M, Malik HS. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 2008;4:e21. doi: 10.1371/journal.pgen.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 26.Lee H, Komano J, Saitoh Y, Yamaoka S, Kozaki T, Misawa T, Takahama M, et al. Zinc-finger antiviral protein mediates retinoic acid inducible gene I-like receptor-independent antiviral response to murine leukemia virus. Proc Natl Acad Sci USA. 2013;110:12379–12384. doi: 10.1073/pnas.1310604110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang N, Dong Q, Li J, Jangra RK, Fan M, Brasier AR, Lemon SM, et al. Viral induction of the zinc finger antiviral protein is IRF3-dependent but NF-κB-independent. J Biol Chem. 2010;285:6080–6090. doi: 10.1074/jbc.M109.054486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of “single protein RING finger” E3 ubiquitin ligases. Bioessays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 29.Carthagena L, Bergamaschi A, Luna JM, David A, Uchil PD, Margottin-Goguet F, Mothes W, et al. Human TRIM gene expression in response to interferons. PLoS One. 2009;4:e4894. doi: 10.1371/journal.pone.0004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajsbaum R, Garcia-Sastre A, Versteeg GA. TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J Mol Biol. 2014;426:1265–1284. doi: 10.1016/j.jmb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu B, Li NL, Shen Y, Bao X, Fabrizio T, Elbahesh H, Webby RJ, et al. The C-terminal tail of TRIM56 dictates antiviral restriction of influenza A and B viruses by impeding viral RNA synthesis. J Virol. 2016;90:4369–4382. doi: 10.1128/JVI.03172-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu B, Li NL, Wang J, Shi PY, Wang T, Miller MA, Li K. Overlapping and distinct molecular determinants dictating the antiviral activities of TRIM56 against flaviviruses and coronavirus. J Virol. 2014;88:13821–13835. doi: 10.1128/JVI.02505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Liu B, Wang N, Lee YM, Liu C, Li K. TRIM56 is a virus- and interferon-inducible E3 ubiquitin ligase that restricts pestivirus infection. J Virol. 2011;85:3733–3745. doi: 10.1128/JVI.02546-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, Inn KS, et al. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38:384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foss S, Watkinson R, Sandlie I, James LC, Andersen JT. TRIM21: a cytosolic Fc receptor with broad antibody isotype specificity. Immunol Rev. 2015;268:328–339. doi: 10.1111/imr.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watkinson RE, McEwan WA, Tam JC, Vaysburd M, James LC. TRIM21 promotes cGAS and RIG-I sensing of viral genomes during infection by antibody-opsonized virus. PLoS Pathog. 2015;11:e1005253. doi: 10.1371/journal.ppat.1005253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McEwan WA, Tam JC, Watkinson RE, Bidgood SR, Mallery DL, James LC. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat Immunol. 2013;14:327–336. doi: 10.1038/ni.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen Y, Li NL, Wang J, Liu B, Lester S, Li K. TRIM56 is an essential component of the TLR3 antiviral signaling pathway. J Biol Chem. 2012;287:36404–36413. doi: 10.1074/jbc.M112.397075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 40.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kane M, Zang TM, Rihn SJ, Zhang F, Kueck T, Alim M, Schoggins J, et al. Identification of interferon-stimulated genes with antiretroviral activity. Cell Host Microbe. 2016;20:392–405. doi: 10.1016/j.chom.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chin KC, Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc Natl Acad Sci USA. 2001;98:15125–15130. doi: 10.1073/pnas.011593298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saitoh T, Satoh T, Yamamoto N, Uematsu S, Takeuchi O, Kawai T, Akira S. Antiviral protein viperin promotes Toll-like receptor 7- and Toll-like receptor 9-mediated type I interferon production in plasmacytoid dendritic cells. Immunity. 2011;34:352–363. doi: 10.1016/j.immuni.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 44.Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, Barber GN, Lin R, et al. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76:5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol Cell Biol. 2011;31:3802–3819. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oshiumi H, Miyashita M, Okamoto M, Morioka Y, Okabe M, Matsumoto M, Seya T. DDX60 is involved in RIG-I-dependent and independent antiviral responses, and its function is attenuated by virus-induced EGFR activation. Cell Rep. 2015;11:1193–1207. doi: 10.1016/j.celrep.2015.04.047. [DOI] [PubMed] [Google Scholar]
- 47.Grunvogel O, Esser-Nobis K, Reustle A, Schult P, Muller B, Metz P, Trippler M, et al. DDX60L is an interferon-stimulated gene product restricting hepatitis C virus replication in cell culture. J Virol. 2015;89:10548–10568. doi: 10.1128/JVI.01297-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goubau D, van der Veen AG, Chakravarty P, Lin R, Rogers N, Rehwinkel J, Deddouche S, et al. Mouse superkiller-2-like helicase DDX60 is dispensable for type I IFN induction and immunity to multiple viruses. Eur J Immunol. 2015;45:3386–3403. doi: 10.1002/eji.201545794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Junt T, Barchet W. Translating nucleic acid-sensing pathways into therapies. Nat Rev Immunol. 2015;15:529–544. doi: 10.1038/nri3875. [DOI] [PubMed] [Google Scholar]
- 50.Savva A, Roger T. Targeting toll-like receptors: promising therapeutic strategies for the management of sepsis-associated pathology and infectious diseases. Front Immunol. 2013;4:387. doi: 10.3389/fimmu.2013.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]