Abstract
Objective
ZNF750, a transcriptional regulator of epidermal differentiation, has been identified as a tumor suppressor in esophageal squamous cell carcinoma (ESCC). The aim of the present study was to investigate the clinical and prognostic significance of ZNF750 expression and to evaluate the effect of ZNF750 knockdown on cell proliferation, migration, and invasion in ESCC.
Methods
A total of 124 patients with ESCC who underwent curative esophagectomy were evaluated in this study. The expression of ZNF750 in surgical specimens was immunohistochemically assessed and used in the analysis of clinicopathological features and overall survival (OS). The molecular role of ZNF750 was investigated by ZNF750 knockdown using small interfering RNA (siRNA) in ESCC cell lines.
Results
Low ZNF750 expression had a significant correlation with positive lymph node metastasis (p = 0.028). Furthermore, there was a significant relationship between low expression of ZNF750 in ESCC and a poor OS, and a multivariate analysis showed that low ZNF750 expression was an independent prognostic factor (p = 0.020). The cell growth, migration, and invasion were significantly increased by downregulation of ZNF750.
Conclusions
The low expression of ZNF750 was significantly associated with a poor prognosis, and ZNF750 expression may, therefore, be a reliable prognostic biomarker in ESCC.
Keywords: ZNF750, Esophageal cancer, Squamous cell carcinoma, Prognostic factor, Biomarker
Introduction
Esophageal squamous cell carcinoma (ESCC) is one of the most aggressive types of gastrointestinal cancers. In recent decades, combination therapy of surgery, chemotherapy, and radiotherapy has been developed, and the outcome of ESCC has improved [1, 2]. However, the prognosis of ESCC is still not desirable [3, 4]; therefore, elucidation of the mechanism underlying the progression of this cancer and the identification of specific molecular markers predicting the prognosis are significant issues regarding the effective management and improvement of the prognosis of ESCC.
Recently, cancer genome research has been extensively conducted for the development of individualized medical treatment. It is, therefore, necessary to clarify the function and examine the clinicopathological significance of cancer-related genes discovered by genome analyses. ZNF750, the gene responsible for seborrhea-like dermatitis with psoriasiform elements, is a remarkable gene shown to have a relationship with ESCC by several whole-exome sequencing studies. This gene is significantly mutated at a frequency of 6–16.7% [5, 6, 7, 8]. ZNF750 was reported to act as a transcriptional regulator of epidermal differentiation, and the expression of ZNF750 induces epidermal differentiation genes and suppresses epidermal progenitor genes [9]. ZNF750 has been shown to function as a tumor suppressor [5, 6, 10], and we previously reported the relationship between ZNF750 expression and chemoradiotherapy response in ESCC patients [11]. We, therefore, consider ZNF750 to be a notable gene in the field of ESCC. However, no reports have described the clinicopathological features and prognostic value of ZNF750 expression in ESCC.
Therefore, in this study, we investigated ZNF750 expression in ESCC using immunohistochemical staining. In addition, we divided the subjects into 2 groups based on their expression level and compared the clinical factors and survival period between the groups. Furthermore, we investigated the molecular role of ZNF750 using human ESCC cell lines.
Materials and Methods
Clinical Specimens and Cell Culture
Primary ESCC specimens were obtained from 124 patients with pathologically diagnosed ESCC at Chiba University Hospital from 1997 to 2010. All patients underwent radical surgery with 3-region lymph node dissection (neck, chest, and abdomen). Patients who received any other preoperative treatments or whose pathological diagnosis was not ESCC were excluded from the present study. The staging classification was assessed using the TNM system (UICC 7th edition) [12]. The patient characteristics are shown in Table 1. Written informed consent was obtained from all patients for research purposes.
Table 1.
Patient characteristics (n = 124)
| Age (minimum-maximum), years | 64 (40–84) |
| Sex (male:female) | 103:21 |
| Pathological T factor | |
| Tis | 1 |
| T1 | 53 |
| T2 | 13 |
| T3 | 49 |
| T4 | 8 |
| Pathological N factor | |
| N0 | 62 |
| N1 | 62 |
| Stage | |
| 0 | 1 |
| I | 33 |
| II | 41 |
| III | 41 |
| IV | 8 |
| Tumor differentiation | |
| Well | 33 |
| Moderate | 70 |
| Poor | 21 |
The TE8 and TE11 human ESCC cell lines were used in this study. Both cell lines were obtained from Cell Resource Center for Biomedical Research, Institute of Development, Aging and Cancer, Tohoku University, Japan. The cells were grown in Dulbecco's Modified Eagle's Medium (Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin sodium, and 100 μg/mL streptomycin in a humidified atmosphere containing 5% CO2 at 37°C.
Immunohistochemistry
The expression of ZNF750 was immunohistochemically assessed using the peroxidase-antiperoxidase complex method in accordance with the previously reported protocol [10]. Anti- human ZNF750 polyclonal antibody (1:200; HPA023012; Sigma-Aldrich, St. Louis, MO, USA) was used as the primary antibody, and anti-mouse/rabbit antibody (EnVision/HRP, anti-mouse/rabbit, K5001; DAKO Japan, Tokyo, Japan) was used as the secondary antibody.
The Evaluation of ZNF750 Immunohistochemical Staining
After ZNF750 immunohistochemical staining, patients were divided into 2 groups as follows. The proportion of positively stained cells was scored as 1 point (0–29%), 2 points (30–59), or 3 points (>60%). The staining intensity was scored as 0 points (no staining), 1 point (weak), 2 points (moderate), or 3 points (strong). The total score was calculated by adding the proportion score and the intensity score. A total score of ≥3 was defined as high expression, and <3 was defined as low expression.
The ZNF750 expression was used in the analysis of the clinicopathological features and of the overall survival (OS). Representative images of ZNF750 expression are shown in Figure 1a. All specimens were assessed by 2 different observers independently.
Fig. 1.
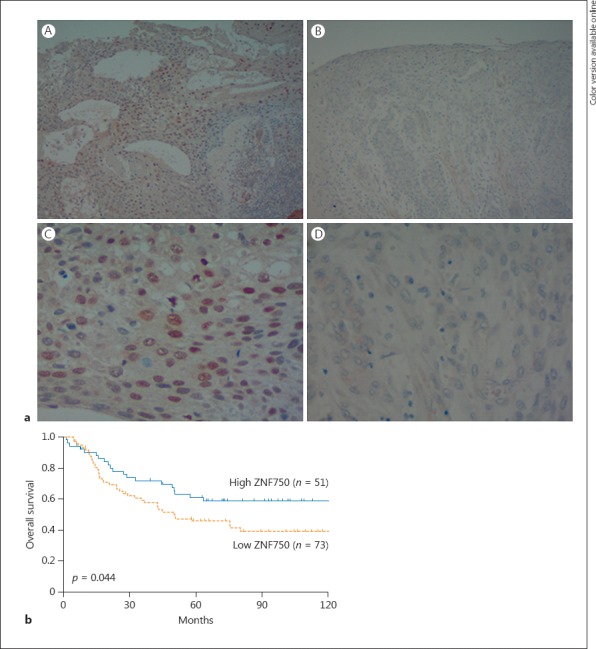
The immunohistochemical analysis of ZNF750 expression in ESCC. a Images for high expression (A and C) and low expression (B and D) are shown (×100 in the upper section, ×400 in the lower section). b A comparison of the overall survival of high- and low-ZNF750 expression groups of ESCC patients. ESCC, esophageal squamous cell carcinoma.
RNA Isolation
Cells were treated with the QIAzol Lysis Reagent (Qiagen, Venlo, The Netherlands) in accordance with the manufacturer's instructions for total RNA extraction. The concentration and purity of extracted RNA was evaluated using a NanoDrop Lite Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Quantitative Reverse-Transcription PCR
Total RNA was converted to cDNA using a QuantiTect Reverse Transcription Kit (Qiagen). Real-time PCR was performed in triplicate using QuantiTect SYBR® Green PCR Kits (Qiagen) and a MyiQ™2 Two-Color Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) in accordance with the manufacturer's protocol. The primers for ZNF750 (assay name: Hs_ZNF750_1_SG) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (assay name: Hs_GAPDH_1_SG) as an internal control, which had been obtained from Qiagen, were used.
Transfection of Small Interfering RNAs
The small interfering RNAs (siRNAs) (3 unique 27mer siRNA duplexes; ZNF750 Cat. No. SR3125578; Origene Technologies, Rockville, MD, USA) and negative control siRNA (Universal scrambled negative control siRNA duplex; Cat. No. SR30004; Origene Technologies) were transfected using the Lipofectamine™ RNAiMAX Transfection Reagent (Thermo Fisher Scientific) in accordance with the manufacturer's protocol.
Cell Proliferation, Migration, and Invasion Assays
Following the transfection of siZNF750 or the negative control, proliferation, migration, and invasion assays were performed in accordance with the previously reported protocol [13]. In brief, cells were seeded in 96-well plates at 5 × 103 cells per well in 100 μL of medium containing 10% FBS. After 72 h, the cell proliferation was evaluated using the Cell Counting Kit-8 (Dojindo, Kumamoto, Japan). In each treatment group, triplicate wells were assessed for cell viability. Cell motility was evaluated using a micropore chamber assay. Cell migration was assessed using a 24-well micropore transparent PET membrane with 8.0-μm pores (BD Biosciences, Bedford, MA, USA), and cell invasion was assessed using a 24-well Matrigel-coated micropore membrane with 8.0-μm pores (BD Biosciences). Cell migration/invasion was evaluated by counting the number of cells that had migrated/invaded through the membrane.
Apoptosis Detection
Apoptosis detection was performed using terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) staining with the In situ Apoptosis Detection Kit (MK500, Takara Bio Inc., Shiga, Japan) after the transfection of siZNF750 or the negative control. TUNEL-positive cells were quantified through manual counting in random microscopic fields.
Statistical Analyses
The correlation between the ZNF750 expression levels and the clinicopathological features was evaluated using the Fisher exact test. The survival rates were assessed using the Kaplan-Meier method and the log-rank test. A Cox proportional hazards model was performed to evaluate the significance of ZNF750 expression as a prognostic factor. The correlation between 2 variables and numerical values was assessed using the Wilcoxon rank sum test or t test. The JMP version 12 software program (SAS Institute Inc., Cary, NC, USA) was used for all statistical analyses, and statistical significance was defined as a p value <0.05.
Results
Relationship between the ZNF750 Expression and Clinicopathological Features
Among 124 cases, 73 (58.9%) had tumors with low ZNF750 expression, and 51 (41.1%) had tumors with high ZNF750 expression. A low expression of ZNF750 was associated with positive lymph node metastasis (p = 0.028). No significant relationship was observed between the expression of ZNF750 and other clinicopathological factors, such as age, sex, tumor depth, stage, and tumor differentiation. However, the proportions of advanced tumor depth, advanced stage, and well-differentiated tumor tended to be higher in the high-expression group (Table 2).
Table 2.
Relation between ZNF750 expression and clinicopathological characteristics
| Characteristics | ZNF750 expression |
p value | |
|---|---|---|---|
| low | high | ||
| Age | |||
| ≤65 years | 40 | 25 | 0.586 |
| >65 years | 33 | 26 | |
| Sex | |||
| Male | 59 | 44 | 0.475 |
| Female | l4 | 7 | |
| Tumor depth | |||
| Tis/T1 | 30 | 24 | 0.582 |
| T2/T3/T4 | 43 | 27 | |
| LN status | |||
| N0 | 30 | 32 | 0.028 |
| N1 | 43 | 19 | |
| Stage | |||
| 0/I | 39 | 35 | 0.098 |
| II/III/IV | 34 | 16 | |
| Tumor differentiation | |||
| Well | 18 | 15 | 0.797 |
| Moderate | 43 | 27 | |
| Poor | 12 | 9 | |
Correlation between the Prognosis and ZNF750 Expression and the Histopathological Features
A survival analysis using the Kaplan-Meier method and the log-rank test showed that a low ZNF750 expression was associated with a significantly poorer OS than a higher expression (5-year survival rate: low, 45.6% vs. high, 60.9%, p = 0.044) (Fig. 1b). A univariate analysis showed that a deeper T stage, positive lymph node metastasis, advanced stage, and low ZNF750 expression were significant prognostic factors. A multivariate analysis revealed that a low ZNF750 expression was an independent prognostic factor for OS in ESCC patients (hazard ratio, 1.922; 95% confidence interval, 1.123–3.389, p = 0.020) (Table 3).
Table 3.
Univariate and multivariate analysis of clinicopathological factors and overall survival in ESCC
| Factors | Univariate analysis, p value | Multivariate analysis |
|
|---|---|---|---|
| hazard ratio (95% CI) | p value | ||
| Age >65 years | 0.097 | 1.678 (0.990 – 2.868) | 0.055 |
| Male | 0.157 | 1.628 (0.783 – 3.961) | 0.232 |
| T2/T3/T4 | <0.001 | 1.865 (0.962 – 3.736) | 0.071 |
| N1 | 0.002 | 0.914 (0.363 – 2.260) | 0.848 |
| Stage II/III/IV | <0.001 | 2.181 (0.799 – 6.059) | 0.132 |
| Poor differentiation | 0.295 | 1.170 (0.587 – 2.160) | 0.633 |
| Low ZNF750 expression | 0.040 | 1.922 (1.123–3.389) | 0.020 |
ESCC, esophageal squamous cell carcinoma; CI, confidence interval.
The Evaluation of the Effect of ZNF750 Knockdown on Proliferation, Migration, Invasion, and Apoptosis in ESCC Cell Lines
The expression of ZNF750 was assessed in ESCC cell lines using TE8 and TE11. After siZNF750 transfection into both cell lines, we confirmed that the levels of ZNF750 mRNA were noticeably decreased (3.4 ± 0.8%, p = 0.001 and 4.5 ± 0.5%, p = 0.010 of control in TE8 cells; 0.8 ± 0.4%, p = 0.008 and 1.2 ± 0.4%, p = 0.034 in TE11 cells) (Fig. 2a). The proliferation assay showed that cell growth was significantly increased by the transfection of siZNF70 in both cell lines (127 ± 3.4%, p = 0.0495 and 160 ± 3.0%, p = 0.0495 of control in TE8 cells; 150 ± 2.1%, p = 0.0495 and 136 ± 3.0%, p = 0.0495 in TE11 cells) (Fig. 2b). In the migration assay, there were significant differences in the acceleration of cell motility between the siZNF750 transfected cells and control cells (305 ± 117.0%, p = 0.021 and 240 ± 43.2%, p = 0.021 of control in TE8 cells; 192 ± 22.9%, p = 0.021 and 204 ± 8.9%, p = 0.021 in TE11 cells) (Fig. 2c). Similarly, the invasion assay revealed that the cell invasion was significantly higher in the siZNF750 transfectants than in the control transfectant (218 ± 42.9%, p = 0.021 and 300 ± 34.8%, p = 0.021 of control in TE8 cells; 267 ± 53.3%, p = 0.021 and 247 ± 85.4%, p = 0.021 in TE11 cells) (Fig. 2c). In addition, the apoptosis assay showed that the apoptotic cells were significantly decreased by the transfection of siZNF750 in both cell lines (33.3% ± 11.2%, p = 0.019 and 15.6 ± 8.5%, p = 0.019 of control in TE8 cells; 16.1 ± 6.5%, p = 0.018 and 38.7 ± 10.5%, p = 0.020 in TE11 cells) (Fig. 2d).
Fig. 2.
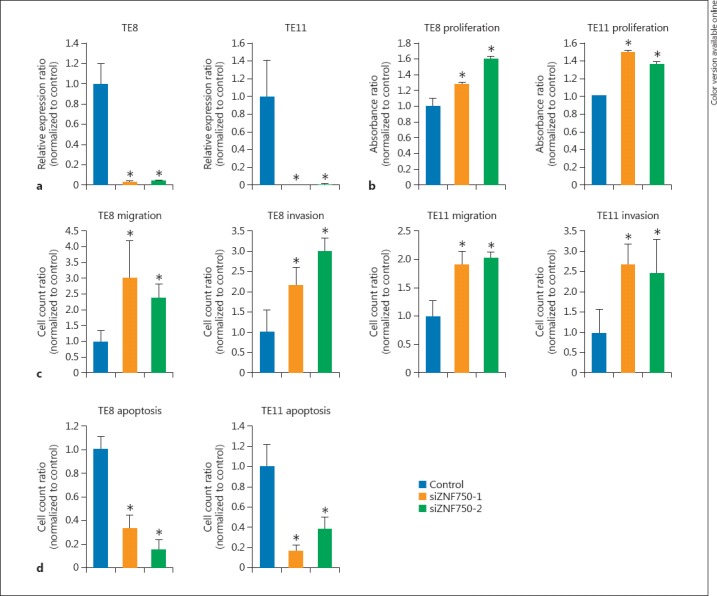
The effects of ZNF750 knockdown on ESCC cell proliferation, migration, and invasion. ESCC cell lines (TE8 and TE11) were transfected with control siRNA or 2 siRNAs (siZNF750-1 and siZNF750-2). a ZNF750 expression in ESCC cell lines after transfection of siZNF750. b Results of the proliferation assay. c Results of the migration/invasion assay. d Results of the apoptosis assay. * p < 0.05. ESCC, esophageal squamous cell carcinoma.
Discussion
The purpose of the present study was to clarify the clinicopathological features and their significance as prognostic factors of ZNF750 expression in ESCC. In this study, a low ZNF750 expression had a significant correlation with positive lymph node metastasis (p = 0.028). Furthermore, there was a significant relationship between the low expression of ZNF750 in ESCC and a poor OS (hazard ratio, 1.922; 95% confidence interval, 1.123–3.389, p = 0.020). We also demonstrated that siZNF750 significantly increased the cell proliferation, migration, and invasion and decreased apoptosis. These results suggest that ZNF750 is a suppressor of tumor progression and that the expression of ZNF750 may be a novel candidate prognostic biomarker for ESCC patients.
Several reports have shown that the expression of ZNF750 is decreased in cancer tissues compared with normal tissues [5, 6, 10], and the mutation of ZNF750 inhibited the activation of differentiation genes and caused functional deterioration [9, 14]. Some studies have shown that the decreased expression of ZNF750 promoted cell growth, migration, and invasion in squamous cell carcinoma [5, 6, 10], and Cohen et al. [15] revealed that the downregulation of ZNF750 decreased apoptosis in human keratinocytes. The present study showed the same results in cell line experiments. This suggests that ZNF750 is involved in suppressing tumor development in ESCC. In addition, Hazawa et al. [10] reported that the low expression of ZNF750 correlated with a poor prognosis in head and neck squamous cell carcinoma and lung squamous cell carcinoma, and we reported this value to be a prognostic factor of ZNF750 in ESCC for the first time.
Differentiation abnormalities play an important role in tumorigenesis. Recent studies have revealed that differentiation-related genes are associated with many cancers [16, 17, 18, 19], including ESCC [20, 21, 22, 23, 24]. Therefore, dysfunction of genes regulating differentiation has a relationship with tumor development in ESCC. For example, KLF4, a transcriptional factor containing zinc finger, has been shown to play an essential role in cell differentiation and proliferation [21, 25, 26, 27]. It was recently reported that KLF4 was a tumor suppressor and prognostic biomarker in ESCC patients [28]. In addition, Zhang et al. [29] showed that KLF4 inhibits the regulation of the epithelial-mesenchymal transition and has a relationship with cisplatin resistance. Several reports have shown that ZNF750, an essential transcriptional regulator of epidermal differentiation, positively regulates KLF4 expression [9, 10, 14]. In other words, ZNF750 is associated with KLF4, a tumor suppressor gene, and is considered to have an important role in treatment resistance. Indeed, we previously revealed that a low ZNF750 expression was associated with poor sensitivity to chemoradiotherapy [11]. In addition, Hu et al. [30] revealed that the upregulation of KLF4 by methylseleninic acid could inhibit cell growth in ESCC cells. These findings suggest that the upregulation of ZNF750 may be an effective treatment strategy for ESCC patients.
In conclusion, we herein demonstrated that the low expression of ZNF750 correlates with positive lymph node metastasis and a poor OS in patients with ESCC. Furthermore, the downregulation of ZNF750 increased the cell growth, migration, and invasion in ESCC cell lines. These findings suggest that ZNF750 may be a useful novel prognostic biomarker and that the regulation of ZNF750 may become an important target in the treatment of ESCC.
Statement of Ethics
This study was conducted in accordance with the Declaration of Helsinki. The present study was approved by the Ethics Committee of Graduate School of Medicine, Chiba University, Chiba, Japan. Informed consent was obtained from all patients for research purposes.
Disclosure Statement
The authors declare no conflicts of interest in association with the present study.
Acknowledgements
This study was supported by a Grant-in-Aid for Scientific Research (grant No. 17K16531) from the Japan Society for the Promotion of Science.
References
- 1.Altorki N, Kent M, Ferrara C, Port J. Three-field lymph node dissection for squamous cell and adenocarcinoma of the esophagus. Ann Surg. 2002;236:177–183. doi: 10.1097/00000658-200208000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medical Research Council Oesophageal Cancer Working Group Surgical resection with or without preoperative chemotherapy in oesophageal cancer: a randomized controlled trial. Lancet. 2002;359:1727–1733. doi: 10.1016/S0140-6736(02)08651-8. [DOI] [PubMed] [Google Scholar]
- 3.Courrech Staal EF, van Coevorden F, Cats A, Aleman BM, van Velthuysen ML, Boot H, et al. Outcome of low-volume surgery for esophageal cancer in a high-volume referral center. Ann Surg Oncol. 2009;6:3219–3226. doi: 10.1245/s10434-009-0700-5. [DOI] [PubMed] [Google Scholar]
- 4.Law SY, Fok M, Cheng SW, Wong J. A comparison of outcome after resection for squamous cell carcinoma and adenocarcinomas of the esophagus and cardia. Surg Gynecol Obstet. 1992;75:107–112. [PubMed] [Google Scholar]
- 5.Zhang L, Zhou Y, Cheng C, Cui H, Cheng L, Kong P, et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet. 2015;96:597–611. doi: 10.1016/j.ajhg.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin DC, Hao JJ, Nagata Y, Xu L, Shang L, Meng X, et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet. 2014;46:467–473. doi: 10.1038/ng.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sawada G, Niida A, Uchi R, Hirata H, Shimamura T, Suzuki Y, et al. Genomic landscape of esophageal squamous cell carcinoma in a Japanese population. Gastroenterology. 2016;150:1171–1182. doi: 10.1053/j.gastro.2016.01.035. [DOI] [PubMed] [Google Scholar]
- 8.Birnbaum RY, Zvulunov A, Hallel-Halevy D, Cagnano E, Finer G, Ofir R, et al. Seborrhea-like dermatitis with psoriasiform elements caused by a mutation in ZNF750, encoding a putative C2H2 zinc finger protein. Nat Genet. 2006;38:749–751. doi: 10.1038/ng1813. [DOI] [PubMed] [Google Scholar]
- 9.Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, et al. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev Cell. 2012;22:669–677. doi: 10.1016/j.devcel.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazawa M, Lin DC, Handral H, Xu L, Chen Y, Jiang YY, et al. ZNF750 is a lineage-specific tumour suppressor in squamous cell carcinoma. Oncogene. 2017;36:2243–2254. doi: 10.1038/onc.2016.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otsuka R, Akutsu Y, Sakata H, Hanari N, Murakami K, Kano M, et al. ZNF750 expression as a novel candidate biomarker of chemoradiosensitivity in esophageal squamous cell carcinoma. Oncology. 2017 doi: 10.1159/000476068. DOI: 10.1159/ 000476068. [DOI] [PubMed] [Google Scholar]
- 12.Sobin LH, Gospodarowicz MK, Wittekind C. TNM Classification of Malignant Tumors. ed 7. Oxford: Wiley-Blackwell; 2010. [Google Scholar]
- 13.Akanuma N, Hoshino I, Akutsu Y, Murakami K, Isozaki Y, Maruyama T, et al. MicroRNA-133a regulates the mRNAs of two invadopodia-related proteins, FSCN1 and MMP14, in esophageal cancer. Br J Cancer. 2014;110:189–198. doi: 10.1038/bjc.2013.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boxer LD, Barajas B, Tao S, Zhang J, Khavari PA. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes Dev. 2014;28:2013–2026. doi: 10.1101/gad.246579.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen I, Birnbaum RY, Leibson K, Taube R, Sivan S, Birk OS. ZNF750 is expressed in differentiated keratinocytes and regulates epidermal late differentiation genes. PLoS One. 2012;7:e42628. doi: 10.1371/journal.pone.0042628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyszkiewicz T, Jarzab M, Szymczyk C, Kowal M, Krajewska J, Jaworska M, et al. Epidermal differentiation complex (locus 1q21) gene expression in head and neck cancer and normal mucosa. Folia Histochem Cytobiol. 2014;52:79–89. doi: 10.5603/FHC.2014.0018. [DOI] [PubMed] [Google Scholar]
- 17.Khammanivong A, Wang C, Sorenson BS, Ross KF, Herzberg MC. S100A8/A9 (calprotectin) negatively regulates G2/M cell cycle progression and growth of squamous cell carcinoma. PLoS One. 2013;8:e69395. doi: 10.1371/journal.pone.0069395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherbet GV. Metastasis promoter S100A4 is a potentially valuable molecular target for cancer therapy. Cancer Lett. 2009;280:15–30. doi: 10.1016/j.canlet.2008.10.037. [DOI] [PubMed] [Google Scholar]
- 19.Lohman FP, Medema JK, Gibbs S, Ponec M, van de Putte P, Backendorf C. Expression of the SPRR cornification genes is differentially affected by carcinogenic transformation. Exp Cell Res. 1997;231:141–148. doi: 10.1006/excr.1996.3458. [DOI] [PubMed] [Google Scholar]
- 20.Wu N, Song Y, Pang L, Chen Z. CRCT1 regulated by microRNA-520 g inhibits proliferation and induces apoptosis in esophageal squamous cell cancer. Tumour Biol. 2016;37:8271–8279. doi: 10.1007/s13277-015-4730-2. [DOI] [PubMed] [Google Scholar]
- 21.He H, Li S, Hong Y, Zou H, Chen H, Ding F, et al. Krüppel-like factor 4 promotes esophageal squamous cell carcinoma differentiation by up-regulating keratin 13 expression. J Biol Chem. 2015;290:13567–13577. doi: 10.1074/jbc.M114.629717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen K, Li Y, Dai Y, Li J, Qin Y, Zhu Y, et al. Characterization of tumor suppressive function of cornulin in esophageal squamous cell carcinoma. PLoS One. 2013;8:e68838. doi: 10.1371/journal.pone.0068838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Ma J, Sunkel B, Luo A, Ding F, Li Y, et al. S100A14: novel modulator of terminal differentiation in esophageal cancer. Mol Cancer Res. 2013;11:1542–1553. doi: 10.1158/1541-7786.MCR-13-0317. [DOI] [PubMed] [Google Scholar]
- 24.Ji J, Zhao L, Wang X, Zhou C, Ding F, Su L, et al. Differential expression of S100 gene family in human esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2004;130:480–486. doi: 10.1007/s00432-004-0555-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 26.Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- 27.Tetreault MP, Yang Y, Travis J, Yu QC, Klein-Szanto A, Tobias JW, et al. Esophageal squamous cell dysplasia and delayed differentiation with deletion of Klf4 in murine esophagus. Gastroenterology. 2010;139:171–181. doi: 10.1053/j.gastro.2010.03.048. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma MQ, Zhang HD, Tang P, Jiang HJ, Chen CG. Association of Kruppel-like factor 4 expression with the prognosis of esophageal squamous cell carcinoma patients. Int J Clin Exp Pathol. 2014;7:6679–6685. [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang P, Hong H, Sun X, Jiang H, Ma S, Zhao S, et al. MicroRNA-10b regulates epithelial-mesenchymal transition by modulating KLF4/Notch1/E-cadherin in cisplatin-resistant nasopharyngeal carcinoma cells. Am J Cancer Res. 2016;6:141–156. [PMC free article] [PubMed] [Google Scholar]
- 30.Hu C, Liu M, Zhang W, Xu Q, Ma K, Chen L, et al. Upregulation of KLF4 by methylseleninic acid in human esophageal squamous cell carcinoma cells: modification of histone H3 acetylation through HAT/HDAC interplay. Mol Carcinog. 2015;54:1051–1059. doi: 10.1002/mc.22174. [DOI] [PubMed] [Google Scholar]


