Abstract
We have generated a site‐directed mutant of the manganese superoxide dismutase SOD‐3 of C.elegans (MnSOD‐3) which modifies the metal specificity of the enzyme. While wild‐type MnSOD‐3 functions with manganese in the active site (3600 U mg−1 of protein) it has little or no activity when iron is incorporated. However, when histidine replaces glutamine 142 in the active site, the enzyme retains 50 % of its activity and becomes cambialistic for its metal cofactor exhibiting very similar specific activity with either manganese or iron.
Keywords: enzymes, iron, manganese, metalloprotein, superoxide dismutase
Introduction
Very little is known about the way most metalloproteins obtain their metal cofactors. Although some metallochaperones exist, in many cases the metal must be not only incorporated into the protein during de novo folding but also needs to be selected from any of the other metals in the immediate environmental vicinity; the process of metalation.1 Mononuclear superoxide dismutases utilise either iron or manganese in their active sites and metalation of these enzymes can be made dependent upon the availability of the metal through provision of the metal in the culture growth medium when they are overexpressed in E.coli.2 The manganese superoxide dismutase (MnSOD) and iron superoxide dismutase (FeSOD) of E.coli are extremely similar in sequence (45 % conservative amino acids) and structure (91 % homology and RMSDCα‐monomer 1.6 Å) and yet to become an active enzyme, each enzyme must bind its cognate metal.3 Each enzyme may fold correctly while incorporating the “wrong” metal but the enzyme does not catalyse the conversion of superoxide to oxygen and hydrogen peroxide, the normal function of SOD.4 Several attempts have been made using site‐directed mutagenesis to change ion specificity with the aim to interconvert SOD ion dependence, but with very little success.3c, 5 Interestingly, there are SODs that do indeed function with either metal and site‐directed mutagenesis has succeeded in adjusting the metal specificity of these, but they remain cambialistic nonetheless.6 One focus of such experiments has been the highly conserved glutamine residue in the active site that forms an intrinsic part of the hydrogen‐bonded network of amino acids and water necessary for full enzyme activity. The main reason for this is that in FeSOD the glutamine is provided by the N‐domain of the protein while in MnSOD the glutamine is contributed by the C‐domain. In either case the Nϵ2 and Oϵ1 atoms occupy almost identical positions in three‐dimensional space.5b, 7 This arrangement does not affect the three histidine residues and the aspartate that also coordinate directly to the metal cofactor, nor does it interfere with the one water (or hydroxyl ion, designated WAT1) directly bound to the metal; a common feature of all SODs. Furthermore, the glutamine, while bonding with the metal‐attached water, continues a hydrogen‐bonding network by binding to a similarly conserved tyrosine at position 34 in the sequence. Various analyses have been performed to identify the amino acid sequences that might form a signature for the discrimination between iron and manganese SODs with some initial success. Today, over three hundred SOD amino acids sequences are known and the boundary has become blurred and more difficult to identify. However, the signature first described by Parker and Blake (1988) to determine an MnSOD is reasonably accurate.8 One notable exception is the arrangement of amino acids found in the FeSOD of M.tuberculosis, in which the normally conserved N‐domain glutamine is replaced by a histidine residue provided by the C‐domain.9 We have therefore tested this intriguing exception by replacing glutamine in a C.elegans MnSOD (MnSOD‐3) with a histidine, generating the site‐directed mutant MnSOD‐3[Q142H]. We show that this single amino acid substitution is sufficient to convert the MnSOD into a cambialistic SOD, which we propose to be an essential element that determines ion specificity while maintaining SOD function.
Results and Discussion
Protein purification and characterization
Wild‐type MnSOD‐3 and site‐directed mutant MnSOD‐3[Q142H] were cultured in minimal media containing either iron or manganese from the start of cell growth, and purified to homogeneity as described previously.10 Metal content was analysed by microwave plasma atomic emission spectroscopy (MP‐AES) and showed that the purified proteins contained the canonical metal (Table 1). It is impossible to keep iron completely out of the growth medium and the Q142H mutant exhibited a higher selectivity for it than did wild type MnSOD‐3 when cultured in manganese‐supplemented media (Table 1).
Table 1.
Metal content and thermal melting analyses of MnSOD‐3 and MnSOD‐3[Q142H] cultured in minimal media containing either iron or manganese.
| Purified protein[a] | Media | Manganese[b] | Iron[b] | T m [°C][c] |
|---|---|---|---|---|
| MnSOD‐3 | Mn | 1.02±0.051 | 0.01±0.001 | 73 |
| MnSOD‐3 | Fe | 0.13±0.006 | 0.92±0.048 | 84 |
| MnSOD‐3[Q142H] | Mn | 0.63±0.031 | 0.20±0.010 | 68 |
| MnSOD‐3[Q142H] | Fe | 0.02±0.001 | 0.93±0.046 | 66 |
[a] Proteins induced from the appropriate plasmid in E.coli SOD‐deficient cells. [b] Metal content indicated in g atoms per subunit. Errors were consistently approx. 5 %. [c] Thermal melting temperatures determined by CD.
Circular dichroism (CD) spectra of the purified proteins indicated no secondary structural changes between mutants and wild type or between iron‐ or manganese‐containing SODs (results not shown). The structural integrity of the SODs was further probed through thermal denaturation experiments to determine their melting temperatures (T m) (Table 1). Although the melting temperatures of the mutant proteins were similar whether they incorporated iron or manganese, we found that iron‐containing wild‐type MnSOD‐3 appeared to be more stable than its manganese counterpart. If this were due to the effect of iron–amino acid bonding in the active site, we would still have expected the same trend to be seen in the Q142H mutant, but we did not.
Enzyme activity
The activity of the SODs was measured by the cytochrome c method and gave 3600±90 U mg−1 of protein for wild‐type MnSOD‐3, similar to previously measured values.10 MnSOD‐3 purified from cultures supplemented with iron and which was shown to contain mostly iron in the active site (Figure 1) yielded a value of only 16 U mg−1 of protein. If any activity in the iron‐substituted MnSOD was due to the manganese present (0.13 g atom per monomer) we would expect an activity of nearly 500. It may be assumed therefore that any residual manganese might be randomly distributed throughout the four subunits of each tetramer. It has previously been assumed that the minimal unit to provide enzyme activity is a fully metalated dimer within a tetramer (see Figure 1). This explains the very low, almost undetectable activity of the iron‐substituted MnSOD‐3, despite some manganese being present. Enzyme activity of the MnSOD‐3[Q142H] mutant was found to be 900±30 U mg−1 of protein when isolated from manganese‐supplemented media and 1179±19 U mg−1 of protein when iron‐substituted. Considering the metalation of each protein (Table 1) these values indicate that the Q142H mutation has converted MnSOD‐3 into a cambialistic enzyme that is active with either manganese or iron in the active site.
Figure 1.
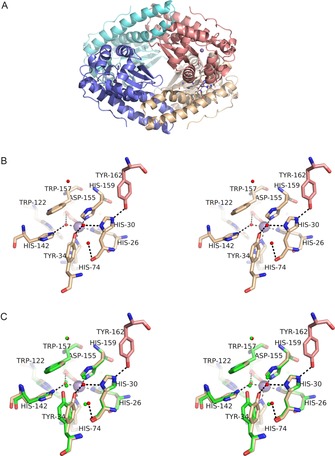
Structure of MnSOD‐3[Q142H]. A) (top) tetrameric assembly of the four SOD monomeric subunits is formed as a dimer of dimers. Each active dimer is shown in blue (left in top Figure) or red (right in top Figure) and illustrates the proximity of the active sites containing metal ions (purple spheres). The tetrameric interface is formed between the hairpin N domain helices of subunits forming two four‐helix bundles typical of many tetrameric SODs. B) (middle) Stereo diagram of the active site of MnSOD‐3[Q142H] shown in approximately the same orientation and colour as A (top). Tyr 162 is derived from a different subunit and hydrogen bonds with His 30 which then hydrogen bonds to Tyr 34 via a solvent molecule (WAT2; solvent molecules shown as small red spheres). Due to the distance between Tyr 34 and His 142 the hydrogen bonding network is discontinuous, and the latter residue can bind only to WAT1 (the metal‐bound solvent) in this orientation (seen in the X‐ray structure). C) (bottom) The same as Figure B but with the addition of residues from wild‐type MnSOD‐3 shown in green. The proximity of Gln 142 NE2 to His 142 NE2 is illustrated as is the repositioning of Tyr 34. Solvent molecules are illustrated as small green spheres two of which are required to link His 30 to Tyr 34 by hydrogen bonds (H bonds not shown).
Structural analysis
We have previously reported the structure of the wild‐type MnSOD‐3 from C.elegans (PDB: 3DC5 and 4X9Q) and its complex with the substrate analogue, azide (PDB: 5AG2).10, 11 These data have reinforced our understanding and the importance of the extensive hydrogen bonded network formed between the external solvent, a number of significant residues and the metal in the active site of the enzyme. Here we report the structure of the cambialistic mutant MnSOD‐3[Q142H] (PDB: 6ELK).
Overall the MnSOD‐3[Q142H] structure was highly similar (RMSDCα‐all‐chains 0.19 and 0.15 Å) to the wild type MnSOD‐3 3DC5 and 4X9Q structures, respectively. As previously reported, there is evidence from the electron density maps of some flexibility in the region of chain A Q40‐A59 with the structure occupying two alternate conformations.11 The predominant density observed was similar to the configuration of 4X9Q. Therefore this conformation was used rather than that seen in 3DC5. Although we can see minor repositioning of some active site residues in the MnSOD‐3[Q142H] structure, the most significant finding is the alteration of the hydrogen‐bonded network (Figure 1). In MnSOD‐3 this is usually formed between H30 and Y34 via two solvent molecules, continues to Q142 and then to WAT1, the solvent ligand bound directly to the metal. The most obvious difference seen in the active site of the Q142H mutant is the movement of Y34. This residue is repositioned by 1.7 Å through an angular movement of 17.2° in comparison with the wild type Y34. The Y34 OH is in the position that is 1.5 Å closer to the H30 ND1. Consequently the requirement of two solvent molecules to form the hydrogen bonding between these atoms is reduced to just one (Figure 1 B). This hydrogen‐bonding arrangement is more like that found in other mononuclear SODs. However, this displacement now places the Y34 OH significantly farther away from the mutated H142, rendering hydrogen bonding impossible. The distance between Y34 OH to H142 NE2 is 4 Å whereas Y34 OH to Q142 NE2 in wild‐type is 2.9 Å, making the two atoms readily available to hydrogen bond. This occurs even though the Q142 NE2 of wild type and the H142 NE2 of the mutant are only 0.5 Å apart in the aligned structures (Figure 1 C). H142 NE2 can form the hydrogen bond with WAT1 being 2.9 Å away, thus connecting H142 with the metal ion centre. Although W122 is twisted slightly (12.2°) away from H142, no other effects of the mutation can be seen. Inter‐subunit distances between metal centres are virtually identical in the two structures (wild‐type and MnSOD‐3[Q142H]) indicating that there is virtually no change in dimeric or tetrameric contacts between subunits.
We can infer from the requirement of protons to form hydrogen peroxide from superoxide substrates that the network of hydrogen bonds connecting solvent to metal must be essential for enzyme function. Y34 may be considered to be one possible source of protonation, but in the mutant Q142 H, communication between the metal centre and Y34 is lost as the hydrogen bonding network cannot connect via H42 and Y34. We therefore investigated this network using molecular dynamics simulations.
Molecular dynamics simulations
In MnSOD‐3[Q142H], the trajectory of a 100 ns simulation revealed an average of three waters in addition to the ubiquitous WAT1; the water or hydroxyl that is coordinated to the manganese cofactor. We observed an extra water between the manganese, Y34 and H30, with respect to the native MnSOD‐3. This additional water is also capable of hydrogen bonding to Y34 and H30. These residues, termed gateway residues, are of significance due to their proximity to the active site funnel through which it is presumed the superoxide substrate passes to reach the active centre. During simulations, this solvent occasionally approaches the manganese to a closest distance of 2.26 Å, making coordination between this water and the cofactor possible. This suggests that a sixth co‐ordination centre may form transiently as suggested for a yeast MnSOD mutant.12 In contrast to the X‐ray determined structure, during the 100 ns simulation the ring of H142 is observed to rotate by up to 180° placing H142 NE2 and H142 ND1 too far to allow for a hydrogen bond with the metal‐bound WAT1 (Figure 2 and Supporting Information). However, it was noted that throughout the trajectory, there were instances when a second water approaches which can form the hydrogen‐bonding network required to link H142 ND1 to the manganese via WAT1. There is, therefore, the potential to form two transient hydrogen bonding networks to the metal centre (via H142 and the gateway residues). The molecular dynamics simulations suggest that solvent movement near and around the active site facilitates the transfer of protons via the hydrogen bonding networks that may form and collapse over time.
Figure 2.
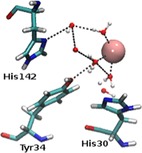
MnSOD‐3[Q142H] second sphere residues, His142, Tyr34 and His30 observed from Molecular Dynamics (MD) simulation. This particular frame of the protein trajectory reveals ND1 and NE2 of His142 in a position that is opposed to what is observed in the X‐ray structure of this protein. Here, ND1 is closer to the inner‐sphere waters. Red and white spheres indicate waters, while manganese is indicated by a pink sphere. Hydrogen bonding is illustrated as black dashes. This Figure was generated by VMD.13 See supporting information for the trajectory.
Electrostatics analysis
The electrostatic potential was calculated by solving linearized Poisson–Boltzmann equation for solvent ionic strength I=0.15 M. Atomic charges of the metal cofactor were derived with the restrained electrostatic potential (RESP) procedure from the electrostatic potential calculated at the B3LYP/def2‐SVP level. The active site surface is mainly positive in character, which is due to the positive formal (+1) charge of the metal cofactor. The surplus charge is not limited to the metal ion as it smears into the ligands coordinated to the metal. Figure 3 shows the results of these calculations for the cambialistic mutant MnSOD‐3[Q142H]. We have observed and analysed similar electrostatics for the wild‐type and iron‐containing MnSOD‐3 and for the manganese‐ and iron‐substituted mutant. We have looked at the overall electrostatic potentials generated at the protein surface as well as within the active site (Figure 3 and Supporting Information) and have found little difference between the proteins.
Figure 3.
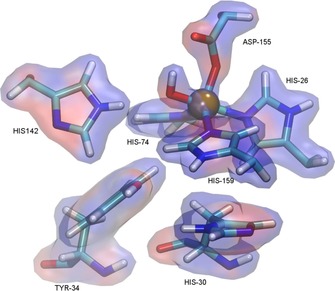
Representation of the active site of MnSOD‐3[Q142H] with solvent accessible surface colour coded according to electrostatic potential. The region exhibiting a negative potential is colored red, whereas highly positive potential region is coloured blue. This Figure was generated by VMD.13.
Conclusion
Metalation is an important event in metalloprotein production, and in the case of the mononuclear SODs, may be considered an aspect of the protein folding problem in general. This is due partly to the fact that the N‐ and C‐domains of the protein must fold independently before capture of the metal and two amino acid ligands to the metal are provided by each domain. This must be the case because the final 18 C‐terminal amino acids are found between the domains and obviously cannot be inserted into position after protein folding is complete. When metal chaperones are not involved, correct metalation is more often accomplished by controlling the metal pool within the cell as proteins are apt to take up the dominant metal (as we show here by manipulating the metal content of our overexpressed SOD).1 When metal homeostasis (for example the control of iron levels in mitochondria) is disturbed, mismetalation may occur and is of direct relevance to the MnSOD described in this study. It is still a matter of some debate as to whether or not mismetalated human MnSOD (i.e. containing iron) is a good or a bad thing for the cell, as on the one hand it removes harmful iron, while on the other it removes essential SOD activity.14 Mismetalation of proteins may occur for a number of reasons and usually results in an inactive protein. Under oxidative stress, mononuclear iron containing enzymes are disabled.15 Here we describe the production of a cambialistic Mn/FeSOD through a single mutation of an MnSOD. We have studied the proteins through physicochemical means and in silico analyses to try to identify the reason for this conversion. Unfortunately there seems to be no simple explanation for the selectivity (during de novo protein folding) or specificity (enzyme activity with a selected metal) of this ubiquitous and important class of enzymes. The possibility of a holistic property of the enzyme as an explanation remains a confounding possibility, but a holistic characteristic is becoming more likely deduced from the increasing number of experimental data.
Experimental Section
Materials and methods
All general‐purpose chemicals were supplied by Sigma–Aldrich (Germany) and VWR (Radnor USA). Bioneer (South Korea) provided the oligonucleotides and DNA sequencing services. The in vitro site‐directed mutagenesis reaction was performed using the Quikchange II XL site‐directed mutagenesis kit from Agilent Technologies (Santa Clara, Ca). Hampton Research (Ca) reagents were used for protein crystallisation.
In vitro site directed mutagenesis
The pTrc99A cDNA clone of C.elegans MnSOD‐3 served as the template for mutagenesis using the following oligonucleotides pairs to replace the glutamine in position 142 by histidine:
Q142H‐forward:
5′CACCTGTGCAAACCACGATCCTTTGGAAGG‐3′
Q142H‐reverse:
5′CCTTCCAAAGGATCGTGGTTTGCACAGGTG3′
The mutation was confirmed by sequencing both DNA strands with the primers:
PKPro‐5 5′CTCGTATAATGTGTGGAATTGTGAGCGG3′
PKTerm‐5 5′CCTGACCCCATGCCGAACTCAGAAG3′.
Protein expression and purification
The E.coli strain OX326A (ΔsodA, ΔsodB),16 transformed with the ptrc‐MnSOD plasmid DNA, was grown in defined minimal media. The media contained 1x M9 salts (Na2PO4 12 g L−1, KH2PO4 6 g L−1, NH4Cl 2 g L−1, NaCl 1 g L−1, CaCl2 6 mg L−1), MgCl2 (1 mmol), fructose (0.2 % w/v), thiamine (5×10−4 % w/v), casamino acids (0.1 % w/v). Cell cultures were supplemented with ampicillin (200 μg mL−1) and MnSO4 (1 mm) or FeSO4 (1 mm) as appropriate. Expression was induced by IPTG (0.1 mm) and cells were grown overnight at 37 °C with aeration. All solutions were prepared with AmberliteTM‐treated deionised water. Proteins were purified to homogeneity as previously described.10
Protein crystallisation, data collection, structure solution and refinement
Crystals of MnSOD‐3[Q142H] were grown at 25 °C via the hanging drop diffusion method with a reservoir liquor of bicine (0.1 m pH 8.8) and ammonium sulfate (2.9 m) from a drop containing reservoir liquor (2 μL) and protein (2 μl, 10 mg mL−1). The crystals were immersed in a cryo‐protectant of glycerol (20 %) and then mounted on loops for data collection. X‐ray diffraction data was collected at 100 K using the RIgaku Micromax 007 X‐ray system. Data was integrated at 1.65 and 1.7 Å (Table 2).
Table 2.
Data collection and refinement statistics for SOD‐3 Q142H (PDB 6ELK).
| MnSOD‐3[Q142H] | |
|---|---|
| Data statistics Source | Rigaku MicroMax‐007 rotating anode |
| Wavelength (Å) | 1.541 |
| Resolution range (Å)[a] | 53.18–1.65 (1.69–1.65) |
| Space group | P41212 |
| Unit‐cell parameters (Å) | a=b=81.5, c=137.8 |
| Completeness (%)[a] | 99.8 (97.3) |
| No. of observed reflections | 581095 |
| No. of unique reflections | 56496 |
| Redundancy | 10.3 (7.0) |
| <I/σ(I)>[a] | 30.1 (2.9) |
| R merge (%)[a,b] | 3.8 (52.9) |
| R pim (%)[a,c] | 1.7 (30.9) |
| CC 1/2 | 0.99 (0.82) |
| Refinement statistics | |
| Resolution range for refinement (Å) | 53.18–1.65 |
| R factor (%) | 18.3 |
| R free (%)[d] | 21.0 |
| No. of protein non‐H atoms | 3172 |
| No. of water molecules | 303 |
| No. of manganese ions | 2 |
| No. of sulfate ions | 6 |
| No. of glycerol molecule | 1 |
| R.m.s.d bond lengths (Å) | 0.008 |
| R.m.s.d bond angles (°) | 1.2 |
| Average overall B factor (Å2) | |
| Protein | 33 |
| Water | 36 |
| Manganese ions | 19 |
| Sulfate ions | 52 |
| Glycerol | 36 |
| Ramachandran analysis, the percentage of residues in the regions of plot (%)[e] | |
| Favoured region | 97.4 |
| Outliers | 0 |
| PDB code | 6ELK |
[a] Values given in parentheses correspond to those in the outermost shell of the resolution range. [b] R merge=∑hkl∑i|I i(hkl)−⟨I(hkl)⟩|/∑hkl∑I i(hkl) [c] R pim=∑hkl{1/[N(hkl)−1]}1/2∑i|I i(hkl)−⟨I(hkl)⟩|/∑hkl∑i I i(hkl) [d] R free was calculated with 5 % of the reflections set aside randomly. [e] Ramachandran analysis using the program MolProbity.17
Molecular dynamic simulations
Two 100 ns Molecular Dynamics (MD) simulations were launched, based on the existing crystal structure of native MnSOD‐3 (4X9Q) and MnSOD Q142H (6ELK) Simulations were carried out using Gromacs 4.6.2 package.18 The AMBER99SB force field was used to describe the protein.19 Four manganese ions were added to both proteins using the Mn2+ parameters as described by Bradbook et al.20 The MnSOD‐3 and Q142 proteins were placed in a cubic box filled with 21 957 and 20613 three‐point transferable intermolecular potential (TIP3P) water molecules, respectively. Eight and four chloride ions were added to neutralise the MnSOD‐3 and Q142H systems, respectively. The final size of the system reached 20621 atoms. The cut‐off van der Waals interactions was set to 1.4 nm. The velocity‐rescale (VR) thermostat and the Parinello‐Rahman were used.21 Particle‐mesh Ewald (PME) method was used to treat electrostatic interactions with a 1 nm cut‐off distance. Periodic boundary conditions were applied to three dimensions of the simulation box. Motion equations were solved numerically at a time step set to 2 fs and the neighbour list was updated every 10 steps. Each solvated system was subjected to a thorough energy minimisation of 5000 steps using a steepest descent minimizer. Thereafter, equilibrium simulations were performed at a constant temperature of 300 K and a constant pressure of 105 Pa. Simulations of 100 ns were performed at a constant temperature of 300 K maintained by the weak‐coupling Scheme. Post‐processing and analysis were carried out using Gromacs and VMD.13
Enzyme assay and metal analysis
Protein concentration was determined by the BCA assay or by absorbance at 280 nm for purified proteins using a molar extinction coefficient for MnSOD‐3 of 43680. Superoxide dismutase activity was measured spectrophotometrically as described by McCord and Fridovich22 and Ysebaert‐Vanneste and Vanneste,23 whereby cytochrome c serves as the detector and xanthine‐xanthine oxidase as the superoxide generator.
The degree of metalation of the purified SOD protein samples was measured using an Agilent Technologies 4100 Microwave‐plasma Atomic Emission Spectrometer (MP‐AES). Samples in KP buffer (between 0.3 and 1.0 mg mL−1) with no additions were used and both iron and manganese content measured directly. Sample analyses were carried out using three replicates at a pump speed of 15 rpm for 15 seconds uptake time. Prior to the metal analysis the MP‐AES torch was heated for 30 minutes and all tubing and the nebuliser were rinsed with a 5 % HNO3 solution. Standard curves were prepared using High Purity Standard CCV‐1 Solution A metal standards solution.
Circular dichroism spectroscopy
The secondary structures of the proteins were assessed by CD measurements using a PiStar spectrometer (Applied Photosystems). Far‐UV (180–260 nm) CD spectra of the proteins (0.3 mg mL−1) in potassium phosphate buffer (50 mm pH 7.8) were recorded in stepwise increments of 0.5 nm at 25 °C in a cuvette of 0.1 cm pathlength with a 2 nm bandwidth. The unfolding transition temperature was followed by CD with a temperature ramp between 20 °C and 90 °C (1 °C per minute) at 222 nm and a 4 nm bandwidth. Temperature was monitored using a probe in the stoppered cuvette.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Supplementary
Acknowledgements
Work reported here was carried out using funding from UoM grants PHBIN02‐1, PHBRP03‐05 and PHBRP03‐17. The European Union Erasmus+ programme (project number: 2016‐1‐PL01‐KA103‐023786) is acknowledged for providing financial support for the mobility traineeship of T.B. This research was supported in part by PL‐Grid Infrastructure. We wish to thank the EU COST association for support through COST actions CM1305 and CM1306.
T. Hunter, R. Bonetta, A. Sacco, M. Vella, P.-M. Sultana, C. H. Trinh, H. B. R. Fadia, T. Borowski, R. Garcia-Fandiño, T. Stockner, G. J. Hunter, Chem. Eur. J. 2018, 24, 5303.
References
- 1. Foster A. W., Osman D., Robinson N. J., J. Biol. Chem. 2014, 289, 28095–28103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beck Y., Bartfeld D., Yavin Z., Levanon A., Gorecki M., Hartman J. R., Nat. Biotechnol. 1988, 6, 930–935. [Google Scholar]
- 3.
- 3a. Hunter G. J., Hunter T., Health 2013, 5, 1719–1729; [Google Scholar]
- 3b. Abreu I. A., Cabelli D. E., Biochim. Biophys. Acta Proteins Proteomics 2010, 1804, 263–274; [DOI] [PubMed] [Google Scholar]
- 3c. Culotta V. C., Yang M., O'Halloran T. V., Biochim. Biophys. Acta Mol. Cell Res. 2006, 1763, 747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. W. F. Beyer, Jr. , Fridovich I., J. Biol. Chem. 1991, 266, 303–308. [PubMed] [Google Scholar]
- 5.
- 5a. Vance C. K., Miller A. F., Biochemistry 1998, 37, 5518–5527; [DOI] [PubMed] [Google Scholar]
- 5b. Schwartz A. L., Yikilmaz E., Vance C. K., Vathyam S., Koder R. L., Miller A. F., J. Inorg. Biochem. 2000, 80, 247–256; [DOI] [PubMed] [Google Scholar]
- 5c. Miller A. F., FEBS Lett. 2012, 586, 585–595; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5d. Osawa M., Yamakura F., Mihara M., Okubo Y., Yamada K., Hiraoka B. Y., Biochim. Biophys. Acta Proteins Proteomics 2010, 1804, 1775–1779. [DOI] [PubMed] [Google Scholar]
- 6.
- 6a. Gabbianelli R., Battistoni A., Capo C., Polticelli F., Rotilio G., Meier B., Desideri A., Arch. Biochem. Biophys. 1997, 345, 156–159; [DOI] [PubMed] [Google Scholar]
- 6b. Sugio S., Hiraoka B. Y., Yamakura F., Eur. J. Biochem. 2000, 267, 3487–3495; [DOI] [PubMed] [Google Scholar]
- 6c. Hiraoka B. Y., Yamakura F., Sugio S., Nakayama K., Biochem. J. 2000, 345, 345–350; [PMC free article] [PubMed] [Google Scholar]
- 6d. Un S., Tabares L. C., Cortez N., Hiraoka B. Y., Yamakura F., J. Am. Chem. Soc. 2004, 126, 2720–2726; [DOI] [PubMed] [Google Scholar]
- 6e. Nakamura T., Torikai K., Uegaki K., Morita J., Machida K., Suzuki A., Kawata Y., FEBS j. 2011, 278, 598–609; [DOI] [PubMed] [Google Scholar]
- 6f. Russo Krauss I. M., Merlino A., Pica A., Rullo R., Bertoni A., Capasso A., Amato M., Riccitiello F., De Vendittis E., Sica F., RSC Adv. 2015, 5, 87876–87887. [Google Scholar]
- 7. Hunter T., Bannister J. V., Hunter G. J., Eur. J. Biochem. 2002, 269, 5137–5148. [DOI] [PubMed] [Google Scholar]
- 8.
- 8a. Parker M. W., Blake C. C., FEBS Lett. 1988, 229, 377–382; [DOI] [PubMed] [Google Scholar]
- 8b. Wintjens R., Gilis D., Rooman M., Proteins Struct. Funct. Bioinf. 2007, 70, 1564–1577. [DOI] [PubMed] [Google Scholar]
- 9.
- 9a. Cooper J. B., McIntyre K., Badasso M. O., Wood S. P., Zhang Y., Garbe T. R., Young D., J. Mol. Biol. 1995, 246, 531–544; [DOI] [PubMed] [Google Scholar]
- 9b. Bunting K., Cooper J. B., Badasso M. O., Tickle I. J., Newton M., Wood S. P., Zhang Y., Young D., Eur. J. Biochem. 1998, 251, 795–803. [DOI] [PubMed] [Google Scholar]
- 10. Trinh C. H., Hunter T., Stewart E. E., Phillips S. E. V., Hunter G. J., Acta Crystallogr. Sect. A 2008, 64, 1110–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hunter G. J., Trinh C. H., Bonetta R., Stewart E. E., Cabelli D. E., Hunter T., Protein Sci. 2015, 24, 1777–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sheng Y., Butler Gralla E., Schumacher M., Cascio D., Cabelli D. E., Valentine J. S., Proc. Natl. Acad. Sci. USA 2012, 109, 14314–14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Humphrey W., Dalke A., Schulten K., J. Mol. Graph. 1996, 14, 33–38. [DOI] [PubMed] [Google Scholar]
- 14.
- 14a. Naranuntarat A., Jensen L. T., Pazicni S., Penner-Hahn J. E., Culotta V. C., J. Biol. Chem. 2009, 284, 22633–22640; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14b. Yamakura F., Kobayashi K., Furukawa S., Suzuki Y., Free Radical Biol. Med. 2007, 43, 423–430. [DOI] [PubMed] [Google Scholar]
- 15. Imlay J. A., J. Biol. Chem. 2014, 289, 28121–28128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Steinman H. M., Mol. Gen. Genet. 1992, 232, 427–430. [DOI] [PubMed] [Google Scholar]
- 17. Chen V. B., Arendall 3rd W. B., Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., Acta Crystallogr. Sect. A 2010, 66, 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J., J. Comput. Chem. 2005, 26, 1701–1718. [DOI] [PubMed] [Google Scholar]
- 19. Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J. L., Dror R. O., Shaw D. E., Proteins Struct. Funct. Bioinf. 2010, 78, 1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bradbrook G. M., Gleichmann T., Harrop S. J., Habash J., Raftery J., Kalb J., Yariv J., Hiller I. H., Helliwell J. R., J. Chem. Soc. Faraday Trans. 1998, 94, 1603–1611. [Google Scholar]
- 21.
- 21a. Bussi G., Donadio D., Parrinello M., J. Chem. Phys. 2007, 126, 014101-014107; [DOI] [PubMed] [Google Scholar]
- 21b. Parrinello M., Rahman A., J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar]
- 22. McCord J. M., Fridovich I., J. Biol. Chem. 1969, 244, 6049–6055. [PubMed] [Google Scholar]
- 23. Ysebaert-Vanneste M., Vanneste W. H., Anal. Biochem. 1980, 107, 86–95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Supplementary


