Duplication of a cytokinin response regulator, followed by the acquisition of a novel expression pattern, generated a Y-encoded sex-determining gene, named Shy Girl in kiwifruit (Actinidia spp).
Abstract
Dioecy, the presence of male and female flowers on distinct individuals, has evolved independently in multiple plant lineages, and the genes involved in this differential development are just starting to be uncovered in a few species. Here, we used genomic approaches to investigate this pathway in kiwifruits (genus Actinidia). Genome-wide cataloging of male-specific subsequences, combined with transcriptome analysis, led to the identification of a type-C cytokinin response regulator as a potential sex determinant gene in this genus. Functional transgenic analyses in two model systems, Arabidopsis thaliana and Nicotiana tabacum, indicated that this gene acts as a dominant suppressor of carpel development, prompting us to name it Shy Girl (SyGI). Evolutionary analyses in a panel of Actinidia species revealed that SyGI is located in the Y-specific region of the genome and probably arose from a lineage-specific gene duplication. Comparisons with the duplicated autosomal counterpart, and with orthologs from other angiosperms, suggest that the SyGI-specific duplication and subsequent evolution of cis-elements may have played a key role in the acquisition of separate sexes in this species.
INTRODUCTION
Among angiosperms, hermaphroditism, or the presence of both types of flower organs in each flower, represents the ancestral and most common state and only a few percent of species have evolved dioecy, i.e., the separation of sexes between male and female individuals (Renner, 2014). Dioecy has evolved multiple independent times across the plant kingdom, and this transition has been accompanied by various lineage-specific adaptive scenarios (Ming et al., 2011; Charlesworth, 2013, 2015; Renner, 2014). Theoretical models for the evolution of dioecy in plants predict the requirement for two mutations: a recessive mutation resulting in male sterility and a dominant female-suppressing factor SuF (Westergaard, 1958; Charlesworth and Charlesworth, 1978; Charlesworth, 2015). In this scenario, the acquisition of SuF requires a gain-of-function mutation with a dominant effect in order to function in heterozygotes, such as in heterogametic males carrying copies of both the X and Y chromosomes. Previous studies on Y chromosome evolution in plants, mostly in Silene latifolia and papaya (Carica papaya), have revealed nonrecombining male-determining regions containing multiple genes (Liu et al., 2004; Ming et al., 2007; Kazama et al., 2016), but the sex-determining genes have not been identified in either species yet. Recent genome assembly in garden asparagus (Asparagus officinalis) has led to the identification of two sex-determining genes: a potential male promoting factor (M) and a potential SuF, named SOFF, hypothesized to have arisen from a gene duplication event followed by the acquisition of a new function, distinct from that of the ancestral gene (Harkess et al., 2017). In diploid persimmon (Diospyros lotus), on the other hand, sex determination is currently hypothesized to be under the control of a pair of paralogous genes called OGI and MeGI (Akagi et al., 2014). MeGI encodes an autosomal transcription factor, and its expression is regulated by the Y-encoded pseudogene OGI. In the presence of OGI, MeGI expression is reduced, resulting in the development of male flowers, as opposed to female flowers, where MeGI expression is higher (Akagi et al., 2014, 2016). The situation in persimmon is reminiscent to that in mammals, where the Y chromosome carries a single male-determining factor (Ming et al., 2011; Bachtrog and Tree of Sex Consortium et al., 2014), although it is possible that a second Y-encoded sex determinant has yet to be uncovered in persimmon.
As exemplified above, the evolution of separate sexes can originate from an initial gene, or whole-genome, duplication event, resulting in a situation more permissive of gain-of-function mutations, possibly required for this transition. Although pseudogenization is the most common outcome after duplication, other scenarios can occur as well, such as neofunctionalization, where one of the duplicates retains its original function while the other evolves a new function, or subfunctionalization, where the original function is split between the duplicates (Moore and Purugganan, 2005). If the same functions are maintained by both duplicates, changes may occur in cis-regulatory regions, possibly establishing novel functionality through restricting expression either in time or location (Moore and Purugganan, 2005; Roulin et al., 2013), or for example, limiting it to one sex. Such regulatory changes are potentially more likely than amino acid changes and may occur rapidly after gene duplication events, especially if the evolution of the duplicated gene is constrained by conflicts caused by pleiotropic effects (Roulin et al., 2013), and influence important lineage-specific traits. For example, functional changes in paralogs have triggered the establishment ripening characteristics in tomato (Solanum lycopersicum) fruits (Tomato Genome Consortium, 2012).
Kiwifruit, a major fruit crop worldwide, belongs to the genus Actinidia. Most Actinidia species are dioecious, even those that are polyploid (Datson and Ferguson, 2011), suggesting that dioecy is probably ancestral to the genus (Chat et al., 2004). In the genus Actinidia, male individuals bear fertile anthers and rudimental carpels, and female individuals bear fertile carpels and regular but functionally sterile anthers. Sex in Actinidia is genetically controlled, with XY males and XX females. Individuals carrying at least one copy of the Y chromosome are male, independent of ploidy level, suggesting that the sex-determining locus includes an active male-determining factor (Gill et al., 1998; Ming et al., 2011; Seal et al., 2012). However, hermaphrodite mutants occasionally appear from both male and female individuals in Actinidia deliciosa (McNeilage, 1991, 1997; Sakellariou et al., 2016), possibly resulting from the action of additional autosomal genetic factors in this species (McNeilage, 1997). Previous cytological analyses and genetic mapping surrounding the sex-determining region suggested that the sex chromosomes are incipient and homomorphic, with a physically small sex-determinant region (He et al., 2005; Fraser et al., 2009; Zhang et al., 2015). A draft of the kiwifruit genome sequence was recently generated from a female cultivar (Huang et al., 2013), but the Y-specific sequence and genomic context surrounding the potential sex determinants are still largely unknown. In this study, we aimed to identify candidate sex-determining genes in kiwifruit, through a combination of de novo whole-genome, transcriptome sequencing, and evolutionary analyses.
RESULTS
Identification of Male-Specific Region of the Y Chromosome in Actinidia
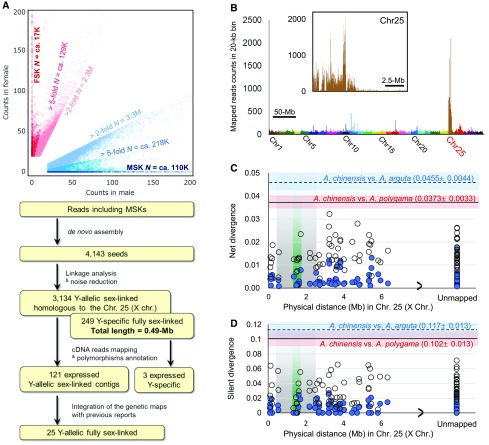
Our first goal was to identify male-specific sequences by comparing a set of genomic sequence reads from male and female Actinidia individuals. Random genomic sequencing libraries were constructed from 20 female and 22 male siblings from a population of F1 sibling trees derived from an interspecific cross between A. rufa and A. chinensis, called the KE population. In this population, flower development and specifically the pattern of differentiation of the sexual organs were similar to those described in most Actinidia species (Figure 1; Supplemental Figure 1). Sequenced reads were obtained using Illumina technology, amounting to a total of 130 to 140× coverage of the estimated A. chinensis genome size (758 Mb) (Huang et al., 2013). Reads from males and females were grouped into separate categories and each cataloged into 30-bp subsequences (k-mers), following the approach previously used to identify Y-linked sequences in persimmon (Akagi et al., 2014). Reads including k-mers found only within male reads (male-specific or MSK), or significantly enriched in the male reads (Figure 2A; Supplemental Figure 2; P < 2.2e−16 with Fisher’s exact test), were used to assemble 4143 potentially sex-linked “seed contigs.” Approximately 53% of the putatively male-specific reads identified by the k-mer analysis mapped to chromosome 25 of the reference genome of an A. chinensis female (Huang et al., 2013) (Figure 2B). Next, we identified single-nucleotide polymorphisms (SNPs) within each of these contigs and characterized how they segregated with sex in our population. Integration of these segregation results with the previous fine genetic mapping of the sex-determining locus (Zhang et al., 2015) indicated that, among the Y-allelic sex-linked 3134 seed contigs that showed significant similarity to the reference chromosome 25 (i.e., the X chromosome) (Figure 2A), only 25 were fully sex-linked. Another 3109 were partially sex-linked (see Supplemental Figure 3 for the definition used in this study and Supplemental Figures 4A and 4B for the fine mapping). On the other hand, an additional 249 contigs were Y-specific (Figure 2A), i.e., they showed no similarity to the X chromosome and female reads did not map to these contigs well, except to the repetitive regions (see Supplemental Figure 3 for the definition used in this study). They also exhibited perfect cosegregation with the plants’ sex, consistent with being located in the male specific region of the Y chromosome, MSY. Together, these 249 contigs accounted for ∼0.49 Mb, suggesting that the MSY region in Actinidia might be a small region flanked by recombining regions.
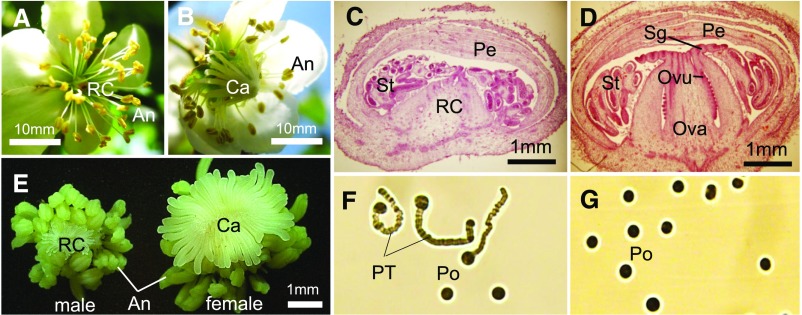
Figure 1.
Characteristics of Flower Sexuality in the KE Population.
(A) and (B) Male (A) and female (B) opened flowers from individuals in the KE population. An, anthers; Ca, carpel; RC, rudimentary carpel.
(C) and (D) Cross sections of male (C) and female (D) flowers in stage 1 (Supplemental Figure 1). Ovu, ovule; Pe, petal; Sg, stigma; St, stamens.
(E) Dissected anthers and carpels in male and female flowers in stage 1.
(F) and (G) Germination of pollen tubes from male (F) and female (G) flowers. Po, pollen grains; PT, pollen tube. These flower phenotypes are consistent within those previously observed in the genus Actinidia.
Figure 2.
Extraction of Male-Specific Sequences for Sex Determinant Discovery.
(A) Distribution of k-mers in genomic and/or transcriptome sequencing reads from the KE population and screening procedure for contigs located on partially or fully sex-linked region and expressed in developing flowers. FSK, female-specific k-mers.
(B) Mapping of reads including MSKs to the draft genome of cv Hong Yang (female, 2n = 2x = 2A + XX = 58); ∼53.7% of reads mapped to chromosome 25 (X chromosome of A. chinensis).
(C) and (D) Distribution of net divergence (C) and silent divergence (D) values between the X- and Y-allelic sequences of genes located within the PAR on chromosome 25 (blue circles). White circles represent interspecific net divergence values between A. chinensis and A. rufa for each gene. The solid black line within the red rectangle and the black dotted line within the blue rectangle represent the average net divergence (C) and average silent divergence (D) rates, respectively, with standard errors, within 20 putative single-copy genes in the Euasterids genome (Wu et al., 2006, Supplemental Table 3), in A. chinensis versus A. polygama and versus A. arguta, respectively. Only the genes expressed significantly in developing flowers (RPKM > 2.0) are included. Gray and green areas represent hypothetical fully sex-linked regions based on our segregation test and a previous report (Zhang et al., 2015) (see Supplemental Figure 4 for details).
Characterization of Y-Encoded Candidate Sex-Determining Genes
Actinidia exhibits cryptic dioecy, where male flowers produce fertile pollen grains and rudimentary carpels, while female flowers produce deceptively normal anthers and nonfunctional pollen grains, presumably because of tapetum (or middle layer) degradation (Supplemental Figure 1; Falasca et al., 2010, 2013). Specifically, sex-specific differences in gynoecium development occur and can be observed early in flower development (defined as stage 1; Supplemental Figure 1). Differential development of the androecium, on the other hand, occurs later, after microspore maturation (defined as stage 2-3; Supplemental Figure 1). To identify candidate sex-determining genes, we conducted transcriptome analyses on 17 male and 17 female young developing flowers sampled at stage 1 (Supplemental Figure 1) from the F1 population described above and searched for differentially expressed genes. Because it required targeting a completely different developmental stage, we did not search for genes involved in the differential development of androecia at this point.
To search for candidate sex determinants, we integrated two complementary approaches: (1) we obtained Illumina mRNA-seq reads from each sample and mapped them to the Y-allelic sex-linked and Y-specific genomic contigs (Figure 2A); and (2) we seeded an assembly using reads that included the putatively male-specific 35-mers identified separately from the cDNA reads (Supplemental Figure 5). These analyses together generated 57 contigs, of which five were located on the Y-allelic fully sex-linked region (Supplemental Figures 4E and 6A and Supplemental Data Set 1A) and two that were Y-specific contigs (Supplemental Figure 6B and Supplemental Data Set 1B). To further check for the presence of potentially expressed genes located within the Y-specific genomic contigs, Illumina mRNA reads were mapped to the 61 genes predicted by AUGUSTUS (Stanke et al., 2004) in the Y-specific contigs. The results were consistent, i.e., the same two genes were expressed. These were annotated as expressed male-specific genes (Supplemental Data Set 2). Despite the fact that they did not exhibit any substantial expression in carpel, the remaining 59 putative genes remain putative candidate genes contributing to sex determination, as discussed later.
The five transcripts in the Y-allelic fully sex-linked contigs did not include any conserved male-specific polymorphisms across A. chinensis, A. rufa, A. polygama, and A. arguta, which would be expected from a potential sex determinant conserved within the Actinidia genus. To further assess whether these contigs could carry sex-determining sequences, we compared nucleotide polymorphisms and divergence between putative X- and Y-alleles (when applicable) and between pairs of Actinidia species for the 57 transcripts located in the Y-allelic sex-linked contigs. Since dioecy is expected to be ancestral to the divergence of Actinidia species, X and Y alleles of candidate sex determining factors are expected to have diverged earlier. All 57 Y-allelic sex-linked genes, including the five fully sex-linked transcripts, exhibited substantially lower divergence (raw silent site divergence, synonymous substitution ratio [Ks], and net divergence) between A. chinensis X-and Y-alleles than between putatively X-allelic sequences of A. chinensis and A. rufa, A. arguta, or A. polygama (Figures 2C and 2D; Supplemental Figure 4E, Supplemental Table 1, and Supplemental Data Set 1A). This result suggests that these X and Y sequences continued to recombine until after the inferred time of the origin of dioecy, making them unlikely candidate sex determinants.
Of the two fully Y-specific contigs (Supplemental Data Sets 1B and 2), one encodes a type-C cytokinin response regulator (Gupta and Rashotte, 2012). We named this gene Shy Girl (SyGI) based on its function (see below). Sequence analysis (see Methods) suggests that this gene originated from a duplication within the Actinidia or Actinidiaceae lineage, ∼20 million years ago (MYA) (Figure 3A; Supplemental Figure 7), with an estimated rate of 2.81 × 10−9 substitutions per synonymous site per year (Shi et al., 2010). Site- and branch-specific tests did not provide any evidence for positive selection after the duplication event for either SyGI or its autosomal counterpart (Achn384741) (Figure 3A), suggesting that the two duplicates might have retained the same protein function. However, their expression patterns differed substantially (Figure 3B). SyGI is expressed in developing flowers, specifically in the surface of rudimentary carpels of male flowers (Supplemental Figure 8), whereas the putative progenitor, Achn384741, is not expressed in any flower organs of either sex but is expressed in young leaves (Figure 3B). The presence of SyGI sequences are also restricted to males in all other Actinidia species tested (Figure 3C), where it was expressed in developing flowers (Figure 3D). The association between sex and the presence of the SyGI sequence was fully conserved, even in polyploid species (Figure 3E), suggesting that the duplication predated the divergence of these species. The other Y-specific contig, Y-encoded FT-like gene (YFT), was expressed faintly in gynoecia (reads per kilobase and million reads [RPKM] = ∼1.0), but its presence was not fully correlated with maleness across all Actinidia species studied (Supplemental Figure 9A). Furthermore, ectopic expression of YFT did not induce any phenotypic change in flower structures (Supplemental Figures 9B and 9C and Supplemental Table 2). In summary, while we cannot exclude the possibility that YFT has acquired a female suppressing function independently in each Actinidia species, these results suggest that SyGI best fits the expected criteria of a likely candidate sex determinant.
Figure 3.
Characterization of the SyGI Gene.
(A) Phylogenetic tree and signs of selective pressure on SyGI-like type-C response regulators (RR) in representative eudicot species. The CRE1-AHK4 histidine kinase, defined as a type-A response regulator, was used as the outgroup. The SyGI-Achn384741 duplication event is estimated to have arisen ∼20 MYA, which postdates the divergence of Actinidiaceae (Ellison et al., 2012).
(B) Expression pattern of SyGI and Achn384741 in flower organs of a male individual from the KE population. SyGI exhibits specific expression in rudimentary carpels. We used rudimentary carpels and normal stamens at stages 1 and 2-3, respectively, which correspond to the earliest stage at which male and female developing structures can be visibly differentiated (Supplemental Figure 1).
(C) Complete male-specific conservation of SyGI in the genomes of a variety of Actinidia species. A. deliciosa is hexaploid, and tetraploid A. arguta and A. macrosperma are shown.
(D) Expression of SyGI and Achn384741 in developing flowers from a variety of Actinidia species. In males, PCR primers detecting both SyGI and Achn384741 only amplified SyGI (see Methods). M, male; F, female.
(E) In higher-ploidy variants of A. arguta, sexuality also cosegregated with the existence of SyGI.
Functional Analysis of SyGI
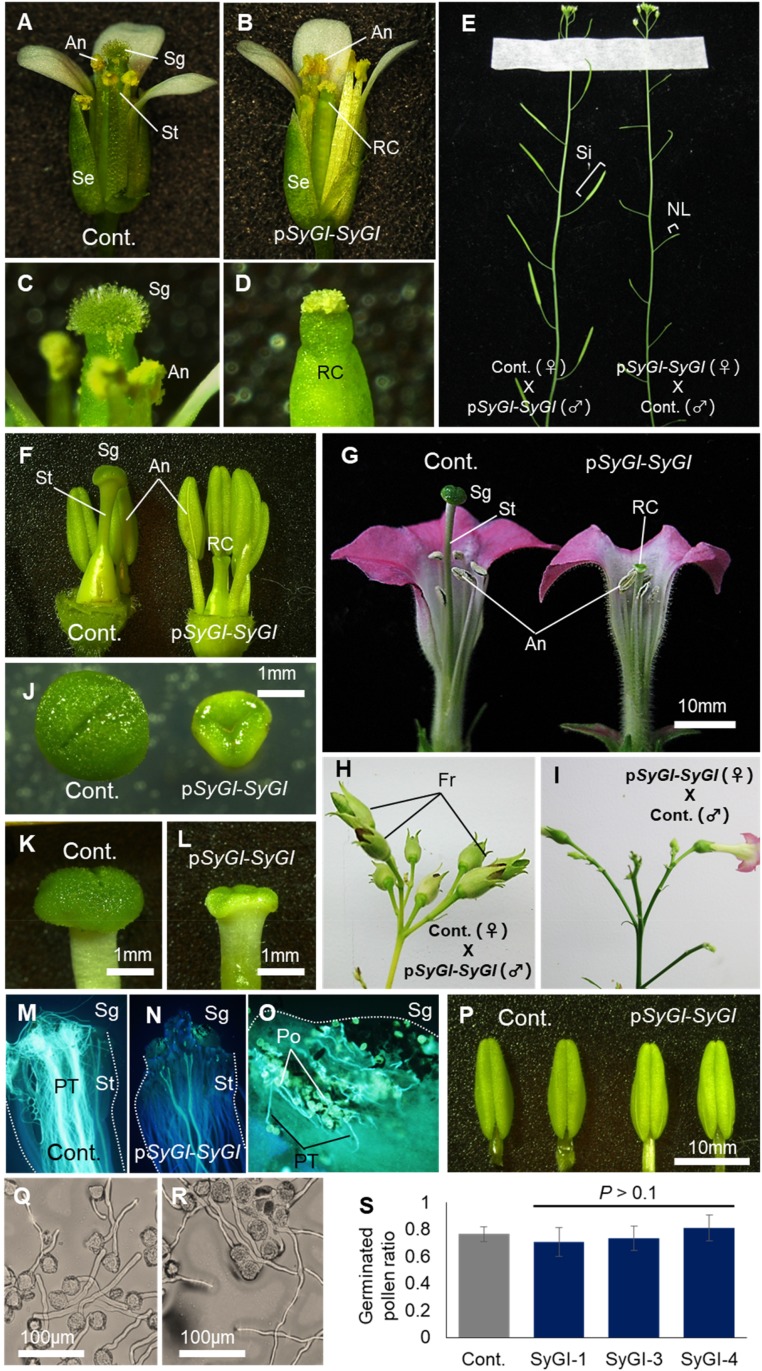
In transgenic kiwifruit, analysis of flower phenotypes is extremely lengthy. Therefore, to validate the function of SyGI, we tested our candidate gene in transgenic lines of two distantly related model plants: Arabidopsis thaliana and Nicotiana tabacum. The transgenic Arabidopsis line expressing SyGI under the control of its native promoter (pSyGI-SyGI) exhibited suppressed carpel development, especially during the stigma elongation stage (Figures 4A to 4E; Supplemental Table 3). The success of reciprocal crosses between wild-type and pSyGI-SyGI plants suggested that SyGI can induce female sterility without impairing male fertility, consistent with its specific expression in male gynoecia in kiwifruit developing flowers. Transgenic N. tabacum lines expressing the same construct (pSyGI-SyGI) produced flowers with rudimentary carpels, sometimes accompanied with pleiotropic effects on other organs, such as altered leaf shape (Figures 4F to 4L; Supplemental Figures 10A to 10M and Supplemental Table 4A). Similar to the observations in Arabidopsis, reciprocal crosses between wild-type and pSyGI-SyGI lines in N. tabacum (Figures 4H and 4I; Supplemental Figure 10R) supported the idea that SyGI acts as a suppressor of female function. In pSyGI-SyGI lines, stigmas were structurally disrupted and wild-type pollen tubes were arrested in the stigma (Figures 4M and 4N; Supplemental Figure 11A). Styles and ovules of pSyGI-SyGI flowers looked structurally similar to those of control plants, including in terms of the cell size (Supplemental Figures 10D to 10K), although these organs were substantially smaller than those of control plants, suggesting reduced cell division rate or pattern. Taken together, these data suggest that SyGI might repress female function by disrupting the ability of the stigma to promote pollen tube growth, possibly via altered cell patterns. In terms of male function, consistent with the observations in Arabidopsis, the pSyGI-SyGI lines exhibited no substantial change in anther structure or pollen germination rates (Figures 4P to 4S; Supplemental Figures 11B and 11C). The SyGI transgene in the N. tabacum pSyGI-SyGI lines showed a peak of expression in young gynoecia, but no expression was detected in androecia or in the other flower organs (Supplemental Figure 12), consistent with and potentially responsible for the gynoecia-specific function of SyGI. Additionally, the expression pattern of SyGI in N. tabacum vegetative organs was slightly different from its endogenous expression in kiwifruit (Supplemental Figure 12). N. tabacum transgenic lines expressing SyGI under the control of the CaMV35S promoter occasionally exhibited inhibited carpel growth (Supplemental Figures 9N to 9R and Supplemental Table 4B), with 3 of the 10 transgenic lines showing irregular carpel development. Inhibition of carpel growth was also induced by ectopic expression of the autosomal copy of the gene Achn384741 controlled by the CaMV35S promoter (Supplemental Figures 9O and 9R and Supplemental Table 4B), where irregularities were found in 5 of the 23 transgenic lines. These results suggest that SyGI and Achn384741 bear similar functions, at least during gynoecium development, which is consistent with the absence of detectable positive selection on their sequences. One of the p35S-SyGI lines exhibited dwarfism (Supplemental Figure 9Q). While it is possible that this is due to insertional effects of the transgene in this particular line, it is consistent with the phenotype of Arabidopsis overexpressing ARR22 (Kiba et al., 2004), an ortholog of SyGI (Supplemental Figure 7), and could further support the hypothesis the protein functions of SyGI and its orthologs/paralogs are conserved.
Figure 4.
Functional Validation of SyGI.
(A) to (D) Phenotypic characterization of representative androgenized Arabidopsis flowers expressing SyGI under the control of its native promoter (pSyGI-SyGI). Compared with control plants ([A] and [C]), mature SyGI-induced plants ([B] and [D]) exhibited rudimentary carpels (RC) with short style (St) and shrunken stigma (Sg), although other organs were unaffected. An, anthers; Se, sepals.
(E) Reciprocal crossing between control and pSyGI-SyGI plants. pSyGI-SyGI plants could function as paternal parent but were sterile as maternal parent. Si, siliques; NL, nonfertile legume.
(F) to (I) Representative androgenized phenotypes in tobacco flowers expressing SyGI under the control of its native promoter. As in Arabidopsis, pSyGI-SyGI lines exhibited rudimentary carpels, although other flower organs were unaffected ([F], 1 week before flowering; [G], mature flower). Reciprocal crossing of control and pSyGI-SyGI lines supported the idea that expression of SyGI leads to repression of female function only ([H] and [I]). Fr, fruits.
(J) to (L) Comparison of stigmas of mature flower from control and pSyGI-SyGI transgenic plants. The observed phenotypes were consistent with pSyGI-SyGI Arabidopsis lines and male Actinidia species.
(M) to (S) aniline blue staining of wild type pollen tubes after pollination to control (M) and pSyGI-SyGI pistils ([N] and [O]). Pollen tubes were arrested in the stigma of pSyGI-SyGI lines ([N] and [O]). Anthers from the pSyGI-SyGI lines were not distinguishable from anthers from control plants (P). Pollen from control (Q) and pSyGI-SyGI (R) flowers germinated normally and showed no significant difference in germination ratio (S).
More in-depth mRNA-Seq analyses of the transcriptome data from young developing A. chinensis flowers supported the idea that SyGI functions as a type-C cytokinin response regulator. Specifically, we identified 223 genes differentially expressed between males and females developing flowers (stage 1) (false discovery rate < 0.01, RPKM > 0.5; Supplemental Data Set 3). Genes in the CLAVATA-WUSCHEL (CLV-WUS) and CUP-SHAPED COTYLEDON-SHOOT MERISTEMLESS (CUC-STM) pathways, both of which contribute to carpel development via cytokinin signaling in Arabidopsis floral meristems (Argueso et al., 2010), were expressed at higher levels in A. chinensis female buds compared with male buds (Supplemental Figure 13). Consistent with this possibility, application of exogenous synthetic cytokinin partially restored gynoecia development in male kiwifruit, with no significant effect on male function (Figure 5; Supplemental Figures 14C, 14F, and 14G), whereas it did not affect gynoecia development in female kiwifruit (Supplemental Figures 14A, 14B, 14D, and 14E). Interestingly, cytokinin has been shown to promote carpel development in several species, seemingly irrespective of the sex determination system (Marsch-Martínez et al., 2012), but not in all cases (Grant et al., 1994). Therefore, while cytokinin is able to restore gynoecia function, it might not reflect complementation of the SyGI function per se.
Figure 5.
Synthetic Cytokinin (CPPU) Treatment Induced Hermaphroditism in Male Actinidia.
Treatment of male individuals from the KE population (A. rufa × A. chinensis) with CPPU at 50 ppm. CPPU-treated male flowers showed incompletely restored stigma (Sg) development (A), while control male plants produced completely rudimentary stigma (RS) (B). An, anthers. Stigma of male flowers (shorter than those of female flowers). Swollen ovary (Ova) of CPPU-treated male flowers, with no ability to bear fertile seeds (C). In A. deliciosa, 50 ppm CPPU treatment induced the formation of small fruits (D) containing ovule-like structures (Ovu) (E).
Evolution of SyGI and Acquisition of Its Specific Expression Pattern
To investigate if the function of SyGI is specific to Actinidia, we compared expression level of the orthologs of SyGI in young developing flowers and carpels from other species, with particular focus on the order Ericales, to which Actinidia belongs. The putative orthologous sequences of SyGI/Achn384741 were identified from the D. lotus (Caucasian persimmon) and Vaccinium corymbosum (blueberry) draft genome sequences (T. Akagi, K. Shirasawa, H. Nagasaki, H. Hirakawa, R. Tao, L. Comai, and I.M. Henry, unpublished results for D. lotus, and Gupta et al. [2015] for V. corymbosum), using BLASTX. While SyGI clearly exhibits a carpel-enriched expression pattern, no substantial other type-C cytokinin RRs from the SyGI family (including Achn384741) are expressed in carpels in other species (Figure 6). This result suggests that SyGI acquired gynoecium-specific expression after the Actinidia-specific duplication event. The silent site divergence estimates for SyGI-Achn384741 across a wide variety of Actinidia species (Ks = 0.1099) and between the duplicates SyGI and Achn384741 (Ks = 0.1108) are almost identical (Figure 7A), indicating that differences in expression patterns between SyGI and Achn384741 might have arisen simultaneously to or very rapidly after the duplication event.
Figure 6.
Evolution of Type-C Response Regulators in Angiosperm and Specific Expression Pattern of SyGI.
Expression patterns of the orthologs/paralogs of SyGI gene in developing flowers (Fl) and in developing carpels (Ca) in D. lotus (Caucasian persimmon), V. corymbosum (blueberry), and A. chinensis (kiwifruit), all members of the Ericales order. SyGI was the only gene to exhibit strong expression in the carpels, suggesting that the ancestral gene was not expressed in the carpel and that SyGI acquired carpel (pistil) or flower expression specifically. The black circle labeled “d” indicates the node representing the SyGI-Achn384741 duplication.
Figure 7.
Evolution of SyGI via Actinidia-Specific Duplication.
(A) Evolutionary relationship between the Shy Girl-Achn384741 duplication event and the timing of divergence between Actinidia species. The black circles labeled “d” and “a” indicate the node of SyGI-Achn384741 duplication and the divergence between Actinidia species, respectively. The white circle labeled “c” indicates the hypothetical timing of the evolution of cis-elements in the Shy Girl promoter sequence. The rate of silent divergence from the current SyGI to the SyGI-Achn384741 duplication (Ks = 0.1108) is only slightly bigger than to the divergence of Actinidia species (Ks = 0.1099). The estimated time between the two (Ks = 0.0009) corresponds to ∼0.16 million years (MY), suggesting very rapid fixation of the proto-SyGI after the hypothetical evolution of the cis-element.
(B) Model for the establishment of SyGI function as a dominant suppressor of feminization. According to the two-mutation model, a male-promoting factor (M/m) could be present in the same haploblock, and the male-sterility mutation might have predated the establishment of duplicated SyGI.
DISCUSSION
In this study, we attempted to uncover the mechanisms underlying the developmental processes resulting in sex determination in dioecious kiwifruit. To that end, we capitalized on a population of interspecific siblings produced by crossing A. rufa and A. chinensis, partly because it has the advantage that all male individuals share the same Y-specific sequences, originating from the A. chinensis male parent, while X allelic sequences from both species were available for detection of the genetic diversity across species. We started by using both genomic and transcriptome data to identify and assemble male-specific sequences (Figure 2). Expressed loci within these sequences were identified based on transcriptome data and only a few loci remained that were fully linked to sexuality. Of those, one Y-specific locus encoding a type-C cytokinin response regulator, which was called SyGI, stood out as being fully male-specific and expressed in developing male flowers, specifically in developing carpels (Figure 3). These data are consistent with SyGI playing the role of a female-suppressor gene in this system. Consistent with this hypothesis, transgenic Arabidopsis and N. tabacum expressing SyGI under its native promoter exhibited underdeveloped carpels (Figure 4).
Previous results had demonstrated that ectopic expression of one type-C cytokinin response regulator, ARR22, in Arabidopsis reduced cytokinin signaling, resulting in the downregulation of genes in the CLV-WUS and CUC-STM pathways (Kiba et al., 2004; Horák et al., 2008). Importantly, the same pathways were downregulated in male kiwifruits (Supplemental Figure 12). In S. latifolia, these pathways were shown to be upregulated in a bisexual mutant that was putatively derived from a SuF-disrupted male, compared with the progenitor non-mutant male, suggesting that the Y-encoded SuF regulates these pathways during the repression of carpel development (Koizumi et al., 2010). Furthermore, in monoecious Oriental persimmon (Diospyros kaki) and in a dioecious wild grapevine (Vitis amurensis), cytokinin signaling can promote carpel development in male flowers (Yonemori et al., 1991; Wang et al., 2013), although cytokinin does not always promote feminization in dioecious plants (Grant et al., 1994). These results suggest that negative regulation of cytokinin signaling by SyGI may therefore inhibit gynoecial development in kiwifruit and that the cytokinin signaling pathways affecting gynoecium development might be a conserved element involved in sexual dimorphism in some dioecious species, despite the fact that the specific mechanism leading to the regulation of the cytokinin pathway might have independent origins and different genetic determinants.
Phylogenetic analyses of SyGI and related sequences within Actinidia (Figure 7A) suggest that SyGI originated from an Actinidia-specific gene duplication event, followed by neofunctionalization, in the form of the acquisition of an expression pattern specific to the developing gynoecium of male flowers (Figures 6 and 7). This suggests that SyGI might play the role of a gain-of-function dominant female-suppressing gene (SuF), the one proposed in the two-mutation model of the evolution of dioecy (Charlesworth and Charlesworth, 1978). Because this duplication results in the production of male flowers but does not affect male fertility, we hypothesize that another mutation causing male sterility also occurred during the evolution of dioecy in Actinidia, generating a male-promoting factor (M), as proposed in the two-mutation model (Charlesworth, 2013, 2015). The two-mutation model would be also supported in this case by the existence of genetically male kiwifruit that bear hermaphroditic flowers, as could be expected from individuals carrying M but not SuF (McNeilage, 1991). If these occasional hermaphrodites originate from recombination between the two sex-determining genes on the MSY region, this would suggest that recombination is not completely suppressed between those loci. Alternatively, it is possible that these individuals bear mutated nonfunctional SuF. No candidate M factor was found in this study, possibly because our transcriptomic analyses only covered the very early flower developmental stages, while the M factor in Actinidia is likely to be expressed in tapetum tissue or surrounding tissues in later developmental stages (Falasca et al., 2010, 2013; Supplemental Figure 1). Analysis of transcriptome data from anthers in stage 2-3, combined with the 61 candidate genes predicted from the Y-specific genomic sequences (Supplemental Table 2), might allow for the identification of the M factor in the future. Hermaphrodite and/or neuter A. deliciosa individuals (McNeilage, 1997) can also provide effective resources to identify the M factor.
In D. lotus (diploid persimmon), also a member of the order Ericales, sex determination involves a Y-encoded small-RNA, OGI, which is the dominant suppressor of the feminizing gene, MeGI (Akagi et al., 2014). While the system described here hinges on different mechanisms and involves factors bearing different functions, one aspect of the evolution of dioecy is common to both systems: a genus-specific duplication event. Specifically, OGI is a product of a Diospyros-specific gene duplication event of the autosomal gene MeGI (Akagi et al., 2014), while SyGI originates from an Actinidia-specific duplication. Interestingly, in Asparagus, the dominant female-suppressing gene, SOFF, is thought to be derived from a lineage-specific gene duplication as well (Harkess et al., 2017). These similarities, in Actinidia, Diospyros, and Asparagus, imply that the establishment of the dominant female suppressor is potentially facilitated by lineage-specific duplication events, potentially more permissive of gain-of-function mutations. Plants have frequently undergone whole genome duplication or paleoploidization events, which can lead to various gene neofunctionalization (Tomato Genome Consortium, 2012; Van de Peer et al., 2017). The timing of the duplication deriving SyGI and Achn384741 (∼20 MYA, Ks = ∼0.11) could correspond to the timing of one of the two Actinidia-specific paleoploidization, called Ad-α (Huang et al., 2013), although a more in-depth synteny analysis between the Y chromosome and autosomes would be indispensable to validate this relationship. This might support the observation that lineage-specific variation in sexual system, especially the transition into dioecy (or dimorphic sex), is associated with frequent (paleo)polyploidization (or large-scale gene duplications) (Ashman et al., 2013; Goldberg et al., 2017). Potentially consistent with this concept, in Fragaria, repeated translocation of a short gene cassette (∼13 kb), is thought to be involved in the establishment of at least three independent sex chromosomes within the species (Tennessen et al., 2017). Our interpretation that lineage-specific duplication is a driving force to derive a sex determinant sheds light on a potential evolutionary pathway to the dominant suppressing factor (SuF) proposed in the two-mutation model. It is also consistent with the idea that, while different plant lineages have evolved dioecy independently, they are using similar evolutionary tools.
METHODS
Plant Materials
Trees from the KE population (Actinidia rufa × Actinidia chinensis) planted in the experimental orchard of the Kagawa University Graduate School of Agriculture (Miki, Kagawa, 34°16′, 134°7′ L/L) (Supplemental Table 5) were used for the Illumina sequencing analyses. A total of 44 trees (20 females and 22 males from the KE population and their parents) were used for MSK identification. To define the developing stages (Supplemental Figure 1), we collected flower buds until anthesis, in April and May 2015 and 2016. For expression profiling by mRNA-seq analysis, developing flowers from 17 of male and 17 female individuals were harvested on April 11, 2015, during the early differentiation stage of the gynoecia (stage 1; Supplemental Figure 1). Flower organs from trees in the KE population were also collected for expression analysis by RT-PCR on 4th and 22nd of April 2016, which corresponded to stage 1 and stage 2-3 of the differentiation of gynoecia and androecia, respectively. For assessment of the candidate gene sequence variation and genetic diversity within Actinidia, male and female individuals from A. chinensis, A. deliciosa, A. rufa, A. hypoleuca, A. polygama, A. kolomikta, and A. argute, and a female individual from A. macrosperma, were used. Information about these Actinidia individuals is summarized in Supplemental Table 5.
Illumina Library Construction and Sequencing
Genomic Libraries
Genomic DNA was extracted from young leaves using Genome-tip 20/G or 100/G (Qiagen), sometimes followed by purification using phenol/chloroform extraction. Approximately 1.5 μg of genomic DNA was used for the construction of each Illumina genomic libraries; the DNA was fragmented using NEBNext dsDNA Fragmentase (New England Biolabs [NEB]) for 40 to 60 min at 37°C and cleaned using Agencourt AMPure XP (Beckman Coulter Genomics) for size selection. To select fragments ranging between 300 and 600 bp, 27 μL of AMPure was added to the 63-μL reaction. After a brief incubation at room temperature, 90 μL of the supernatant was transferred to a new tube and 20 μL water and 30 μL AMPure were added. After a second brief incubation at room temperature, the supernatant was discarded and the DNA was eluted from the beads in 20 μL of water, as recommended. Next, DNA fragments were subjected to end repair using NEB’s End Repair Module Enzyme Mix, and A-base overhangs were added with Klenow (NEB), as recommended by the manufacturer. A-base addition was followed by AMPure cleanup using 1.8:1 (v/v) AMPure reaction. Barcoded NEXTflex adaptors (Bioo Scientific) were ligated at room temperature using NEB Quick Ligase (NEB) following the manufacturer’s recommendations. To remove contamination of self-ligated adapter dimers, libraries were size-selected using AMPure in 0.8:1 (v/v) AMPure:reaction volume to select for adapter-ligated DNA fragments at least 400-bp long. Half of the eluted DNA was enriched by PCR reaction using Prime STAR Max (Takara) at the following PCR conditions: 30 s at 98°C, 10 cycles of 10 s at 98°C, 30 s at 65°C, and 30 s at 72°C, and a final extension step of 5 min at 72°C. Enriched libraries were purified with AMPure (0.7:1 [v/v] AMPure to reaction volume), and quality and quantity were assessed using the Agilent BioAnalyzer (Agilent Technologies) and Qubit fluorometer (Invitrogen). Libraries were sequenced using Illumina’s HiSeq 2500 or HiSeq 4000 (150-bp paired-end reads), according to the manufacturer’s instructions.
mRNA Libraries
Young flower buds and carpel samples corresponding to the early differentiation stages of male or female primordia (stage 1) were harvested on April 10, 2015 and April 5, 2016, respectively. Total RNA was extracted using the Plant RNA Reagent (Invitrogen) and purified by phenol/chloroform extraction. Five to ten micrograms of total RNA were processed in preparation for Illumina Sequencing, according to a previous report (Akagi et al., 2014). In brief, mRNA was purified using the Dynabeads mRNA purification kit (Life Technologies). Next, cDNA was synthesized via random priming using Superscript III (Life Technologies) followed by heat inactivation for 5 min at 65°C. Second-strand cDNA was synthesized using the second-strand buffer (200 mM Tris-HCl, pH 7.0, 22 mM MgCl2, and 425 mM KCl), DNA polymerase I (NEB), and RNaseH (NEB) with incubation at 16°C for 2.5 h. Double-stranded cDNA was purified using AMPure with a 0.7:1 (v/v) AMPure to reaction volume ratio. The resulting double-stranded cDNA was subjected to fragmentation and library construction, as described above, for genomic library preparation. Ten cycles of PCR enrichment were performed using the method described above. The constructed libraries were sequenced on Illumina’s HiSeq 4000 sequencer (150-bp paired-end reads).
All Illumina sequencing was conducted at the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, and the raw sequencing reads were processed using custom Python scripts developed in the Comai laboratory and available online (http://comailab.genomecenter.ucdavis.edu/index.php/Barcoded_data_preparation_tools), as previously described (Akagi et al., 2014). In brief, reads were split based on index information and trimmed for quality (average Phred sequence quality > 20 over a 5-bp sliding window) and adaptor sequence contamination. A read length cutoff of 35 bp was applied to mRNA reads. All samples used to generate Illumina sequences are listed in Supplemental Table 6.
Screening of the Expressed Candidate Sex Determinants
Cataloguing of k-mers and Construction of Polymorphic Contigs
Sex-specific or sex-biased sequences and polymorphisms were identified using custom python scripts (https://github.com/Comai-Lab/kmer-extract-by-trigger-site), as described previously (Akagi et al., 2014). In brief, all 30-bp k-mers (words) starting with “A” were selected from all genomic reads, whereas for the mRNA-seq data, all 35-bp k-mers starting with “A” were collected. The set of k-mers that met a minimum total count threshold of 20 were retained. To identify the sex-specific/biased k-mers, k-mer counts were compared between male and female reads. Fully male-specific k-mers (MSKs), which were completely absent in the female reads, were identified and used to select all pair-ended reads containing at least one of the them. Enrichment of MSKs was examined with Fisher’s exact test in R, comparing the numbers of MSKs and female-specific k-mers against the expected numbers of k-mers randomly extracted from the male and female reads.
De novo assembly was performed using the CLC assembler and all 150-bp paired-end reads that included at least one MSK (Figure 2A; Supplemental Figure 2). In the gRNA-seq and mRNA-seq analyses, 4143 and 1376 seed contigs were generated, respectively. To assess linkage to the sex determinants, all genomic reads from each individual of the KE population were aligned to these contigs using the Burrows-Wheeler Aligner (BWA) (Li and Durbin, 2009) and default parameters (http://bio-bwa.sourceforge.net/) or allowing up to 12-nucleotide (∼8%) mismatches. The number of reads mapping to each reference sequences was recorded from the alignment file produced by the sequence alignment/map (SAM) tool (Li et al., 2009) (http://samtools.sourceforge.net/). Next, informative polymorphisms were identified with custom Python scripts (http://comailab.genomecenter.ucdavis.edu/index.php/Mpileup). For SNPs and short indels, only those observed in multiple individuals (n > 3) were defined as informative.
The paired-end reads including MSKs were also mapped to the kiwifruit (A. chinensis, cv Hong Yang, female) reference genome sequence (Huang et al., 2013) (http://bioinfo.bti.cornell.edu/cgi-bin/kiwi/home.cgi) with BWA using default parameters (http://bio-bwa.sourceforge.net/). The number of mapped reads per 50-kb bin was recorded using a custom python script (http://comailab.genomecenter.ucdavis.edu/index.php/Bin-by-sam) according to a previous study (Henry et al., 2014). Enrichment of the reads mapped to the X chromosome (Chr. 25) was assessed with Fisher’s exact test in R, by comparing the numbers of reads including MSKs mapped to each chromosome to the total number of reads mapped to each chromosome.
Linkage Test and Definition in the gRNA-Seq-Derived Contigs
Linkage tests were performed in three steps using the informative polymorphisms detected in the mapped reads. First, after default mapping using BWA, and mapping allowing up to 10% nucleotide mismatches, we classified the contigs into four categories: putative Y-specific contigs, putative Y-allelic sex-linked contigs, repetitive contigs, and “other” contigs. Contigs including a >300-bp region to which no female reads mapped were defined as Y-specific. These contigs are expected to be mostly located on the MSY (Supplemental Figure 3). The contigs bearing significant SNPs and/or short indels (<300 bp), and which cosegregated with sexuality in >90% of the KE population, were defined as putative Y-allelic sex-linked contigs. Here, we detected male-specific polymorphisms only in the nucleotide sequences with appropriate mapping coverage (15 < n < 120, expected coverage for Y = ∼32 in pooled male), both in male and female reads. For further verification, the SNP haplotype of each contig was confirmed by visualization of polymorphisms using the Integrative Genomics Viewer (IGV) version 2.3 (Robinson et al., 2011), according to a previous report (Akagi et al., 2014). If a contig exhibited at least two independent Y-specific polymorphisms in pooled female reads, it was labeled as recombined with respect to the sex determination locus. Contigs were labeled “repetitive” when the coverage of mapped reads from the male reads was at least 5-fold higher than the expected coverage for Y (∼32). The other contigs were defined as “other” (or “pending”). Here, we identified 249 Y-specific contigs, 3134 Y-allelic sex-linked contigs, 211 repetitive contigs and 549 other contigs (Supplemental Figure 2).
To further examine the 3134 sex-linked contigs, we assessed recombination in each individual, as previously described (Akagi et al., 2014). Briefly, all informative polymorphisms present in each contig were pooled to derive the X/Y genotype of each individual (XY or XX) in the following way. The haplotype of each contig was manually determined using the IGV. For each contig and each individual, the number of reads containing X- or Y-specific polymorphisms were recorded. Individuals with six or more independent reads containing an X-specific polymorphism, but no read containing Y-specific polymorphisms, were labeled as homozygous XX for that particular contig. If the genotype of each individual in that particular contig was completely consistent with sex in the 42 segregated lines, we defined that as “fully sex-linked contig.” If not, we defined it as “partially sex-linked contig” (Supplemental Figure 3).
Third, to anchor the sex-linked contigs to the reference X chromosome (Chr. 25), the sequences of the Y-allelic sex-linked contigs were subjected to BLASTN homology searches, allowing up to 10% mismatches in 100-bp bin. Next, the anchored contigs were pooled by 100-kb bins in the X chromosome, to define the recombination break points in our population. From the estimated sex determinant locus, the first bin where >70% of the anchored contigs were partially sex-linked was defined as the break points. Finally, we integrated information about the physical location of some markers to locate the previous genetic mapping of the sex determinant on the X chromosome by Zhang et al. (2015) (Supplemental Figure 4).
Identification of the Candidate Expressed Genes
mRNA-seq reads from 17 male and 17 female individuals from the KE population were used to identify the genes substantially expressed in developing flowers. The mRNA 150-bp paired-end reads were fragmented to 50 bp and mapped to the 249 Y-specific and 3134 Y-allelic sex-linked genomic contigs. RPKM values were calculated in the region where transcript reads were mapped. We defined the contigs in which RPKM > 2.0, as “expressed contigs” resulting in 121 and 3 expressed polymorphic contigs from the sex-linked and Y-specific contigs, respectively (Supplemental Figure 6).
For the 1376 mRNA-seq-derived contigs described above, recombination mapping was performed to define a subset of sex-linked contigs. gRNA-seq reads, which were fragmented to 50 bp, were mapped to these contigs to check linkage to the sex determination locus using BWA and default parameters. The informative polymorphisms were visualized using IGV and used for linkage test. Next, we classified the contigs into the same four categories mentioned above for the genomic contigs. RPKM values were calculated in the Y-allelic sex-linked and Y-specific contigs to discard subtly expressed contigs (RPKM < 2.0). Finally, 88 and 4 candidate contigs were retained as expressed Y-allelic sex-linked and Y-specific contigs, respectively (Supplemental Figure 6).
To combine the candidate genes obtained using both methods, the contigs exhibiting >99% nucleotide identity and overlapping over at least 50 bp with each other were defined as belonging to the same gene, as shown in Supplemental Figure 6.
To further identify potential expressed genes within the Y-specific genomic contigs, we used AUGUSTUS (Stanke et al., 2004) for gene prediction from the sequences of the Y-specific genomic contigs, using default parameters and without prediction of alternatively spliced isoforms. The mRNA reads from developing flowers and carpels from four male and four female individuals from the KE population were mapped to the 61 predicted genes using BWA and default parameters. The read counts per gene were generated from the aligned SAM files using a custom R script.
Genetic Diversities of the Allelic Sex-Linked Genes in Actinidia
The Y-allelic sex-linked contigs (Supplemental Data Set 1A) of the candidate genes were aligned to the reference genome sequences of A. chinensis (Huang et al., 2013) to obtain the corresponding X-allelic sequences. To compare X-Y with interspecies divergence, putative X-alleles from A. rufa were also constructed from the Illumina reads. Potential transposable elements identified by BLASTN using the TAIR/nr database were removed, and the coding frame was assigned after integration of alternative splicing and isoforms based on putative functional orthologous sequences in model plants (such as Arabidopsis thaliana, grape [Vitis vinifera], and tomato [Solanum lycopersicum]). The X- and Y-allelic nucleotide sequences were aligned using MAFFT ver. 7 (Katoh and Standley, 2013), with the L-INS-i model and by SeaView ver. 4 for manual pruning. The resulting alignments were subjected to DnaSP 5.1 (Librado and Rozas, 2009) to calculate the number of segregation sites (S), Jukes and Cantor corrected values of synonymous (Ks), nonsynonymous (Ka) substitutions, and the index of evolutionary speed (Ka/Ks ratio) (Supplemental Data Set 1A). To compare the timing of divergence between the X- and Y-alleles and between the X alleles of A. chinensis and A. rufa, we compared Ks value and all-site divergence of each allelic gene pair retaining at least 90 bp of aligned sequence. The timing of divergence between A. chinensis and A. polygama or A. arguta was estimated using Ks and all-site divergence of 20 putatively single-copied genes in these species (Supplemental Table 3). These sequences were aligned using MAFFT ver. 7 under the L-INS-i model followed by manual revision using SeaView ver. 4 and subjected to DnaSP 5.1 to analyze informative SNPs.
For the Y-specific contigs (Supplemental Figure 3) of the candidate genes, conservation of male specificity in the genus Actinidia was examined using male and female individuals from eight Actinidia species (Supplemental Table 5). Genomic DNAs were extracted from young leaves using Genome-tip 20/G or 100/G (Qiagen) followed by purification with phenol/chloroform extraction. Multiple partial regions of the candidates were amplified with primer sets designed from the A. chinensis sequences using PrimerSTAR GXL (TaKaRa). Two of the male-specific candidates, SyGI and YFT, showed high similarity to Achn384741 (Chr. unknown) and Achn077261 (Chr. 24), respectively. Thus, the full-length genomic regions covering the 5′ promoters of these two genes were amplified with multiple primer sets designed from the sequences from A. chinensis and then the amplified PCR products in other Actinidia species were sequenced. For SyGI, we designed an optimized primer set, specifically amplifying the SyGI from conserved in Actinidia species (Supplemental Table 7).
Expression Profiling
The mRNA-seq reads from 17 male and 17 female individuals from the KE population were aligned to the reference sequences of A. chinensis (cv Hong Yang, female) using BWA with default parameters. The read counts per contig were generated from the aligned SAM files using a custom R script. Differential expression between male and female individuals was analyzed in R (version 3.0.1) using the R package DESeq (Anders and Huber, 2010) (version 1.14; http://bioconductor.org/packages/release/bioc/html/DESeq.html). We conducted DESeq analysis using 17 biological replicates from male and female individuals, with the following parameters: method=‘per-condition’ and sharingMode=‘gene-est-only’. A false discovery rate threshold of 0.01 was used to identify differentially expressed genes.
Total RNA was also extracted from each flower organ to assess the specific expression pattern by RT-PCR. Carpels at stage 1, anthers at stage 2-3, and petals and sepals at stage 2-3 were collected on April 4th and April 22nd from the KE population and on April 22nd and May 12th from A. polygama and A. arguta, respectively. cDNA was synthesized from 500 ng of total RNA using the ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo), and 4-fold diluted cDNA was used as a template for RT-PCR analysis on SyGI and Achn384741 (Supplemental Table 7). Gene-specific amplification was confirmed by direct sequencing of the amplicons. For the reference gene, primer pairs specific to a gene encoding an abundant actin (Achn107351) were used.
To compare the expression patterns of type-C cytokinin RRs nested in the SyGI/Achn384741 family within the Ericales, the orthologous sequences of SyGI/Achn384741 from Diospyros lotus (Caucasian persimmon) and Vaccinium corymbosum (blueberry) were identified from the draft genome sequences (T. Akagi, K. Shirasawa, H. Nagasaki, H. Hirakawa, R. Tao, L. Comai, and I.M. Henry, unpublished results for D. lotus, and Gupta et al. [2015], https://www.vaccinium.org/tools/blast, for V. corymbosum). Developing carpels and flowers were sampled from D. lotus cv Kunsenshi (female) and V. corymbosum cv O’Neil at a stage corresponding to stage 1 of Actinidia development. These samples were subjected to RNA extraction using Plant RNA Reagent. cDNA was synthesized from 500 ng of total RNA using the ReverTra Ace qPCR RT Master Mix with gDNA Remover and 4-fold diluted cDNA was used as a template for RT-PCR analysis (Supplemental Table 7). For the reference genes, primer pairs specific to a gene encoding an abundant actin (DkAct) (Akagi et al., 2009) and ubiquitin (Vashisth et al., 2011) were used for Diospyros and Vaccinium, respectively.
In Situ RNA Hybridization
RNA in situ hybridization was performed as previously described (Esumi et al., 2007), with minor modifications. Young flower samples, around stage 1, were fixed in FAA (1.8% formaldehyde, 5% acetic acid, and 50% ethanol), subsequently displaced by a 10 to 30% sucrose solution series before being sliced by a cryostat (Leica CM1520) using Cryofilm, according to Kawamoto’s film method (Kawamoto, 2003). The tissues were sliced into ∼10-μm sections and mounted on Frontier-coated glass slides (Matsunami Glass Ind.). The tissue sections were rehydrated in an ethanol series and incubated in a Proteinase K solution (700 units/mL Proteinase K, 50 mM EDTA, and 0.1 M Tris-HCl, pH 7.5) for 30 min at 37°C, followed by acetylation with acetic anhydride (0.25% acetic anhydride in 0.1 M triethanolamine solution) for 10 min. Full-length SyGI cDNA sequences were cloned into the pGEM-T Easy vector (Promega) to synthesize DIG-labeled antisense RNA probes using the DIG-labeling RNA synthesis kit (Roche). The probe solution including RNaseOUT (Thermo Fisher Scientific) was applied to the slides and covered with Parafilm. Hybridization was performed at 48°C for >16 h. For detection, 0.1% Anti-Digoxigenin-AP Fab fragments (Sigma-Aldrich) was used as the secondary antibody to stain with NBT/BCIP solutions.
Evolutionary Analysis of SyGI
The putative full-length sequences of SyGI and Achn384741 from A. chinensis, ARR22 and ARR24 from Arabidopsis, and 89 putative type-C cytokinin response regulator proteins with significant sequence similarity to SyGI (e−19 cut off) were identified from the genomes of 30 angiosperms using BLASTp in Phytozome (JGI release version 11.0, https://phytozome.jgi.doe.gov/pz/portal.html). CRE1/AHK4, which encodes a well-known type-A cytokinin response regulator from Arabidopsis, was used as the outgroup gene. Alignment analyses on amino acid sequences were conducted using MAFFT ver. 7 with L-INS-i model, followed by manual revision using SeaView ver. 4. The evolutionary topology was examined using the maximum likelihood method (ML) by Mega v. 5 (Tamura et al., 2011) with WAG+I+G model and 1000 replications for bootstraps (number of discrete gamma categorization = 3). All sites, including missing and gap data, were used for the construction of phylogenic trees, and the nearest neighbor interchange technique was used. Bootstraps were shown on the branches as 1/10 of the calculated values (Supplemental Figure 7). The phylogenetic tree using the entire gene sets included many branches that were not statistically supported, mainly because of the lack of substitutions in the properly aligned region. The orthologous genes in three representative species, grape, tomato, and Arabidopsis, were subjected to in-codon frame alignment using Pal2Nal (Suyama et al., 2006) and MAFFT ver. 7. The aligned nucleotide sequences were used for construction of evolutionary topology using ML method by Mega, with 3-parameter (+I+G) model with 1000 replications for bootstrap. Based on these alignments and topology, the codon-based detection of branch- and site-specific positive selection test was performed using PAML (Yang, 1997). The statistical significance of positive selection on the foreground branches was evaluated using the likelihood ratio test of the null hypothesis that Ka/Ks = 1. Site-specific positive selection was assessed by Empirical Bayes analysis.
For assessment of genetic diversity and selective pressure in SyGI and Achn384741, in the Actinidia genus, the full-length cDNA sequence from SyGI and Achn384741 from various Actinidia species were subjected to in-codon frame alignment by MAFFT ver. 7 and SeaView, followed by a ML approach using Mega v. 6 to construct an evolutionary topology. The ancestral sequences of SyGI was inferred using Mega and GSVIVT0102634009 from V. vinifera, which is the closest ortholog of SyGI/Achn384741 in the registered angiosperm genomes, as the outgroup gene. To estimate the divergence time between SyGI and Achn384741, synonymous substitution ratio (Ks) was calculated using DnaSP v 5.1, as previously reported from Actinidia (Shi et al., 2010). Here, we adopted an estimated rate of 2.81 × 10−9 substitutions per synonymous site per year.
Transformation and Phenotyping of Transgenic Plants
The full lengths of the SyGI and Achn384741 transcript sequences were amplified by PCR using PrimeSTAR Max (TaKaRa) from cDNA from carpel and leaves from A. chinensis sel. FCM1, respectively (Supplemental Table 5). The amplicons were cloned into the pGWB2 vector (Nakagawa et al., 2007) to place the genes under the control of the CaMV35S promoter. The full-length SyGI genomic sequences, including the ∼1500-bp 5′ promoter region, were also amplified from genomic DNA of A. chinensis sel. FCM1 (Supplemental Table 5) to connect to the pPLV2 (De Rybel et al., 2011) vector when the gene is under the control of the inserted native promoter. We constructed pGWB2-SyGI and pGWB2-Achn384741 using the Gateway system (Invitrogen) using pENTR/D-TOPO cloning kit and LR clonase. pPLV2-native promoter (pSyGI)-SyGI was constructed using the In-Fusion Cloning kit (Clontech) and the underlined sequences of the primers listed above as overlap with pPLV2 digested by HpaI.
Arabidopsis ecotype Columbia-0 was grown under white light (400–750 nm) with 16-h-light and 8-h-dark cycles at 21°C until transformation. The binary construct was introduced into Agrobacterium tumefaciens strain GV3101 (pMP90) using the helper vector pSOUP by electroporation. Next, Arabidopsis wild-type plants were transformed using the flower-dipping method, according to the previous protocol (Zhang et al., 2006). Screening of transgenic plants was conducted on Murashige and Skoog media containing 50 μg/mL kanamycin. Tobacco plants (Nicotiana tabacum) cv Petit Havana SR1 were grown in vitro under white light (400–750 nm) with 16-h-light and 8-h-dark cycles at 22°C until transformation. The binary construct was introduced into the Agrobacterium strain EHA101 using the helper vector pSOUP by electroporation. Young petioles and leaves of tobacco plants were transformed by the leaf disk method. Transgenic plants were selected on Murashige and Skoog medium supplemented with 100 μg/mL kanamycin. Pollen tube germination was assessed 3 h after placing the pollen grains on 15% sucrose/0.005% boric acid/1.0% agarose media at 25°C. The pollen germination ratio was counted as average percentages in batches of 200 pollen grains from the first three flowers. Pollen grains exhibiting pollen tubes longer than the length of the grain were considered as “germinated grains.” To investigate pollen tube growth in the style of the transgenic plants, the styles were fixed 48 h after pollination in formalin-acetic-ethanol (1:1:8) and then cleared and softened in a 10 n sodium hydroxide solution. The styles were squashed and pollen tubes were stained in a 0.1% solution of aniline blue dye dissolved in 0.1 n K3PO4.
Treatment of Actinidia Species with Synthetic Cytokinins
Ten parts per million (ppm) or 50 ppm of N-(2-chloro-4-pyridyl)-N′-phenylurea was administered to two male individuals of the KE population (KE-17, 23) at stage 2, which were maintained at Kagawa University, and to two individuals of A. deliciosa cv Tomuri at stage 2-3, which were planted on 20-liter pots in a greenhouse at Kyoto University. Approximately 25 to 50 mL of the solutions was used to directly spray each tree.
Accession Numbers
All sequence data generated in the context of this manuscript have been deposited in the appropriate DDBJ database: Illumina reads for gRNA-seq and mRNA-seq in the Short Read Archive database (DRA accession DRA005439, BioProject ID PRJDB5449) and the Y-specific genomic contigs set derived from MSK (IDs LC218452-LC218700) and the SyGI/Achn384741 promoter/genic sequences in Actinidia species (IDs LC260493-LC260502).
Supplemental Data
Supplemental Figure 1. Development of male and female flowers in the KE population.
Supplemental Figure 2. Pipeline for the identification of Y-specific and Y-allelic sex-linked contigs in the KE population.
Supplemental Figure 3. Definition of the MSY, and Y-allelic sex-linked and Y-specific contigs.
Supplemental Figure 4. Mapping of MSK-derived reads to chromosome 25 (X chromosome) and definition of the fully sex-linked region within the X chromosome.
Supplemental Figure 5. Identification of male-specific k-mers in mRNA-seq reads
Supplemental Figure 6. Integration of the expressed candidate genes identified from MSK-derived contigs using gRNA-seq and mRNA-seq approaches.
Supplemental Figure 7. Evolutionary tree of Shy Girl and Achn384741 orthologs in angiosperms.
Supplemental Figure 8. RNA in situ hybridization using SyGI antisense probes, in male flowers around stage 1.
Supplemental Figure 9. Conservation of male specificity of the YFT across Actinidia species and its functional analysis.
Supplemental Figure 10. Phenotypic variation in pSyGI-SyGI, pCaMV35S-SyGI, and pCaMV35S-Achn384741 N. tabacum transformed lines.
Supplemental Figure 11. Pollen tube growth in the styles of pSyGI-SyGI transgenic lines.
Supplemental Figure 12. Expression patterns of SyGI under the control of its native promoter in N. tabacum.
Supplemental Figure 13. Several cytokinin signaling related genes exhibit higher expression in developing female flowers of Actinidia.
Supplemental Figure 14. Phenotypic variation in Actinidia after treatment with synthetic cytokinin (CPPU).
Supplemental Table 1. Rates of interspecific silent and net divergences between A. chinensis and A. arguta/A. polygama.
Supplemental Table 2. Characterization of the pCaMV35S-YFT and pYFT-YFT in Arabidopsis transformed lines.
Supplemental Table 3. Characterization of the pSyGI-SyGI Arabidopsis transformed lines.
Supplemental Table 4. Characterization of the pSyGI-SyGI, pCaMV35S-SyGI, and pCaMV35S-Achn384741 N. tabacum transformed lines.
Supplemental Table 5. Plant materials.
Supplemental Table 6. Indexes and annotation of the Illumina sequences.
Supplemental Table 7. Primers used in this study.
Supplemental File 1. Text file of the alignment corresponding to the phylogenetic analysis in Supplemental Figure 7.
Supplemental Data Set 1. Annotations of the integrated candidate expressed contigs.
Supplemental Data Set 2. Expression patterns of the genes predicted in the Y-specific genes.
Supplemental Data Set 3. Genes differentially expressed between male and female developing flowers of Actinidia.
Acknowledgments
We thank Luca Comai (UC Davis Department of Plant Biology and Genome Center) for bioinformatics support and Minori Sonoda (Graduate School of Agriculture, Kyoto University) for experimental support. A part of the plant materials of this work were originally provided from Kagawa Prefectural Agricultural Experiment Station. Some of this work was performed at the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley supported by NIH S10 OD018174 Instrumentation Grant. This work was supported by the Japan Science and Technology Agency (PRESTO to T.A.), by the Japan Society for Promotion of Science through a Grant-in-Aid for Scientific Research on Innovative Areas No. J16H06471 (to T.A.), and by a National Science Foundation IOS award under Grant 1457230 (to I.M.H.).
AUTHOR CONTRIBUTIONS
T.A., I.K., and R.T. conceived the study. T.A. designed the experiments. T.A., H.O., and T.M. conducted the experiments. T.A. and H.O. analyzed the data. I.M.H. participated in the interpretation of the results. K.B. and I.K. initiated and maintained the plant materials. T.A. and I.M.H. drafted the manuscript. All authors approved the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Akagi T., Ikegami A., Tsujimoto T., Kobayashi S., Sato A., Kono A., Yonemori K. (2009). DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol. 151: 2028–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T., Henry I.M., Tao R., Comai L. (2014). Plant genetics. A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science 346: 646–650. [DOI] [PubMed] [Google Scholar]
- Akagi T., Henry I.M., Kawai T., Comai L., Tao R. (2016). Epigenetic regulation of the sex determination gene MeGI in polyploid persimmon. Plant Cell 28: 2905–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso C.T., Raines T., Kieber J.J. (2010). Cytokinin signaling and transcriptional networks. Curr. Opin. Plant Biol. 13: 533–539. [DOI] [PubMed] [Google Scholar]
- Ashman T.L., Kwok A., Husband B.C. (2013). Revisiting the dioecy-polyploidy association: alternate pathways and research opportunities. Cytogenet. Genome Res. 140: 241–255. [DOI] [PubMed] [Google Scholar]
- Bachtrog D., et al.; Tree of Sex Consortium (2014). Sex determination: why so many ways of doing it? PLoS Biol. 12: e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. (2013). Plant sex chromosome evolution. J. Exp. Bot. 64: 405–420. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. (2015). Plant contributions to our understanding of sex chromosome evolution. New Phytol. 208: 52–65. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D. (1978). A model for the evolution of dioecy and gynodioecy. Am. Nat. 112: 975–997. [Google Scholar]
- Chat J., Jáuregui B., Petit R.J., Nadot S. (2004). Reticulate evolution in kiwifruit (Actinidia, Actinidiaceae) identified by comparing their maternal and paternal phylogenies. Am. J. Bot. 91: 736–747. [DOI] [PubMed] [Google Scholar]
- Datson P.M., Ferguson A.R. (2011). Actinidia. Wild Crop Relatives: Genomic and Breeding Resources, Tropic. Subtropic. Fruits, Kole C., ed (Berlin, Heidelberg: Springer; ), pp. 1–20. [Google Scholar]
- De Rybel B., van den Berg W., Lokerse A., Liao C.Y., van Mourik H., Möller B., Peris C.L., Weijers D. (2011). A versatile set of ligation-independent cloning vectors for functional studies in plants. Plant Physiol. 156: 1292–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison A.M., Butler E.D., Hicks E.J., Naczi R.F., Calie P.J., Bell C.D., Davis C.C. (2012). Phylogeny and biogeography of the carnivorous plant family Sarraceniaceae. PLoS One 7: e39291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esumi T., Tao R., Yonemori K. (2007). Relationship between floral development and transcription levels of LEAFY and TERMINAL FLOWER 1 homologs in Japanese pear (Pyrus pyrifolia Nakai) and quince (Cydonia oblonga Mill.). J. Jpn. Soc. Hortic. Sci. 76: 294–304. [Google Scholar]
- Falasca G., Franceschetti M., Bagni N., Altamura M.M., Biasi R. (2010). Polyamine biosynthesis and control of the development of functional pollen in kiwifruit. Plant Physiol. Biochem. 48: 565–573. [DOI] [PubMed] [Google Scholar]
- Falasca G., D’Angeli S., Biasi R., Fattorini L., Matteucci M., Canini A., Altamura M.M. (2013). Tapetum and middle layer control male fertility in Actinidia deliciosa. Ann. Bot. 112: 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser L.G., Tsang G.K., Datson P.M., De Silva H.N., Harvey C.F., Gill G.P., Crowhurst R.N., McNeilage M.A. (2009). A gene-rich linkage map in the dioecious species Actinidia chinensis (kiwifruit) reveals putative X/Y sex-determining chromosomes. BMC Genomics 10: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G.P., et al. (1998). Development of sex-linked PCR markers for gender identification in Actinidia. Theor. Appl. Genet. 97: 439–445. [Google Scholar]
- Goldberg E.E., Otto S.P., Vamosi J.C., Mayrose I., Sabath N., Ming R., Ashman T.L. (2017). Macroevolutionary synthesis of flowering plant sexual systems. Evolution 71: 898–912. [DOI] [PubMed] [Google Scholar]
- Grant S., et al. (1994). Genetics of sex determination in flowering plants. J. Genet. Dev. 15: 214–230. [Google Scholar]
- Gupta S., Rashotte A.M. (2012). Down-stream components of cytokinin signaling and the role of cytokinin throughout the plant. Plant Cell Rep. 31: 801–812. [DOI] [PubMed] [Google Scholar]
- Gupta V., Estrada A.D., Blakley I., Reid R., Patel K., Meyer M.D., Andersen S.U., Brown A.F., Lila M.A., Loraine A.E. (2015). RNA-Seq analysis and annotation of a draft blueberry genome assembly identifies candidate genes involved in fruit ripening, biosynthesis of bioactive compounds, and stage-specific alternative splicing. Gigascience 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkess A., et al. (2017). The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat. Commun. 8: 1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Li J.-Q., Cai Q., Wang Q. (2005). The cytology of Actinidia, Saurauia and Clematoclethra (Actinidiaceae). Bot. J. Linn. Soc. 147: 369–374. [Google Scholar]
- Henry I.M., Dilkes B.P., Tyagi A., Gao J., Christensen B., Comai L. (2014). The BOY NAMED SUE quantitative trait locus confers increased meiotic stability to an adapted natural allopolyploid of Arabidopsis. Plant Cell 26: 181–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horák J., Grefen C., Berendzen K.W., Hahn A., Stierhof Y.D., Stadelhofer B., Stahl M., Koncz C., Harter K. (2008). The Arabidopsis thaliana response regulator ARR22 is a putative AHP phospho-histidine phosphatase expressed in the chalaza of developing seeds. BMC Plant Biol. 8: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., et al. (2013). Draft genome of the kiwifruit Actinidia chinensis. Nat. Commun. 4: 2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T. (2003). Use of a new adhesive film for the preparation of multi-purpose fresh-frozen sections from hard tissues, whole-animals, insects and plants. Arch. Histol. Cytol. 66: 123–143. [DOI] [PubMed] [Google Scholar]
- Kazama Y., Ishii K., Aonuma W., Ikeda T., Kawamoto H., Koizumi A., Filatov D.A., Chibalina M., Bergero R., Charlesworth D., Abe T., Kawano S. (2016). A new physical mapping approach refines the sex-determining gene positions on the Silene latifolia Y-chromosome. Sci. Rep. 6: 18917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T., Aoki K., Sakakibara H., Mizuno T. (2004). Arabidopsis response regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol. 45: 1063–1077. [DOI] [PubMed] [Google Scholar]
- Koizumi A., Yamanaka K., Nishihara K., Kazama Y., Abe T., Kawano S. (2010). Two separate pathways including SlCLV1, SlSTM and SlCUC that control carpel development in a bisexual mutant of Silene latifolia. Plant Cell Physiol. 51: 282–293. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Liu Z., et al. (2004). A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427: 348–352. [DOI] [PubMed] [Google Scholar]
- Marsch-Martínez N., Ramos-Cruz D., Irepan Reyes-Olalde J., Lozano-Sotomayor P., Zúñiga-Mayo V.M., de Folter S. (2012). The role of cytokinin during Arabidopsis gynoecia and fruit morphogenesis and patterning. Plant J. 72: 222–234. [DOI] [PubMed] [Google Scholar]
- McNeilage M.A. (1991). Sex expression in fruiting male vines of kiwifruit. Sex. Plant Reprod. 4: 274–278. [Google Scholar]
- McNeilage M.A. (1997). Progress in breeding hermaphrodite kiwifruit cultivars and understanding the genetics of sex determination. Acta Hortic. 444: 73–78. [Google Scholar]
- Ming R., Wang J., Moore P.H., Paterson A.H. (2007). Sex chromosomes in flowering plants. Am. J. Bot. 94: 141–150. [DOI] [PubMed] [Google Scholar]
- Ming R., Bendahmane A., Renner S.S. (2011). Sex chromosomes in land plants. Annu. Rev. Plant Biol. 62: 485–514. [DOI] [PubMed] [Google Scholar]
- Moore R.C., Purugganan M.D. (2005). The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8: 122–128. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Renner S.S. (2014). The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. Am. J. Bot. 101: 1588–1596. [DOI] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulin A., Auer P.L., Libault M., Schlueter J., Farmer A., May G., Stacey G., Doerge R.W., Jackson S.A. (2013). The fate of duplicated genes in a polyploid plant genome. Plant J. 73: 143–153. [DOI] [PubMed] [Google Scholar]
- Sakellariou M.A., et al. (2016). Agronomic, cytogenetic and molecular studies on hermaphroditism and self-compatibility in the Greek kiwifruit (Actinidia deliciosa) cultivar ‘Tsechelidis’. J. Hortic. Sci. Biotechnol. 91: 2–13. [Google Scholar]
- Seal A.G., Ferguson A.R., de Silva H.N., Zhang J.L. (2012). The effect of 2n gametes on sex ratios in Actinidia. Sex. Plant Reprod. 25: 197–203. [DOI] [PubMed] [Google Scholar]
- Shi T., Huang H., Barker M.S. (2010). Ancient genome duplications during the evolution of kiwifruit (Actinidia) and related Ericales. Ann. Bot. 106: 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Steinkamp R., Waack S., Morgenstern B. (2004). AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 32: W309–W312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34: W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J.A., Wei N., Straub S., Govindarajulu R., Liston A., Ashman T.-L. (2017). Repeated translocation of a gene cassette drives sex chromosome turnover in strawberries. bioRxiv doi/10.1101/163808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y., Mizrachi E., Marchal K. (2017). The evolutionary significance of polyploidy. Nat. Rev. Genet. 18: 411–424. [DOI] [PubMed] [Google Scholar]
- Vashisth T., Johnson L.K., Malladi A. (2011). An efficient RNA isolation procedure and identification of reference genes for normalization of gene expression in blueberry. Plant Cell Rep. 30: 2167–2176. [DOI] [PubMed] [Google Scholar]
- Wang Z., et al. (2013). Bisexual flower ontogeny after chemical induction and berry characteristics evaluation in male Vitis amurensis Rupr. Sci. Hortic. (Amsterdam) 162: 11–19. [Google Scholar]
- Westergaard M. (1958). The mechanism of sex determination in dioecious flowering plants. Adv. Genet. 9: 217–281. [DOI] [PubMed] [Google Scholar]
- Wu F., Mueller L.A., Crouzillat D., Pétiard V., Tanksley S.D. (2006). Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant clade. Genetics 174: 1407–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (1997). PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Yonemori K., Sugiura A., Kameda K., Yomo Y. (1991). Regularity and modification of sex expression in monoecious persimmon. HortScience 26: 777. [Google Scholar]
- Zhang Q., Liu C., Liu Y., VanBuren R., Yao X., Zhong C., Huang H. (2015). High-density interspecific genetic maps of kiwifruit and the identification of sex-specific markers. DNA Res. 22: 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.S., Niu Q.W., Chua N.H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646. [DOI] [PubMed] [Google Scholar]