The locus underlying the panicle apical abortion1 mutant in rice encodes a putative aluminum-activated malate transporter (OsALMT7) that is essential for maintaining sink size and grain yield by mediating malate transport in rice panicles.
Abstract
Panicle size is a critical determinant of grain yield in rice (Oryza sativa) and other grain crops. During rice growth and development, spikelet abortion often occurs at either the top or the basal part of the panicle under unfavorable conditions, causing a reduction in fertile spikelet number and thus grain yield. In this study, we report the isolation and functional characterization of a panicle abortion mutant named panicle apical abortion1-1 (paab1-1). paab1-1 exhibits degeneration of spikelets on the apical portion of panicles during late stage of panicle development. Cellular and physiological analyses revealed that the apical spikelets in the paab1-1 mutant undergo programmed cell death, accompanied by nuclear DNA fragmentation and accumulation of higher levels of H2O2 and malondialdehyde. Molecular cloning revealed that paab1-1 harbors a mutation in OsALMT7, which encodes a putative aluminum-activated malate transporter (OsALMT7) localized to the plasma membrane, and is preferentially expressed in the vascular tissues of developing panicles. Consistent with a function for OsALMT7 as a malate transporter, the panicle of the paab1-1 mutant contained less malate than the wild type, particularly at the apical portions, and injection of malate into the paab1-1 panicle could alleviate the spikelet degeneration phenotype. Together, these results suggest that OsALMT7-mediated transport of malate into the apical portion of panicle is required for normal panicle development, thus highlighting a key role of malate in maintaining the sink size and grain yield in rice and probably other grain crops.
INTRODUCTION
Rice (Oryza sativa) is the major staple food for more than half of the world’s population. Grain yield in rice is mainly determined by the number of panicles, number of grains per panicle, and grain weight, all of which are typical quantitative traits (Xing and Zhang, 2010). Panicle architecture, characterized by its size and branching pattern, determines the number of spikelets and, thus, the number of grains per panicle. Large panicles with more branches and spikelets (and, thus, higher grain number per panicle) have been preferred in breeding programs for new rice types with higher yield (Khush, 2000). Thus, understanding the molecular genetic mechanisms of panicle development and identification of superior alleles for large panicles are of great interest to both plant biologists and plant breeders.
The rice panicles are initiated following the switch from the vegetative to reproductive growth. At the transition, the shoot apical meristem is converted into an inflorescence meristem, which further initiates the primary branch meristems (BMs) and forms the main axis of the inflorescence. Subsequently, the primary BMs produce secondary BMs and spikelet meristems (SMs). SMs are also initiated from the secondary BMs and finally form spikelets (Ikeda et al., 2004; Tanaka et al., 2013). At maturity, rice panicle architecture, and thus the number of spikelets per panicle, is determined mainly by the length of the main axis and the length and number of primary and secondary branches. Over the past two decades, a number of genes regulating panicle development have been identified and functionally characterized. For example, SMALL PANICLE, REDUCED CULM NUMBER1 (RCN1), LAX PANICLE1 (LAX1), and LAX2 are involved in the initiation of BMs and SMs (Nakagawa et al., 2002; Komatsu et al., 2003a; Oikawa and Kyozuka, 2009; Tabuchi et al., 2011). ABERRANT PANICLE ORGANIZATION1 (APO1), APO2, and TAWAW1 are involved in maintaining the identity of BMs by preventing precocious conversion of BMs to SMs (Ikeda et al., 2005, 2007; Ikeda-Kawakatsu et al., 2012; Yoshida et al., 2013). FRIZZY PANICLE is required to ensure initiation and identity of floral organs by inhibiting the continued formation of axillary meristems from differentiated SMs (Komatsu et al., 2001, 2003b). Notably, the above genes encode different types of transcription factors. In addition, GRAIN NUMBER1, which encodes a cytokinin oxidase/dehydrogenase, regulates cytokinin accumulation in inflorescence meristems and plays an important role in determining the final number of SMs during panicle development (Ashikari et al., 2005). DENSE AND ERECT PANICLE1 encodes a phosphatidylethanolamine binding protein-like domain protein that regulates meristematic activity and, thus, panicle morphology and grain number per panicle (Delhaize et al., 1993, 2007; Ashikari et al., 2005; Huang et al., 2009). Recently, miR156 was found to negatively regulate panicle branch number by downregulating the expression of SOUAMOSA PROMOTER BINDING PROTEIN-LIKE14 (Jiao et al., 2010; Miura et al., 2010). Elucidating the functions of these genes and their regulatory relationships will contribute to breeding of new elite rice cultivars with “ideal” plant architecture and higher grain yield.
During rice growth and development, panicle abortion frequently occurs at either the top or basal parts of the panicle, particularly under unfavorable climatic conditions (malnutrition, extreme temperatures, shading, and water stress) (Senanayake et al., 1991; Saha et al., 1998; Yao et al., 2000; Kobayasi et al., 2001; Kato et al., 2008). Molecular genetic studies have identified several quantitative trait loci responsible for panicle apical abortion in rice (Cheng et al., 2011; Tan et al., 2011). In addition, a few genes involved in panicle degeneration have also been reported. SHORT PANICLE1 was reported to control the spikelet abortion at the basal part of panicles, likely through regulating nitrate transport (Li et al., 2009). ABERRANT SPIKELET AND PANICLE1 encodes a TOPLESS-related transcriptional corepressor; its loss-of-function mutant exhibits pleiotropic phenotypes including spikelet abortion at the middle and basal portions of panicle (Yoshida et al., 2012). Mutation in TUTOU1, which encodes a SCAR-like protein modulating actin organization, causes a pleiotropic phenotype including panicle apical abortion (Bai et al., 2015). Despite this progress, the molecular and genetic mechanisms underlying panicle abortion are still poorly understood.
In this study, we report the isolation and characterization of a spikelet abortion mutant named panicle apical abortion1-1 (paab1-1) that exhibits degeneration of spikelets at the tops of panicles during the late stage of panicle development. Our data suggest that OsALMT7 (an aluminum-activated malate transporter) is responsible for panicle apical abortion in paab1-1 and plays an essential role in maintaining panicle size and grain yield by mediating malate transport in rice.
RESULTS
paab1-1 Exhibits an Apical Abortion Phenotype during Late Panicle Development
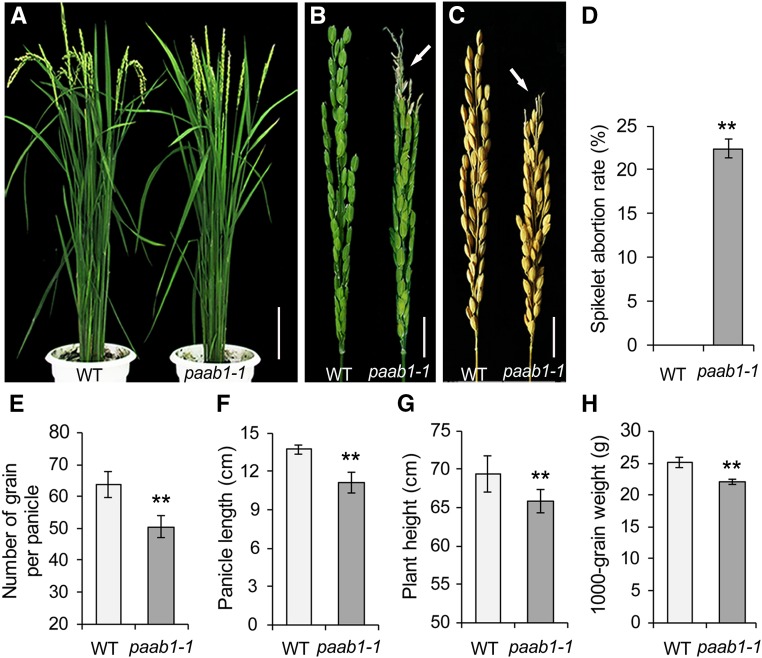
To understand the genetic and molecular mechanism underlying panicle development in rice, we identified a mutant, named panicle apical abortion1-1, from a tissue culture-derived population of the japonica rice cv Kitaake. The paab1-1 mutant plants were morphologically similar to the wild type before the heading stage but showed severely aborted spikelets at the apical portion of each panicle at the heading stage (Figures 1A and 1B). The spikelet abortion rate, as the percentage of aborted spikelets in a panicle, was ∼22% on average, and as a result, the number of grains per mature panicle dropped by ∼20% compared with the wild type (Figures 1C to 1E). In addition, the plant height of the paab1-1 mutant was slightly lower than the wild type, largely due to a reduction in panicle length (Figures 1F and 1G). Moreover, the overall grain weight per panicle was reduced in paab1-1 (Figure 1H). A more detailed analysis based on dividing the paab1-1 panicle into three portions indicated that the spikelets and grains of the middle portion were smaller, whereas the basal spikelets and grains were normal in size compared with the wild type (Supplemental Figures 1A to 1F). Thus, paab1-1 is a typical spikelet abortion mutant with a significant reduction in the sink size and grain yield.
Figure 1.
Phenotypic Chatracterization of paab1-1.
(A) Gross morphology of an adult paab1-1 plant, showing the erect panicles due to aborted spikelets at the top as compared with the wild type (WT).
(B) A representative paab1-1 panicle showing the aborted apical portion. The arrow indicates the degenerated spikelets.
(C) Comparison of a representative paab1-1 mature panicle with the wild type, showing reduced panicle length due to the aborted tip in paab1-1.
(D) to (H) Comparison of spikelet abortion rate (D), number of grain per panicle (E), panicle length (F), plant height (G), and 1000-grain weight (H) between the wild type and paab1-1. All data shown are mean ± se (n = 15). Asterisks represent statistically significant differences from the wild type, as determined by Student’s t test. **P < 0.01.
Bars = 10 cm in (A) and 2 cm in (B) and (C).
To investigate when the apical spikelet degeneration starts to occur during panicle development, we divided the developmental course for panicles into six stages according to the panicle length (∼1, ∼3, ∼5, ∼7, ∼10, and ∼13 cm) (Figures 2A to 2F). In the variety Kitaake, the 1-cm stage represents a tiny panicle that already has differentiated spikelets, whereas the panicle at the 13-cm stage has almost reached the final size, although it is still inside the flag leaf sheath (Figures 2A, 2F, and 2G). During the development of panicles, the size of the spikelets also increases and reaches a maximum at the 13-cm stage (Figures 2H to 2M). Parallel observations showed that there were no apparent differences in the color and size of the panicles, spikelets, and anthers between the wild-type and paab1-1 plants until the 10-cm stage, but the apical spikelet degeneration phenotype became clearly visible at the 13-cm stage in paab1-1 (Figures 2A to 2F and 2H to 2M), as indicated by the white and smaller spikelets and shrunk anthers (Supplemental Figures 2A and 2B). These aberrant spikelets became shriveled after heading, and finally aborted (Figure 2N). To define the developmental defect more precisely, we examined the spikelet morphology at early development stages by scanning electron microscopy. The scanning electron microscopy observations revealed no difference in floral primordia between paab1-1 and the wild type (Supplemental Figures 3A to 3F). We then performed histological analysis on apical spikelets at late stages. The lemma, palea, lodicules, stamens, and pistil in a paab1-1 spikelet all were morphologically similar to those in the wild type at the 10-cm stage (Supplemental Figures 4A, 4B, 4E, and 4F). However, the floral organs in paab1-1, but not in the wild type, collapsed as the apical spikelets developed further (Supplemental Figures 4C, 4D, 4G, and 4H). Thus, apical abortion of the paab1-1 panicle occurred during late panicle development (after 10 cm in length).
Figure 2.
Degeneration of Top Spikelets Occurs at Late Stages of Panicle Development in paab1-1.
(A) to (G) Representative images of wild-type (left) and paab1-1 (right) developing panicles, showing different stages as indicated by panicle length: 1 cm (A), 3 cm (B), 5 cm (C), 7 cm (D), 10 cm (E), 13 cm (F), and final size (G). White arrows indicate the degenerating spikelets in (F) and (G).
(H) to (N) Representative spikelets from the top of the corresponding panicles shown in (A) to (G).
Note that degeneration of apical panicle becomes visible in paab1-1 when the panicle reaches the 13-cm stage. Bars = 1 cm in (A) to (G) and 3 mm in (H) to (N).
Cell Death Occurs in the Apical Spikelets of paab1-1 Panicles
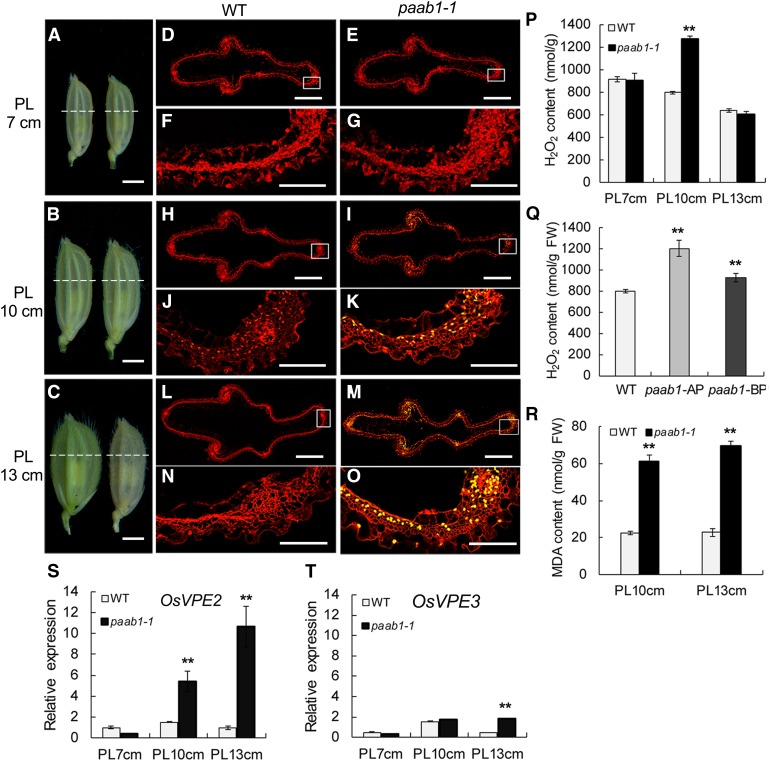
To further examine the cellular changes associated with the panicle apical abortion phenotype in paab1-1, we performed transmission electron microscopy (TEM) analysis of the apical spikelets at the 7-, 10-, and 13-cm stages, respectively. There were no obvious alterations in term of the organization and organelle content in paab1-1 cells at the 7-cm stage, but less defined organelles with reduced contents were seen at the 10-cm stage. Breakdown of organelles was clear at the 13-cm stage compared with the wild type (Supplemental Figures 5A to 5F). Consistent with the findings from the TEM analysis, a terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay, which detects nuclear DNA fragmentation, showed signals in paab1-1 apical spikelet hull cells at the 10-cm stage (Figures 3B, 3I, and 3K), and the signals became more intense at the 13-cm stage (Figures 3C, 3M, and 3O). In addition, an electrophoresis analysis also showed clear DNA fragmentation in the aborted spikelets, further confirming that DNA degradation occurred in the aborted apical spikelets in paab1-1 (Supplemental Figure 6). On the contrary, neither positive TUNEL signals nor obvious DNA fragments were detected in the wild-type spikelet cells of the corresponding stages (Figures 3D, 3F, 3H, 3J, 3L, and 3N). These results collectively suggest that cell degeneration in the paab1-1 apical spikelets starts to occur between the 7- and 10-cm stages, before spikelet abortion becomes visible.
Figure 3.
Cell Death-Related Events Are Triggered in paab1-1.
(A) to (C) Apical spikelets of the wild-type (left) and paab1-1 (right) panicles at the 7-, 10-, and 13-cm stages (PL 7 cm, PL 10 cm, and PL 13 cm) were used for cell death analysis. The dotted white lines indicate the sites of cross sections for TUNEL assays.
(D) and (E) TUNEL assay of apical spikelet hull cells at the 7-cm stage. No DNA fragmentation signal was detected in both the wild type (D) and paab1-1 (E).
(F) and (G) Magnified views of the boxed areas in (D) and (E), respectively.
(H) and (I) TUNEL assay of apical spikelet hull cells at the 10-cm stage. DNA fragmentation signal was detected in the paab1-1 mutant (H), but not in the wild type (I).
(J) and (K) Magnified views of the boxed areas in (H) and (I), respectively.
(L) and (M) TUNEL assay of apical spikelet hull cells at the 13-cm stage. There was no DNA fragmentation signal detected in the wild type (L), but intensive signals were seen in paab1-1 (M).
(N) and (O) Magnified views of the boxed areas in (L) and (M), respectively.
(P) Measurement of H2O2 content in panicles of the 7-, 10-, or 13-cm stages in the wild type and paab1-1, showing higher accumulation of H2O2 only at the 10-cm stage in paab1-1. The calculation was based on fresh weight of the whole panicles.
(Q) H2O2 content measurement in the wild-type panicles, compared with apical and lower portions of 10-cm young panicles of paab1-1. AP, apical portion; LP, lower portion; FW, fresh weight.
(R) Higher MDA content in the paab1-1 apical spikelets at PL10 cm and PL13 cm.
(S) and (T) Expression level of OsVPE2 (S) and OsVPE3 (T) in the top regions of wild-type and paab1-1 panicles at the 7-, 10-, or 13-cm stage, relative to the internal control (rice UBIQUITIN gene).
All data in (P) to (T) are presented as means ± se of three independent biological replicates. **P < 0.01, analyzed by the Student’s t test compared with the wild type. Bars = 2 mm in (A) to (C), 0.5 mm in (D), (E), (H), (I), (L), and (M), and 0.1 mm in (F), (G), (J), (K), (N), and (O).
It has been well documented that excessive accumulation of H2O2 can trigger cell death (Van Breusegem and Dat, 2006). We therefore measured H2O2 content and found that there was a H2O2 blast in the paab1-1 panicle at the 10-cm stage and that this change was located in the apical portion (Figures 3P and 3Q). High reactive oxygen species levels lead to oxidative damage of cellular components, such as membrane lipids; and malondialdehyde (MDA), an end product of lipid peroxidation, is regarded as an indicator of the production of reactive oxygen species (Chen and Murata, 2002). We measured the MDA content and found that MDA levels were elevated in the apical portions of 10- and 13-cm paab1-1 panicles compared with the wild type (Figure 3R). We further examined the expression level of OsVPE2 and OsVPE3, two representative genes associated with programmed cell death (PCD; Deng et al., 2011), and found that expression of OsVPE2 was significantly increased in paab1-1 at the 10-cm stage and even higher at the 13-cm stage, whereas a significant elevation was not seen until the 13-cm stage for OsVPE3 (Figures 3S and 3T). Those results suggest that overaccumulation of H2O2 in the apical portion of the paab1-1 panicle may trigger PCD at a stage not later than 10 cm in length.
Cloning and Characterization of the Causal Gene OsALMT7
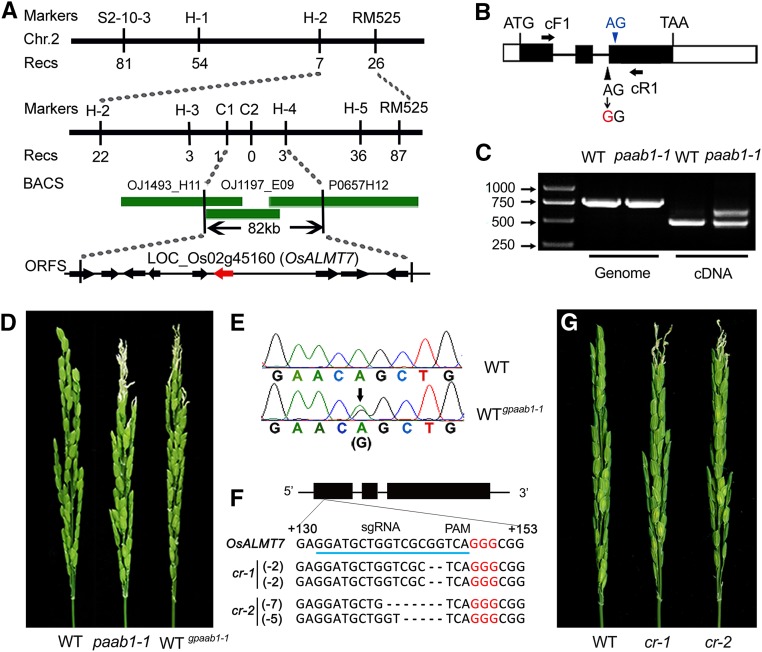
To isolate the causal gene for the observed mutant phenotype, we constructed an F2 mapping population by crossing paab1-1 with the japonica cultivar IRAT129. All the F1 individuals showed the panicle apical abortion phenotype, indicating that the mutation is dominant. We selected 1340 individuals with typical apical abortion from 2300 field-grown F2 plants to map paab1-1. The paab1-1 locus was initially mapped to the long arm of chromosome 2 between the markers S2-10-3 and RM525. After fine mapping, the mutation was further narrowed down to an 82-kb genomic region between the markers C1 and H-4, in which nine open reading frames were annotated (Figure 4A). Sequence comparison between paab1-1 and the wild type revealed a single nucleotide substitution of A to G in the predicted splicing site between the second intron and the third exon of LOC_Os02g45160 (referred to hereafter as OsALMT7; Liu et al., 2017) (Figure 4B). Consistent with the expected defect in splicing of the second intron, RT-PCR analysis detected two transcripts in the paab1-1 mutant. One was 157 bp longer than that in the wild type due to retention of the second intron. In addition, a smaller transcript caused by alternative splicing of the second intron was detected (Figures 4B and 4C). Both transcripts detected in paab1-1 had frame shifts starting from the Ala at position 164 and were terminated prematurely (Supplemental Figure 7). RT-qPCR analysis showed that OsALMT7 is expressed at a higher level in the paab1-1 mutant than in the wild-type plants (Supplemental Figure 8).
Figure 4.
Map-Based Cloning of paab1-1.
(A) Fine mapping of paab1-1. The molecular markers and numbers of recombinants are indicated above and below the filled bars, respectively. The candidate open reading frame is highlighted in red.
(B) Genomic structure of the LOC_Os02g45160 (OsALMT7) gene. The white and filled boxes indicate untranslated regions and exons, respectively, and the black lines indicate introns. A base substitution in paab1-1 is highlighted in red and an alternative splicing site in blue. cF1 and cR1 are a pair of primers for PCR analysis in (C).
(C) RT-PCR analysis showing the presence of two abnormally sized transcripts in paab1-1. The larger transcript contains the 2nd intron due to mutation of the splicing site. The slightly smaller transcript is caused by the utilization of an alternative downstream splicing site as indicated by green in (B). The genomic DNA was used as a control.
(D) Genetic confirmation of the OsALMT7 gene. Wild-type plants transformed with a genomic fragment with the base substitution amplified from paab1 (WTgpaab1-1) resembled the paab1-1 plants.
(E) Verification of coexistence of the wild type and mutated OsALMT7 in the WTgpaab1-1 plants by sequencing.
(F) Deletion mutation at the target site in two representative knockout lines generated by the CRISPR/Cas9 technology. The cr-1 plant is a homozygous mutant carrying a 2-bp deletion on both homochromosomes and cr-2 is a biallelic mutant carrying a 5-bp deletion on one chromosome and a 7-bp deletion on another. The filled bars indicate exons and lines indicate introns of OsALMT7. The sgRNA target sequence is underlined in blue and the PAM motif is highlighted in red letters.
(G) Representative panicles of knockout mutants showing the apical abortion phenotype.
To verify whether the mutation of OsALMT7 is responsible for panicle apical abortion in paab1-1, a 6-kb genomic fragment isolated from paab1-1, including the entire coding region of LOC_Os02g45160 and its flanking sequences, was introduced into the wild type. All 22 positive transformants (T0) displayed the typical panicle apical abortion phenotype, mimicking the paab1-1 phenotype (Figures 4D and 4E). This observation confirmed the identity of OsALMT7 for panicle apical abortion and verified the dominant nature of the OsALMT7 mutation in paab1-1. In addition, we isolated an allelic mutant, named paab1-2, which carried a point mutant in the third exon of OsALMT7 (resulting an amino acid change from Ala to Phe at position 195) and displayed a similar panicle apical abortion phenotype to paab1-1 (Supplemental Figures 7, 9A, and 9B). We also transformed a CRISPR-Cas9 construct targeting the first exon of OsALMT7 into the wild type (Kitaake) and identified 48 knockout plants (with frame shift). Strikingly, all the knockout plants exhibited the apical panicle abortion phenotype (Figures 4F and 4G). Similarly, RNA interference (RNAi) knockdown plants of OsALMT7 in four japonica variety backgrounds (Kittake, Zhonghua 11, Asominori, and Nipponbare) all exhibited the apical panicle abortion phenotype (Supplemental Figures 10A to 10H). RT-qPCR analysis showed that expression of OsALMT7, but not that of its two closest homologs OsALMT8 and OsALMT9, was significantly downregulated in the RNAi plants (Supplemental Figures 11A and 11B). Collectively, these results demonstrate that the mutation in OsALMT7 is responsible for panicle apical abortion and that the mutant phenotype observed in paab1-1 is likely caused by a dominant-negative mutation.
The predicted OsALMT7 protein consists of 488 amino acid residues and contains a conserved ALMT domain. Secondary structure prediction showed that OsALMT7 has seven potential transmembrane helices (Supplemental Figures 12A to 12C). BLAST searches of databases revealed that there are a number of ALMT orthologous proteins in land plants and that the rice ALMT family includes nine members. Phylogenetic analysis indicated that OsALMT7 is most closely related to AtALMT10 (with 45.6% amino acid identity), an uncharacterized Arabidopsis family member, but is more distantly related to other functionally characterized members, such as AtALMT1 (30.6% identity), AtALMT6 (22.2%), AtALMT9 (24.8%), AtALMT12 (26%), and TaALMT1 (37.4%) (Supplemental Figure 13). This bioinformatic information implies that OsALMT7 may execute a distinct function from those previously characterized homologs.
OsALMT7 Is Preferentially Expressed in Vascular Tissue and Localized to the Plasma Membrane
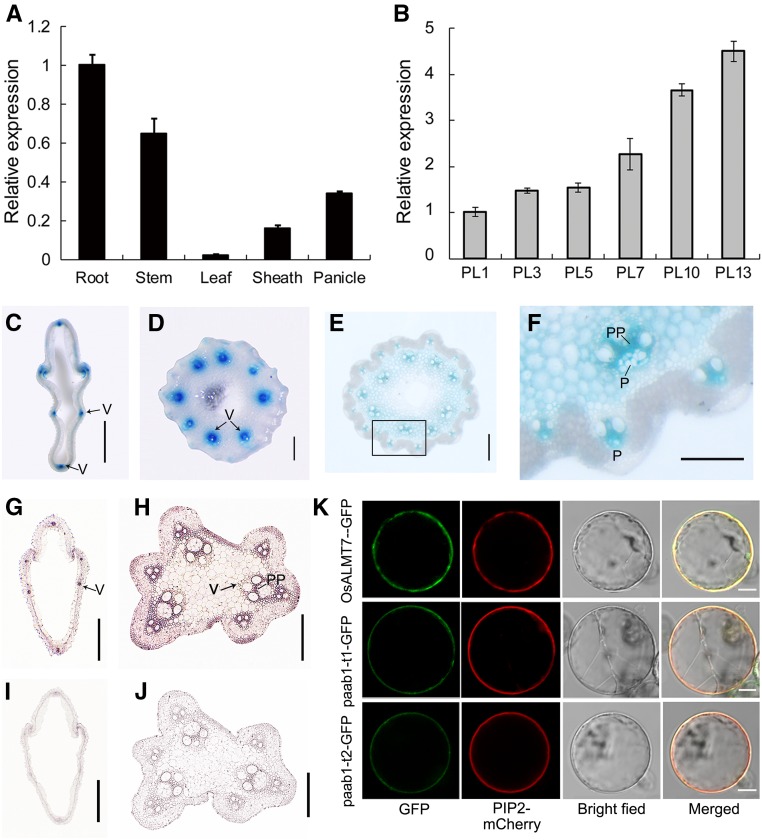
To elucidate the function of OsALMT7, we first analyzed the expression pattern of OsALMT7 by RT-qPCR analysis. Expression of OsALMT7 was detected in all rice organs analyzed, with relatively higher expression in the roots, stems, and panicles and lower expression in other organs including leaves and leaf sheathes (Figure 5A). A detailed analysis focusing on the panicle indicated that OsALMT7 expression increased continuously during panicle development (Figure 5B). The expression pattern of OsALMT7 was further evaluated in plants transformed with a GUS reporter gene driven by a 2614-bp promoter sequence of OsALMT7. We observed GUS activity in various organs examined, with the strongest staining in the roots, stems, and panicles and faint staining in leaves and leaf sheaths, similar to the RT-qPCR results (Supplemental Figures 14A to 14E). Since organ defects were observed only on the apical part of the panicle in paab1-1, we further analyzed OsALMT7 expression in the panicle at the 10-cm stage. GUS staining in transverse sections of spikelet hulls and rachises of the panicle revealed strong promoter activity of OsALMT7 in the vascular tissues (Figures 5C to 5E). A close-up observation showed that the GUS signals were mainly distributed in the phloem and phloem parenchyma cells of vascular tissues (Figure 5F). To verify this observation, an RNA in situ hybridization approach was employed. Strong signal was detected again in the vascular tissues of the spikelet hull and rachis (Figures 5G to 5J). The expression pattern of OsALMT7 is consistent with its putative function as an anion transporter required for normal panicle development, likely involved in phloem loading of malate.
Figure 5.
Expression Pattern and Subcellular Localization of the OsALMT7 Protein.
(A) Relative expression of OsALMT7 in various tissues, including root, stem, leaf blade, leaf sheath, and panicle at the 7-cm stage (PL7). Rice UBIQUITIN was used as an internal control. Data are presented as mean ± se (n = 3).
(B) Relative expression of OsALMT7 in developing panicles at the 1-, 3-, 5-, 7-, 10-, and 13-cm stages (PL1 to PL13) before heading. Rice UBIQUITIN was used as an internal control. Data are presented as mean ± se (n = 3).
(C) to (F) Promoter activity of OsALMT7 as shown by GUS staining. Vascular tissue-preferred expression was seen in spikelet hull (C) and rachises at basal (D) and middle (E) parts of the panicles carrying the OsALMT7-GUS fusion. The squared area in (E) was magnified to show a vascular tissue (F). P, phloem; PP, phloem parenchyma cells. Bars = 0.5 mm in (C), 0.2 mm in (D) and (E), and 0.1 mm in (F).
(G) to (J) mRNA in situ hybridization of OsALMT7 on transverse sections of panicle spikelet hull (F) and rachis (G). Abundant OsALMT7 transcripts were detected in vascular tissues of the panicle. OsALMT7 sense probe was used as a negative control (I) and (J). V, vascular tissue; PP, phloem parenchyma cells. Bars = 0.5 mm in (G) and (I) and 0.2 mm in (H) and (J).
(K) Plasma membrane localization of OsALMT7 in rice protoplasts. The OsALMT7-GFP fusion protein was transiently coexpressed with PIP2-mCherry, a plasma membrane marker in rice protoplasts (upper row). cDNAs corresponding to the two mutant versions of transcripts detected in paab1-1 were also fused to GFP (paab1-t1-GFP and paab1-t2-GFP) and coexpressed with PIP2-mCherry in rice protoplasts (middle and lower rows). From left to right: image of GFP (green), mCherry (red), protoplast, and merged GFP and mCherry. Bars = 5 μm.
To test if OsALMT7 is localized to the plasma membrane as annotated (http://harrier.nagahama-i-bio.ac.jp/sosui/), OsALMT7 was fused to the GFP reporter gene driven by the CaMV35S promoter. The resulting construct together with PIP2-mCherry, a plasma membrane marker (Lee et al., 2009), was transiently expressed in rice leaf sheath protoplasts. OsALMT7 was indeed colocalized with PIP2 on the plasma membrane in the protoplasts (Figure 5K). We also generated transgenic plants carrying the OsALMT7-GFP fusion protein transgene in the wild-type background. Plasma membrane localization was observed for OsALMT7-GFP fusion protein in the transgenic root cells (Supplemental Figures 15A to 15C). Notably, the predicted mutant products of paab1-1 still retained the first five transmembrane helices (Supplemental Figure 7) and were also localized to the plasma membrane (Figure 5K). Moreover, OsALMT7-GFP was localized to the plasma membrane in plasmolyzed onion epidermal cells (Supplemental Figures 15D to 15G). To verify if the OsALMT7-GFP fusion protein was biologically functional, we cotransformed the CRISPR-Cas9 construct together with an OsALMT7ʹ-GFP fusion construct carrying synonymous base substitutions at the target site into the variety Zhonghua 11 (Supplemental Figure 16A). The knockout plants generated with the CRISPR-Cas9 construct alone displayed panicle apical abortion, whereas all the 13 knockout (with frame shift in the internal OsALMT7) double transformants were normal in panicle development, suggesting that plasma membrane-localized OsALMT7ʹ-GFP fusion protein is biologically functional (Supplemental Figures 16B to 16D).
OsALMT7 Functions as an Aluminum-Independent Malate Transporter
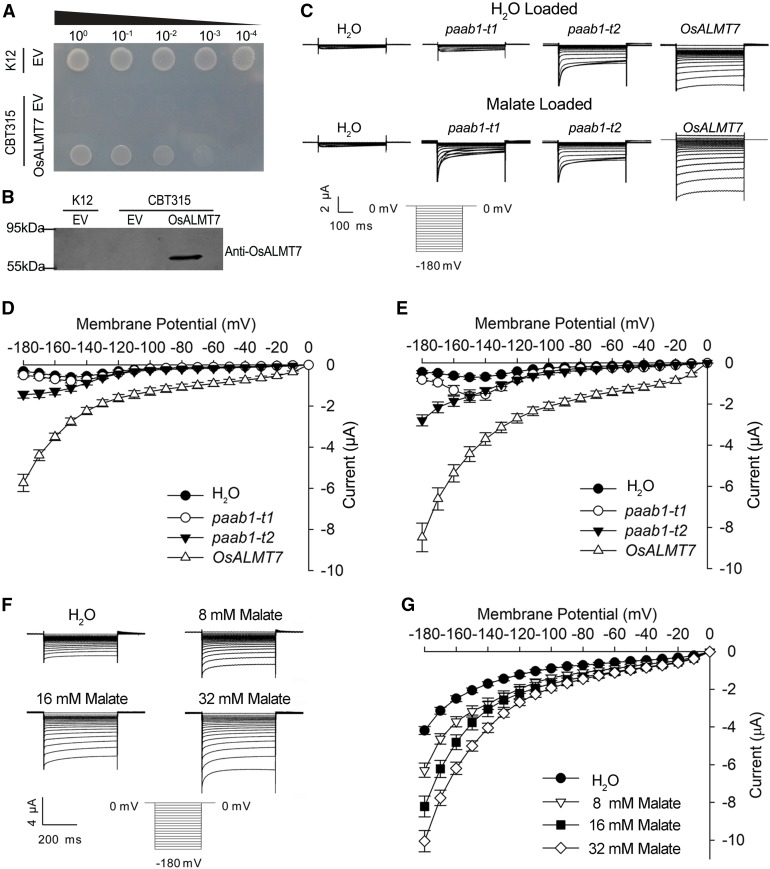
Several members of the ALMT family have been shown to function as anion channels and mediate malate transport (Barbier-Brygoo et al., 2011). To examine whether OsALMT7 exerts a similar biochemical function, an OsALMT7 cDNA fragment was cloned into the vector pKK223-3 and then heterologously expressed in the Escherichia coli strain CBT315. CBT315 is a mutant strain defective in dicarboxylate transporter and is incapable of utilizing malate as the carbon source (Jeong et al., 2004). Growth of the transformed strain was partially restored on the M9 medium when malate was used as the sole carbon source, suggesting that OsALMT7 could mediate malate transport in E. coli (Figures 6A and 6B).
Figure 6.
sALMT7 Mediates Malate Transport in E. coli and X. laevis Oocytes Systems.
(A) and (B) Partial suppression of the growth defect of the E. coli dicarboxylate transporter mutant CBT315 by expression of OsALMT7. CBT315 transfected with the empty vector (EV) or the OsALMT7 gene (OsALMT7) and its wild type (K12) transfected with the EV only were grown on M9 agar medium with 10 mM malate (pH 6.6) as the sole carbon source. From left to right: transfected cells without dilution (100), diluted by 10× (10−1), 100× (10−2), 1000× (10−3), or 10,000× (10−4). Pictures were taken 3 d after inoculation. Immunoblot analysis showing the expression of the OsALMT7 protein in CBT315 is in (B).
(C) Current recordings in X. laevis oocytes expressing OsALMT7 and the paab1-1 mutants. Whole-cell currents were recorded in oocytes injected with different cRNAs: OsALMT7, paab1-t1, paab1-t2, and with water as a control. The oocytes were preloaded with either water (upper panel) or malate (bottom panel). The holding potential was set to 0 mV and voltage protocols, as well as time and current scale bars for the recordings, are shown. paab1-t1 and paab1-t2 represent two cDNAs corresponding the two mutant versions of transcripts detected in paab1-1.
(D) and (E) The current-voltage (I-V) relationship of the steady state currents in oocytes preloaded with water (D) or malate (E). The data are derived from the current recordings as those shown in (C) and presented as mean ± se (n = 10 for each cRNA).
(F) OsALMT7-mediated currents recorded from X. laevis oocytes preloaded with a range of malate concentrations. Whole-cell currents were recorded in oocytes expressing OsALMT7 at different intracellular malate concentrations (8, 16, and 32 mM). The oocytes preloaded with water were used as a control. The holding potential was set to 0 mV and voltage pluses were stepped between −180 mV and 0 mV in 10-mV increments. The time and current scale bars for the recordings are shown.
(G) The current-voltage (I-V) relationship of the steady state currents in oocytes with different intracellular malate concentrations. The data are derived from the current recordings as those shown in (F) and presented as mean ± se (n = 10 for each malate concentration).
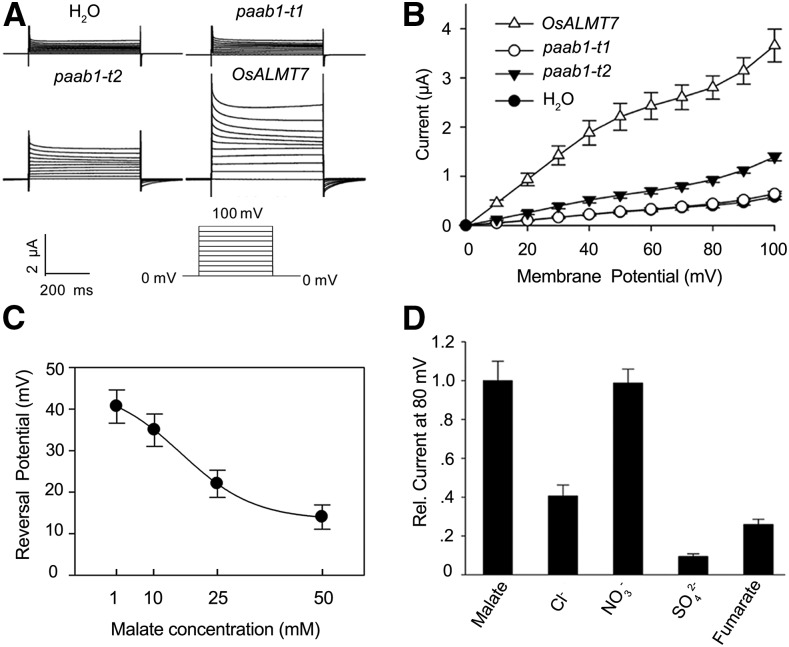
We further determined the electrophysiological properties of OsALMT7 by heterologously expressing it in Xenopus laevis oocytes. The two-electrode voltage clamp method was used to measure OsALMT7-mediated malate current in oocytes (Wagner et al., 2000). We observed that larger inward currents (anion efflux) appeared in the oocytes injected with the wild-type OsALMT7-cRNA than in the control oocytes injected with water or either of the two mutant cRNAs corresponding to the transcripts detected in paab1-1 (paab1-t1 or paab1-t2) and that the differences were greater when the oocytes were preloaded with malate than with water (Figures 6C to 6E). Moreover, the degree of enhanced inward currents by preloading of malate was concentration dependent (Figures 6F and 6G). In a separate assay where malate was added in the bathing solution, greater outward current (anion influx) was detected from the oocytes expressing OsALMT7 than from those expressing paab1-t1 or paab1-t2 or from the control cells injected with water only (Figures 7A and 7B). Reversal potential measurements in oocytes expressing OsALMT7 showed that increasing malate concentration in bath lead to negative shifts in the reversal potential (Figure 7C), which is indicative of malate permeation. Since inward current was recorded at hyperpolarization membrane potential and outward current was recorded at depolarization membrane potential, inside-out single channel current was recorded in oocytes to detect the directivity of OsALMT7 in oocytes. As shown in Supplemental Figure 17, at 180 mV command voltage, which means −180 mV membrane potential in the plasma membrane, inward currents were detected showing the efflux of malate. Furthermore, at −60 mV command voltage, which means 60 mV membrane potential in the plasma membrane, outward currents were detected showing the influx of malate. Taken together, these results confirmed that OsALMT7 functions as an anion channel capable of mediating transport of malate across the membrane. As ion channels are well known to form oligomers to perform transport activity (Middleton et al., 1996; Xicluna et al., 2007) and the predicted protein product of paab1-1 is localized to the plasma membrane like the wild-type protein, we speculated that the truncated mutant protein may interfere with the normal function of the wild-type protein, thus causing a dominant-negative effect at a posttranslational level.
Figure 7.
OsALMT7-Mediated Outward Currents and Anion Selectivity of OsALMT7 in X. laevis Oocytes.
(A) Recordings of OsALMT7-mediated outward currents in X. laevis oocytes. Whole-cell currents were recorded in oocytes injected with different cRNAs: OsALMT7, paab1-t1 and paab1-t2, and with H2O as a control. The outside solution was added with malate at a final concentration of 25 mM. The holding potential was set to 0 mV and voltage protocols, as well as time and current scale bars for the recordings are shown.
(B) The current-voltage (I-V) relationship of the steady state outward currents in oocytes. The data are derived from the current recordings as those shown in (A) and presented as mean ± se (n = 15 for each cRNA).
(C) Reversal potential of OsALMT7-expressing oocytes preloaded with 46 nL of 100 mM malate was recorded in bath solutions with 1, 10, 25 or 50 mM malate. The reversal potential was valued through cross points of the I-V curves and X-axes (I=0). The data are presented as mean ± se (n = 12 for each concentration).
(D) Anion selectivity of OsALMT7 in X. laevis oocytes. The outside solution was added with Cl-, NO3-, SO42- or fumarate, and with malate as a control. The relative current is the D-value from OsALMT7-expressing oocytes subtracted with that from control oocytes. Current for malate was set to 1. Calculation was based on currents measured at 80 mV only. The data are presented as mean ± se (n = 10 for each anion).
In addition to malate, some ALMT transporters were reported to translocate other organic anions such as fumarate or inorganic anions such as Cl−, NO3−, and SO42− (Kovermann et al., 2007; Piñeros et al., 2008a, 2008b). To examine the selectivity of OsALMT7 between malate and other anions, we replaced malate with fumarate, chloride, nitrate, or sulfate in the bathing solution and measured the corresponding outward currents in oocytes. The oocytes injected with OsALMT7-cRNA were permeable to fumarate, nitrate, chloride, or sulfate relative to the control injected with water, and the permeability to nitrate and malate was similar, as indicated by the outward currents (Figure 7D; Supplemental Figures 18A to 18C). Thus, OsALMT71 may also transport other anions in addition to malate, at least in X. laevis oocytes.
Several members of the ALMT family are involved in aluminum (Al) tolerance by mediating malate exudation from roots to soil (Sasaki et al., 2004; Hoekenga et al., 2006; Ye et al., 2017). To test whether OsALMT7 is involved in Al tolerance, we first tested if Al enhances the OsALMT7-mediated malate transport in the two-electrode voltage clamp assay. The addition of Al to the bathing solution had no significant effect on the inward currents in the oocytes preloaded with malate and injected with different versions of OsALMT7-cRNAs (Supplemental Figures 19A and 19B). Next, we treated rice seedlings with Al and found that the paab1-1 plants had similar sensitivity as the wild type (Supplemental Figure 20A). In addition, our RT-qPCR analysis indicated that OsALMT7 expression was not induced by Al (Supplemental Figure 20B). These results together suggest that OsALMT7 is an aluminum-independent malate transporter and is not associated with Al tolerance.
paab1-1 Is Defective in Malate Transport in the Apical Spikelets
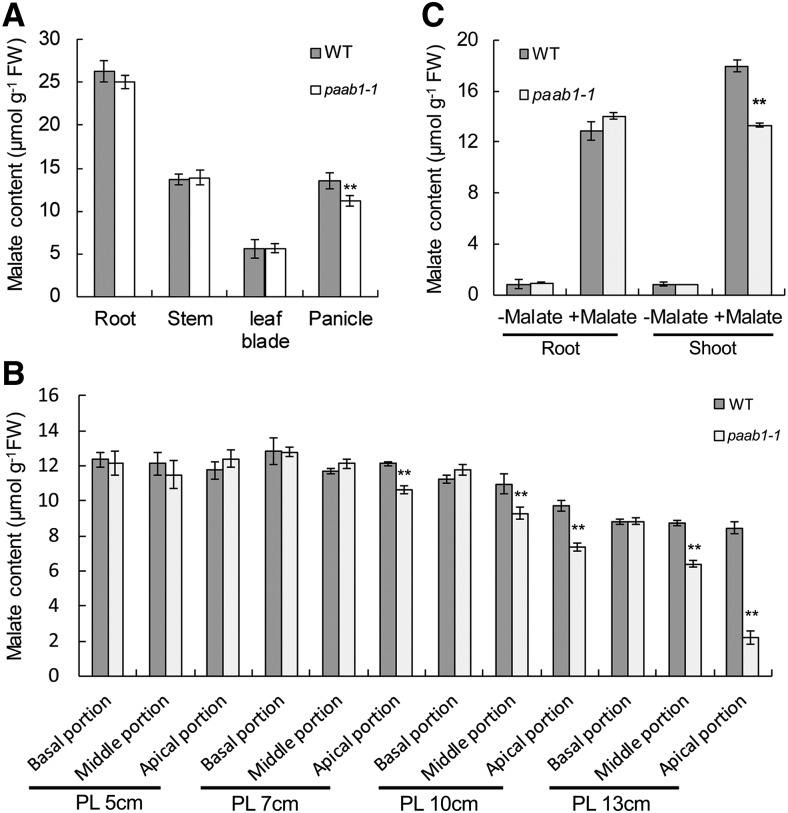
Since OsALMT7 functions as a malate transporter, we next investigated if the apical spikelet abortion in paab1-1 is associated with any change in malate level. Our malate content analysis indicated that there was a significant drop in the malate level of the panicle, but not of the root, stem, and leaf blade of paab1-1, compared with the wild type (Figure 8A). More detailed analysis revealed that malate content started to decrease in the apical portion at the 7-cm stage, and the decrease became more dramatic and extended to the middle portion during further panicle development, but no significant differences were detected in the lower portion of panicle between paab1-1 and the wild type (Figure 8B). No difference in nitrate level was detected between paab1-1 and the wild type (Supplemental Figure 21), suggesting that other transporters might be responsible for transporting nitrate in the panicle of paab1-1. To further investigate if malate transport is impaired in other tissues of paab1-1, we grew rice seedlings in a hydroponic solution supplemented with 0.3 mM malate for 24 h and measured the malate content in the roots and shoots. The malate content was lower in the shoots but not in the roots of paab1-1, compared with the wild type (Figure 8C). This result implies that transport of malate might be impaired in paab1-1, thus causing spikelet degeneration in the apical portion of panicle.
Figure 8.
Malate Content Is Decreased in the paab1-1 Panicle.
(A) Malate content in the indicated tissues sampled from field plants at the PL10 cm stage. Whole panicles were used for this analysis.
(B) Malate content in different panicle portions at different developmental stages. The panicle was divided into basal, middle, and apical portions to measure gradients in changes of malate content.
(C) Malate transport assay in seedlings grown in hydroponic solution. Five-day-old rice seedlings were exposed to the hydroponic solution added with 0 or 0.3 mM malate for 24 h before sampling.
All data are presented as means ± se (n = 3) of three independent biological replicates. Asterisks above bars indicate significant difference from the wild type (**P < 0.01) analyzed by the Student’s t test.
Injection of Malate into Developing Panicles Alleviates Apical Spikelet Degeneration in paab1-1
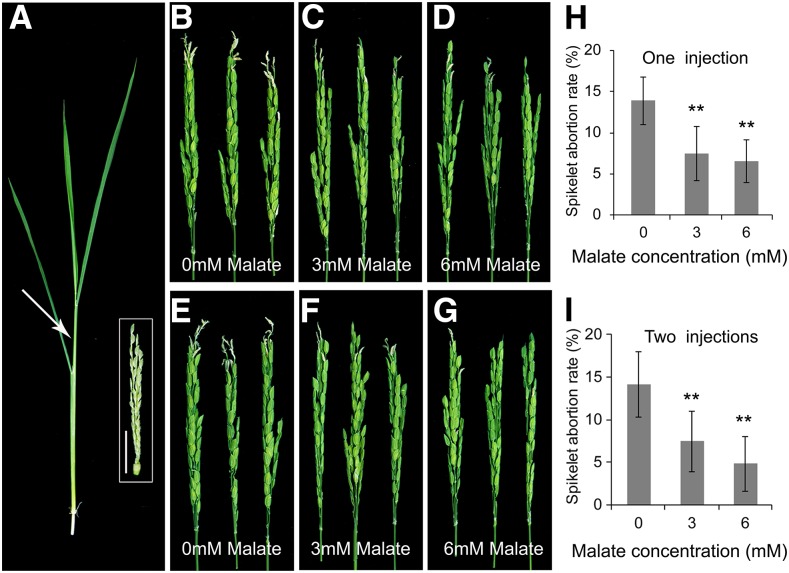
To further test the notion that decreased malate might be the trigger for apical panicle abortion in paab1-1, we first injected a top site of the wild-type panicle at approximately the 7-cm stage with water or with 1.5 mM malate and observed that the injections did not pose any visible injury or phenotypic change to the subsequent growth of the panicles (Supplemental Figures 22A to 22C). Then we injected 1.5 mM malate into the paab1-1 panicles at a similar stage. The paab1-1 plants injected at this stage still developed aborted apical spikelets, not different from the control injected with water only (Supplemental Figures 22D and 22E). When 3 mM malate was injected into younger paab1-1 plants with panicles of ∼5 cm in length, however, the apical spikelet abortion phenotype was alleviated significantly (Figures 9A to 9C and 9H). Increase of malate concentration to 6 mM further rescued the apical spikelets in paab1-1 (Figures 9D and 9H). A second injection 4 d after the first had a mild improvement in maintaining the panicle growth in paab1-1 (Figures 9E to 9G and 9I). Together, those results support the conclusion that malate deficiency is the underlying cause of spikelet apical degeneration in paab1-1.
Figure 9.
Injection of Malate to paab1-1 Plants Alleviates Apical Panicle Abortion.
(A) A representative tiller, with the panicle at about the 5-cm stage, used for malate injection. Arrow indicates an approximate injection site. The dissected panicle is shown in the white box. Bar = 1 cm.
(B) to (D) Representative paab1-1 panicles after injection with 0 (water control), 3, or 6 mM malate.
(E) to (G) Representative paab1-1 panicles after two sequential injections with 0, 3, or 6 mM malate. The second injection was made 4 d after the first injection with the same malate concentrations.
(H) and (I) Spikelet abortion rate of paab1-1 panicles injected with malate for one (H) or two sequential times (I).
Photographs in (B) to (G) were taken after the panicles emerged out of the flag leaf sheaths. All data shown in (H) and (I) are mean ± se (n = 10). Asterisks indicate significant difference from 0 mM malate, as determined by the Student’s t test at P < 0.01.
DISCUSSION
Panicle Apical Abortion in paab1-1 Reduces Panicle Size and Is Accompanied by Programmed Cell Death
Panicle abortion is a widespread physiological defect that reduces grain yield in rice and other cereal crops (Sheehy et al., 2001; Yamagishi et al., 2004). However, the genetic and molecular mechanisms governing panicle abortion have remained poorly characterized. In this study, we isolated a new panicle abortion mutant named paab1-1, which exhibits spikelet degeneration at the apical portion of panicle, and consequently a severe reduction in panicle size and grain yield (Figures 1B and 1E). In addition, growth of spikelets in the middle portion of paab1-1 panicle is also affected, resulting in reduced grain size (Supplemental Figure 1). Notably, the defects in both the apical and middle portions are observed after the panicles have already formed, suggesting that OsALMT7 is required for maintaining the development of differentiated panicles. Detailed observation revealed that panicle abortion becomes visible at the stage of 10 cm in length (Figures 2F and 2M). Furthermore, we found that there is a H2O2 burst and increased MDA levels in the aborted spikelets, accompanied by organelle degeneration, DNA degradation, and enhanced PCD-related gene expression (Figure 3). Together, these results indicate that the phenotype of paab1-1 is accompanied by programmed cell death in the apical spikelets.
OsALMT7 Encodes an ALMT Transporter Independent of Aluminum Activation
The family of aluminum-activated malate transporters has been shown to perform a range of physiological functions related to guard cell regulation, mineral nutrition transport, grain metabolism, and malate accumulation during tomato fruit ripening, besides being involved in aluminum tolerance (Hoekenga et al., 2006; Piñeros et al., 2008b; Sasaki et al., 2010; Xu et al., 2015; Ye et al., 2017). For example, The Arabidopsis thaliana members AtALMT6 and AtALMT9 mediate malate currents across the vacuole and regulate stomata movement (Meyer et al., 2011; De Angeli et al., 2013). TaALMT1 and AtALMT1 have been reported to mediate malate excretion from the plasma membrane of root cells into rhizosphere to form nontoxic complexes and detoxify Al3+ accumulation in soil (Sasaki et al., 2004; Hoekenga et al., 2006). In this study, we show that OsALMT7 encodes an aluminum-activated malate transporter. The rice genome contains nine members of the ALMT family (Delhaize et al., 2007) and functional analysis has been reported for only one of these genes (Liu et al., 2017). Our phylogenetic analysis revealed that OsALMT7 belongs to the same clade as TaALMT1, AtALMT1, and ZmALMT1 (Supplemental Figure 13). However, in contrast with TaALMT1 and AtALMT1, we found that OsALMT7 can mediate transport of malate across the plasma membrane independent of Al3+ activation (Figures 6 and 7; Supplemental Figure 19). As most anion channels mediate anion transport depending on the membrane potential and anion concentration on both sides of the plasma membrane, and the membrane potential of most plant cells is negative (hyperpolarized potential) under normal physiological conditions, anion channels generally favor the function of anion efflux from the cytosol. However, some cells such as xylem cells show depolarized membrane potential in plants (Wegner and Zimmermann, 2004; Wegner et al., 2011). Given that OsALMT7 mediates large inward/outward currents in oocytes (Figures 6C to 6E, 7A and 7B), we speculate that OsALMT7 may mediate influx or efflux in spikelet cells on the panicle, depending on the membrane potential and cytosolic and extracellular malate concentration.
OsALMT7 Is Required for Malate Supply and Sustained Growth of the Panicle during Late Development Stages
Panicle outgrowth in rice is a rapid elongation process, accompanied by maturation of reproductive organs that must be supported by higher rates of nutrient and energy supply. As such, abortion of spikelets is often seen, particularly under unfavorable environmental conditions (Yao et al., 2000; Itoh et al., 2005; Smith and Stitt, 2007; Kato et al., 2008). In support of this notion, nitrogen nutrition has also been shown to be required for spikelet development on the panicle (Fu et al., 2011). In addition, the maize (Zea mays) tassel-less1 mutant, which has a defect in boron transport and thus insufficient boron supply to the rapidly growing reproductive tissue and developing organs, shows a small and tip-barren ear phenotype (Durbak et al., 2014).
In this study, we demonstrated that malate is a key metabolite required to support sustained growth of the panicle during late development and that this process requires OsALMT7 based on the following observations. First, we found that the accumulation of malate is reduced in the aborted portions of paab1-1 panicles, compared with the apical portions of the wild type (Figure 8B). Second, we showed that injection of malate can effectively alleviate the aborted spikelets phenotype in paab1-1 (Figure 9). Third, we found that OsALMT7 can mediate transport of malate across the plasma membrane independent of Al3+ activation (Figures 6 and 7; Supplemental Figure 19). As malate is a central metabolite in the plant cell, involved in the mitochondrial tricarboxylic acid (TCA) and glyoxylate cycles in plant species (Martinoia and Rentsch, 1994; Fernie and Martinoia, 2009; Sweetman et al., 2009), one possibility might be that a deficiency of malate exerts a negative influence on energy generation and storage, biosynthesis of intermediates in TCA and glyoxylate cycle, or cellular pH status, which in turn impair other cellular activities such as import of sucrose and other nutrients, nitrogen reduction, and photorespiration. Therefore, further investigation of paab1-1 on expression level of TCA enzyme genes, content of TCA intermediates, nitrogen and sucrose level, and photorespiration activity may provide useful clues to understanding the link between low malate and PCD. Alternatively, the charge movement caused by uptake of anion uptake (in this case, malate) and the associated changes in electroneutrality or pH cannot be excluded. Moreover, it has been shown that malate protects plants from aluminum toxicity by its efflux from root cells to rhizosphere (Hoekenga et al., 2006) and that malate plays an important role in regulating guard cell aperture by maintaining cellular osmotic pressure in guard cells (Mathieu et al., 1986; Meyer et al., 2010). A recent study showed that altered expression of OsALMT4 in rice disrupted the distribution of certain minerals including Mn and B nutrition (Liu et al., 2017). OsALMT7 might also play a role in regulating mineral nutrition balance in rice. Furthermore, as OsALMT7 is also permeable to nitrate and defects in nitrate transport have been shown to cause a small-panicle phenotype (Li et al., 2009), possible effects of OsALMT7 on these cellular processes and their relationship with panicle development remain to be investigated in future studies.
METHODS
Plant Materials and Growing Conditions
The paab1-1 mutant was isolated from a tissue culture-derived population of the japonica cultivar Kitaake. The F2 mapping population was derived from a cross between paab1-1 and the japonica cultivar IRAT129. At the heading stage, F2 plants with the paab1-1 phenotype were used for gene mapping. Rice (Oryza sativa) plants were grown in the experimental field at the Chinese Academy of Agricultural Sciences during the natural growing season. Transgenic plants were grown in pots in a greenhouse under standard growth conditions.
For aluminum treatment experiment, the wild-type and paab1-1 seeds were soaked in water overnight at 30°C in the dark and then germinated on wet filter paper at 30°C for 2 d. The germinating seeds were transferred to a net floating on a 0.5 mM CaCl2 solution (pH 4.5). After 3 d, the seedlings were exposed to the same solution but supplemented with 0, 50, or 100 μM AlCl3 for 3 d. Sensitivity of the plants to Al was determined by measuring their root length. To examine whether Al has an effect on the expression level of OsALMT7, 6-d-old seedlings of the wild type and paab1-1 were exposed to 0 or 50 μM AlCl3 for 6 h. The roots and shoots treated with or without Al were harvested for RNA extraction.
TUNEL Assay
Apical spikelet hulls of the wild type and paab1-1 at different developmental stages were collected and fixed in the FAA fixation solution (containing an 18:1:1[v/v] mixture of formalin, 70% ethanol, and acetic acid) for 24 h. The hulls were then dehydrated through an ethanol series (90, 70, and 50%) and embedded in paraffin (Sigma-Aldrich). Tissue cross sections (10 μm in thickness) were cut with a rotary microtome and hydrated through an ethanol series (100, 85, 70, and 50%) and treated with proteinase K in phosphate buffer (pH 7.4). The TUNEL assay was performed with a Dead End Fluorometric TUNEL Kit (Promega) according to the manufacturer’s instructions. The green fluorescence of fluorescein (TUNEL signal) and red fluorescence of propidium iodide were analyzed at 488 nm (excitation) and 520 nm (detection), and 488 nm (excitation) and 610 nm (detection), respectively, under a confocal laser scanning microscope (LSM 700; Carl Zeiss).
Scanning Electron Microscopy and TEM
For scanning electron microscopy, flowers at different developmental stages were fixed in 2.5% glutaraldehyde and 0.1 M phosphate buffer at 4°C overnight. Following ethanol dehydration, samples were critical point dried, sputter coated with gold in an E-100 ion sputter, and observed using a scanning electron microscope (S3400N; Hitachi).
For TEM, apical spikelets of the wild type and paab1-1 at different developmental stages (7, 10, and 13 cm in panicle length) were collected and fixed with 2.5% glutaraldehyde in a 0.1 M phosphate buffer at 4°C overnight. After being rinsed with 0.1 M phosphate buffer, they were incubated in a solution containing 1% (w/v) osmium tetraoxide for 4 h at room temperature. The samples were subsequently dehydrated through a graded series of ethanol and then embedded in acrylic resin (London Resin Company). Ultrathin sections (50–70 nm) were double stained with 2% (w/v) uranyl acetate and 2.6% (w/v) lead citrate aqueous solutions and observed with a JEM-1230 transmission electron microscope (JEOL) at 80 kV.
DNA Laddering Analysis
Apical spikelets of the wild type and paab1-1 at different developmental stages (7, 10, and 13 cm) were collected and ground in liquid nitrogen. The DNA was exacted by the CTAB method as previously described (Dellaporta et al., 1983). Then, the DNA concentration was estimated using NanoDrop (ND-1000). For each sample, 2 µg DNA was loaded per lane and separated on a 2% (w/v) agarose gel by electrophoresis. Fragmented DNA was captured under UV light with a GEL Doc XR Imager.
Quantitative Measurement of H2O2 Content
H2O2 was extracted from panicles of different developmental stages according to the previously described method (Rao et al., 2000). Reactions for quantifying H2O2 content were performed using a hydrogen peroxide assay kit (Beyotime) according to the manufacturer’s instructions. The H2O2 content was measured instantly with a spectrometer (SpectraMax Plus384) at a wavelength of 560 nm. The measurements were performed in triplicate and the data were analyzed by the variance (ANOVA), and means were compared using a Student’s t test.
MDA Content Analysis
MDA accumulation has been considered an indicator of lipid peroxidation and cell death (Hodges et al., 1999). For determination of MDA content, panicle samples (0.2 g fresh weight) were homogenized in 5 mL of phosphate buffer (pH 7.8) and centrifuged at 3,000g for 10 min. The supernatant was mixed with an equal volume of 5% (w/v) trichloroacetic acid containing 0.5% (w/v) thiobarbituric acid. The mixture was incubated at 95°C for 15 min and then immediately cooled in an ice bath. After centrifugation at 3,000g for 10 min, the absorbance of the supernatant was measured at 450, 532, and 600 nm, respectively. Finally, the MDA content was calculated according to the equation 6.45 × (OD532 − OD600) − 0.56 × OD450 (Lin et al., 2012). The MDA content analysis was performed with three independent replicates on which five different plants were used.
Map-Based Cloning of paab1-1
Map-based cloning was performed using more than 1300 mutant individuals selected from the F2 mapping population. We first developed molecular markers according to sequence polymorphisms between Kitaake and IRAT129 including SSR, InDel, CAPS, and dCAPS. Primers used for generating those markers are listed in Supplemental Table 1. The paab1-1 locus was mapped to an interval between the markers H-2 and RM525 on the long arm of chromosome 2. Fine mapping anchored the mutation site to an 82-kb genomic region between the markers C1 and H-4. This region spans the BAC clones OJ1493-H11, OJ1197-E09, and P0657H12. The primers used for amplifying the candidate gene LOC_Os02g45160 are listed in Supplemental Table 1.
Constructs for Complementation Test, RNAi, and CRISPR
For the genetic complementation test, a 6-kb genomic fragment covering the entire coding region of LOC_Os02g45160, plus 1705 bp upstream sequence and 997 bp downstream sequence, was amplified by PCR from paab1-1 with the primer pair p2300-OsALMT7-F and p2300-OsALMT7-R and inserted into the binary vector pCAMBIA2300 using the restriction endonucleases SacI and SalI. To generate the OsALMT7 RNAi construct, two inverted repeats of 325 bp were amplified from the wild type with the primer pair RNAi-F and RNAi-R. The PCR fragments were sequentially cloned into the pCUbi1390-ΔFAD2 vector in the sense and antisense orientations using the XhoI/KpnI and BamHI/XbaI sites to create the RNAi construct pUbi-dsRNAi OsALMT7. To create OsALMT7 CRISPR lines, an 18-bp sgRNA targeting the first exon of OsALMT7 was cloned into the CRISPR-Cas9 expression vector according to the method previously described (Miao et al., 2013). The resulting construct was introduced into the Kitaake (wild type) by Agrobacterium tumefaciens-mediated transformation as described (Hiei and Komari, 2008). The primers for creating the above constructs are listed in Supplemental Table 2. The CRISPR transgenic plants were genotyped by PCR amplification and DNA sequencing.
RNA Extraction and RT-qPCR Analysis
Total rice RNA was extracted using a RNA prep pure kit (Zymo Research) and treated with DNaseI according to the manufacturer’s instructions. The first-strand cDNA was synthesized from 2 μg of total RNA with oligo(dT) as the primer, using a reverse transcription kit (TaKaRa). Subsequently, the first-strand cDNA was used for PCR amplification with the primer pair cF1 and cR1 to detect unusual splicing caused by the base pair substitution mutation (Supplemental Table 2).
RT-qPCR was performed using ABI7500HT Fast real-time PCR system with the SYBR Premix Ex Taq (TaKaRa; RR041A), following the manufacturer’s instructions. The rice UBIQUITIN gene was used as an internal control. Gene expression level was determined from three independent replicates, each consisting of five plants for tissue samples, and three technical replicates per tissue sample were analyzed. Gene-specific primers are shown in Supplemental Table 2.
GUS Staining Assay
For promoter activity analysis, a 2614-bp genomic fragment upstream of the ATG start codon was PCR-amplified from wild-type genomic DNA with the primer pair promoter-F and promoter-R (Supplemental Table 2) and fused to the GUS reporter gene in the binary vector pCAMBIA1305. The resulting construct pOsALMT7 promoter-GUS was introduced into the wild type by the Agrobacterium-mediated transformation method. Histochemical staining of GUS activity in the transgenic plants was performed as previously described (Jefferson, 1987). Images were captured using Leica Application Suite 3.3.0 software.
Subcellular Localization of OsALMT7
To investigate the subcellular localization of OsALMT7, the coding sequence of OsALMT7 was amplified with the primer pair pAN580-P1F and pAN580-P1R from the wild-type plant and then cloned into the pAN580 vector to generate an N-terminal fusion with GFP under control of the CaMV 35S promoter, resulting in the pAN580-OsALMT7-GFP construct. cDNAs corresponding to the two mutant versions of transcripts were amplified with the primer pairs (pAN580-P2F/pAN580-P2R and pAN580-P3F/pAN580-P3R; Supplemental Table 2) from paab1-1 and cloned into pAN580, generating pAN580-paab1-t1-GFP and pAN580-paab1-t2-GFP, respectively. Each of those constructs was then transiently coexpressed with PIP2-mCherry, a plasma membrane marker, in rice protoplasts according to the protocol described previously (Bart et al., 2006; Lee et al., 2009). Stable transgenic plants carrying the OsALMT7-GFP fusion was also produced by the Agrobacterium-mediated method. OsALMT7ʹ-GFP fusion construct was created with three synonymous base substitutions at the sgRNA targeted site and then was cotransformed into the variety Zhonghua 11 together with the OsALMT7 CRISPR-Cas9 construct. GFP fluorescence in the rice protoplasts and roots of transgenic plants was visualized with a confocal laser scanning microscope (LSM 700).
In addition, the construct pAN580-OsALMT7-GFP and the membrane marker were heterologously expressed in onion epidermal cells by the particle bombardment method (Bio-Rad PDS-1000) according to the protocol previously described (Von Arnim, 2007). To induce plasmolysis, cells were treated with 1 M mannitol for 15 min. The GFP signal in the epidermal cells was visualized with a confocal laser scanning microscope (LSM 700).
Phylogenetic Analysis of ALMT Proteins
Amino acid sequences homologous to OsALMT7 were downloaded from the National Center for Biotechnology Information website (http://blast.ncbi.nlm.nih.gov/). Multiple sequence alignments of these homologs (Supplemental Data Set 1) were performed using ClustalX 2.0 and the phylogenetic tree was constructed using the neighbor-joining method (MEGA5 software).
Complementation Test of OsALMT7 in Escherichia coli
Functional complementation test of OsALMT7 in E. coli was conducted according to the method described previously (Lee et al., 2008). The wild-type strain K12 (CGSC4401) and its dicarboxylate transport mutant strain CBT315 (CGSC5269) were obtained from The E. coli Genetic Resources Center at Yale University (http://cgsc.biology.yale.edu). The OsALMT7 cDNA fragment was cloned into the EcoRI site of the pKK223-3 vector with the primer pair pkk223-OsALMT7-F and pkk223-OsALMT7-R (Supplemental Table 2) under the control of the tac promoter, forming the construct pKK223-OsALMT7.Then, the mutant stain CBT315 was transformed with the construct pKK223-OsALMT7. The empty vector (pKK223-3) was transformed into CBT315 as a negative control and into K-12 as a positive control. All strains were grown on M9 medium supplemented with 10 mM l-malic acid as the sole carbon source (pH adjusted to 6.6 with NaOH) at 37°C for 3 d. To confirm OsALMT7 expression in the CBT315 strain, immunoblotting was performed using standard protocols with anti- OsALMT7 antisera (Abmart; 15222), and the signal was detected by an enhanced HRP-DAB substrate kit (PA110; Tiangen).
In Vitro Transcription and Expression in Xenopus laevis Oocytes
For functional analysis in oocytes, the coding sequences of OsALMT7, paab1-t1, and paab1-t2 were cloned into the pGEMHE vector (Liman et al., 1992). The cRNAs were prepared in vitro using the T7 RiboMAX large-scale production system (Promega). Oocytes were isolated and maintained in the ND96 solution prior to the injections. The oocytes were injected with or without (control) 25 ng of cRNA in 50 nL water and incubated at 17°C in a modified Barth’s solution containing 88 mM NaCl, 1.0 mM KCl, 0.91 mM CaCl2, 0.33 mM Ca(NO3)2, 0.82 mM MgSO4, 2.4 mM NaHCO3, and 10 mM HEPES-NaOH (pH 7.5). Electrophysiological experiments were performed 2 d after cRNA injection, as described previously (Xu et al., 2006).
Electrophysiological Measurements in X. laevis Oocytes
Whole-cell recordings from oocytes were performed under constant perfusion at room temperature (22°C) with a Gene-Clamp 500B amplifier (Axon Instruments) using the conventional two-electrode voltage-clamp technique. The recording electrodes were filled with 3 M KCl. The inward currents were recorded in oocytes that were preloaded with 50 nL water or 200 mM Na-malate. For the recording involved in different intracellular Na-malate concentrations, oocytes were injected with 50 nL of 100, 200, and 400 mM Na-malate to have intracellular Na-malate concentration of 8, 16, and 32 mM, respectively. The main bath solution consisted of 96 mM NaCl, 1.8 mM KCl, 1.8 mM CaCl2, and 0.1 mM LaCl3, with or without 0.1 mM AlCl3 (pH 4.5). The holding potential was set to 0 mV and voltage test pluses were stepped between −180 and 0 mV (in 10 mV increments). The bath solutions used for outward current recording and anion selectivity experiment contained 25 mM NaCl, 25 mM NaNO3, 25 mM Na2SO4, 25 mM Na-malate, or Na-fumarate, added to the same solution (100 mM Na-gluconate, 100 mM Ca-gluconate, 100 mM Mg-gluconate, 100 mM K-gluconate, and 10 mM MES, pH 4.5). The voltage test pluses were stepped between 0 and 100 mV (in 10-mV increments). The current-voltage (I/V) relationships were constructed by measuring the current amplitude at the end of test pulses. Reversal potential of oocytes expressing OsALMT7 was measured in bath solutions with 1, 10, 25, or 50 mM malate, after preloading with 46 nL of 100 mM malate in oocytes. Reversal potential was valued through cross points of the I-V curves and x axes (I = 0). In addition, inside-out single-channel recording in oocytes was performed following the method (Maksaev and Haswell, 2015). The pipette solution contains 25 mM malate, 5 mM Mg-gluconate, 0.5 MgCl2, 10 mM MES/Tris (pH 7.2), and d-mannitol (Π = 220 mosmol kg−1). The bath solution contains 35 mM malate, 5 mM Mg-gluconate, 0.5 MgCl2, 10 mM MES/Tris (pH 7.2), and d -mannitol (Π = 220 mosmol kg−1).
Malate Content Analysis
For malate transport assay, 5-d-old seedlings were transferred into a hydroponic solution containing 0 or 0.3 mM malate and treated for 24 h. The roots and shoots were collected and rinsed three times with deionized water. Malate content was measured according to a previously described enzymatic method (Delhaize et al., 1993). Briefly, samples (0.2 g fresh weight) obtained from seedlings after malate treatment and panicles at different elongation stages were homogenized in 3 mL water and incubated at 75°C for 15 min. After centrifugation, 0.2 mL of the supernatant was taken and mixed with 2.4 mL buffer (0.1 M glycine, 0.1 M glutamate, 2 mM NAD, and 2 μL of 1KU GOT). Then, the mix was preincubated for 10 min to obtain a stable absorbance reading at 340 nm, followed by addition of 5 μL 5KU malate dehydrogenase to activate the reaction for 10 min. The absorbance readings before and after the addition of malate dehydrogenase were recorded and used for calculation of NADH production and malate content. The malate content analysis was performed with three independent replicates and each sample included five plants.
Nitrate Content Analysis
Nitrate contents in different portions of panicles were determined according to the salicylic acid method (Vendrell and Zupancic, 1990). Specifically, samples (0.1 g fresh weight) were frozen with liquid nitrogen, ground to powder, and then suspended in 1 mL of deionized water, followed by boiling at 100°C for 20 min. After centrifugation at 15,000g for 10 min, 0.1 mL supernatant was transferred into a new tube. Then, 0.4 mL of salicylic acid-sulfate acid (5 g salicylic acid in 100 mL sulfate acid) was added into the tube. After thorough mixing, the reactions were incubated at room temperature for 30 min, and then 9.5 mL of 8% NaOH solution was added. The reaction was cooled down to room temperature before the absorbance was measured at 410 nm. Nitrate concentration was determined according to a standard curve made with KNO3 at concentrations between 10 and 100 mg/L. Finally, the nitrate content in samples was calculated using the following equation: Y = CV/W (Y, nitrate content; C, nitrate concentration; V, total volume of extracted sample; W, weight of sample). The nitrate content analysis was performed with three independent replicates and each sample included five plants.
Malate Injections to Developing Panicles
Field-grown wild-type and paab1-1 plants were used for the malate injection experiments. First, tillers of the wild-type plants with panicles at approximately the 7-cm stage were tested by injecting either distilled water or 1.5 mM malate (pH 5.2) to examine if the injection method and exogenous supply of malate have any effect on growth of the young panicles. Injection to tillers was performed with a 1-mL plastic syringe at a site slightly above the top of a panicle. A volume of 0.5 mL was injected to each of the chosen tillers. Subsequently, the paab1-1 tillers with panicles at either the 7- or 5-cm stage were injected with 0.5 mL of water or 3 and 6 mM malate, respectively, in the same way. In addition, sequential injections (4 d after the first injection) were applied to some of the tillers. Ten tillers were used for each treatment. When the treated panicles emerged out of the flag leaf sheaths, the spikelet abortion rate was calculated and then analyzed by the Student’s t test.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: OsALMT7, AK108963; OsVPE2, AK067597; OsVPE3,AK070079; and UBIQUITIN, AK059011.
Supplemental Data
Supplemental Figure 1. Impaired Spikelet Growth and Reduced Grain Size in Middle Portion of the paab1-1 Panicle.
Supplemental Figure 2. Degenerating Flower Organs in Top Spikelets of paab1-1 Panicle.
Supplemental Figure 3. Scanning Electron Microscopy of Apical Spikelets in the Wild Type and paab1-1 at Early Stages.
Supplemental Figure 4. Histological Analysis of Apical Spikelets in the Wild Type and paab1-1.
Supplemental Figure 5. Microscopic Observation of the Apical Spikelet Hulls from Panicles of Three Selected Developmental Stages.
Supplemental Figure 6. DNA Fragmentation in the paab1-1 Apical Spikelets.
Supplemental Figure 7. Alignment of the Amino Acid Sequences of OsALMT7 in Wild-Type, paab1-1, and paab1-2 mutants.
Supplemental Figure 8. Reverse-Transcription Quantitative PCR Analysis of OsALMT7 and Mutant Transcripts in the Wild Type and paab1-1 Mutant, Respectively.
Supplemental Figure 9. Characterization of the OsALMT7 Allelic Mutant paab1-2.
Supplemental Figure 10. Phenotypic Observations of OsALMT7-RNAi Transgenic Lines in Four japonica Backgrounds.
Supplemental Figure 11. Relative Expression of OsALMT7 Homologs in OsALMT7 Knockdown Plants.
Supplemental Figure 12. Transmembrane Helix Prediction of OsALMT7.
Supplemental Figure 13. Phylogenetic Tree of ALMT Proteins.
Supplemental Figure 14. Promoter Activity Analysis of OsALMT7 Using GUS as a Reporter.
Supplemental Figure 15. OsALMT7 Is Localized in the Plasma Membranes of Transgenic Rice and Onion Epidermal Cells.
Supplemental Figure 16. Plasma Membrane Localization and Functional Evaluation of the OsALMT7-GFP Fusion Protein.
Supplemental Figure 17. Inside-out Single Channel Currents from Oocytes Expressing OsALMT7 and Water (Control) at Two Membrane Potentials.
Supplemental Figure 18. OsALMT7-Mediated Outward Currents Recorded from Oocytes.
Supplemental Figure 19. OsALMT7-Mediated Inward Currents Are Not Activated by Al.
Supplemental Figure 20. Al Sensitivity Is Not Changed in paab1-1.
Supplemental Figure 21. Nitrate Content Is Not Changed in the paab1-1 Panicle.
Supplemental Figure 22. Injection of Malate to paab1-1 Plants with Panicles at 7 cm Stage Did Not Alleviate the Apical Panicle Abortion.
Supplemental Table 1. Molecular Marker Primers Used in Map-Based Cloning.
Supplemental Table 2. Primers Used for Plasmid Construction and Functional Analysis
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 13.
Acknowledgments
We thank Weihua Wu (China Agricultural University) for help with electrophysiological experiments. This research was supported by the grants from the National Key Research and Development Program of China (2016YFD0100403), the National Transgenic Science and Technology Program (2016ZX0800938B), and the National Natural Science Foundation of China (91535302 and 31401466).
AUTHOR CONTRIBUTIONS
J.W. supervised the project. Z.C., C.W., and J.W. designed the research. Y.H., Y.L., S.L., J.C., and H.Z. performed research. Y.H., Y.L., J.M., and M.W. analyzed data. Z.C. provided the plant material. J.L., J.T., S.Z., Y.R., and C.W. provided technical assistance. C.L. and J.W. cultivated the transgenic plants in the field. X.Z. and X.G. generated the transgenic plants. Y.H. wrote the article. Z.C., C.W., and H.W. revised the article.
References
- Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., Angeles E.R., Qian Q., Kitano H., Matsuoka M. (2005). Cytokinin oxidase regulates rice grain production. Science 309: 741–745. [DOI] [PubMed] [Google Scholar]
- Bai J., et al. (2015). Rice TUTOU1 encodes a suppressor of cAMP receptor-like protein that is important for actin organization and panicle development. Plant Physiol. 169: 1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier-Brygoo H., De Angeli A., Filleur S., Frachisse J.M., Gambale F., Thomine S., Wege S. (2011). Anion channels/transporters in plants: from molecular bases to regulatory networks. Annu. Rev. Plant Biol. 62: 25–51. [DOI] [PubMed] [Google Scholar]
- Bart R., Chern M., Park C.J., Bartley L., Ronald P.C. (2006). A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.H., Murata N. (2002). Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Curr. Opin. Plant Biol. 5: 250–257. [DOI] [PubMed] [Google Scholar]
- Cheng Z.J., Mao B.G., Gao S.W., Zhang L., Wang J.L., Lei C.L., Zhang X., Wu F.Q., Guo X.P., Wan J. (2011). Fine mapping of qPAA8, a gene controlling panicle apical development in rice. J. Integr. Plant Biol. 53: 710–718. [DOI] [PubMed] [Google Scholar]
- De Angeli A., Zhang J., Meyer S., Martinoia E. (2013). AtALMT9 is a malate-activated vacuolar chloride channel required for stomatal opening in Arabidopsis. Nat. Commun. 4: 1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E., Ryan P.R., Randall P.J. (1993). Aluminum tolerance in wheat (Triticum aestivum L.) (II. Aluminum-stimulated excretion of malic acid from root apices). Plant Physiol. 103: 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E., Gruber B.D., Ryan P.R. (2007). The roles of organic anion permeases in aluminium resistance and mineral nutrition. FEBS Lett. 581: 2255–2262. [DOI] [PubMed] [Google Scholar]
- Dellaporta S., Wood J., Hicks J. (1983). A plant DNA minipreparation: Version II. Plant Mol. Biol. Report. 1: 19–21. [Google Scholar]
- Deng M., et al. (2011). Bcl-2 suppresses hydrogen peroxide-induced programmed cell death via OsVPE2 and OsVPE3, but not via OsVPE1 and OsVPE4, in rice. FEBS J. 278: 4797–4810. [DOI] [PubMed] [Google Scholar]
- Durbak A.R., Phillips K.A., Pike S., O’Neill M.A., Mares J., Gallavotti A., Malcomber S.T., Gassmann W., McSteen P. (2014). Transport of boron by the tassel-less1 aquaporin is critical for vegetative and reproductive development in maize. Plant Cell 26: 2978–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie A.R., Martinoia E. (2009). Malate. Jack of all trades or master of a few? Phytochemistry 70: 828–832. [DOI] [PubMed] [Google Scholar]
- Fu J., Huang Z., Wang Z., Yang J., Zhang J. (2011). Pre-anthesis non-structural carbohydrate reserve in the stem enhances the sink strength of inferior spikelets during grain filling of rice. Field Crops Res. 123: 170–182. [Google Scholar]
- Hiei Y., Komari T. (2008). Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 3: 824–834. [DOI] [PubMed] [Google Scholar]
- Hodges D.M., DeLong J.M., Forney C.F., Prange R.K. (1999). Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207: 604–611. [DOI] [PubMed] [Google Scholar]
- Hoekenga O.A., et al. (2006). AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 9738–9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Qian Q., Liu Z., Sun H., He S., Luo D., Xia G., Chu C., Li J., Fu X. (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41: 494–497. [DOI] [PubMed] [Google Scholar]
- Ikeda K., Sunohara H., Nagato Y. (2004). Developmental course of inflorescence and spikelet in rice. Breed. Sci. 54: 147–156. [Google Scholar]
- Ikeda K., Nagasawa N., Nagato Y. (2005). ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Dev. Biol. 282: 349–360. [DOI] [PubMed] [Google Scholar]
- Ikeda K., Ito M., Nagasawa N., Kyozuka J., Nagato Y. (2007). Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J. 51: 1030–1040. [DOI] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K., Maekawa M., Izawa T., Itoh J., Nagato Y. (2012). ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 69: 168–180. [DOI] [PubMed] [Google Scholar]
- Itoh J., Nonomura K., Ikeda K., Yamaki S., Inukai Y., Yamagishi H., Kitano H., Nagato Y. (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46: 23–47. [DOI] [PubMed] [Google Scholar]
- Jefferson R. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Report. 5: 387–405. [Google Scholar]
- Jeong J., Suh S., Guan C., Tsay Y.F., Moran N., Oh C.J., An C.S., Demchenko K.N., Pawlowski K., Lee Y. (2004). A nodule-specific dicarboxylate transporter from alder is a member of the peptide transporter family. Plant Physiol. 134: 969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., Dong G., Zeng D., Lu Z., Zhu X., Qian Q., Li J. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42: 541–544. [DOI] [PubMed] [Google Scholar]
- Kato Y., Hirotsu S., Nemoto K., Yamagishi J. (2008). Identification of QTLs controlling rice drought tolerance at seedling stage in hydroponic culture. Euphytica 160: 423–430. [Google Scholar]
- Khush G.S. (2000). New plant type of rice for increasing the genetic yield potential. In Rice Breeding and Genetics, Nanda J.S., ed (Boca Raton, FL: Science Publishers; ), pp. 99–108. [Google Scholar]
- Kobayasi K., Yamane K., Imaki T. (2001). Effects of Non-Structural Carbohydrates on Spikelet Differentiation in Rice. Plant Prod. Sci. 4: 9–14. [Google Scholar]
- Komatsu K., Maekawa M., Ujiie S., Satake Y., Furutani I., Okamoto H., Shimamoto K., Kyozuka J. (2003b). LAX and SPA: major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA 100: 11765–11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Maekawa M., Shimamoto K., Kyozuka J. (2001). The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev. Biol. 231: 364–373. [DOI] [PubMed] [Google Scholar]
- Komatsu M., Chujo A., Nagato Y., Shimamoto K., Kyozuka J. (2003a). FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130: 3841–3850. [DOI] [PubMed] [Google Scholar]
- Kovermann P., Meyer S., Hörtensteiner S., Picco C., Scholz-Starke J., Ravera S., Lee Y., Martinoia E. (2007). The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J. 52: 1169–1180. [DOI] [PubMed] [Google Scholar]
- Lee H.K., Cho S.K., Son O., Xu Z., Hwang I., Kim W.T. (2009). Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21: 622–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Choi Y., Burla B., Kim Y.Y., Jeon B., Maeshima M., Yoo J.Y., Martinoia E., Lee Y. (2008). The ABC transporter AtABCB14 is a malate importer and modulates stomatal response to CO2. Nat. Cell Biol. 10: 1217–1223. [DOI] [PubMed] [Google Scholar]
- Li S., Qian Q., Fu Z., Zeng D., Meng X., Kyozuka J., Maekawa M., Zhu X., Zhang J., Li J., Wang Y. (2009). Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J. 58: 592–605. [DOI] [PubMed] [Google Scholar]
- Liman E.R., Tytgat J., Hess P. (1992). Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron 9: 861–871. [DOI] [PubMed] [Google Scholar]
- Lin A., Wang Y., Tang J., Xue P., Li C., Liu L., Hu B., Yang F., Loake G.J., Chu C. (2012). Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 158: 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhou M., Delhaize E., Ryan P.R. (2017). Altered expression of a malate-permeable anion channel, OsALMT4, disrupts mineral nutrition. Plant Physiol. 175: 1745–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksaev G., Haswell E.S. (2015). Expressing and characterizing mechanosensitive channels in Xenopus oocytes. Methods Mol. Biol. 1309: 151–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia E., Rentsch D. (1994). Malate compartmentation: responses to a complex metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45: 447–467. [Google Scholar]
- Mathieu Y., Guern J., Pean M., Pasquier C., Beloeil J.C., Lallemand J.Y. (1986). Cytoplasmic pH regulation in Acer pseudoplatanus cells: II. Possible mechanisms involved in pH regulation during acid load. Plant Physiol. 82: 846–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S., De Angeli A., Fernie A.R., Martinoia E. (2010). Intra- and extra-cellular excretion of carboxylates. Trends Plant Sci. 15: 40–47. [DOI] [PubMed] [Google Scholar]
- Meyer S., Scholz-Starke J., De Angeli A., Kovermann P., Burla B., Gambale F., Martinoia E. (2011). Malate transport by the vacuolar AtALMT6 channel in guard cells is subject to multiple regulation. Plant J. 67: 247–257. [DOI] [PubMed] [Google Scholar]
- Miao J., Guo D., Zhang J., Huang Q., Qin G., Zhang X., Wan J., Gu H., Qu L.J. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23: 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton R.E., Pheasant D.J., Miller C. (1996). Homodimeric architecture of a ClC-type chloride ion channel. Nature 383: 337–340. [DOI] [PubMed] [Google Scholar]
- Miura K., Ikeda M., Matsubara A., Song X.J., Ito M., Asano K., Matsuoka M., Kitano H., Ashikari M. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42: 545–549. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Shimamoto K., Kyozuka J. (2002). Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 29: 743–750. [DOI] [PubMed] [Google Scholar]
- Oikawa T., Kyozuka J. (2009). Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 21: 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeros M.A., Cançado G.M.A., Kochian L.V. (2008a). Novel properties of the wheat aluminum tolerance organic acid transporter (TaALMT1) revealed by electrophysiological characterization in Xenopus oocytes: functional and structural implications. Plant Physiol. 147: 2131–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeros M.A., Cançado G.M., Maron L.G., Lyi S.M., Menossi M., Kochian L.V. (2008b). Not all ALMT1-type transporters mediate aluminum-activated organic acid responses: the case of ZmALMT1-an anion-selective transporter. Plant J. 53: 352–367. [DOI] [PubMed] [Google Scholar]
- Rao M.V., Lee H., Creelman R.A., Mullet J.E., Davis K.R. (2000). Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12: 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A., Sarkar R.K., Yamagishi Y. (1998). Effect of time of nitrogen application on spikelet differention and degeneration of rice. Bot. Bull. Acad. Sin. 39: 119–123. [Google Scholar]
- Sasaki T., Yamamoto Y., Ezaki B., Katsuhara M., Ahn S.J., Ryan P.R., Delhaize E., Matsumoto H. (2004). A wheat gene encoding an aluminum-activated malate transporter. Plant J. 37: 645–653. [DOI] [PubMed] [Google Scholar]
- Sasaki T., Mori I.C., Furuichi T., Munemasa S., Toyooka K., Matsuoka K., Murata Y., Yamamoto Y. (2010). Closing plant stomata requires a homolog of an aluminum-activated malate transporter. Plant Cell Physiol. 51: 354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake N., De Datta S.K., Naylor R.E.L., Tomson W.J. (1991). Lowland rice apical development: stages and cultivar differences detected by electron microscopy. Agron. J. 83: 1013–1023. [Google Scholar]
- Sheehy J.E., Dionora M.J.A., Mitchell P.L. (2001). Spikelet numbers, sink size and potential yield in rice. Field Crops Res. 71: 77–85. [Google Scholar]
- Smith A.M., Stitt M. (2007). Coordination of carbon supply and plant growth. Plant Cell Environ. 30: 1126–1149. [DOI] [PubMed] [Google Scholar]
- Sweetman C., Deluc L.G., Cramer G.R., Ford C.M., Soole K.L. (2009). Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry 70: 1329–1344. [DOI] [PubMed] [Google Scholar]
- Tabuchi H., et al. (2011). LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23: 3276–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.J., Sun Y.J., Xu H.S., Yu S.B. (2011). Identification of quantitative trait locus and epistatic interaction for degenerated spikelets on the top of panicle in rice. Plant Breed. 130: 177–184. [Google Scholar]
- Tanaka W., Pautler M., Jackson D., Hirano H.Y. (2013). Grass meristems II: inflorescence architecture, flower development and meristem fate. Plant Cell Physiol. 54: 313–324. [DOI] [PubMed] [Google Scholar]
- Van Breusegem F., Dat J.F. (2006). Reactive oxygen species in plant cell death. Plant Physiol. 141: 384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendrell P.F., Zupancic J. (1990). Determination of soil nitrate by transnitration of salicylic acid. Commun. Soil Sci. Plant Anal. 21: 1705–1713. [Google Scholar]
- Von Arnim A. (2007). Subcellular localization of GUS- and GFP-tagged proteins in onion epidermal cells. Cold Spring Harb. Protoc. 2007: pdb prot4689. [DOI] [PubMed] [Google Scholar]
- Wagner C.A., Friedrich B., Setiawan I., Lang F., Bröer S. (2000). The use of Xenopus laevis oocytes for the functional characterization of heterologously expressed membrane proteins. Cell. Physiol. Biochem. 10: 1–12. [DOI] [PubMed] [Google Scholar]
- Wegner L.H., Zimmermann U. (2004). Bicarbonate-induced alkalinization of the xylem sap in intact maize seedlings as measured in situ with a novel xylem pH probe. Plant Physiol. 136: 3469–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner L.H., Stefano G., Shabala L., Rossi M., Mancuso S., Shabala S. (2011). Sequential depolarization of root cortical and stelar cells induced by an acute salt shock - implications for Na(+) and K(+) transport into xylem vessels. Plant Cell Environ. 34: 859–869. [DOI] [PubMed] [Google Scholar]
- Xicluna J., Lacombe B., Dreyer I., Alcon C., Jeanguenin L., Sentenac H., Thibaud J.B., Chérel I. (2007). Increased functional diversity of plant K+ channels by preferential heteromerization of the shaker-like subunits AKT2 and KAT2. J. Biol. Chem. 282: 486–494. [DOI] [PubMed] [Google Scholar]
- Xing Y., Zhang Q. (2010). Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61: 421–442. [DOI] [PubMed] [Google Scholar]
- Xu J., Li H.D., Chen L.Q., Wang Y., Liu L.L., He L., Wu W.H. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360. [DOI] [PubMed] [Google Scholar]
- Xu M., Gruber B.D., Delhaize E., White R.G., James R.A., You J., Yang Z., Ryan P.R. (2015). The barley anion channel, HvALMT1, has multiple roles in guard cell physiology and grain metabolism. Physiol. Plant. 153: 183–193. [DOI] [PubMed] [Google Scholar]
- Yamagishi J., Miyamoto N., Hirotsu S., Laza R.C., Nemoto K. (2004). QTLs for branching, floret formation, and pre-flowering floret abortion of rice panicle in a temperate japonica x tropical japonica cross. Theor. Appl. Genet. 109: 1555–1561. [DOI] [PubMed] [Google Scholar]
- Yao Y., Yamamoto Y., Yoshida T., Nitta Y., Miyazaki A. (2000). Response of differentiated and degenerated spikelets to top-dressing, shading and day/night temperature treatments in rice cultivars with large panicles. Soil Sci. Plant Nutr. 46: 631–641. [Google Scholar]
- Ye J., Wang X., Hu T., Zhang F., Wang B., Li C., Yang T., Li H., Lu Y., Giovannoni J.J., Zhang Y., Ye Z. (2017). An InDel in the promoter ofAl-ACTIVATED MALATE TRANSPORTER9 selected during tomato domestication determines fruit malate contents and aluminum tolerance. Plant Cell 29: 2249–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., et al. (2013). TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc. Natl. Acad. Sci. USA 110: 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., Ohmori Y., Kitano H., Taguchi-Shiobara F., Hirano H.Y. (2012). Aberrant spikelet and panicle1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J. 70: 327–339. [DOI] [PubMed] [Google Scholar]