Abstract
Purpose of Review
The opportunities afforded through the recent advent of genome-editing technologies have allowed investigators to more easily study a number of diseases. The advantages and limitations of the most prominent genome-editing technologies are described in this review, along with potential applications specifically focused on cardiovascular diseases.
Recent Findings
The recent genome-editing tools using programmable nucleases, such as zinc-finger nucleases, transcription activator-like effector nucleases, and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9), have rapidly been adapted to manipulate genes in a variety of cellular and animal models. A number of recent cardiovascular disease-related publications report cases in which specific mutations are introduced into disease models for functional characterization and for testing of therapeutic strategies.
Summary
Recent advances in genome-editing technologies offer new approaches to understand and treat diseases. Here, we discuss genome editing strategies to easily characterize naturally occurring mutations and offer strategies with potential clinical relevance.
Keywords: Genome editing, Cardiovascular disease, ZFN, TALEN, CRISPR/Cas9, iPSC
Introduction
Cardiovascular diseases (CVD) continue to account for the main cause of death annually and are responsible for more than $300 billion in direct and indirect costs [1]. Although considerable efforts have aimed at alleviating the burden of CVD, significant advances are still needed as CVD broadly encompasses both hereditary and acquired diseases. We are now evolving past the age of “one size fits most therapeutic treatment options and into a period of personalized medicine. Vast deposits of genome-sequencing data have become increasingly available from individuals with diseases or disease-correlated traits, but much less functional laboratory follow-up has been completed to understand how these genetic differences contribute to the progression of disease. Recent genome-editing technologies have advanced our ability to rapidly study candidate genes of interest in both cellular and animal models. In this review, we highlight several of the most recent technologies used for genome editing. We also describe their use in different in vitro and in vivo model systems and focus our discussion on how recent advances in genome-editing technology may contribute to ultimately understanding and treating CVD.
Tools for Genome Editing
Prior to genome editing, technologies using viral transgene expression to restore gene function or RNA interference (RNAi) to repress dysfunctional genes were regularly used to alter target gene expression for functional analysis [2–4]. These genetic therapeutic technologies have been successfully used to treat a number of disorders [4, 5], albeit with strong limitations such as poor specificity and safety concerns due to potential mutagenesis at the insertion site [6–8]. Further limitations include RNAi as only being useful for knocking down genes and its inability to fully silence expression.
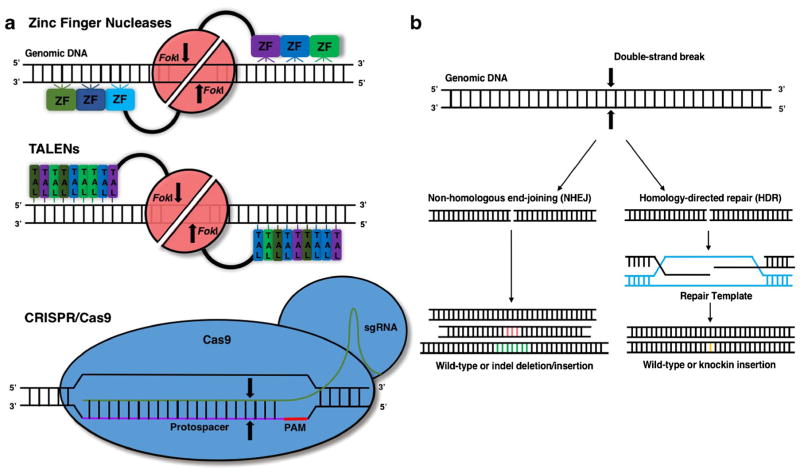
In recent years, genome-editing tools using programmable nucleases have drastically opened up the possibility for restoring genes by inserting protective mutations, correcting deleterious frameshift mutations, and disrupting genes for total knockdown. The most prominent genome-editing technologies, zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9) (Fig. 1a) introduce directed changes in the genome and are explored in more detail in this section. These technologies take advantage of the endogenous DNA repair machinery involved in DNA breaks, such as non-homologous end-joining (NHEJ) and homology-directed repair (HDR) (Fig. 1b) [9]. When a double-stranded break (DSB) occurs, it is most frequently repaired by NHEJ, whereby the DSB ends are directly rejoined without the presence of a repair template. However, since this process is error-prone, insertion or deletion of DNA sequences (indels) can be introduced at the site of the DSB that cause a frameshift and/or truncation of the targeted area; if this occurs in a coding region, the resultant encoded protein may have a deleterious mutation or truncation, and the protein could be rendered nonfunctional. In the other repair pathway of HDR, an exogenous DNA repair template that is either a double-strand DNA vector or a single-strand DNA oligonucleotide is introduced to incorporate specific changes in the genomic DNA sequence. Of note, HDR is limited to proliferating cells and is only active in the S and G2 phases of the cell cycle. Although NHEJ occurs much more frequently than HDR, genetic or chemical components that either inhibit NHEJ or activate HDR may be used to bias the repair outcome [10–12]. For example, design optimization of single-stranded DNA donors to be asymmetric in length and complementary to the non-target strand released first during dissociation of CRISPR-Cas9 from cleaved DNA was reported to substantially increase HDR frequencies [12].
Fig. 1.
General overview of genome-editing tools. a The general architecture of zinc finger nucleases (ZFN), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9) are shown here. For ZFNs, the DNA-binding zinc-finger domains (ZF) recognize 3–4 basepairs (bp) that account for a total recognition of a unique 9–18 bp DNA sequence. TALENs rely on components called TAL repeats that contain a highly conserved 33–35 amino acid repeat with differing, variable amino acids in the 12th and 13th position that can specifically recognize one of the four DNA bases. Both ZFNs and TALENs require a cleavage domain comprising the nuclease domain of the bacterial enzyme, FokI, that dimerizes upon DNA targeting. For the CRISPR/ Cas9 system, a Cas9 nuclease complexed with a synthetic “guide” RNA about 100 nucleotides in length can target and introduce mutations at a specific genomic site. The first 20 nucleotides or so of the guide RNA (protospacer) will recognize and hybridize to a complementary sequence on either stand of a DNA molecule immediately next to a protospacer-adjacent motif (PAM). All of these technologies introduce a double-strand break, as denoted by arrows. b When a DNA double-stranded break occurs, it is repaired by either non-homologous end-joining (NHEJ) or homology-directed repair (HDR). Since NHEJ is error-prone, repair can result in deletions or insertions (indels) at the break site, potentially resulting in frameshifts. During HDR, if a repair template is introduced that contains a mutation, that mutation will be permanently introduced into the genome upon repair
Other tools such as meganucleases [13], adenoviruses [14], and adeno-associated viruses (AAVs) [15] have been used, albeit with lower adaptability and efficiency. With the recent advent of a number of these technologies, it is highly likely that more genome-editing technologies will become available in the near future.
Zinc-Finger Nucleases
ZFNs are engineered, chimeric proteins that are designed in pairs to recognize DNA sequences flanking the genome editing site of interest. The architecture of ZFNs include two protein domains tethered together: (1) a DNA-binding domain from the class of zinc-finger transcription factors, and (2) a cleavage domain comprising the nuclease domain of the bacterial enzyme, FokI. The DNA-binding domain usually includes between three and six zinc-finger domains, with each tandem zinc-finger recognizing 3–4 basepair (bp) that accounts for a total recognition of a unique 9–18 bp DNA sequence. After targeting to the flanking sequences, the two ZFN nuclease domains dimerize and generate a DSB, with subsequent genome editing [16–18].
Although ZFNs offer advantages in flexibility over the classical genome editing strategies like RNAi or viral transgene expression, investigators considering genome editing should understand the accompanying challenges. First, initial engineering of ZFNs to target a specific DNA sequence may prove quite challenging since assembling tandem zinc-finger domains that target a specific DNA sequence is technically challenging, potentially expensive, and normally requires extensive optimization [19]. Furthermore, ZFNs are limited in their site selection, since they generally cannot be designed to target guanine-poor sequences [4].
Transcription Activation-Like Effector Nucleases
TALENs are conceptually similar in their architecture to ZFNs, each comprising a DNA-binding domain fused to a FokI nuclease domain. However, TALENs differ in their binding of DNA by relying on components called TAL repeats; these repeats contain a highly conserved 33–35 amino acid repeat with differing, variable amino acids in the 12th and 13th position [termed the repeat variable diresidue (RVD)] that can specifically recognize one of the four DNA bases [20, 21]. Since RVDs differ in their affinity toward specific nucleotides, tandem TAL repeats can be engineered to target a specific DNA sequence [22–25]. Therefore, TALENs are comparatively easier to engineer than ZFNs. Furthermore, TALENs do not have the same limitations as ZFNs in their ability to target sequences, having numerous dimeric target sequences that on average occur every 3 bp [23–25].
The largest limitation of TALENs is the need to screen and optimize an extensive number of potential pairs since a significant proportion of TALENs do not have robust activity at the desired target sites, especially near highly methylated regions [23, 26, 27]. TALENs also have a preference for the 5′ targeted base being a thymine [4].
Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-Associated 9
CRISPR-Cas9 systems were first identified as a mechanism for bacterial adaptive immunity and have since been developed as a highly promising genome-editing tool [28–30]. A Cas9 nuclease complexed with a synthetic “guide RNA about 100 nucleotides in length can target and introduce mutations at a specific genomic site. The first 20 nucleotides or so of the guide RNA, termed the protospacer, will recognize and hybridize to a complementary sequence on either stand of a DNA molecule immediately next to a protospacer-adjacent motif (PAM) [28, 30, 31]. The first characterized CRISPR/ Cas9 system from Streptococcus pyogenes (SpCas9) is still the most widely used; however, other systems have been adapted from additional bacterial species in the last few years [28, 30–35]. Since investigators need only change the protospacer in the guide RNA in order to target different genomic sites, CRISPR/Cas9 systems for genome editing are particularly easy for investigators to establish in their laboratories.
However, CRISPR/Cas9 systems are not without their limitations; editing requires the presence of a PAM, such as an NGG motif for SpCas9 binding, which only occurs approximately every 8 bp in the genome. This requirement may be burdensome when a precise mutation needs to be introduced and there is no nearby PAM. Recent work has aimed at overcoming this limitation by changing PAM specificity to nonconanical sequences including but not limited to NGA and NGCG sequences [36, 37]. Similar to other genome-editing technologies outlined in this review, the CRISPR/ Cas9 system also suffers from a possibly serious limitation—the potential for “off-target effects from DSBs throughout the genome, including disruption of non-targeted genes, the potential for oncogenesis, and altered cell survival and proliferative capacity. Alterations to CRISPR/Cas9 have demonstrated reduced off-target effects and improved on-target specificity, including conversion of Cas9 protein to a nickase [38, 39], fusing catalytically inactive Cas9 to a FokI nuclease domain [40], and mutagenesis to create “high-fidelity forms of Cas9 [41–43].
Using Genome Editing to Create In Vitro Disease Models
Although in vitro models such as cultured-transformed cell lines are widely used since they are relatively easy to maintain and manipulate, these models do not necessarily accurately reflect human physiology. In vivo models that do, such as nonhuman primates, are extremely costly and difficult to maintain. Therefore, there is a great need for developing model systems that more accurately reflect human physiology and are still cost-effective and easy to maintain.
One such attractive in vitro system uses human pluripotent stem cell (hPSC) lines. Harvested cells from patients can be reprogrammed into induced pluripotent stem cell (iPSC) lines and differentiated into a number of different cell types for functional analysis. However, it can be problematic to compare iPSC lines from different individuals as they are extremely varied in their genetic backgrounds, epigenetics, and ability to differentiate into the desired cell type. Creation of matched, isogenic cell lines using genome editing to introduce only a specific alteration at the region of interest avoids these concerns, allowing for more rigorous characterization of the functional consequence of the genetic alteration. Generation of isogenic hPSC lines using ZFNs, TALENs, or CRISPR/ Cas9 have already furthered our knowledge of cardiomyopathies [44•, 45, 46], lipid metabolism [47•, 48, 49], valvular disease [50], and electrophysiological disorders [51, 52].
Using Genome Editing to Create In Vivo Disease Models
Since existing genome-editing tools are relatively efficient, strains of genetically modified animals, with the mouse model as a leading example, can be generated in the course of weeks, whether by knockout or knockin of specific mutations. Traditionally, genetically modified mice are produced by introducing the desired genetic change in mouse embryonic stem cells. Antibiotic selection is first used to promote homologous recombination that introduces the desired alteration, and correctly targeted stem cells are then injected into blastocysts for production of chimeric mice. Homozygous alleles are then obtained in future generations through breeding. Recently, CRISPR/Cas9 systems have been used to more efficiently edit a specific DNA target by directly injecting the genome-editing machinery into single-cell mouse embryos [53•, 54]. A target gene in this system can be disrupted by NHEJ with either one or more guide RNAs, or a knock-in mouse can be generated if a repair template is injected in parallel with the CRISPR/Cas9 machinery. If a conditional knockout system is preferred, loxP sites flanking the target gene can be introduced using repair templates [54]. To date, CVD has been studied in a variety of generated mammalian models, including but not limited to rats [55–60], rabbits [61–63], and pigs [64, 65]. In zebrafish models alone, cardiac development [66, 67], cardiac regeneration [68], vascular development [69, 70], and inherited cardiomyopathy [71] have been explored.
Somatic in vivo genome editing has also been demonstrated in adult animals with CRISPR/Cas9 delivery via viral vectors and lipid nanoparticles [34, 72•, 73]. To efficiently deliver genetic material in adult animals, viral vectors such as adenovirus and adeno-associated virus (AAV) can be used. AAVs are the preferred method of delivery for clinical use since adenovirus has been shown to induce the innate immune response [74, 75] and lentivirus can lead to oncogenesis [76], but AAVs are limited in their “cargo size capacity to approximately 4.8 kb. Unfortunately, SpCas9 delivered with a promoter and polyadenylation sequence, along with a guide RNA expression cassette, is too large to fit into a single AAV, and so recent work has aimed at overcoming this limitation. One solution is the production of a transgenic mouse that expresses Cas9 in one or more tissues; delivery of a guide RNA cassette via AAV can rapidly produce a knockdown mouse [77]. For example, a cardiac muscle-specific Cas9 transgenic mouse was created in order to study diseases such as hypertrophic cardiomyopathy [78]. Alternative approaches to effectively deliver Cas9 by AAVs have included using smaller Cas9 proteins, such as from Staphylococcus aureus (SaCas9) [34], as well as dividing SpCas9 into two pieces, delivered via two AAVs, followed by spontaneous reassembly in vivo through intein-mediated splicing [79, 80].
Genome Editing for Therapeutic Benefit
A number of genes have been established to be causal for CVD, some of which encode proteins that are potential therapeutic targets. However, even if a drug that can successfully target a protein is available, it typically must be taken on a daily basis for the entirety of a patient’s life. Furthermore, many associated proteins are not easily targeted by traditional small molecular therapeutics, i.e., are not “druggable. In principle, genome editing offers an entirely new approach for the prevention or treatment of CVD, as with a single application it can permanently change the expression of a target protein that could result in a lifelong therapeutic benefit. In some cases, interventions to disrupt CVD-promoting genes may be useful; in other cases, correction of a disease-promoting mutation may be the optimal strategy.
One example of using genome editing as a therapeutic tool to disrupt a gene that promotes CVD has been the targeting of the gene encoding proprotein convertase subtilisin/kexin type 9 (PCSK9) [81–83]. When PCSK9 is dysregulated, such as through gain-of-function mutations in the PCSK9 gene, the protein impairs low-density lipoprotein (LDL)-cholesterolclearance by acting as an antagonist to the LDL receptor, thereby promoting hypercholesterolemia. Conversely, naturally occurring loss-of-function mutations reduce risk of myocardial infarction by markedly reducing blood cholesterol levels [84]. Due to this observation, two antibody-based therapies targeting PCSK9 havebeen developed and recently approved; however, these therapies must be delivered by injection every few weeks [85–88]. Testing the concept of permanent reduction of cholesterol levels, one study used adenovirus to deliver CRISPR/SpCas9 targeting mouse Pcsk9 to the liver in adult mice, resulting in 90% reduction of blood PCSK9 levels and 40% reduction of blood cholesterol levels [72•]. Similar work using AAV to deliver CRISPR/SaCas9 targeting Pcsk9 in mice showed similar results [34]. More recently, the targeting of human PCSK9 in liver-humanized mice generated by primary human hepatocyte transplantation resulted in 50% reduction in circulating human PCSK9 levels, establishing the efficacy of CRISPR/ Cas9 in human hepatocytes in vivo [89].
A number of types of CVD such as inherited cardiomyopathies and rhythm disorders are largely caused by single, dominant mutations that result in dysfunctional protein function. Using genome editing, these diseases could be addressed by (1) disruption of the mutant allele by NHEJ to remove the dominant effect of the allele, (2) correcting the mutant allele with HDR in the presence of a repair template, or (3) inserting extra copies of the wild-type allele with HDR to dilute the effect of the mutant allele. The first approach, involving disruption of the mutant allele, has been partially validated through the use of RNAi in a mouse model of hypertrophic cardiomyopathy [90]. Stringent specificity toward the mutant allele, with sparing of the wild-type allele, is imperative for this approach since silencing of the working gene could prove deleterious. The second and third approaches may be exceedingly difficult to achieve, since there is essentially no HDR in non-proliferative adult cardiomyocytes. However, recent studies in mouse models correcting type 1 tyrosinaemia [73] and ornithine transcarbamylase deficiency [91] suggest that HDR-based therapies might be successful if used in very young patients or even in utero, when the heart is still growing and cardiomyocytes are still proliferating.
While promising for future therapeutic applications, several challenges and limitations remain as to the use of genome-editing tools in human patients. The predominant concern that has garnered much attention from the scientific community has been the potential for off-target mutagenesis. Indeed, it would be detrimental to introduce a mutation that causes oncogenesis, promotes a different disease than the one being treated, or reduces cellular fitness. Even if a gene is permanently silenced without off-target mutagenesis, silencing of the gene may inadvertently cause dysregulation of other complex biological processes to which it had unknown contributions. Furthermore, other limitations of genome editing such as lack of on-target editing efficiency, difficulty in targeting specific tissues in vivo, and the potential for toxicity or immune responses upon delivery require more characterization before genome editing can be routinely used as therapy in humans.
That said, although it will be some years before specific genome-editing therapies to prevent or treat CVD will be approved for use in patients, it is already clear that genome editing will be an important tool in the therapeutic armamentarium. Indeed, treatment of HIV infection in patients via disruption of the CCR5 gene in autologously transplanted T cells has already achieved a measure of success in clinical trials [92••], and a number of other genome-editing therapies are in active development.
Conclusion
The recent advent of a number of genome-editing technologies has allowed for a rapid advancement in the generation of cellular and animal models for functional characterization of diseases, as well as opening the door to a new class of therapeutics. Whether investigators are looking to disrupt disease-associated genes or correct pathogenic mutations, genome-editing technologies are already proving to be transformational. Whether as a research tool or as a therapeutic modality, genome editing will undoubtedly contribute to preventing and alleviating symptoms in patients suffering from CVD.
Footnotes
Compliance with Ethical Standards
Conflict of Interest Alexandra Chadwick and Kiran Musunuru declare they have no conflicts of interest.
Human and Animal Rights and Informed Consent All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, et al. Heart disease and stroke statistics—2013 update: a report from the American Heart Association. Circulation. 2013;127(1):e6–245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kay MA. State-of-the-art gene-based therapies: the road ahead. Nat Rev Genet. 2011;12(5):316–28. doi: 10.1038/nrg2971. [DOI] [PubMed] [Google Scholar]
- 3.Vaishnaw AK, Gollob J, Gamba-Vitalo C, Hutabarat R, Sah D, Meyers R, et al. A status report on RNAi therapeutics. Silence. 2010;1(1):14. doi: 10.1186/1758-907X-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21(2):121–31. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powell SK, Rivera-Soto R, Gray SJ. Viral expression cassette elements to enhance transgene target specificity and expression in gene therapy. Discov Med. 2015;19(102):49–57. [PMC free article] [PubMed] [Google Scholar]
- 6.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457(7228):426–33. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiemann K, Rossi JJ. RNAi-based therapeutics-current status, challenges and prospects. EMBO Mol Med. 2009;1(3):142–51. doi: 10.1002/emmm.200900023. doi:10. 1002/emmm.200900023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9(1):57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 9.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4(6):435–45. doi: 10.1038/nrm1127. doi:10.1038/ nrm1127. [DOI] [PubMed] [Google Scholar]
- 10.Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33(5):543–8. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 11.Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34(3):339–44. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 13.Grizot S, Smith J, Daboussi F, Prieto J, Redondo P, Merino N, et al. Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res. 2009;37(16):5405–19. doi: 10.1093/nar/gkp548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki K, Mitsui K, Aizawa E, Hasegawa K, Kawase E, Yamagishi T, et al. Highly efficient transient gene expression and gene targeting in primate embryonic stem cells with helper-dependent adenoviral vectors. Proc Natl Acad Sci U S A. 2008;105(37):13781–6. doi: 10.1073/pnas.0806976105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan IF, Hirata RK, Wang PR, Li Y, Kho J, Nelson A, et al. Engineering of human pluripotent stem cells by AAV-mediated gene targeting. Mol Ther. 2010;18(6):1192–9. doi: 10.1038/mt.2010.55. doi:10.1038/mt. 2010.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–46. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 17.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25(11):1298–306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 18.Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci U S A. 2007;104(9):3055–60. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5(5):374–5. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326(5959):1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 21.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326(5959):1509–12. doi: 10.1126/science.1178811. doi:10.1126/science. 1178811. [DOI] [PubMed] [Google Scholar]
- 22.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39(12):e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30(5):460–5. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32(6):569–76. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller JC, Zhang L, Xia DF, Campo JJ, Ankoudinova IV, Guschin DY, et al. Improved specificity of TALE-based genome editing using an expanded RVD repertoire. Nat Methods. 2015;12(5):465–71. doi: 10.1038/nmeth.3330. [DOI] [PubMed] [Google Scholar]
- 26.Bultmann S, Morbitzer R, Schmidt CS, Thanisch K, Spada F, Elsaesser J, et al. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res. 2012;40(12):5368–77. doi: 10.1093/nar/gks199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, et al. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol. 2013;31(3):251–8. doi: 10.1038/nbt.2517. [DOI] [PubMed] [Google Scholar]
- 28.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–2. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 32.Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013;110(39):15644–9. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10(11):1116–21. doi: 10.1038/nmeth.2681. doi:10.1038/ nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–91. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirano H, Gootenberg JS, Horii T, Abudayyeh OO, Kimura M, Hsu PD, et al. Structure and Engineering of Francisella novicida Cas9. Cell. 2016;164(5):950–61. doi: 10.1016/j.cell.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523(7561):481–5. doi: 10.1038/nature14592. doi:10.1038/ nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Ge X, Yang F, Zhang L, Zheng J, Tan X, et al. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci Rep. 2014;4:5405. doi: 10.1038/srep05405. doi:10.1038/ srep05405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–9. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31(9):833–8. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32(6):577–82. doi: 10.1038/nbt.2909. doi:10.1038/ nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84–8. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279–84. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–5. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20(6):616–23. doi: 10.1038/nm.3545. doi:10.1038/nm.3545. • This study was one of the first to use genome editing in human pluripotent stem cells for the purpose of cardiovascuar disease modeling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, et al. HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349(6251):982–6. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karakikes I, Stillitano F, Nonnenmacher M, Tzimas C, Sanoudou D, Termglinchan V, et al. Correction of human phospholamban R14del mutation associated with cardiomyopathy using targeted nucleases and combination therapy. Nat Commun. 2015;6:6955. doi: 10.1038/ncomms7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12(2):238–51. doi: 10.1016/j.stem.2012.11.011. doi:10. 1016/j.stem.2012.11.011. • This study was one of the first to use genome editing in human pluripotent stem cells for the purpose of cardiovascuar disease modeling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maetzel D, Sarkar S, Wang H, Abi-Mosleh L, Xu P, Cheng AW, et al. Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann-Pick Type C patient-specific iPS cells. Stem Cell Rep. 2014;2(6):866–80. doi: 10.1016/j.stemcr.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta RM, Meissner TB, Cowan CA, Musunuru K. Genome-edited human pluripotent stem cell-derived macrophages as a model of reverse cholesterol transport—brief report. Arterioscler Thromb Vasc Biol. 2016;36(1):15–8. doi: 10.1161/ATVBAHA.115.305956. doi:10.1161/ATVBAHA. 115.305956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Theodoris CV, Li M, White MP, Liu L, He D, Pollard KS, et al. Human disease modeling reveals integrated transcriptional and epigenetic mechanisms of NOTCH1 haploinsufficiency. Cell. 2015;160(6):1072–86. doi: 10.1016/j.cell.2015.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Liang P, Lan F, Wu H, Lisowski L, Gu M, et al. Genome editing of isogenic human induced pluripotent stem cells recapitulates long QT phenotype for drug testing. J Am Coll Cardiol. 2014;64(5):451–9. doi: 10.1016/j.jacc.2014.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang M, D’Aniello C, Verkerk AO, Wrobel E, Frank S, Wardvan Oostwaard D, et al. Recessive cardiac phenotypes in induced pluripotent stem cell models of Jervell and Lange-Nielsen syndrome: disease mechanisms and pharmacological rescue. Proc Natl Acad Sci U S A. 2014;111(50):E5383–92. doi: 10.1073/pnas.1419553111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–8. doi: 10.1016/j.cell.2013.04.025. doi:10.1016/j.cell.2013.04.025. • This study established the high degree of efficacy of CRISPR/Cas9 in mouse embryos, foreshadowing its efficacy in embryos from a wide variety of species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. Onestep generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–9. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacob HJ, Lazar J, Dwinell MR, Moreno C, Geurts AM. Gene targeting in the rat: advances and opportunities. Trends Genet. 2010;26(12):510–8. doi: 10.1016/j.tig.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moreno C, Hoffman M, Stodola TJ, Didier DN, Lazar J, Geurts AM, et al. Creation and characterization of a renin knockout rat. Hypertension. 2011;57(3):614–9. doi: 10.1161/HYPERTENSIONAHA.110.163840. doi:10.1161/ HYPERTENSIONAHA.110.163840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Endres BT, Priestley JR, Palygin O, Flister MJ, Hoffman MJ, Weinberg BD, et al. Mutation of Plekha7 attenuates salt-sensitive hypertension in the rat. Proc Natl Acad Sci U S A. 2014;111(35):12817–22. doi: 10.1073/pnas.1410745111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larcher T, Lafoux A, Tesson L, Remy S, Thepenier V, Francois V, et al. Characterization of dystrophin deficient rats: a new model for Duchenne muscular dystrophy. PLoS One. 2014;9(10):e110371. doi: 10.1371/journal.pone.0110371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J, Xie D, Huang J, Lv F, Shi D, Liu Y, et al. Cold-inducible RNA-binding protein regulates cardiac repolarization by targeting transient outward potassium channels. Circ Res. 2015;116(10):1655–9. doi: 10.1161/CIRCRESAHA.116.306287. [DOI] [PubMed] [Google Scholar]
- 60.Zhu X, Fang J, Jiang DS, Zhang P, Zhao GN, Zhu X, et al. Exacerbating pressure overload-induced cardiac hypertrophy: novel role of adaptor molecule Src homology 2-B3. Hypertension. 2015;66(3):571–81. doi: 10.1161/HYPERTENSIONAHA.115.05183. doi:10.1161/HYPERTENSIONAHA.115. 05183. [DOI] [PubMed] [Google Scholar]
- 61.Yang D, Xu J, Zhu T, Fan J, Lai L, Zhang J, et al. Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. J Mol Cell Biol. 2014;6(1):97–9. doi: 10.1093/jmcb/mjt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji D, Zhao G, Songstad A, Cui X, Weinstein EJ. Efficient creation of an APOE knockout rabbit. Transgenic Res. 2015;24(2):227–35. doi: 10.1007/s11248-014-9834-8. [DOI] [PubMed] [Google Scholar]
- 63.Niimi M, Yang D, Kitajima S, Ning B, Wang C, Li S, et al. ApoE knockout rabbits: a novel model for the study of human hyperlipidemia. Atherosclerosis. 2016;245:187–93. doi: 10.1016/j.atherosclerosis.2015.12.002. doi:10.1016/j. atherosclerosis.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, Du Y, Shen B, Zhou X, Li J, Liu Y, et al. Efficient generation of gene-modified pigs via injection of zygote with Cas9/sgRNA. Sci Rep. 2015;5:8256. doi: 10.1038/srep08256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Umeyama K, Watanabe K, Watanabe M, Horiuchi K, Nakano K, Kitashiro M, et al. Generation of heterozygous fibrillin-1 mutant cloned pigs from genome-edited foetal fibroblasts. Sci Rep. 2016;6:24413. doi: 10.1038/srep24413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ota S, Hisano Y, Ikawa Y, Kawahara A. Multiple genome modifications by the CRISPR/Cas9 system in zebrafish. Genes Cells. 2014;19(7):555–64. doi: 10.1111/gtc.12154. [DOI] [PubMed] [Google Scholar]
- 67.Kotani H, Taimatsu K, Ohga R, Ota S, Kawahara A. Efficient multiple genome modifications induced by the crRNAs, tracrRNA and Cas9 protein complex in zebrafish. PLoS One. 2015;10(5):e0128319. doi: 10.1371/journal.pone.0128319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cao J, Navis A, Cox BD, Dickson AL, Gemberling M, Karra R, et al. Single epicardial cell transcriptome sequencing identifies Caveolin 1 as an essential factor in zebrafish heart regeneration. Development. 2016;143(2):232–43. doi: 10.1242/dev.130534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524(7564):230–3. doi: 10.1038/nature14580. doi:10.1038/ nature14580. [DOI] [PubMed] [Google Scholar]
- 70.Novodvorsky P, Watson O, Gray C, Wilkinson RN, Reeve S, Smythe C, et al. klf2ash317 mutant zebrafish do not recapitulate morpholino-induced vascular and haematopoietic phenotypes. PLoS One. 2015;10(10):e0141611. doi: 10.1371/journal.pone.0141611. doi:10.1371/journal.pone. 0141611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zou J, Tran D, Baalbaki M, Tang LF, Poon A, Pelonero A, et al. An internal promoter underlies the difference in disease severity between N- and C-terminal truncation mutations of Titin in zebrafish. Elife. 2015;4:e09406. doi: 10.7554/eLife.09406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115(5):488–92. doi: 10.1161/CIRCRESAHA.115.304351. doi:10.1161/ CIRCRESAHA.115.304351. • This study established the high degree of efficacy of CRISPR/Cas9 in the mouse liver. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34(3):328–33. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muruve DA. The innate immune response to adenovirus vectors. Hum Gene Ther. 2004;15(12):1157–66. doi: 10.1089/hum.2004.15.1157. doi:10.1089/hum.2004.15. 1157. [DOI] [PubMed] [Google Scholar]
- 75.Gregory SM, Nazir SA, Metcalf JP. Implications of the innate immune response to adenovirus and adenoviral vectors. Futur Virol. 2011;6(3):357–74. doi: 10.2217/fvl.11.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Themis M, Waddington SN, Schmidt M, von Kalle C, Wang Y, Al-Allaf F, et al. Oncogenesis following delivery of a nonprimate lentiviral gene therapy vector to fetal and neonatal mice. Mol Ther. 2005;12(4):763–71. doi: 10.1016/j.ymthe.2005.07.358. [DOI] [PubMed] [Google Scholar]
- 77.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–55. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carroll KJ, Makarewich CA, McAnally J, Anderson DM, Zentilin L, Liu N, et al. A mouse model for adult cardiac-specific gene deletion with CRISPR/Cas9. Proc Natl Acad Sci U S A. 2016;113(2):338–43. doi: 10.1073/pnas.1523918113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Truong DJ, Kuhner K, Kuhn R, Werfel S, Engelhardt S, Wurst W, et al. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. 2015;43(13):6450–8. doi: 10.1093/nar/gkv601. doi:10.1093/nar/ gkv601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, et al. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods. 2016;13(10):868–74. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Seidah NG. Proprotein convertase subtilisin kexin 9 (PCSK9) inhibitors in the treatment of hypercholesterolemia and other pathologies. Curr Pharm Des. 2013;19(17):3161–72. doi: 10.2174/13816128113199990313. [DOI] [PubMed] [Google Scholar]
- 82.Lambert G, Charlton F, Rye KA, Piper DE. Molecular basis of PCSK9 function. Atherosclerosis. 2009;203(1):1–7. doi: 10.1016/j.atherosclerosis.2008.06.010. doi:10.1016/ j.atherosclerosis.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 83.Poirier S, Mayer G, Poupon V, McPherson PS, Desjardins R, Ly K, et al. Dissection of the endogenous cellular pathways of PCSK9-induced low density lipoprotein receptor degradation: evidence for an intracellular route. J Biol Chem. 2009;284(42):28856–64. doi: 10.1074/jbc.M109.037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354(12):1264–72. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 85.Shan L, Pang L, Zhang R, Murgolo NJ, Lan H, Hedrick JA. PCSK9 binds to multiple receptors and can be functionally inhibited by an EGF-A peptide. Biochem Biophys Res Commun. 2008;375(1):69–73. doi: 10.1016/j.bbrc.2008.07.106. [DOI] [PubMed] [Google Scholar]
- 86.Duff CJ, Scott MJ, Kirby IT, Hutchinson SE, Martin SL, Hooper NM. Antibody-mediated disruption of the interaction between PCSK9 and the low-density lipoprotein receptor. Biochem J. 2009;419(3):577–84. doi: 10.1042/BJ20082407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ni YG, Condra JH, Orsatti L, Shen X, Di Marco S, Pandit S, et al. A proprotein convertase subtilisin-like/kexin type 9 (PCSK9) C-terminal domain antibody antigen-binding fragment inhibits PCSK9 internalization and restores low density lipoprotein uptake. J Biol Chem. 2010;285(17):12882–91. doi: 10.1074/jbc.M110.113035. doi:10.1074/jbc.M110. 113035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Graham MJ, Lemonidis KM, Whipple CP, Subramaniam A, Monia BP, Crooke ST, et al. Antisense inhibition of proprotein convertase subtilisin/kexin type 9 reduces serum LDL in hyperlipidemic mice. J Lipid Res. 2007;48(4):763–7. doi: 10.1194/jlr.C600025-JLR200. [DOI] [PubMed] [Google Scholar]
- 89.Wang X, Raghavan A, Chen T, Qiao L, Zhang Y, Ding Q, et al. CRISPR-Cas9 targeting of PCSK9 in human hepatocytes in vivo-brief report. Arterioscler Thromb Vasc Biol. 2016;36(5):783–6. doi: 10.1161/ATVBAHA.116.307227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jiang J, Wakimoto H, Seidman JG, Seidman CE. Allele-specific silencing of mutant Myh6 transcripts in mice suppresses hypertrophic cardiomyopathy. Science. 2013;342(6154):111–4. doi: 10.1126/science.1236921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34(3):334–8. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370(10):901–10. doi: 10.1056/NEJMoa1300662. doi:10.1056/ NEJMoa1300662. •• This was the first clinical trial to demonstrate the efficacy of therapeutic genome editing in human patients. [DOI] [PMC free article] [PubMed] [Google Scholar]