Abstract
A key player in the intracellular trafficking network is cytoplasmic dynein, a protein complex that transports molecular cargo along microtubules. Vertebrate dynein’s movement becomes strikingly enhanced upon interacting with dynactin and a cargo-adapter, such as BicaudalD2. However, the mechanisms responsible for increased transport are not well understood, largely due to limited structural information. We used cryo-electron tomography to visualize the three-dimensional structure of the microtubule-bound dynein-dynactin complex from Mus musculus, and show that the dynactin-cargo-adapter complex binds two dimeric dyneins. This configuration imposes spatial and conformational constraints on both dynein dimers, positioning the four motor domains in close proximity and oriented towards the microtubule minus-end. We propose that grouping multiple dyneins onto a single dynactin scaffold promotes collective force production, increased processivity, and favors unidirectional movement, suggesting mechanistic parallels to axonemal dynein. These findings provide structural insights into a previously unknown mechanism for dynein regulation.
Introduction
Precise spatial and temporal delivery of components to specific locations within a cell requires tightly regulated trafficking across a vast microtubule (MT) network1. A key player in intracellular trafficking is cytoplasmic dynein-1 (hereafter dynein), which transports molecular cargo towards MT minus ends. Dynein functions as a multi-subunit complex of dimerized “heavy chains” (DHCs), containing a carboxy- (C)-terminal “motor” domain and an amino- (N)-terminal “tail” region that contains a dimerization domain and attachment sites for several non-catalytic subunits. The dynein motor is distinct from other cytoskeletal motors, composed of an AAA+ ATPase ring interrupted by a coiled-coil stalk with a globular microtubule-binding domain (MTBD)2,3. Notably, purified vertebrate dynein exhibits limited, diffusive movement on MTs. Long-range, minus end-directed movement requires the association of dynactin, a megadalton-sized multi-subunit cofactor, as well as one of various cargo adaptors, such as the N-terminal fragment of BicaudalD2 (BICD2N)4,5. Mutations that disrupt these dynein-cofactor interactions are associated with a variety of neurological pathologies6. Although the manner by which BICD2N structurally mediates interactions between the dynein tail 2 and dynactin has been elucidated by cryo-EM7, a fundamental question remains: How do interactions with the dynein tail confer unidirectional processivity on the dynein motor domains (MDs)?
Results
Structure determination of microtubule-bound dynein-dynactin-BICD2 complex
To understand how dynein is harnessed to yield processive movement, we isolated dynein-dynactin-BICD2N (DDB) complexes bound to microtubules (DDB-MT) from mouse brain tissue following methods previously described8. The sparse, non-periodic decoration of DDB complexes bound to MTs, as well as the ice thickness required to completely embed intact complexes in all orientations around the MTs, precluded the application of traditional single particle cryo-EM methodologies. Thus, we used cryo-ET and subtomogram averaging to determine the 3D structure of this massive, extremely flexible, and asymmetric complex (Fig. 1, Supplementary Figs. 1 and 2, and Supplementary Data Movie 1).
Figure 1.
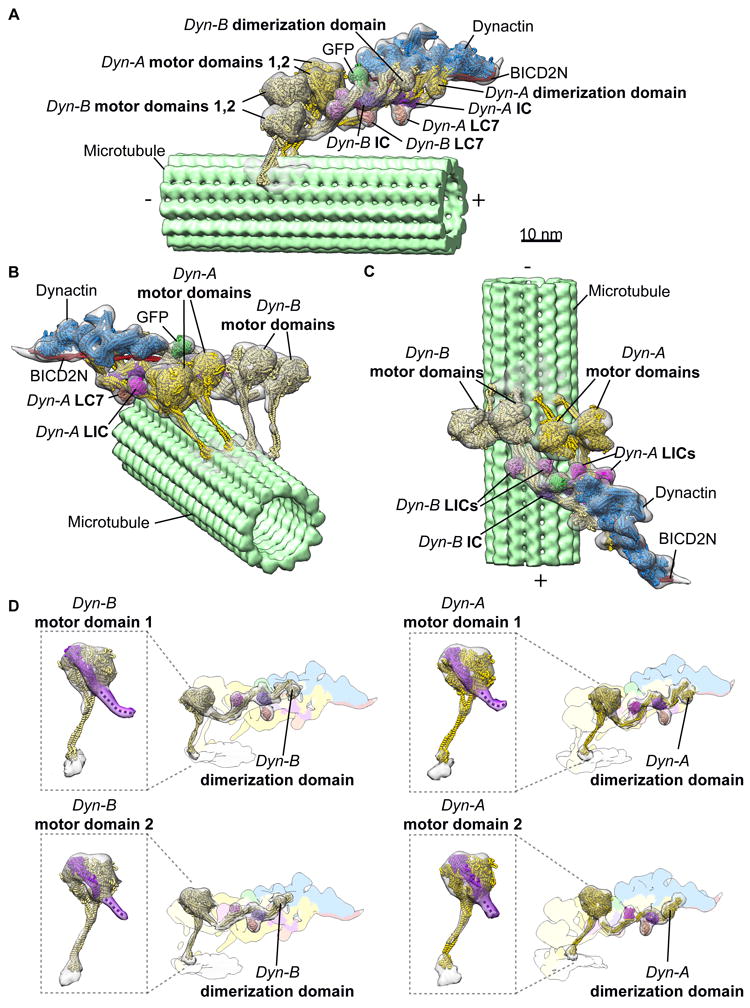
3D organization of the microtubule-bound dynein-dynactin-BICD2N complex. (a-c) Three views of the subtomogram average (gray transparent density) of the MT-DDB complex are shown, with fitted atomic models of dynein dimer-1 (Dyn-A; yellow), dynein dimer-2 (Dyn-B; light yellow), dynactin (blue), BICD2N (red), associated chains (purple, salmon, magenta), and the BICD2N GFP tag (green), and a microtubule model (light green) PDBs used in fitting listed in Methods. (d) Cryo-EM density of each dynein motor domain (boxed region) shows the linker arm (purple) in the post-powerstroke conformation, consistent with AMPPNP binding. Cryo-EM density for each dynein HC and associated subunits with docked models, with the remainder of the cryo-EM density colored according to component composition (all coloring as in a-c).
To facilitate the 3D reconstruction of this complex, we developed an assisted alignment procedure into the RELION subtomogram averaging workflow9, followed by focused refinement of the individual components (dynein tails-dynactin-BICD2N (TDB), and each pair of dynein motors) (Supplementary Figs. 3 and 4, see Methods). The resulting structures were merged in UCSF Chimera10 to obtain the final reconstruction of the intact DDB-MT complex (Fig. 1 and Supplementary Fig. 5, see Methods).
BICD2N mediates the association of two dynein dimers with a single dynactin
The overall organization of the DDB-MT resembles previous structures7,8, but a striking new feature emerged: the presence of two complete dimeric dynein densities bound to dynactin (Fig. 1). The details of the reconstruction were sufficient to visualize the entirety of the four DHCs from the dynactin-bound N-terminus to the C-terminal MDs, and to confirm the post-power-stroke conformation of the motor linker domain11,12, which is consistent with the presence of AMPPNP during the isolation procedure (Fig. 1d). The four MDs are positioned in a row, ~17nm from the MT surface, with weak density attributable to the stalk contacting the corresponding MT. Additionally, the structure displays densities for several other dynein subunits, including the light intermediate chain (LIC), light chain 7 (LC7), and intermediate chain (IC) (Fig. 1), in positions that are consistent with previous studies2,7,8. The majority of the dynein subunits could be rigid-body docked into the DDB-MT structure using individual chains from previous atomic models of human cytoplasmic dynein-1 complexes (EMD-5NW4, 5NVS), only short segments proximal to the MDs had to be repositioned to fit the cryo-EM density. The resulting model of the complete DDB-MT complex shows the organization of two dynein dimers (Dyn-A and Dyn-B) with associated dynein subunits, one dynactin-BICD2N complex, as well as the GFP-tag at the N-terminus of BICD2N (Fig. 1a).
The observation that the dynactin-BICD2N assembly binds to two dynein dimers in the presence of MTs is unexpected because prior motility assays and structural studies concluded that only one dynein dimer was present in the dynactin-BICD2N complex2,4,5,7 (Fig. 1, Supplementary Fig. 6a and d). Manual inspection of the individual raw subtomograms revealed that over 97% of the dynactin densities were associated with four dynein MDs (Fig. 2a, Supplementary Fig. 2b). Importantly, 3D refinement improved the quality of the dynactin density, and also revealed poorly resolved density corresponding to four MDs (Supplementary Fig. 6b). Focused 3D classification on the region surrounding the dynein MDs did not yield any well-resolved 3D classes containing a single dynein dimer (Supplementary Fig. 6c), reinforcing our conclusion. Furthermore, comparison of our reconstruction with previously determined 2D averages of negatively stained DDB-MT complexes8 revealed highly correlated structural features (Supplementary Fig. 6d), suggesting that two dynein dimers were associated with a single dynactin in our earlier 2D averages of DDB-MT complex8. Prior 2D analyses likely hindered the visualization of the four dynein MDs due to flattening effects of the methodology, compounded by the inability to discern overlapping motor densities from a projection image.
Figure 2. Association of two dyneins with dynactin in the presence of cargo adaptor proteins.
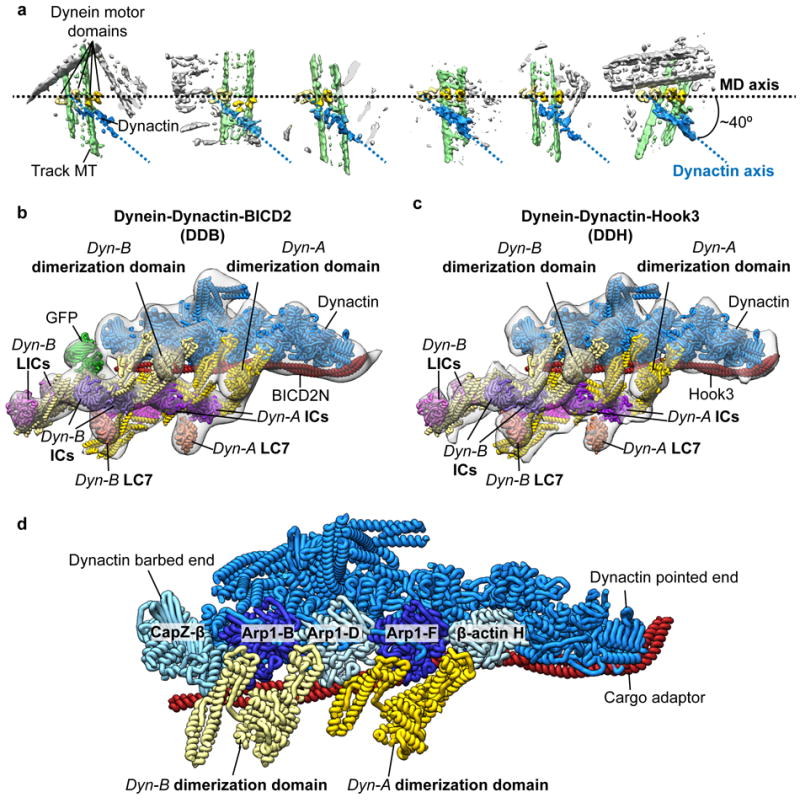
(a) Raw subtomograms show that dynein dimers (motor domains (MDs) colored in two shades of yellow) associate with a single dynactin (blue) in Dyn-adaptor-MT complexes. The MDs are arranged horizontally (axis represented by black dotted line) showing that the dynactin is oriented at a ~40° relative to the MD to the axis. The DDB-associated MT is colored green, non-associated MTs are colored gray. (b-c) Subtomogram averages (gray transparent density) of the dynactin-dynein tail-cargo adaptor portion of the DDH-MT (b) and DDB-MT (c) complexes with docked atomic models of dynein tails (colored as in Fig. 1). Both complexes present a similar overall architecture with two dimeric dyneins bound to a single dynactin. (d) A pseudo-atomic model of the dynactin-dynein tail-cargo adaptor complex shows interactions between two dimeric dynein tails and the dynactin filament. The tail of Dyn-A binds to dynactin across Arp1-F subunit with one heavy chain binding at the interface between ß-actin H and Arp1-F, and the other chain binds at the interface between Arp1-F and Arp1-D. The tail of Dyn-B binds across Arp1-B subunit of dynactin with one heavy chain binding at the interface between Arp1-B and D subunits and the other between Arp1-B and CapZ-β. (e) Subtomogram average (gray transparent density) of the DDB-MT complex with fitted atomic models (as shown in Fig. 1) shows that spacing between ATPase rings and microtubule-binding domains (MTBDs) is ~12nm and ~6nm, respectively. The position of the MDs relative to the entire DDB-MT complex is shown in the inset (right panel).
The ability of BICD2N to facilitate binding of two dynein dimers to a single dynactin complex may explain why a subset of DDB complexes exhibited extreme run lengths (>50 μm) in motility assays5. Furthermore, recent single molecule experiments show that DDB complex velocities on MTs distribute into two populations, with one exhibiting twice the velocity of the other13. Additionally, recent structural studies have shown that dynactin-BICD2N is capable of binding two dimeric dyneins14. Together, these data suggest that regulatory mechanisms exist that influence the DDB’s dynein:dynactin stoichiometry, and that perhaps inclusion of AMPPNP in our brain lysate, which immobilizes DDB complexes on MTs for structural analyses, may have induced a dynein conformation that favors the observed 2:1 stoichiometry.
Hook3 also recruits two dynein dimers to dynactin
To assess whether the recruitment of two dynein dimers is unique to the BICD2N scaffold, we isolated dynein-dynactin complexes bound to MTs in the presence of another cargo adaptor, an N-terminal fragment of Hook3, which was also shown to endow dynein-dynactin with processive motility5,15,16. Strikingly, the subtomogram average of the resulting dynein-dynactin-Hook3 (DDH) complex again revealed two tail domains interacting with dynactin and EM density attributable to two sets of dynein’s accessory subunits (LC, IC, LIC) (Fig. 2c, Supplementary Fig. 7). The fact that the structures of the DDH and DDB are largely indistinguishable (Fig. 2b, c, Supplementary Fig. 7) suggests that recruitment of two dynein molecules to the dynactin-cargo adaptor complex is a widely conserved mechanism for inducing processive motility.
Our 3D reconstruction illustrates how one dynactin-adaptor complex can accommodate two dynein dimers. The previously determined TDB structure showed the dynein tail to be bound to two clefts along dynactin’s Arp filament: one between Arp1-D and F, and the other between Arp1-F and β-actin H7. We observe identical interactions here (Fig. 2b, c). The second dynein tail binds the Arp1 filament in a highly similar fashion, interacting with two adjacent clefts near the barbed-end of dynactin, one between the Arp1-D and B and the other between Arp1-B and the CapZ- dimer (Fig. 2 b, c, d). The fact that neither our study, nor previous studies, observe complexes in which dynein straddles the clefts in the center of the Arp filament (i.e., on either side of Arp1-D) suggests that the dynactin-cargo adaptor interface has evolved to maximize dynein occupancy on dynactin.
Motor domains are positioned for processive motility
In contrast to previous structural studies of isolated DDB complexes7,8,14, our structure reveals the spatial organization of the dynein MDs (MDs) relative to the dynactin complex. Formation of the stable TDB complex appears to constrain the dynein motors in a semi-parallel organization on the microtubule (Fig. 1, Fig. 2a). The MDs are visible, but poorly resolved in the cryo-EM reconstruction prior to focused refinement of these domains (Supplementary Fig. 6b), suggesting that the TDB architecture constrains the lateral positioning of the motors around the MT, yet allows sufficient flexibility to facilitate the conformational changes necessary for dynein motility. Focused refinement of the MDs shows that they are equidistantly spaced ~12 nm apart, with all four MTBDs projecting towards the MT minus end (Fig. 1b,c, Fig. 2e). Interestingly, there is some variability in the transverse angle at which the MD pairs attach to the MT axis, which limits our ability to resolve individual tubulin dimers on the MT lattice (Fig. 1, Fig. 2a). Regardless, the spacing between the MTBDs is consistent with the MT helical protofilament spacing, suggesting that the four MDs associate with four distinct but adjacent MT protofilaments (Fig. 2e). Notably, interactions of the dynein tail with dynactin’s helical Arp filament yield a conspicuous “skewed” organization in which dynactin is oriented approximately 40° relative to the linear array of dynein motors (Fig. 2a).
To confirm that the dynein MD configuration on MTs is promoted by the dynactin-adaptor complex, we used cryo-ET to visualize dynein dimers bound to MTs in the absence of dynactin and adaptors. Manual inspection of 229 sub-volumes showed that isolated dynein dimers bind the MT surface individually, with their MDs at a range of distances from one another (Supplementary Fig. 8), hindering our ability to generate a 3D average of these complexes. Despite this complication, our results suggest that in the absence of cofactors, individual cytoplasmic dynein complexes bind individually to the MT, with the two MDs positioned at variable distances from one another. Thus, not only does the dynactin-cargo adaptor complex recruit multiple dyneins, it positions their MDs in an array highly compatible with unidirectional processive movement. This is consistent with prior work showing that association of a single dynein with dynactin results in a dramatic reorganization of dynein from an auto-inhibited conformation to one that is capable of productive minus-end movement2.
Discussion
In addition to positioning the dynein MDs for processive motility, dynactin can also serve as a scaffold for collective force production. Vertebrate dynein motors have been shown to work collectively to generate forces that far exceed those produced by an individual dynein motor17,18, and this multi-motor coordination may be required to carry out high-load transport processes, such as nuclear positioning, mitotic spindle rotation, and organelle trafficking. A well-characterized example of teamwork among dynein motors can be found in cilliary and flagellar axonemes, where axonemal dyneins are known to work in huge ensembles to accomplish large-scale, synchronized, cilliary and flagellar motility19.
We wondered if the dynein configuration observed in our structures showed any similarities to axonemal dynein. Axonemal dyneins contain a C-terminal MD that is similar to cytoplasmic dynein, but have evolved a distinct N-terminal tail to accommodate its cellular function20. Intriguingly, the spatial organization and 3D shape of not only the dynein MDs, but also much of the tail domain in the DDB-MT structure is strikingly similar to that of sea urchin sperm flagella outer dynein arms in the post-powerstroke state (Fig. 3a, Supplementary Fig. 9)21. In both structures, dynein tails exhibit a flexible “kink” domain projecting from the linker arm, which extends away from the MD to associate with an elongated, filamentous structure – a microtubule doublet in the case of axonemal dynein, and dynactin’s actin-like filament in the case of cytoplasmic dynein (Fig. 3b). This leads us to hypothesize that cytoplasmic and axonemal dyneins utilize a similar mechanism for coordinating the activity of multiple dynein motors, in which parallel arrangement of the MDs relative to the MT allows the conformational change associated with ATP hydrolysis to propel the MTBD more effectively toward the MT minus end 21. Another non-mutually exclusive possibility is that the second motor might increase the duty ratio of the entire complex by providing an additional attachment to the MT lattice and reducing the probability of complex dissociation from the MT during movement. Such an effect could enhance complex processivity, as has been observed in assays that multimerize motors on an artificial scaffolding, such as a bead or DNA chassis22-24. Given that a range of duty ratios are observed in different types of axonemal dyneins25, further study will be required to investigate the extent of the structural parallels between axonemal and cytoplasmic dynein, and if there exist additional regulatory commonalities or evolutionary divergences. However, due to the technical challenges in structurally characterizing this large, conformationally heterogeneous complex, investigating these proposed mechanisms of dynein motility will require a combination of more sophisticated reconstitution systems with improved cryo-ET methodologies.
Figure 3.
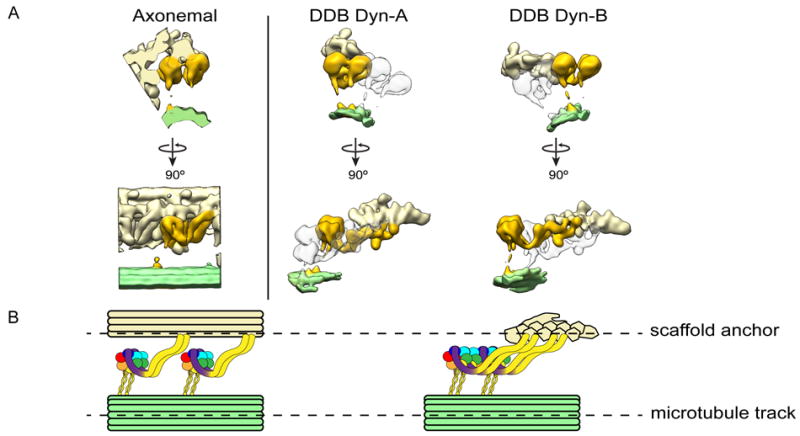
Organizational and mechanistic commonalities between axonemal dynein and cytoplasmic dynein, suggesting a model for processivity. (a) Orthogonal views of the axonemal dynein subtomogram average (EMD-575721) are shown in the left panel. Axonemal dynein (golden) associates with a MT doublet scaffold (light yellow) through its tail and another MT doublet (green) through the MT binding stalk of the motor. The right side panel shows the organization of cytoplasmic dyneins in dynein-dynactin-cargo adaptor-MT complexes. Each of the two dimeric dyneins (Dyn-A and Dyn-B) are highlighted in gold and associate with the dynactin scaffold (light yellow) via the tails and to MT surface (light green) through the MT binding stalk of the motors. (b) Similarities between the overall organization of multiple axonemal dyneins in axoneme (left) and two cytoplasmic dyneins in Dyn-cargo adaptor-MT complexes (right) are shown using diagrammatic representations. Each AAA+ domain with the dynein motor domain is colored uniquely, with linker arm colored purple. In both systems, multiple dyneins are associated with a filamentous scaffold (a MT doublet or dynactin) via N-terminal tail interactions. The dynein motors associate with MT tracks through the binding stalk. In this way, both axonemal and cytoplasmic dyneins integrate into scaffolds to work in teams.
In conclusion, the MT-dynactin-dynein-adaptor complexes presented here provide a platform that integrates decades of biochemical and biophysical studies on the unusual behavior of this large, highly conserved, minus end-directed motor protein, while posing further interesting questions regarding the underlying mechanisms of dynein-mediated intracellular transport.
Online Methods
Purification of MT-bound complexes
The cargo adaptor proteins GFP-BICD2N (a.a. 25-400) and SNAPf-Hook3 (a.a. 1-552) were recombinantly expressed and purified as described previously5,8. MT-bound DDB complexes were prepared from mouse brain tissue as described previously8. Isolation of MT-bound DDH complexes was performed using the same MT-DDB protocol, with a minor modification to incorporate aspects of a protocol established by Amos26 to enrich DDH complex on MTs. We initially removed bulk tubulin from the lysate by adding 6 μM Taxol and 0.2 mM GTP, performing one round of MT polymerization, and then pelleting and discarding the polymerized MTs and MAPs by centrifugation. In order to prevent endogenous dynein from associating with the MTs prior to pelleting, 0.5mM Mg2+-ATP was added to the lysate. This resulted in lysate having a higher dynein-to-tubulin ratio. The remaining tubulin in lysate was then polymerized by adding 10 μM Taxol and 1 mM GTP, and 4mM Mg2+-AMPPNP and 500nM of Hook3 was added to promote engagement of the DDH complexes to the MTs.
MT-engaged dynein was prepared from mouse brain using similar procedures as described for MT-DDH complex, but to prevent the association of endogenous dynactin with dynein, the lysate was not supplemented with recombinant cargo adaptor proteins. The protocol for this work was approved by the TSRI IACUC office under protocol 14-0013.
Grid preparation for cryo-EM analysis
All samples were prepared for cryo imaging in a similar manner. The complex-bound MT pellets were diluted 20 fold with PMEE buffer supplemented with 1mM GTP, 4mM Mg2+-AMPPNP, and 20μM Taxol at room temperature. 5nm colloidal gold (Ted Pella) were pretreated with BSA to prevent aggregation as described previously27. Immediately before freezing, samples were diluted 60 to 120-fold and mixed with the pre-treated colloidal gold (optimal dilution for each sample was determined by screening the cryo-EM grids at a range of concentrations). 4 μl aliquots of sample were applied to freshly plasma-cleaned (75% argon / 25% oxygen mixture) UltrAuFoil grids (Quantifoil) containing holes 1.2 μm in diameter spaced 1.3 μm apart. Plunge freezing was performed using a custom-built manual plunging device. The grid was manually blotted from the side opposite to which the sample was applied with a Whatman 1 filter paper for 5-7 s to remove excess sample. After blotting, the grid with remaining sample was immediately vitrified by plunge freezing into a liquid-ethane slurry. The entire procedure was carried out in a cold room maintained at 4°C and >90% relative humidity.
Cryo-ET data acquisition
Tilt series for DDB-MT and DDH-MT samples were collected using a Thermo Fisher Titan Krios electron microscope operating at 300 keV and equipped with a Gatan K2 Summit direct electron detector. Data acquisition was performed using the UCSF tomography package28 implemented within the Leginon automated data acquisition software29. Tilt series were acquired using a sequential tilting scheme, starting at 0° and increasing to +59° at 1° increments, then returning to 0° and increasing to -59° at 1° increments. Each tilt series was collected with a nominal defocus value that was randomly set to between 6-8 μm for the DDB-MT data set, and 2-5 μm DDH-MT data set. Each tilt was acquired as movies in counting mode using a dose rate of 5.3 e-/pixel/s, with a per-frame exposure time of 80 ms and a dose of 0.09 e-/Å2. The total cumulative dose for each tilt series was 114 e-/Å2, and was distributed throughout the tilts based on the cosine of the tilt angle to account for changing sample thickness with increasing tilt. 154 and 126 tilt series were collected for DDB-MT and DDH-MT samples at a nominal magnification of 14,000X, giving a calibrated pixel size of 2.13 Å/pixel at the detector level.
Tilt series for the dynein-MT sample were collected on a Thermo Fisher Arctica electron microscope operating at 200 keV and equipped with a Gatan K2 Summit direct electron detector operating in movie mode, as described above. The total cumulative dose and dose distribution for each tilt series was same as described for DDB-MT and DDH-MT data sets. Data were collected using the Leginon package29 with an alternating tilt scheme30. A total of 58 tilt series were collected at a nominal magnification of 17,500X, giving a calibrated pixel size of 2.33 Å/pixel at the detector level.
Tomogram reconstruction
Image processing and tomogram reconstructions were performed in similar fashion for all samples. Movie frames for each tilt were translationally aligned to account for beam-induced motion and drift using the GPU frame alignment program MotionCorr31. A frame offset of 7 and a B-factor of 2000 pixels was used for frame alignment. The raw tilts were initially Fourier-binned by a factor of 2. All micrographs were aligned using the 5nm gold beads as fiducial markers, and further binned by a factor of 2 (final pixel size of 8.52 Å/pixel for DDB-MT and DDH-MT datasets, and 9.32 Å/pixel for dynein-MT dataset) for reconstruction in the IMOD package32. Tomograms were reconstructed using simultaneous iterative reconstruction technique (SIRT) with seven iterations in IMOD provided sufficient contrast for the purposes of particle selection. Tomograms were also reconstructed by weighted back projection (WBP) for the purposes of subtomogram averaging.
Subtomogram averaging and data processing
Intact DDB-MT, DDH-MT, or dynein-MT complexes were completely embedded in vitreous ice, such that they fully encircled the MTs, providing all possible views of the complexes attached to MTs (i.e. top, side, and bottom “views” of MT-bound dynein complexes). Sub-volumes containing DDB-MT, DDH-MT, or dynein-MT were manually picked from SIRT-reconstructed tomograms with the EMAN2 single-particle tomography boxer program33. Complexes above or below the MTs when viewed along the z-axis were readily identifiable when examining the z-slices above or below the MT. Picked coordinates for each sub-volume were imported into the RELION 1.4 subtomogram averaging workflow9. 502 and 303 sub-volumes were extracted from the WBP reconstructions of the DDB-MT and DDH-MT datasets, respectively. Sub-volumes were extracted using a cube-size of 96 voxels for the DDB-MT and 84 voxels for the DDH-MT dataset. Reference-free 3D classification in RELION did not yield any structures resembling dynein or dynactin complexes, and instead predominantly produced averages of MTs. Attempts to remove signal from MTs by applying binary masks did not improve our ability to resolve the MT-bound complexes. To overcome this issue, we developed an assisted 3D subtomogram averaging procedure (Supplementary Fig. 2), wherein we manually docked the available reconstruction of the dynein tail-dynactin-BICD2N (TBD) complex (EMDB 28607) into the DDB-MT or DDH-MT sub-volumes using UCSF Chimera10. The docked densities provided the rotational and translational parameters to generate initial subtomogram averages of the DDB and DDH complexes. These initial averages contained recognizable molecular features consistent with the previously published TDB structure (2AFU)7 (Supplementary Fig. 2b). To better resolve different components (dynein tail-adaptor-dynactin region and dynein motors) of the DDB and DDH complexes, focused 3D refinements were performed using 3D ellipsoidal binary masks corresponding to the individual sub-regions, and the same particles were used for refinement of the individual subunit (i.e. no particles were excluded when refining individual components) (Supplementary Fig. 2c). For each component, 3D refinement was performed in RELION using the initial alignment parameters, with a HEALPix order of 3, an angular step size of 7.5°, and an offset range of 5 pixels. All particles were used in the focused 3D refinement to produce better-resolved reconstructions of the individual components of the MT-bound dynein complexes. These refinements resulted in better-defined sub-regions of the MT-DDB and MT-DDH complexes (Supplementary Fig. 2c). The final resolutions of these reconstructions are conservatively estimated to be ~38 Å (by Fourier Shell Correlation at a 0.5 cutoff) (Supplementary Figs. 4, 6a). Composite reconstructions of the DDB-MT and DDH-MT complexes were generated by aligning and stitching together the focused reconstructions using the “vop maximum” function in UCSF Chimera10, which retains the maximum voxel values of overlapping volumes. The following crystal structures and atomic models of individual components were rigid-body docked into the final, combined DDB-MT structure using UCSF chimera10: dynein AMPPNP-bound AAA+ ATPase motor domains (4W8F)12; dynein motor domain stalk (3VKG)34; dynein motor microtubule-binding domain (MTBD) in high affinity state (3J1T)35; human cytoplasmic dynein-1 heavy chain and associated subunits (IC, LIC, LC7) bound to Dynactin and N-terminal GFP-BICD2N (5NW4, 5NVS)2; and green fluorescent protein (GFP) (1GFL)36.
Although the presence of an additional tail dimer and appearance of four dynein motors in the DDB subtomogram average, as well as the absence of GFP in the DDH reconstruction, all serve as internal controls that preclude the introduction of model bias into our refinement procedure, we performed additional control experiments to rule out this possibility. We first tested the ability of our sub-volumes to reproduce the well-resolved dynein tail-adaptor-dynactin region after focused refinement of the MDs. Focused refinement of the motors results in misalignment of the dynein tail-adaptor-dynactin region, resulting in poorly-resolved dynactin density. Re-refining this region using an ellipsoidal binary mask reproduces the dynactin with well-resolved structural features. Next, we docked the TDB complex (EMDB 28607) into the sub-volumes using randomly assigned Euler angles and performed the same refinement strategy outlined above. This was repeated using three unique seeds for randomization, and in each case the resulting subtomogram did not yield a recognizable complex (Supplementary Fig. 3b).
229 dynein-MT sub-volumes were extracted from the WBP tomograms with a cube-size of 96 voxels. As with the DDB and DDH datasets, ab-initio 3D classification mostly resulted in MT averages, and did not yield any recognizable dynein structures. We attempted to perform an assisted alignment approach, which involved placing spherical markers on the individual dynein MDs using IMOD32. However, due to the variability the inter-motor spacing and disordered arrangement of the dyneins relative to the MTs, we were unable to produce a 3D subtomogram average of MT-bound dimeric dynein. The spherical markers in 3D tomograms were used to measure the 3D inter-motor distances shown in Supplementary Fig. 7. Only motors that were visibly connected as a dimer were used for MD-MD distance measurements.
Data Availability
Reconstructed maps of DDB-MT and DDH-MT were deposited in EM Data Bank with accession IDs EMD-7000 and EMD-7001, respectively. The datasets that support the findings of this study are also available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank J. C. Ducom at The Scripps Research Institute High Performance Computing for computational support, and B. Anderson at The Scripps Research Institute electron microscopy facility for microscope support. We also thank E. Mattson for input on the manuscript. D.G. is supported by a National Sciences Foundation predoctoral fellowship. G.C.L. is supported as a Searle Scholar, a Pew Scholar, and by the National Institutes of Health (NIH) DP2EB020402. R.M. is supported by the National Institutes of Health (NIH) R00 grant R00NS089428. T.S. is supported by the Johns Hopkins Krieger School of Arts and Sciences. Computational analyses of EM data were performed using shared instrumentation funded by NIH S10OD021634 to G.C.L. R.J.M. provided purified SNAPf-Hook3.
Footnotes
Author Contributions:
D.A.G. and S.C. prepared the MT-bound complexes and performed all electron microscopy data collection. D.A.G, S.C., Y.X, and G.C.L. performed the image analyses. All authors contributed to the experimental design and assembly of the manuscript.
Competing Financial Interests Statement:
The authors declare no competing financial interests.
References
- 1.Welte MA. Bidirectional transport along microtubules. Curr Biol. 2004;14:R525–37. doi: 10.1016/j.cub.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 2.Zhang K, et al. Cryo-EM Reveals How Human Cytoplasmic Dynein Is Auto-inhibited and Activated. Cell. 2017;169:1303–1314 e18. doi: 10.1016/j.cell.2017.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt H, Carter AP. Review: Structure and mechanism of the dynein motor ATPase. Biopolymers. 2016;105:557–67. doi: 10.1002/bip.22856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 2014;33:1855–68. doi: 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 2014;345:337–41. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoang HT, Schlager MA, Carter AP, Bullock SL. DYNC1H1 mutations associated with neurological diseases compromise processivity of dynein-dynactin-cargo adaptor complexes. Proc Natl Acad Sci U S A. 2017;114:E1597–E1606. doi: 10.1073/pnas.1620141114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urnavicius L, et al. The structure of the dynactin complex and its interaction with dynein. Science. 2015;347:1441–6. doi: 10.1126/science.aaa4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowdhury S, Ketcham SA, Schroer TA, Lander GC. Structural organization of the dynein-dynactin complex bound to microtubules. Nat Struct Mol Biol. 2015;22:345–7. doi: 10.1038/nsmb.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bharat TA, Scheres SH. Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nat Protoc. 2016;11:2054–65. doi: 10.1038/nprot.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–7. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-A crystal structure. Nat Struct Mol Biol. 2012;19:492–7, S1. doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhabha G, et al. Allosteric communication in the dynein motor domain. Cell. 2014;159:857–68. doi: 10.1016/j.cell.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez PA, Ackermann BE, Vershinin M, McKenney RJ. Differential effects of the dynein-regulatory factor Lissencephaly-1 on processive dynein-dynactin motility. J Biol Chem. 2017;292:12245–12255. doi: 10.1074/jbc.M117.790048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urnavicius L, et al. Cryo-EM shows how dynactin recruits two dyneins for faster movement. bioRxiv. 2017 doi: 10.1038/nature25462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroeder CM, Vale RD. Assembly and activation of dynein-dynactin by the cargo adaptor protein Hook3. J Cell Biol. 2016;214:309–18. doi: 10.1083/jcb.201604002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olenick MA, Tokito M, Boczkowska M, Dominguez R, Holzbaur EL. Hook Adaptors Induce Unidirectional Processive Motility by Enhancing the Dynein-Dynactin Interaction. J Biol Chem. 2016;291:18239–51. doi: 10.1074/jbc.M116.738211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rai AK, Rai A, Ramaiya AJ, Jha R, Mallik R. Molecular adaptations allow dynein to generate large collective forces inside cells. Cell. 2013;152:172–82. doi: 10.1016/j.cell.2012.11.044. [DOI] [PubMed] [Google Scholar]
- 18.Rai A, et al. Dynein Clusters into Lipid Microdomains on Phagosomes to Drive Rapid Transport toward Lysosomes. Cell. 2016;164:722–34. doi: 10.1016/j.cell.2015.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wemmer KA, Marshall WF. Flagellar motility: all pull together. Curr Biol. 2004;14:R992–3. doi: 10.1016/j.cub.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 20.Ishikawa T. Structural biology of cytoplasmic and axonemal dyneins. J Struct Biol. 2012;179:229–34. doi: 10.1016/j.jsb.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 21.Lin J, Okada K, Raytchev M, Smith MC, Nicastro D. Structural mechanism of the dynein power stroke. Nat Cell Biol. 2014;16:479–85. doi: 10.1038/ncb2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torisawa T, et al. Autoinhibition and cooperative activation mechanisms of cytoplasmic dynein. Nat Cell Biol. 2014;16:1118–24. doi: 10.1038/ncb3048. [DOI] [PubMed] [Google Scholar]
- 23.Xu J, Shu Z, King SJ, Gross SP. Tuning multiple motor travel via single motor velocity. Traffic. 2012;13:1198–205. doi: 10.1111/j.1600-0854.2012.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derr ND, et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science. 2012;338:662–5. doi: 10.1126/science.1226734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotani N, Sakakibara H, Burgess SA, Kojima H, Oiwa K. Mechanical properties of inner-arm dynein-f (dynein I1) studied with in vitro motility assays. Biophys J. 2007;93:886–94. doi: 10.1529/biophysj.106.101964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 26.Amos LA. Brain dynein crossbridges microtubules into bundles. J Cell Sci. 1989;93(Pt 1):19–28. doi: 10.1242/jcs.93.1.19. [DOI] [PubMed] [Google Scholar]
- 27.Iancu CV, et al. Electron cryotomography sample preparation using the Vitrobot. Nat Protoc. 2006;1:2813–9. doi: 10.1038/nprot.2006.432. [DOI] [PubMed] [Google Scholar]
- 28.Suloway C, et al. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol. 2009;167:11–8. doi: 10.1016/j.jsb.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 30.Hagen WJ, Wan W, Briggs JA. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J Struct Biol. 2017;197:191–198. doi: 10.1016/j.jsb.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–90. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–6. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 33.Galaz-Montoya JG, Flanagan J, Schmid MF, Ludtke SJ. Single particle tomography in EMAN2. J Struct Biol. 2015;190:279–90. doi: 10.1016/j.jsb.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kon T, et al. The 2.8 A crystal structure of the dynein motor domain. Nature. 2012;484:345–50. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- 35.Redwine WB, et al. Structural basis for microtubule binding and release by dynein. Science. 2012;337:1532–1536. doi: 10.1126/science.1224151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang F, Moss LG, Phillips GN., Jr The molecular structure of green fluorescent protein. Nat Biotechnol. 1996;14:1246–51. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Reconstructed maps of DDB-MT and DDH-MT were deposited in EM Data Bank with accession IDs EMD-7000 and EMD-7001, respectively. The datasets that support the findings of this study are also available from the corresponding author upon request.


