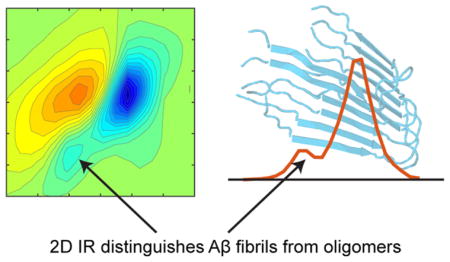
Abstract
We use two-dimensional IR (2D IR) spectroscopy to explore fibril formation for the two predominant isoforms of the β-amyloid (Aβ1–40 and Aβ1–42) protein associated with Alzheimer’s disease. Two-dimensional IR spectra resolve a transition at 1610 cm−1 in Aβ fibrils that does not appear in other Aβ aggregates, even those with predominantly β-sheet-structure-like oligomers. This transition is not resolved in linear IR spectroscopy because it lies under the broad band centered at 1625 cm−1, which is the traditional infrared signature for amyloid fibrils. The feature is prominent in 2D IR spectra because 2D lineshapes are narrower and scale nonlinearly with transition dipole strengths. Transmission electron microscopy measurements demonstrate that the 1610 cm−1 band is a positive identification of amyloid fibrils. Sodium dodecyl sulfate micelles that solubilize and disaggregate preaggregated Aβ samples deplete the 1625 cm 1 band but do not affect the 1610 cm 1 band, demonstrating that the 1610 cm−1 band is due to very stable fibrils. We demonstrate that the 1610 cm−1 transition arises from amide I modes by mutating out the only side-chain residue that could give rise to this transition, and we explore the potential structural origins of the transition by simulating 2D IR spectra based on Aβ crystal structures. It was not previously possible to distinguish stable Aβ fibrils from the less stable β-sheet-rich oligomers with infrared light. This 2D IR signature will be useful for Alzheimer’s research on Aβ aggregation, fibril formation, and toxicity.
Graphical Abstract
INTRODUCTION
β-Amyloid (Aβ) plaques are a hallmark of Alzheimer’s disease (AD), and similar β-sheet-rich plaques are associated with over 20 human amyloid diseases.1 Aβ is produced as a mixture of 38–43 residue isoforms, with the 40 (Aβ1–40) and 42 (Aβ1–42) residue species predominating in humans. Although both spontaneously form amyloid fibrils in vitro, Aβ1–42 is more aggregation-prone and associated with disease states.2–4 Therapeutics capable of clearing Aβ plaques can facilitate recovery from AD symptoms in mice,5,6 and reduced endogenous clearance of Aβ correlates with the onset of AD.7–9 There is also evidence that nonfibrillar oligomers may be the most neurotoxic species.10,11 The characterization of Aβ aggregates and fibrils represents major areas of focus in Alzheimer’s research, as accurate structural characterization is needed to understand the factors that influence and inhibit aggregation. Toward this end, we demonstrate a method to spectroscopically differentiate stable Aβ fibrils from other β-sheet-rich oligomers, a distinction that is difficult or impossible to make with other standard methods in the absence of electron microscopy.
Aβ aggregation is most commonly characterized by circular dichroism (CD), thioflavin-T (ThT) fluorescence, and infrared (IR) spectroscopy. CD provides an estimate of relative β-sheet content, although all β-sheets typically give similar spectra. ThT fluorescence is often used to identify amyloid fibrils, but the dye is also known to bind to other β-sheet-rich aggregates,12 induce amyloid formation,13 be insensitive to major fibril restructuring,14 and fail to produce signal in the presence of amyloid.15 IR spectroscopy resolves the β-sheets of amyloids from the β-sheets of soluble proteins according to their frequencies,16,17 but typically does not distinguish Aβ amyloid fibrils from other, possibly off-pathway, oligomers and aggregates that have high β-sheet content.
Two-dimensional IR (2D IR) spectroscopy offers advantages over these other methods. In particular, the 2D IR signals scale nonlinearly with the absorption coefficient,18–20 resulting in narrower line widths and improved spectral resolution. In a standard IR spectrum, strong transitions can be obscured by more numerous weak transitions, whereas in a 2D IR spectrum, strong transitions tend to dominate because of the nonlinear scaling. β-Sheets of amyloid fibrils have very strong amide I transitions because many backbone carbonyls vibrate in union, and thus become enhanced over random coil and other less regular structures. In this report, we resolve a transition at 1610 cm−1 that allows clear positive identification of very stable Aβ amyloid fibrils in aggregated Aβ samples, distinguishing fibrils from other β-sheet-rich oligomers.
The idea that the structures of intermediates formed en route to amyloid formation differ from the β-sheet structure of the fibrils is not a new concept. There is evidence for transient α-helical intermediates in the aggregation of Aβ, α-synuclein, and amylin.21–26 More germane to the present study are the observations of restructuring of β-sheet structures present in intermediates preceding amyloid fibril formation. For example, there is evidence for antiparallel structure in intermediates of α-synuclein,27 the SH3-domain,28,29 and Aβ30 (though we do not observe the standard signature of antiparallel β-sheet structure in the oligomers observed here). Crystal structures for toxic “cylindrin” intermediates in the aggregation of αB-crystallin have also been reported, and a segment of the Aβ sequence was noted to be compatible with the cylindrin oligomer structure.31 Using 2D IR and isotope labeling, our group has demonstrated the presence of a transient β-sheet in amylin that is ultimately part of a disordered loop in the fiber.32–34 Thus, it appears that oligomers of amyloidogenic proteins may often adopt a different structure than that of the amyloid fibril. In this study, we find that we can exploit the differences in β-sheet sheet structure, even in the absence of isotope labels, to distinguish β-sheet-rich Aβ oligomers from Aβ fibrils.
MATERIALS AND METHODS
Two-Dimensional IR Spectroscopy
The 2D IR spectrometer used has been described previously.35–38 A ca. 3.2 W regenerative amplifier (Spectra Physics, Solstice) outputs 800 nm, 100 fs pulses at a rate of 1 kHz, which pump an optical parametric amplifier (OPA, Light Conversion, TOPAS). Signal (1417 nm) and idler (1845 nm) pulses undergo difference-frequency generation to produce 100 fs mid-IR pulses centered at 1600 cm−1, with bandwidth of approximately 150 cm−1 full width at half-maximum. The mid-IR pulses are split into pump (95%) and probe (5%) beams using a CaF2 wedge. The pump beam passes through a horizontal fully reflective germanium acousto-optic modulator pulse shaper described previously.38 The pulse shaper generates a collinear pair of compressed pump pulses with variable delays that are scanned to generate the pump axis of the 2D IR spectra. Four-frame phase cycling is used to subtract background and suppress pump scatter.37 Pump and probe beams are spatially overlapped and focused at the sample using parabolic mirrors. The temporal delay (t2) between the pump pulse pair and probe beam is controlled using a motorized delay stage (Newport) and set to zero for the experiments in this article. The probe beam is spatially dispersed onto a 64-element mercury–cadmium–telluride detector array (Infrared Associates), which proves ca. 3 cm−1 resolution along the probe axis.
Sample Preparation
Aβ1–40 and Aβ1–42 were purchased from Anaspec. Proteins were dissolved in deuterated hexafluoroisopropanol to deuterate exchangeable sites and to promote disaggregation of the protein samples, and the hexafluoroisopropanol was removed by lyophilization prior to addition of aqueous buffers. Concentration measurements for each protein were determined in aqueous solution and made via 280 nm extinction coefficients using a NanoDrop 2000 (Thermo Scientific). The A280 coefficient used for both proteins was 1490 M−1 cm−1.39 Proteins aggregated spontaneously upon dissolution in D2O buffers at pD 7.5 or 2.0 as indicated in the text. Protein samples are placed between a pair of 2 mm thick CaF2 windows separated by a 56 μm Teflon spacer for 2D IR experiments. Transmission electron microscopy (TEM) images were collected on a Phillips CM120 at the UW-Madison Medical School Electron Microscopy Facility.
Simulations of 2D IR Spectra
Two-dimensional IR vibrational spectra were simulated using transition dipole coupling40–42 to compute the couplings between amide I modes in structures obtained from the protein data bank (PDB). Couplings between nearest neighbor residues were computed separately from dihedral angles using the map of Jansen et al.43 Diagonal disorder (full width at half-maximum (FWHM) 25 cm−1) was added to the local mode frequencies. The computed stick spectra were convoluted with a two-dimensional Gaussian lineshape (FWHM 18 cm−1 along the diagonal) elongated along the diagonal to simulate the 2D IR lineshape at t2 = 0 waiting time, and the local mode frequency was set to 1668 cm−1 such that the 1625 cm−1 transition observed experimentally matched the frequency of the higher-frequency mode for the calculated structures. We note that the need for a higher local mode frequency (1668 cm−1) than might be expected for an amide I transition is due to the Jansen map local mode site shifts being too far to the red for the dihedral angles associated with β-sheets. All of the calculations were performed in Matlab using a custom Matlab program called coupled oscillator spectrum simulator (COSMOSS).44 COSMOSS is an open-source spectral simulation software available on the GitHub: https://github.com/JJ-Ho/COSMOSS.
RESULTS AND DISCUSSION
Two-Dimensional IR Spectra and TEM Images of Aβ Oligomers and Fibrils
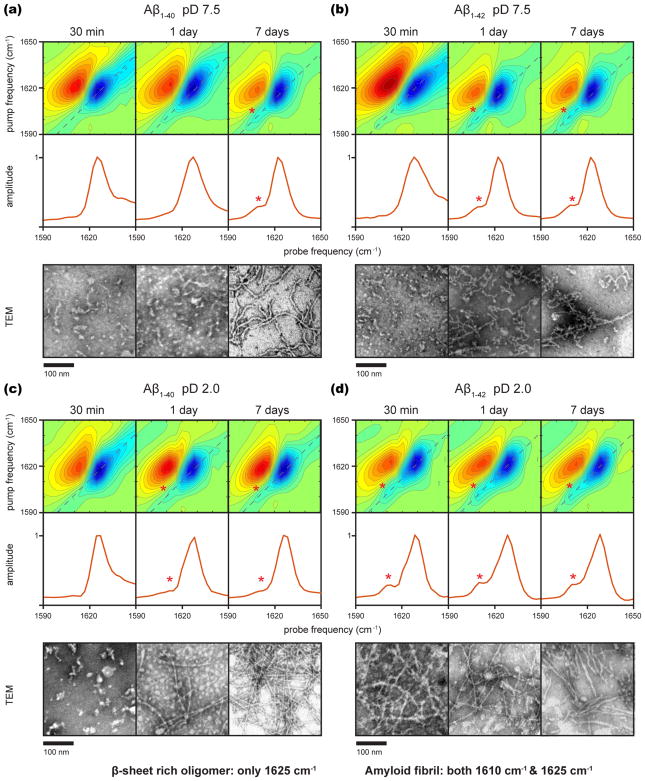
Two-dimensional IR spectra of Aβ1–40 and Aβ1–42 are collected over the course of 7 days along with corresponding TEM images to assess the presence of amyloid fibrils. Data are collected at both pD 7.5 and 2.0. At pD 2.0, Aβ is known to form longer, straighter fibrils that are more similar to those observed in human brain tissue. Low pD also speeds the kinetics, and we used 0.04% NaN3 in the pD 2.0 solution such that our results could be directly compared to earlier 2D IR studies on Aβ1–40 that used the same solvent.45–48 At micromolar concentrations, aggregation of Aβ can take days or weeks;2 our experiments are performed at 1 mM so that the aggregation is complete in 7 days or less. The absorption spectra collected at 1 mM are similar to those previously reported at 100–500 μM45–50 (see Figure S1 for linear spectra from this study), although we note that non-isotope labeled amide I bands are not always sensitive to details of amyloid β-sheets.
Figure 1 shows the representative TEM images and the 2D IR spectra at 30 min, 1 day, and 7 days after initiating aggregation by dissolving lyophilized samples in buffer, along with diagonal cuts through the 2D spectra. In all of the samples, the TEM images at 7 days exhibit fibrils, as expected. In contrast, the TEM images collected 30 min after aggregation is initiated show small amorphous structures consistent with many previous reports of Aβ oligomers and protofibrils. The TEM images at 1 day are either those of oligomers/protofibrils, fibrils, or a mixture, depending on the Aβ isoform and pD. The corresponding 2D IR spectra for all of the samples exhibit a strong set of peaks at 1625 cm−1. These features are from the amide I modes (backbone carbonyl vibrations) of β-sheet transitions that are commonly used to monitor aggregation in the IR studies on Aβ,16 as expected from earlier work on protofibrils/oligomers and fibrils of Aβ. In samples that contain oligomers/protofibrils, random coil features are also observed in the 1640–1670 cm−1 region. The 1625 cm−1 β-sheet feature is always the dominant feature because the 2D intensities scale with the transition dipole strength to the fourth power,18–20 whereas linear spectra scale with the second power, as mentioned above.
Figure 1.
Two-dimensional IR spectra of 1 mM Aβ aggregated for 30 min, 1 day, and 7 days, diagonal slices through the fundamental transition, and representative TEM images for the following samples: (a) Aβ1–40 in pD 7.5 20 mM Tris buffer, (b) Aβ1–42 in pD 7.5 20 mM Tris buffer, (c) Aβ1–40 in pD 2.0 0.04% NaN3 solution, and (d) Aβ1–42 in pD 2.0 0.04% NaN3 solution.
Interestingly, in samples that contain fibrils according to TEM, we also observe a second pair of transitions at 1610 cm−1. This 1610 cm−1 transition is observed for both Aβ1–40 and Aβ1–42. The transition is marked with an asterisk in the diagonal cuts and 2D spectra in the spectra where it is observed. It only appears in samples where fibrils are observed in the TEM images and appears to correlate with the relative amounts of fibril versus amorphous structures, although quantifying morphologies with TEM is very difficult. In the 7 day samples, the intensity is about 25% of the main amide I β-sheet peak at 1625 cm−1, except for Aβ1–40 at pD = 2, where it is about 10%. Figure S2 shows fits to the diagonal traces of the equilibrated samples after 1 week, which are consistent with these relative intensities. In principle, the 2D lineshapes could provide additional information, though we do not fit them here because there is no significant difference in inhomogeneous broadening between the two bands. Though low pD speeds equilibration of the samples to their final state, Aβ1–40 at low pD still shows less intensity at 1610 cm−1 when equilibrated. It thus appears that the 1610 cm−1 transition is indicative of amyloid fibrils and can be used to positively identify the presence of Aβ fibrils. This band is not resolved in the broader linear infrared spectra of Aβ fibrils, as it is not visible in published linear spectra of Aβ fibrils,49–51 and we also do not observe it in linear spectra (Figure S1 shows the FT-IR spectra under the same conditions used in Figure 1).
The 1610 cm−1 Absorption is an Amide I Mode
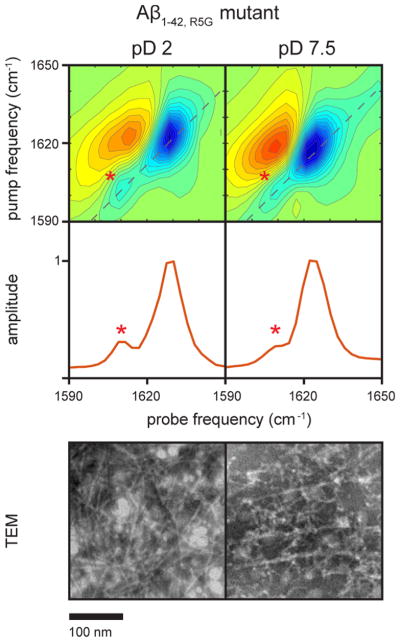
The amide I transitions of most amyloid forming proteins fall between 1610 and 1630 cm−1.16 Thus, the 1610 cm−1 transition could be created by a backbone vibrational mode (i.e., an amide I mode), but it would be among the lowest frequency amyloid transitions reported. Some side chains also have absorbances that fall in this region. In the Aβ sequence, only the arginine at residue 5 could exhibit an absorption in this region.52 To test if arginine is the origin of this transition, we collected the spectra of the R5G mutant fibrils under the same conditions as above. Shown in Figure 2 are the TEM and 2D IR spectra of the mutant fibrils generated in pD 2 and 7.5 buffers after several days of incubation in buffer. Consistent with the observations for the native sequences, aggregation occurs more quickly at pD 2. In both samples, long fibrils are formed and the amyloid β-sheet transition is also observed at 1625 cm−1, indicating that the R5G mutation has little effect on the structure of the fibrils. In addition, the 1610 cm−1 transition is observed.
Figure 2.
Two-dimensional IR spectra and diagonal slices of 1 mM Aβ1–42,R5G collected after 6 days of aggregation in pD 2.0 0.04% NaN3 solution and after 14 days of aggregation in pD 7.5 20 mM Tris buffer and corresponding TEM images. Fibril formation in the pD 7.5 buffer took longer than in the pD 2.0 buffer. Similar to the 2D IR spectra of Aβ1–40 and Aβ1–42 fibrils shown in Figure 1, a transition is observed at ca. 1610 cm−1.
We also note that the anharmonic shift of the 1610 cm−1 transition (which is the frequency difference between the blue fundamental transition and the corresponding red sequence band transition) is about 14 cm−1, similar to that of the 1625 cm−1 transition. Anharmonic shifts less than ca. 20 cm−1 are typical of delocalized vibrational modes.53–55 Thus, we conclude that the 1610 cm−1 mode is created by a backbone amide I vibration with an atypically low frequency.
Stability of the 1610 cm−1 Species toward Sodium Dodecyl Sulfate (SDS) Independently Confirms It Represents Amyloid Fibrils
We next investigated the correlation of the 1610 cm−1 mode with Aβ stability, as measured by the ability of SDS micelles to disrupt the structures present in our aggregated samples. SDS is known to disaggregate/solubilize nonfibrillar β-sheet-rich oligomers but not amyloid fibrils and to stabilize a nonaggregated, partially helical state.56–58 The behavior of SDS toward aggregates can cause inaccurate results when studying oligomer sizes,59–62 but it is useful for distinguishing amyloid fibrils from other partially aggregated structures.57,58 Thus, the addition of SDS to preformed Aβ aggregates is an additional assessment of the presence of amyloid, allowing us to determine if the 1610 cm−1 mode corresponds to protofibrils/oligomers versus the presence of amyloid, independent of the TEM measurements in Figures 1 and 2.
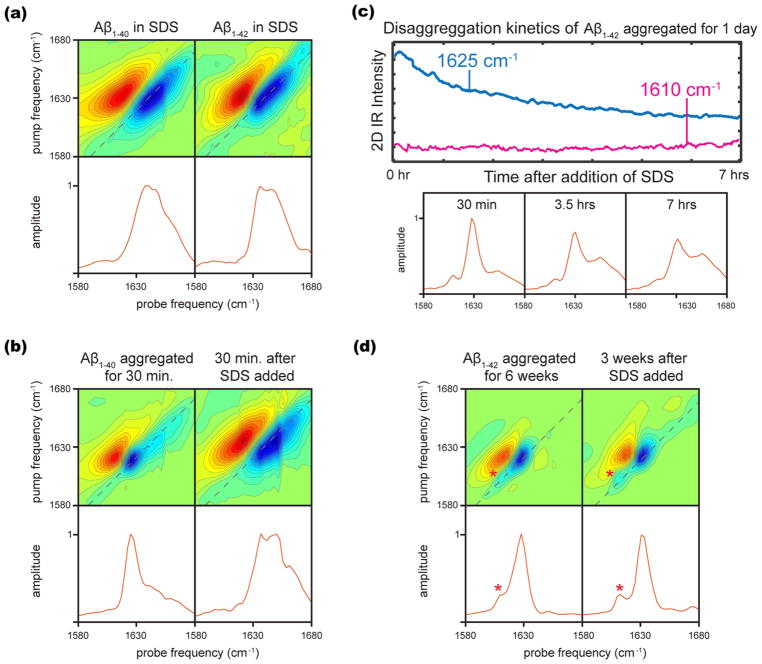
SDS micelles are 3.2–4.2 nm in diameter63 and have an aggregation number of ~62, which means that there are an average of 0.62 Aβ molecules per micelle for a 1 mM Aβ sample in 100 mM SDS.64 Figure 3a shows the 2D IR spectra of lyophilized Aβ1–40 and Aβ1–42 reconstituted in 100 mM SDS without any earlier incubation. Two maxima are observed for each Aβ isoform. The maxima for Aβ1–40 are at 1639 and 1647 cm−1, whereas the maxima for Aβ1–42 are at 1636 and 1647 cm−1. The spectra also show a higher frequency shoulder representing the disordered peptide structures. The transitions at 1647 cm−1 are consistent with α-helical structure as reported from the NMR studies, and the lower frequency transition in each may correspond to a second distinct α-helical structure, or it is possible that a fraction of the protein still adopts a nonamyloid β-sheet conformation under these conditions, as this frequency is also consistent with the range expected for native β-sheets.16 Comparison to the Aβ spectra in the absence of micelles confirms the absence of extended, amyloid-like β-sheet aggregates.
Figure 3.
(a) Two-dimensional IR spectra and diagonal slices of 1 mM Aβ1–40 and Aβ1–42 reconstituted directly into 100 mM SDS. (b) Two-dimensional IR spectra and diagonal slices of Aβ1–40 aggregated for 30 min in pD 7.5 Tris buffer and 30 min after the addition of an equal volume 100 mM SDS. (c) Kinetic traces from rapid scan 2D IR in the β-sheet and α-helical regions for Aβ1–42 aggregated for 1 day in pD 2.0 NaN3 solution, and diagonal slices at various disaggregation times scaled to the maximum intensity. (d) Two-dimensional IR spectra of Aβ1–42 aggregated for 6 weeks in pD 2.0 NaN3 solution and 3 weeks after the addition of an equal volume 100 mM SDS.
To study the SDS-induced disruption of aggregates (disaggregation) in samples that have already been allowed to incubate in buffer (thus preforming aggregates), 1 mM solutions of Aβ1–40 and Aβ1–42 are allowed to incubate in buffer for some time, at which point, an equal volume of 100 mM SDS is added to the solution and 2D IR spectra are collected. These conditions again result in a ratio of 0.62 Aβ molecules per micelle. Figure 3b shows the 2D IR spectra of Aβ1–40 allowed to aggregate in pD 7.5 buffer for 30 min, at which point SDS micelles are added and the 2D IR spectrum is collected 30 min later. Consistent with our expectations from Figure 1, no 1610 cm−1 transition is present after 30 min of aggregation, indicating no fibrils are present in the sample. Within 30 min of adding SDS, the sample is completely disaggregated, judged by the comparison of the 2D spectrum in Figure 3b (right) to those in Figure 3a. Specifically, the peak frequency blue shifts to well above the characteristic β-sheet rich/amyloid region, the line width broadens significantly, and the anharmonicity increases. The fact that SDS has disrupted the β-sheet structure indicates that only nonamyloid oligomers were present and have been disassembled.
In additional experiments, we allow 1 mM Aβ1–42 to aggregate for 24 h in pD 2.0 solution, allowing the fibrils time to mature, and monitor the kinetics of their disaggregation by SDS micelles. In this case, we did observe the 1610 cm−1 transition, and TEM images (refer to Figure 1) indicate that fibrils form under these conditions. We use rapid-scan 2D IR to track the course of disaggregation, and kinetic traces are shown in Figure 3c for the 1625 cm−1 (both oligomers and fibrils) and 1610 cm−1 (fibril only) transitions. Representative diagonal slices through the 2D spectra are shown below the kinetic traces; 30 min into disaggregation, both the 1625 and 1610 cm−1 transitions are observed. As disaggregation proceeds, the β-sheet transition partially decays, along with a rise in the higher-frequency region at ca. 1635–1650 cm−1. The 1610 cm−1 transition does not decay, indicating that the 1610 cm−1 transitions correspond to mature amyloid species that are not disaggregated by SDS micelles. These observations in Figure 3c are representative of our observations in general; SDS is not able to disaggregate the amyloid species corresponding to the 1610 cm−1 transition.
We are also interested in whether we can prepare samples so strongly aggregated that SDS micelles cannot disaggregate them to any significant extent. We allow Aβ1–42 to aggregate for 6 weeks in pD 2.0 solution. These amyloid fibrils are very stable toward SDS micelle disaggregation, showing very little disaggregation even 3 weeks after SDS micelles have been added (Figure 3d).
The observations in SDS provide further evidence that the 1610 cm−1 transition is associated with stable amyloid fibrils. Because the 1625 cm−1 mode is always present when there is also a 1610 cm−1 mode, they must both be coming from the same fibers, not polymorphs each with a single distinct absorption frequency. It is likely that multiple polymorphs are present, and that one (or more) give(s) rise to transitions at both 1610 and 1625 cm−1, whereas others may only exhibit transitions at 1625 cm−1. We investigate this further with simulations below.
Simulating the 2D IR Spectra of Aβ Fibrils
To test the possible structural origins for the 1610 cm−1 mode, we simulate the spectra using structures of Aβ fibrils taken from the protein data bank (PDB). Our goal with these simulations is to establish whether published Aβ structures can produce the 1610 cm−1 2D IR peaks observed experimentally. The 1625 cm−1 mode has been simulated many times in the past with model β-sheets.53,65–69 Because 2D IR spectra are very sensitive to short-range couplings, we opted to simulate our Aβ 2D IR spectra using the crystal structures formed from fragments of Aβ, as X-ray structures generally provide the highest structural resolution (ca. 2 Å). Accordingly, we calculated the 2D IR spectra for several published Aβ crystal structures, and show the results from two structures here (Figure 4b). These two structures are taken from the coordinates of PDB file 2y3k, which is the crystal structure of Aβ fragment 35–42.70 This PDB file contains coordinates for four geometries of strands that are only slightly different from one another, but all fall within the crystal structure accuracy. On average, the carbonyl groups for the structure on the left are about 5° more parallel to their nearest interstrand neighbors, and the strands are spaced ca. 0.3 Å closer together than those on the right. Even though these differences are small, 2D IR spectra are very sensitive to local structure. The simulated spectrum of the left structure has its strongest (A⊥, named for the orientation of the transition dipole relative to the β-strands) transition at 1611 cm−1, whereas the A transition of the right structure is at 1625 cm−1 (details of the simulations are found in the Materials and Methods section). To reproduce the experimental data, the spectra are added in a 1:4 ratio (vide infra). The 2D spectrum and diagonal slice are shown in Figure 4c. This 14 cm−1 difference is caused by the couplings due to differences in dihedral angles, interstrand spacings, and relative amide I mode orientations. The 1611 cm−1 absorption is created by interstrand couplings (βISC) that are about 5 cm−1 larger than those for the structure on the right, associated with more parallel β-strands and shorter distances between residues in adjacent strands. The couplings between neighboring residues are very sensitive to the relative orientation and distance between the local mode transition dipoles,40–42 such that the narrower interstrand distances and more aligned amide oscillators (mentioned above) of the structure on the left lead to the 5 cm−1 average change in interstrand couplings. Because the frequency difference scales as 2*βISC, these couplings account for 10 cm−1 out of 14 cm−1 of the frequency difference between the structures. Couplings between nearest neighbors and other residues account for the remainder of the frequency difference. We note that, across amyloid species, these couplings depend on the structure of the fibril and not necessarily its age or maturity.20 Figure S3 shows the individual simulated spectra, and Figure S4 shows the principle eigenstate responsible for the IR intensity for each in the case with no diagonal disorder. Although a doorway mode analysis71,72 could be performed for the case with diagonal disorder turned on, a molecular eigenstate picture is appropriate for describing the spectra of these highly ordered β-sheet structures in which a single mode is responsible for nearly all of the IR intensity. Figure S5 shows an overlay of the simulated diagonal trace with the experimental data. A recent study has also examined the effects of varying coupling constants in simulated amyloid β-sheets on the frequencies and intensities of the amide I transitions.20 Experimentally, solvent exposure may also play a role in shifting the frequencies of the individual β-sheet regions.
Figure 4.
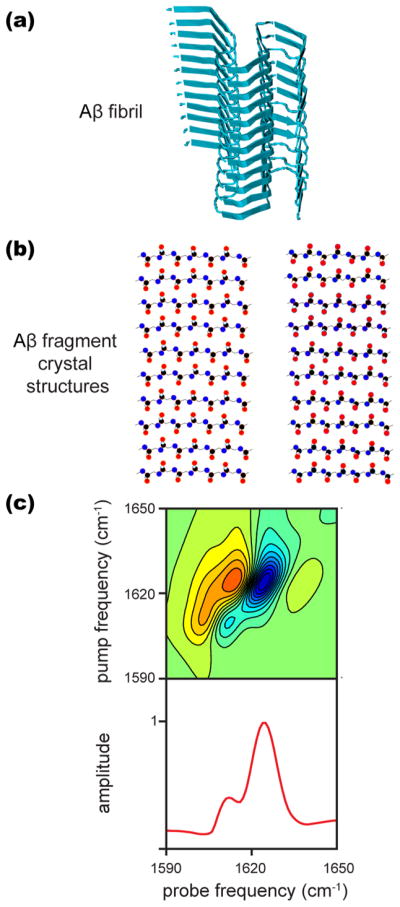
(a) ssNMR structure of an Aβ fibril obtained from PDB structure 2mxu.73 (b) Atomic structures of Aβ35–42 fragments from PDB file 2y3k from which β-sheets were generated as described in the text. (c) The 2D IR spectrum simulated as a mixture of the two structures from panel (b) added in a 1:4 ratio to simulate the spectrum observed experimentally. Note that the local mode transition frequency was shifted to best match the experimental data, though the difference in frequency is unaffected by this shift. Details of the calculations are given in Materials and Methods section.
Amyloid fibrils often contain multiple β-sheets. Each β-sheet within a fibril will have its own A⊥ mode and corresponding intensity and frequency. Coupling between β-sheets is a minor perturbation because apposing β-sheets have small vibrational coupling, and adjacent sheets are separated by loops and turns.74 Figure 4a shows a structure for a full-length Aβ fibril to illustrate the arrangements of multiple β-sheets within a fibril (PDB: 2mxu73). With this idea in mind, we added together the 2D IR spectra for the two structures in Figure 4b in a 1:4 ratio to produce the spectrum shown in Figure 4c. Figure 4c is very similar to that observed experimentally for Aβ fibrils (Figures 1 and 2). The addition of the spectra is intended to model distinct β-sheet regions in a full-length Aβ fibril, each having its own A⊥ mode. Thus, we conclude that 1610 cm−1 is a marker for amyloid fibers and that our experimental results are consistent with an Aβ fibril that has multiple β-sheet regions.
We note that previously reported 2D IR spectra45–48 of Aβ1–40 contained a transition at 1610 cm−1, although it is not assigned nor mentioned in the article, which focuses on isotopic labeling (see Figure 4 of ref 45). To our knowledge, 2D IR spectra of Aβ1–42 have not been previously reported.
SUMMARY AND CONCLUSIONS
In summary, we have used 2D IR spectroscopy to reveal a vibrational transition that allows us to differentiate samples containing Aβ fibrils from those containing other β-sheet-rich structures, such as oligomers and protofibrils, as verified by TEM and SDS disaggregation. As has been noted previously,59–62 the observation that SDS disaggregates β-sheet-rich Aβ oligomers carries broad relevance for Aβ and other amyloid aggregation research, where SDS is commonly used to characterize sizes and relative abundances of protein aggregates and fibrils by gel electrophoresis or other methods. Our evidence indicates that the 1610 cm−1 transition stems from a delocalized amide I mode, and spectral modeling of amyloid structures suggests the lower transition frequency of this mode can be explained by differences in β-sheet structure of different regions of the fibril. It is worth noting that similar observations could exist for other amyloid species, as amyloid fibrils are characterized by specific fibril structures that involve multiple β-sheets;75–78 the enhanced ability of 2D IR spectroscopy to resolve overlapping lineshapes may continue to prove powerful in this regard. The ability to spectrally distinguish Aβ fibrils from β-sheet-rich oligomers carries broad implications for AD research into the aggregation, toxicity, and fibril formation of Aβ. For example, infrared spectroscopy has recently been demonstrated to be a powerful tool for the early prediction and detection of AD by monitoring the amide I spectra of Aβ extracted from human blood and cerebrospinal fluid using antibodies,79,80 and the additional structural sensitivity of 2D IR could allow direct detection of fibrils in antibody-extracted Aβ samples.
Supplementary Material
Acknowledgments
J.P.L. is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation. K.L.R. acknowledges support through an NSF Graduate Research Fellowship (award DGE-1256259).
Funding
This work was funded by the National Institute of Health NIDDK under award number 79895.
Footnotes
Notes
The authors declare the following competing financial interest(s): M.T.Z. is co-owner of PhaseTech Spectroscopy, Inc., which sells mid-IR and visible pulse shapers and 2D spectrometers.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpcb.7b10765.
FT-IR spectra (Figure S1); fit of two summed Gaussian functions to the spectra of each Aβ isoformaggregated for 1 week at pD 2 and 7.5 (Figure S2); individual simulated 2D IR spectra and diagonal traces (Figure S3); eigenvector coefficients (Figure S4); diagonal trace of the simulated spectrum (Figure S5) (PDF)
References
- 1.Chiti F, Dobson CM. Protein Misfolding, Functional Amyloid, and Human Disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Jarrett JT, Berger EP, Lansbury PT. The Carboxy Terminus of the β-Amyloid Protein Is Critical for the Seeding of Amyloid Formation: Implications for the Pathogenesis of Alzheimer’s Disease. Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 3.Iwatsubo T, Odaka A, Suzuki N, Mizusawa H, Nukina N, Ihara Y. Visualization of Aβ42(43) and Aβ40 in Senile Plaques with End-Specific Aβ Monoclonals: Evidence That an Initially Deposited Species Is Aβ 42(43) Neuron. 1994;13:45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- 4.Selkoe DJ. Alzheimer’s Disease: Genes, Proteins, and Therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 5.Cramer PE, Cirrito JR, Wesson DW, Lee CYD, Karlo JC, Zinn AE, Casali BT, Restivo JL, Goebel WD, James MJ, Brunden KR, Wilson DA, Landreth GE. ApoE-Directed Therapeutics Rapidly Clear β-Amyloid and Reverse Deficits in AD Mouse Models. Science. 2012;335:1503–1506. doi: 10.1126/science.1217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HY, Kim HV, Jo S, Lee CJ, Choi SY, Kim DJ, Kim Y. EPPS Rescues Hippocampus-Dependent Cognitive Deficits in APP/PS1 Mice by Disaggregation of Amyloid-β Oligomers and Plaques. Nat Commun. 2015;6(8997) doi: 10.1038/ncomms9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yasojima K, Akiyama H, McGeer EG, McGeer PL. Reduced Neprilysin in High Plaque Areas of Alzheimer Brain: A Possible Relationship to Deficient Degradation of β-Amyloid Peptide. Neurosci Lett. 2001;297:97–100. doi: 10.1016/s0304-3940(00)01675-x. [DOI] [PubMed] [Google Scholar]
- 8.Cook DG, Leverenz JB, McMillan PJ, Kulstad JJ, Ericksen S, Roth RA, Schellenberg GD, Jin LW, Kovacina KS, Craft S. Reduced Hippocampal Insulin-Degrading Enzyme in Late-Onset Alzheimer’s Disease Is Associated with the Apolipoprotein E-ε4 Allele. Am J Pathol. 2003;162:313–319. doi: 10.1016/s0002-9440(10)63822-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deane R, Sagare A, Hamm K, Parisi M, Lane S, Finn MB, Holtzman DM, Zlokovic BV. apoE Isoform-Specific Disruption of Amyloid β Peptide Clearance from Mouse Brain. J Clin Invest. 2008;118:4002–4013. doi: 10.1172/JCI36663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardy J, Selkoe DJ. The Amyloid Hypothesis of Alzheimer’s Disease: Progress and Problems on the Road to Therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 11.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common Structure of Soluble Amyloid Oligomers Implies Common Mechanism of Pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 12.Lindgren M, Sörgjerd K, Hammarström P. Detection and Characterization of Aggregates, Prefibrillar Amyloidogenic Oligomers, and Protofibrils Using Fluorescence Spectroscopy. Biophys J. 2005;88:4200–4212. doi: 10.1529/biophysj.104.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D’Amico M, Di Carlo MG, Groenning M, Militello V, Vetri V, Leone M. Thioflavin T Promotes Aβ(1–40) Amyloid Fibrils Formation. J Phys Chem Lett. 2012;3:1596–1601. doi: 10.1021/jz300412v. [DOI] [PubMed] [Google Scholar]
- 14.Middleton CT, Marek P, Cao P, Chiu C-c, Singh S, Woys AM, de Pablo JJ, Raleigh DP, Zanni MT. Two-Dimensional Infrared Spectroscopy Reveals the Complex Behaviour of an Amyloid Fibril Inhibitor. Nat Chem. 2012;4:355–360. doi: 10.1038/nchem.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cloe AL, Orgel JPRO, Sachleben JR, Tycko R, Meredith SC. The Japanese Mutant Aβ (ΔE22-Aβ1–39) Forms Fibrils Instantaneously, with Low-Thioflavin T Fluorescence: Seeding of Wild-Type Aβ1–40 into Atypical Fibrils by ΔE22-Aβ1–39. Biochemistry. 2011;50:2026–2039. doi: 10.1021/bi1016217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zandomeneghi G, Krebs MRH, McCammon MG, Fändrich M. FTIR Reveals Structural Differences between Native β-Sheet Proteins and Amyloid Fibrils. Protein Sci. 2004;13:3314–3321. doi: 10.1110/ps.041024904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang H, Yang S, Kong J, Dong A, Yu S. Obtaining Information about Protein Secondary Structures in Aqueous Solution Using Fourier Transform IR Spectroscopy. Nat Protoc. 2015;10:382–396. doi: 10.1038/nprot.2015.024. [DOI] [PubMed] [Google Scholar]
- 18.Grechko M, Zanni MT. Quantification of Transition Dipole Strengths Using 1D and 2D Spectroscopy for the Identification of Molecular Structures via Exciton Delocalization: Application to α-Helices. J Chem Phys. 2012;137(184202) doi: 10.1063/1.4764861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunkelberger EB, Grechko M, Zanni MT. Transition Dipoles from 1D and 2D Infrared Spectroscopy Help Reveal the Secondary Structures of Proteins: Application to Amyloids. J Phys Chem B. 2015;119:14065–14075. doi: 10.1021/acs.jpcb.5b07706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomont JP, Ostrander JS, Ho J-J, Petti MK, Zanni MT. Not All β-Sheets Are the Same: Amyloid Infrared Spectra, Transition Dipole Strengths, and Couplings Investigated by 2D IR Spectroscopy. J Phys Chem B. 2017:8935. doi: 10.1021/acs.jpcb.7b06826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jayasinghe SA, Langen R. Lipid Membranes Modulate the Structure of Islet Amyloid Polypeptide. Biochemistry. 2005;44:12113–12119. doi: 10.1021/bi050840w. [DOI] [PubMed] [Google Scholar]
- 22.Knight JD, Hebda JA, Miranker AD. Conserved and Cooperative Assembly of Membrane-Bound α-Helical States of Islet Amyloid Polypeptide. Biochemistry. 2006;45:9496–9508. doi: 10.1021/bi060579z. [DOI] [PubMed] [Google Scholar]
- 23.Apostolidou M, Jayasinghe SA, Langen R. Structure of α-Helical Membrane-Bound Human Islet Amyloid Polypeptide and Its Implications for Membrane-Mediated Misfolding. J Biol Chem. 2008;283:17205–17210. doi: 10.1074/jbc.M801383200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williamson JA, Loria JP, Miranker AD. Helix Stabilization Precedes Aqueous and Bilayer Catalyzed Fiber Formation in Islet Amyloid Polypeptide. J Mol Biol. 2009;393:383–396. doi: 10.1016/j.jmb.2009.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang Z, Hess SK, Heinrich F, Lee JC. Molecular Details of α-Synuclein Membrane Association Revealed by Neutrons and Photons. J Phys Chem B. 2015;119:4812–4823. doi: 10.1021/jp512499r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camargo DCR, Tripsianes K, Buday K, Franko A, Göbl C, Hartlmüller C, Sarkar R, Aichler M, Mettenleiter G, Schulz M, Böddrich A, Erck C, Martens H, Walch AK, Madl T, Wanker EE, Conrad M, de Angelis MH, Reif B. The Redox Environment Triggers Conformational Changes and Aggregation of hIAPP in Type II Diabetes. Sci Rep. 2017;7(44041) doi: 10.1038/srep44041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Celej MS, Sarroukh R, Goormaghtigh E, Fidelio GD, Ruysschaert JM, Raussens V. Toxic Prefibrillar α-Synuclein Amyloid Oligomers Adopt a Distinctive Antiparallel β-Sheet Structure. Biochem J. 2012;443:719–726. doi: 10.1042/BJ20111924. [DOI] [PubMed] [Google Scholar]
- 28.Zurdo J, Guijarro JI, Jiménez JL, Saibil HR, Dobson CM. Dependence on Solution Conditions of Aggregation and Amyloid Formation by an SH3 domain1. J Mol Biol. 2001;311:325–340. doi: 10.1006/jmbi.2001.4858. [DOI] [PubMed] [Google Scholar]
- 29.Zurdo J, Guijarro JI, Dobson CM. Preparation and Characterization of Purified Amyloid Fibrils. J Am Chem Soc. 2001;123:8141–8142. doi: 10.1021/ja016229b. [DOI] [PubMed] [Google Scholar]
- 30.Sarroukh R, Cerf E, Derclaye S, Dufrêne YF, Goormaghtigh E, Ruysschaert JM, Raussens V. Transformation of Amyloid β(1–40) Oligomers into Fibrils Is Characterized by a Major Change in Secondary Structure. Cell Mol Life Sci. 2011;68:1429–1438. doi: 10.1007/s00018-010-0529-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laganowsky A, Liu C, Sawaya MR, Whitelegge JP, Park J, Zhao M, Pensalfini A, Soriaga AB, Landau M, Teng PK, Cascio D, Glabe C, Eisenberg D. Atomic View of a Toxic Amyloid Small Oligomer. Science. 2012;335:1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buchanan LE, Dunkelberger EB, Tran HQ, Cheng PN, Chiu CC, Cao P, Raleigh DP, de Pablo JJ, Nowick JS, Zanni MT. Mechanism of IAPP Amyloid Fibril Formation Involves an Intermediate with a Transient β-Sheet. Proc Natl Acad Sci US A. 2013;110:19285–19290. doi: 10.1073/pnas.1314481110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luca S, Yau WM, Leapman R, Tycko R. Peptide Conformation and Supramolecular Organization in Amylin Fibrils: Constraints from Solid-State NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serrano AL, Lomont JP, Tu L-H, Raleigh DP, Zanni MT. A Free Energy Barrier Caused by the Refolding of an Oligomeric Intermediate Controls the Lag Time of Amyloid Formation by hIAPP. J Am Chem Soc. 2017:16478. doi: 10.1021/jacs.7b08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shim SH, Strasfeld DB, Ling YL, Zanni MT. Automated 2D IR Spectroscopy Using a Mid-IR Pulse Shaper and Application of This Technology to the Human Islet Amyloid Polypeptide. Proc Natl Acad Sci US A. 2007;104:14197–14202. doi: 10.1073/pnas.0700804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strasfeld DB, Ling YL, Shim SH, Zanni MT. Tracking Fiber Formation in Human Islet Amyloid Polypeptide with Automated 2D-IR Spectroscopy. J Am Chem Soc. 2008;130:6698–6699. doi: 10.1021/ja801483n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Middleton CT, Woys AM, Mukherjee SS, Zanni MT. Residue-Specific Structural Kinetics of Proteins through the Union of Isotope Labeling, Mid-IR Pulse Shaping, and Coherent 2D IR Spectroscopy. Methods. 2010;52:12–22. doi: 10.1016/j.ymeth.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh A, Serrano AL, Oudenhoven TA, Ostrander JS, Eklund EC, Blair AF, Zanni MT. Experimental Implementations of 2D IR Spectroscopy through a Horizontal Pulse Shaper Design and a Focal Plane Array Detector. Opt Lett. 2016;41:524–527. doi: 10.1364/OL.41.000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jan A, Hartley DM, Lashuel HA. Preparation and Characterization of Toxic Aβ Aggregates for Structural and Functional Studies in Alzheimer’s Disease Research. Nat Protoc. 2010;5:1186–1209. doi: 10.1038/nprot.2010.72. [DOI] [PubMed] [Google Scholar]
- 40.Krimm S, Abe Y. Intermolecular Interaction Effects in the Amide I Vibrations of β Polypeptides. Proc Natl Acad Sci US A. 1972;69:2788–2792. doi: 10.1073/pnas.69.10.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheam TC, Krimm S. Transition Dipole Interaction in Polypeptides: Ab Initio Calculation of Transition Dipole Parameters. Chem Phys Lett. 1984;107:613–616. [Google Scholar]
- 42.Torii H, Tasumi M. Ab Initio Molecular Orbital Study of the Amide I Vibrational Interactions between the Peptide Groups in Di-and Tripeptides and Considerations on the Conformation of the Extended Helix. J Raman Spectrosc. 1998;29:81–86. [Google Scholar]
- 43.Jansen TlC, Dijkstra AG, Watson TM, Hirst JD, Knoester J. Modeling the Amide I Bands of Small Peptides. J Chem Phys. 2006;125(044312) doi: 10.1063/1.2218516. [DOI] [PubMed] [Google Scholar]
- 44.Ho J-J. Coupled OScillator MOdel Spectrum Simulator. https://gitlab.com/jjho/COSMOSS.
- 45.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. Two-Dimensional Infrared Spectra of Isotopically Diluted Amyloid Fibrils from Aβ40. Proc Natl Acad Sci US A. 2008;105:7720–7725. doi: 10.1073/pnas.0802993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. 2D IR Provides Evidence for Mobile Water Molecules in β-Amyloid Fibrils. Proc Natl Acad Sci US A. 2009;106:17751–17756. doi: 10.1073/pnas.0909888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Falvo C, Zhuang W, Kim YS, Axelsen PH, Hochstrasser RM, Mukamel S. Frequency Distribution of the Amide-I Vibration Sorted by Residues in Amyloid Fibrils Revealed by 2D-IR Measurements and Simulations. J Phys Chem B. 2012;116:3322–3330. doi: 10.1021/jp2096423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma J, Komatsu H, Kim YS, Liu L, Hochstrasser RM, Axelsen PH. Intrinsic Structural Heterogeneity and Long-Term Maturation of Amyloid β Peptide Fibrils. ACS Chem Neurosci. 2013;4:1236–1243. doi: 10.1021/cn400092v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cerf E, Sarroukh R, Tamamizu-Kato S, Breydo L, Derclaye S, Dufrêne YF, Narayanaswami V, Goormaghtigh E, Ruysschaert JM, Raussens V. Antiparallel β-Sheet: A Signature Structure of the Oligomeric Amyloid β-Peptide. Biochem J. 2009;421:415–423. doi: 10.1042/BJ20090379. [DOI] [PubMed] [Google Scholar]
- 50.Cerf E, Ruysschaert J-M, Goormaghtigh E, Raussens V. [accessed Oct 23, 2017];ATR–FTIR, a New Tool to Analyze the Oligomeric Content of Aβ Samples in the Presence of Apolipoprotein E isoforms. 2017 https://www.hindawi.com/journals/jspec/2010/916373/abs/
- 51.Fraser PE, Nguyen JT, Inouye H, Surewicz WK, Selkoe DJ, Podlisny MB, Kirschner DA. Fibril Formation by Primate, Rodent, and Dutch-Hemorrhagic Analogs of Alzheimer Amyloid β-Protein. Biochemistry. 1992;31:10716–10723. doi: 10.1021/bi00159a011. [DOI] [PubMed] [Google Scholar]
- 52.Barth A. The Infrared Absorption of Amino Acid Side Chains. Prog Biophys Mol Biol. 2000;74:141–173. doi: 10.1016/s0079-6107(00)00021-3. [DOI] [PubMed] [Google Scholar]
- 53.Demirdöven N, Cheatum CM, Chung HS, Khalil M, Knoester J, Tokmakoff A. Two-Dimensional Infrared Spectroscopy of Antiparallel β-Sheet Secondary Structure. J Am Chem Soc. 2004;126:7981–7990. doi: 10.1021/ja049811j. [DOI] [PubMed] [Google Scholar]
- 54.Ling YL, Strasfeld DB, Shim SH, Raleigh DP, Zanni MT. Two-Dimensional Infrared Spectroscopy Provides Evidence of an Intermediate in the Membrane-Catalyzed Assembly of Diabetic Amyloid. J Phys Chem B. 2009;113:2498–2505. doi: 10.1021/jp810261x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hamm P, Zanni M. Concepts and Methods of 2D Infrared Spectroscopy. Cambridge University Press; 2011. [Google Scholar]
- 56.Shao H, Jao S-c, Ma K, Zagorski MG. Solution Structures of Micelle-Bound Amyloid β-(1–40) and β-(1–42) Peptides of Alzheimer’s Disease. J Mol Biol. 1999;285:755–773. doi: 10.1006/jmbi.1998.2348. [DOI] [PubMed] [Google Scholar]
- 57.Kushnirov VV, Alexandrov IM, Mitkevich OV, Shkundina IS, Ter-Avanesyan MD. Purification and Analysis of Prion and Amyloid Aggregates. Methods. 2006;39:50–55. doi: 10.1016/j.ymeth.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 58.Rostagno A, Ghiso J. Isolation and Biochemical Characterization of Amyloid Plaques and Paired Helical Filaments. Curr Protoc Cell Biol. 2009:3.33.1. doi: 10.1002/0471143030.cb0333s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bitan G, Fradinger EA, Spring SM, Teplow DB. Neurotoxic Protein Oligomers—What You See Is Not Always What You Get. Amyloid. 2005;12:88–95. doi: 10.1080/13506120500106958. [DOI] [PubMed] [Google Scholar]
- 60.Hepler RW, Grimm KM, Nahas DD, Breese R, Dodson EC, Acton P, Keller PM, Yeager M, Wang H, Shughrue P, Kinney G, Joyce JG. Solution State Characterization of Amyloid β-Derived Diffusible Ligands. Biochemistry. 2006;45:15157–15167. doi: 10.1021/bi061850f. [DOI] [PubMed] [Google Scholar]
- 61.Watt AD, Perez KA, Rembach A, Sherrat NA, Hung LW, Johanssen T, McLean CA, Kok WM, Hutton CA, Fodero-Tavoletti M, Masters CL, Villemagne VL, Barnham KJ. Oligomers, Fact or Artefact? SDS-PAGE Induces Dimerization of β-Amyloid in Human Brain Samples. Acta Neuropathol. 2013;125:549–564. doi: 10.1007/s00401-013-1083-z. [DOI] [PubMed] [Google Scholar]
- 62.Pujol-Pina R, Vilaprinyó-Pascual S, Mazzucato R, Arcella A, Vilaseca M, Orozco M, Carulla N. SDS-PAGE Analysis of Aβ Oligomers Is Disserving Research into Alzheimeŕs Disease: Appealing for ESI-IM-MS. Sci Rep. 2015;5(14809) doi: 10.1038/srep14809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duplâtre G, Marques MFF, da Graça Miguel M. Size of Sodium Dodecyl Sulfate Micelles in Aqueous Solutions as Studied by Positron Annihilation Lifetime Spectroscopy. J Phys Chem. 1996;100:16608–16612. [Google Scholar]
- 64.Turro NJ, Yekta A. Luminescent Probes for Detergent Solutions. A Simple Procedure for Determination of the Mean Aggregation Number of Micelles. J Am Chem Soc. 1978;100:5951–5952. [Google Scholar]
- 65.Kubelka J, Keiderling TA. Differentiation of β-Sheet-Forming Structures: Ab Initio-Based Simulations of IR Absorption and Vibrational CD for Model Peptide and Protein β-Sheets. J Am Chem Soc. 2001;123:12048–12058. doi: 10.1021/ja0116627. [DOI] [PubMed] [Google Scholar]
- 66.Paul C, Wang J, Wimley WC, Hochstrasser RM, Axelsen PH. Vibrational Coupling, Isotopic Editing, and β-Sheet Structure in a Membrane-Bound Polypeptide. J Am Chem Soc. 2004;126:5843–5850. doi: 10.1021/ja038869f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheatum CM, Tokmakoff A, Knoester J. Signatures of β-Sheet Secondary Structures in Linear and Two-Dimensional Infrared Spectroscopy. J Chem Phys. 2004;120:8201–8215. doi: 10.1063/1.1689637. [DOI] [PubMed] [Google Scholar]
- 68.Lee C, Cho M. Local Amide I Mode Frequencies and Coupling Constants in Multiple-Stranded Antiparallel β-Sheet Polypeptides. J Phys Chem B. 2004;108:20397–20407. [Google Scholar]
- 69.Kim J, Huang R, Kubelka J, Bouř P, Keiderling TA. Simulation of Infrared Spectra for β-Hairpin Peptides Stabilized by an Aib-Gly Turn Sequence: Correlation between Conformational Fluctuation and Vibrational Coupling. J Phys Chem B. 2006;110:23590–23602. doi: 10.1021/jp0640575. [DOI] [PubMed] [Google Scholar]
- 70.Colletier JP, Laganowsky A, Landau M, Zhao M, Soriaga AB, Goldschmidt L, Flot D, Cascio D, Sawaya MR, Eisenberg D. Molecular Basis for Amyloid-β Polymorphism. Proc Natl Acad Sci US A. 2011;108:16938–16943. doi: 10.1073/pnas.1112600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Torii H, Tasumi M. Three-dimensional Doorway-state Theory for Analyses of Absorption Bands of Many-oscillator Systems. J Chem Phys. 1992;97:86–91. [Google Scholar]
- 72.Torii H, Tasumi M. Application of the Three-dimensional Doorway-state Theory to Analyses of the amide-I Infrared Bands of Globular Proteins. J Chem Phys. 1992;97:92–98. [Google Scholar]
- 73.Xiao Y, Ma B, McElheny D, Parthasarathy S, Long F, Hoshi M, Nussinov R, Ishii Y. Aβ(1–42) Fibril Structure Illuminates Self-Recognition and Replication of Amyloid in Alzheimer’s Disease. Nat Struct Mol Biol. 2015;22:499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strasfeld DB, Ling YL, Gupta R, Raleigh DP, Zanni MT. Strategies for Extracting Structural Information from 2D IR Spectroscopy of Amyloid: Application to Islet Amyloid Polypeptide. J Phys Chem B. 2009;113:15679–15691. doi: 10.1021/jp9072203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tycko R. Solid-State NMR Studies of Amyloid Fibril Structure. Annu Rev Phys Chem. 2011;62:279–299. doi: 10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tuttle MD, Comellas G, Nieuwkoop AJ, Covell DJ, Berthold DA, Kloepper KD, Courtney JM, Kim JK, Barclay AM, Kendall A, Wan W, Stubbs G, Schwieters CD, Lee VMY, George JM, Rienstra CM. Solid-State NMR Structure of a Pathogenic Fibril of Full-Length Human α-Synuclein. Nat Struct Mol Biol. 2016;23:409–415. doi: 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Colvin MT, Silvers R, Ni QZ, Can TV, Sergeyev I, Rosay M, Donovan KJ, Michael B, Wall J, Linse S, Griffin RG. Atomic Resolution Structure of Monomorphic Aβ42 Amyloid Fibrils. J Am Chem Soc. 2016;138:9663–9674. doi: 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wälti MA, Ravotti F, Arai H, Glabe CG, Wall JS, Böckmann A, Güntert P, Meier BH, Riek R. Atomic-Resolution Structure of a Disease-Relevant Aβ(1–42) Amyloid Fibril. Proc Natl Acad Sci US A. 2016;113:E4976–E4984. doi: 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nabers A, Ollesch J, Schartner J, Kötting C, Genius J, Hafermann H, Klafki H, Gerwert K, Wiltfang J. Amyloid-β-Secondary Structure Distribution in Cerebrospinal Fluid and Blood Measured by an Immuno-Infrared-Sensor: A Biomarker Candidate for Alzheimer’s Disease. Anal Chem. 2016;88:2755–2762. doi: 10.1021/acs.analchem.5b04286. [DOI] [PubMed] [Google Scholar]
- 80.Nabers A, Ollesch J, Schartner J, Kötting C, Genius J, Haußmann U, Klafki H, Wiltfang J, Gerwert K. An Infrared Sensor Analysing Label-Free the Secondary Structure of the Aβ Peptide in Presence of Complex Fluids. J Biophotonics. 2016;9:224–234. doi: 10.1002/jbio.201400145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.