Abstract
The purpose of this study is to evaluate the ability of a machine learning algorithm to classify in vivo magnetic resonance images (MRI) of human articular cartilage for development of osteoarthritis (OA). Sixty-eight subjects were selected from the Osteoarthritis Initiative (OAI) control and incidence cohorts. Progression to clinical OA was defined by the development of symptoms as quantified by the Western Ontario and McMaster Universities Arthritis (WOMAC) questionnaire three years after baseline evaluation. Multi-slice T2–weighted knee images, obtained through the OAI, of these subjects were registered using a nonlinear image registration algorithm. T2 maps of cartilage from the central weight bearing slices of the medial femoral condyle were derived from the registered images using the multiple available echo times and were classified for “progression to symptomatic OA” using the machine learning tool, weighted neighbor distance using compound hierarchy of algorithms representing morphology (WND-CHRM). WND-CHRM classified the isolated T2 maps for the progression to symptomatic OA with 75% accuracy.
Statement of clinical significance
Machine learning algorithms applied to T2 maps have the potential to provide important prognostic information for the development of OA.
Keywords: osteoarthritis, MRI, pattern recognition, classification, registration, segmentation
Introduction
Osteoarthritis (OA) is a highly prevalent disease associated with articular cartilage degeneration1, 2. Magnetic resonance imaging (MRI) is a noninvasive and multi-contrast modality that can detect subtle morphologic changes in articular cartilage and is an effective tool for diagnosing established OA3, 4. Computer-aided diagnostic tools have the potential to assess earlier disease status and are currently the focus of significant MRI research5-8. Specifically, image content descriptors (features) have been shown to be informative in detecting subtle differences between textures and intensities between healthy and pathological cartilage in both the in vitro and in vivo setting6, 8.
Recently, we evaluated the ability for a machine learning algorithm, weighted neighbor distance using compound hierarchy of algorithms representing morphology (WND-CHRM)9, to classify MRIs of human osteochondral plugs of normal and pathologic cartilage based on Osteoarthritis Research Society International (OARSI) histological scores8, 10. The features computed from WND-CHRM were used to develop a multivariable least-squares regression model to predict OARSI scores and classify OA cartilage with up to 86% accuracy in various MR contrast modalities. Our previous in vitro results indicate the ability to non-invasively assign individual subjects to a degree of OA pathology8. One essential difference between this previous study of explants and analyses performed in the in vivo setting is the requirement for reproducible tissue segmentation in the latter.
Related work has been performed using in vivo data from the Osteoarthritis Initiative (OAI)11. Urish et al. found that texture analysis of cartilage MRI relaxation time (T2) was effective at differentiating subjects at risk for developing OA symptoms6. In that study, they used a rigid registration algorithm to align double echo in the steady state (DESS) images to T2 parameter maps. Using a global active statistical shape model with a local active contour model, they performed semi-automated segmentation of the DESS images and applied the mask to T2 parameter maps for feature extraction. They achieved approximately 70% classification accuracy in predicting the symptomatic progression of OA as defined by the Western Ontario and McMaster Universities Arthritis (WOMAC) questionnaire12. While these results are promising, the analysis was restricted to texture features extracted from the raw image. However, additional families of features computed on multiple image transforms may provide further information about early cartilage degeneration.
It is of additional importance to note that the intra-subject registration method used in Reference 6 does not provide the ability to evaluate localized cartilage segments with spatial correspondence between subjects of different groups. Therefore, the development of 3D nonlinear image registration methods is of particular interest for application to large clinical datasets, such as the OAI, so that highly specific regions of cartilage can be isolated and directly compared between subjects. In addition, improved methods for inter-subject registration would provide the ability to accurately evaluate longitudinal changes to localized regions. Once images are registered to a universal target, automated segmentation of knee structures becomes straightforward. Hence, as we demonstrate in this study, nonlinear registration prior to segmentation has the potential to overcome the various limitations of other previously established automated segmentation algorithms13-15, which have been reported to overestimate the articular surface, resulting in partial volume effects with subchondral bone, and exhibit substantial variation among methods16.
Here, we evaluate the ability of a machine learning algorithm to classify in vivo MRI of human articular cartilage for the development of OA. Our approach is based on an automated segmentation of articular cartilage in T2-weighted (T2W) images through a nonlinear inter-subject registration scheme applied to data from the OAI. The registered T2 parameter maps of localized regions of segmented cartilage from the weight-bearing region of the medial femoral condyles were classified by WND-CHRM for symptomatic OA progression, according to a change in three-year WOMAC score.
Materials and Methods
Patient cohort
Data were obtained from the OAI database, available for public access at http://oai.epi-ucsf.org. The specific OAI imaging datasets explored here included 0.E.1 and 0.C.2. The kXR SQ (BU) clinical dataset [versions 0.6 and 5.5] was used for obtaining the Kellgren and Lawrence (KL) grades for each participant17. The PhysExam dataset [versions 0.2.2 and 5.2.1] was also used to assess the WOMAC score12 at the time of baseline MR screening and 36 months later. A total of 68 patients were selected from the OAI prospective cohort; 28 were from the non-exposed control sub-cohort and 40 were from the incidence sub-cohort (Fig. 1). In this study, subjects assigned to the “non-progression” group were selected from the non-exposed control sub-cohort (n = 122) and defined at baseline and at 36 months by a WOMAC score ≤10 with a KL score < 2 (Fig. 1). The 20 patients from the incidence sub-cohort were selected based on the initial criteria of a WOMAC score ≤ 10, but with a change in WOMAC score of > 10 within 36 months from baseline, and with minimal baseline radiographic signs of OA defined as a KL < 2 (Fig. 1). Here, we define this group as “symptomatic progression of OA”.
Figure 1.
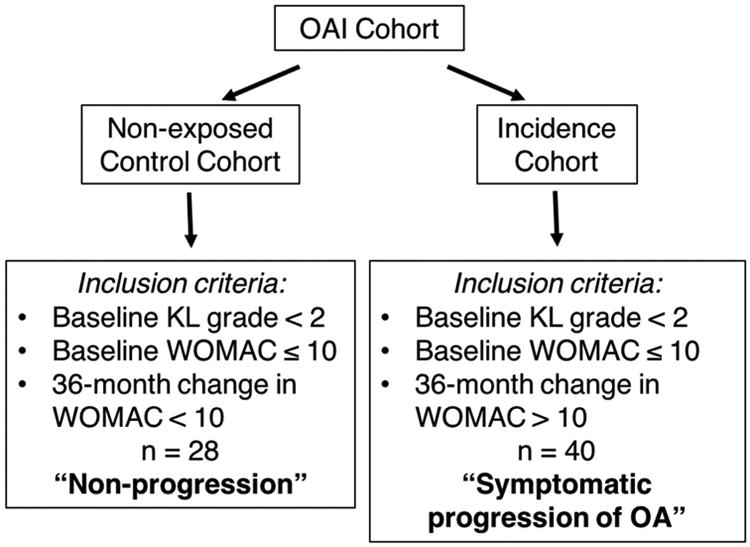
Outline of study design. The non-progression group was randomly selected from the OAI Control cohort based on criteria of baseline KL grade < 2, baseline WOMAC ≤ 10 and a 36-month change in WOMAC < 10 (n = 28). The symptomatic progression of OA group was randomly selected from the OAI Incidence cohort based on the criteria of baseline KL grade < 2, baseline WOMAC ≤ 10 and a 36-month change in WOMAC > 10 (n = 40). MRIs from these 68 patients were then processed and used for classification analysis.
MRI acquisition
The T2W images were obtained from the OAI database (http://oai.epi-ucsf.org). Briefly, MRI of the knee joint was performed on whole body MAGNETOM Trio 3 tesla Multisite scanners (Siemens, Erlangen, Germany) using a standard extremity bird-cage coil11. Sagittal 2D T2W images were acquired using a multi-spin-echo sequence. Seven images with echo time (TE) increasing linearly from 10 to 70 ms were acquired with repetition time = 2700 ms, field of view = 120 × 120 mm2, matrix = 384 × 384, and 27 slices of 3 mm thickness. Total acquisition time was ∼ 11 minutes. Full acquisition details are given in Reference 11.
Image registration
All images were first filtered to achieve noise reduction through application of a multi-spectral non-local means filter, an edge preserving filter, which averages pixel intensities across multiple T2 weightings according to their intensity similarity18. A nonlinear image registration technique using the Automatic Image Registration (A.I.R) toolbox (http://bishopw.loni.ucla.edu/air5/)19-24, implemented in Matlab (MathWorks, Natick, MA, USA), was optimized and applied for inter-subject registration of T2W images obtained at different TE (Fig. 2). A control patient with a BMI of 25.7 and KL grade of zero was selected as the target image to which all subjects' T2W images were registered.
Figure 2.
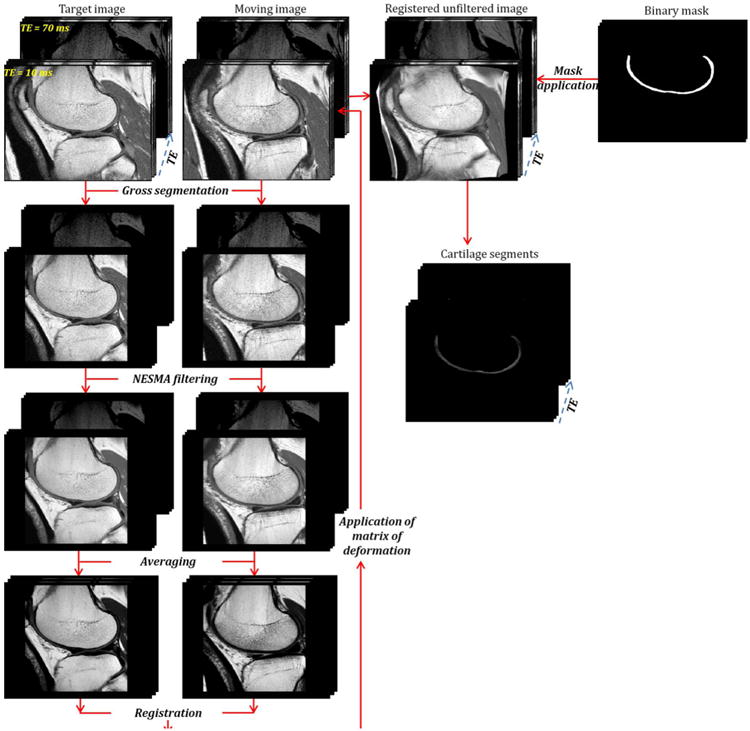
Flow chart of processing steps leading from the unfiltered and unregistered moving image to the cartilage segments. The target and unregistered images were cropped to reduce the field of view and to suppress regions of fat and muscle. Then, a multispectral non-local filter was applied to these images. After filtering, both the multi-spectral target and unregistered filtered weighted images were each averaged across TE. The averaged filtered unregistered image was then registered to the averaged filtered target image. The calculated matrix of deformation derived from this registration was directly applied to the original unfiltered and unregistered T2W image at each TE. A 3D cartilage mask was generated through manual segmentation of each slice from the target image, which was then systematically applied to each of the registered images to isolate cartilage segments from all subjects.
Image registration was performed using a reference image obtained by simple averaging across the images obtained with different TEs in order to maximize signal-to-noise and contrast. The A.I.R. toolbox incorporates variants of nonlinear polynomial spatial transformation models ranging from first (12 parameters) to twelfth (1365 parameters) order. After evaluation of all parameter models (Fig. 3), registration was performed using the tenth order model to optimize accuracy and computational efficiency. The calculated deformation matrix was applied to the original unfiltered T2W images for each TE in order to create the registered image using linear interpolation in all three directions (Fig. 2).
Figure 3.
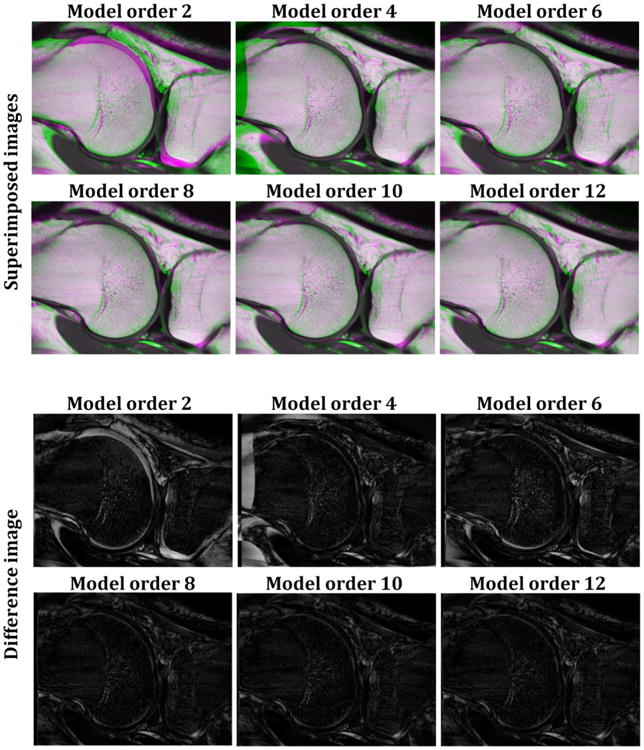
Examples of the quality of registration for six different orders of the nonlinear polynomial transformation model. For each model order, performance of the registration was evaluated through superimposed and difference images between target and registered images. The green and purple colors in the superimposed images indicate misregistered regions. Both the overlap and difference between target and registered images decrease as the order of the model increases from second to eighth order, with no improvement seen for higher order corrections.
Registration validation
The quality of registration was evaluated both qualitatively and quantitatively. The qualitative evaluation entailed a visual inspection of the superimposed target and registered images at each TE. The quantitative evaluation was performed by computing the difference image between the target and registered images at each TE.
In order to ensure that the registration process did not introduce bias, we also compared T2 maps in the cartilage regions of the unregistered images before and after registration. T2 parameter maps were generated from all seven weighted images taking into account the expectation value of the Rician probability density function, as appropriate for magnitude MR images25, 26. In addition, we used the extended phase graph algorithm27, 28 to correct for stimulated echoes caused by radio frequency pulse imperfections29, 30. All numerical calculations were performed using Matlab.
Cartilage segmentation
A cartilage mask was generated through a single manual segmentation of the medial femoral cartilage in each slice of the target T2 map image. The cartilage mask was systematically applied to the each of the registered images to isolate the desired segments from all subjects. Three central weight-bearing slices from the medial femoral condyle were used for classification analysis.
Classification Analysis
WND-CHRM has previously been described in detail9, 31. Briefly, the WND-CHRM algorithm extracts a generic set of numerical image content descriptors, including textures, which measure spatial variation in intensity for several directions and resolutions, statistical distribution of pixel intensity values from polynomial decomposition of the raw image, as well as several image transforms, such as Fourier, Chebyshev, and wavelet, of the raw image. Further, these features are implemented as numerical decompositions of the image and its transforms according to pre-described procedures. The physical meaning of these descriptors is therefore in terms of image characteristics and features. However, there are no specific correspondences between tissue state and these image features. This is a fact often seen in machine learning applications, in which the physical meaning of classifiers takes a back seat to the structure of the images themselves. This results in a final set of 2,919 features (WND-CHRM version 1.60). The features are then ranked by their Fisher discriminant, defined here as the ratio of variance of class means from the pooled mean to the mean of within-class variances32. Because not all features are equally informative and many represent noise9, 32, only the top 100 were used for classification.
A leave-one-out analysis was performed where the class probabilities were computed using the weighted neighbor distance classifier (WND5)31. In the WND5 classifier, the feature vectors from the training images are arranged in a (weighted) feature space, where each feature value is multiplied by its corresponding Fisher discriminant as defined above. The relative probability of a test image belonging to a certain class is calculated as the mean of the inverse fifth power of the Fisher-score weighted distances, r, between the test image and all training images of that class. Each image feature is assigned a Fisher score, Wf :
where Wf is the Fisher score of feature f, N is the total number of classes, T̄f is the mean of the values of feature f among the images allocated for training, and
and
 are the mean and variance of the values of feature f among all training images of class c. All variances are computed after the values of feature f are standardized to the interval [0,1].
are the mean and variance of the values of feature f among all training images of class c. All variances are computed after the values of feature f are standardized to the interval [0,1].
A leave-one-out cross validation analysis was performed such that training sets were constructed with equal numbers of images from the non-progression and symptomatic progression of OA classes. Each sample was in turn excluded from the training set and designated as a test image for assignment to the class with the highest probability. Classification results are reported as sensitivity (proportion of correctly classified symptomatic progression of OA images), specificity (proportion of correctly assigned non-progression images) and accuracy (proportion of correctly assigned samples).
Results
Patient Cohort
Table 1 shows the mean characteristics of the patients selected from the OAI Control and Incidence cohorts. All subjects in the control and symptomatic progression of OA groups had baseline KL grades of zero. The control group had a 36-month change in WOMAC of -0.1, indicating lack of progression. The symptomatic progression of OA group had a change in WOMAC of 25, indicating clear progression of OA symptoms over the 36-month follow-up period. The mean age and BMI of the symptomatic progression of OA group were slightly higher than in the control group (Table 1).
Table 1. Patient characteristics.
| Non-progression group | |||
| Mean age (years) | Mean baseline KL | Mean change in WOMAC | Mean BMI |
| 56 | 0 | -0.1 | 25 |
| Symptomatic progression of OA group | |||
| Mean age (years) | Mean baseline KL | Mean change in WOMAC | Mean BMI |
| 59 | 0 | 25 | 29 |
Image Registration
Performance of the registration as a function of the order of the nonlinear polynomial transformation model is shown in Figure 3. Before registration, the superimposition of the target (green) and the moving (purple) images showed several misaligned regions (Fig. 3). For each model order, the quality of registration was evaluated through superimposition of the target and registered images (Fig. 3, top two rows), and by calculating the difference between the target and registered images (Fig. 3, bottom two rows). The green and purple colors in the superimposed images in Figure 3 indicate mis-registered regions. As shown, both the misregistered regions and the difference between target and registered images decrease as the order of the model increases from second to eighth, with no improvement seen for higher order corrections.
Figure 4 shows a representative example of a T2 map of cartilage calculated before and after registration. In all cases, calculated T2 values were similar, within 10% with no systematic trends, before and after registration in all regions of cartilage, indicating that no significant bias was introduced during the proposed registration process.
Figure 4.
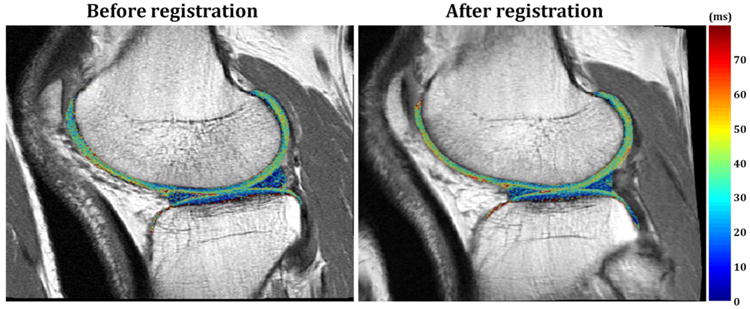
Representative T2 maps calculated before and after registration in knee cartilage regions. T2 maps were superimposed on their corresponding T2W images before and after registration. The T2 values are shown to be minimally changed before and after registration, indicating that the registration methods did not introduce bias to the T2 maps.
Classification and regression analysis
Table 2 shows the classification accuracy of WND-CHRM applied to T2 maps of medial femoral condyle cartilage. The sensitivity, specificity and accuracy were 74%, 76% and 75%, respectively. The most informative features for classification, according to their rank ordered Fisher scores, were the Zernike coefficients and Chebyshev statistics computed from the Fourier transform, as well as the Haralick textures computed from the wavelet (Symlet 5, level 1) transform. Other informative features included multi-scale histograms from several of the image transforms, as well as Zernike coefficients and Haralick textures computed from the Fourier transform.
Table 2.
WND-CHRM classification accuracy of isolated T2 maps of cartilage. Intervals based on 95% confidence using the Normal Approximation method.
| Sensitivity | 0.74 ± 1.3% |
| Specificity | 0.76 ± 1.3% |
| Accuracy | 0.75 ± 0.9% |
Discussion
The weight-bearing regions, specifically the central portion of the medial femoral condyle, of the knee are the most susceptible to changes in cartilage thickness associated with early OA33, 34. Consequently, this region of cartilage may be expected to show subtle pattern differences before the frank transition to OA. T2 relaxation time as a quantitative metric is not the best marker for OA, as there is no highly-sensitive reference standard with which to compare35. However, since T2 relaxation is related to the presence of free water molecules, which slow down the loss of transverse magnetization, T2 maps provide for a direct measurement of water content in cartilage35. In OA, as cartilage matrix begins to break down and become more permeable to water, there is an elevation of T2 relaxation time. Thus in turn, T2 provides for an indirect assessment of collagen content and orientation, which are important indicators for early OA, and is used clinically to evaluate disease status36, 37. In this study, we demonstrate the ability for a state-of-the-art machine learning algorithm, WND-CHRM, to classify T2 maps of the central weight-bearing slices of cartilage within the medial femoral condyle for the progression to symptomatic OA, defined as a change in WOMAC score of >10 three years from baseline evaluation.
WND-CHRM classification is based on inherent image texture and intensity information, rather than measurements such as cartilage volume or thickness. The pattern recognition component of these methods relies on aligned images from which to extract information. Therefore, we developed a new approach for automated articular cartilage segmentation using nonlinear inter-subject registration of T2W images. The application of nonlinear registration provided the ability to automate cartilage segmentation within a large dataset and to isolate corresponding slices of cartilage from the medial femoral condyles between patients. From this, we were able to apply WND-CHRM to successfully evaluate image features from standardized regions within the knee. We found that image features computed by WND-CHRM were correlated with patient-reported ground truth data, that is, WOMAC score. We demonstrated that WND-CHRM was able to classify T2 maps of cartilage, obtained prior to clinical OA, to predict the development of clinical OA with 75% accuracy. This represents a substantial improvement as compared to the result obtained using conventional classification analysis based on T2 according to the Euclidean distance metric (EDM). The EDM showed sensitivity, specificity and accuracy of only 50% using the same dataset, indicating lack of any prognostic ability.
The most informative features for classification provided information about pixel intensities and textures. Specifically, low frequency features including Zernike and Haralick features, Chebyshev statistics and multi-scale histograms measure small variations in pixel values and provide information about texture over large scale areas with smooth intensity transitions31. The variation in pixel intensity patterns derived from these groups of texture features have been previously shown to correlate with the biochemical and biomechanical alterations of OA cartilage in the hip joint38. Further, we found that these low frequency features, which measure subtle variations over regions spanning many pixels, were most informative for evaluating cartilage. Since the T2 maps used in this study included only cartilage, the images had relatively low intensity variation, and in turn, high contrast features, such as edge and object statistics did not provide useful information for classification.
We note that in our previous study, we used WND-CHRM features computed on MRIs of osteochondral plugs to construct a multiple linear regression model for classification based on OARSI score8. We found that the accuracy of binary classification based on several MR measurements, including the diffusion weighted image with b = 999 s/mm2 and the T2W image with TE = 50 ms, improved through use of regression, which establishes relationships throughout the entire dataset for classification of single samples. In Reference 8, the predictors, were the descriptive values assigned to particular features, while the outcome variables, were the histologic OARSI score for each sample. Yet, when similar analyses were performed in the present study using change in WOMAC scores as the outcome variables, we found no such improvement. We attribute this difference in findings to the ground truth information associated with the images; in our previous study, each osteochondral plug had a corresponding OARSI score, which is an objective histological measurement for OA and corresponds to degradation status reflected in the images; These plugs [from the previous study] were obtained from patients with severe OA undergoing knee arthroplasty, in contrast to the present study where the patients did not have clinical OA at baseline. Thus the relationships between MR outcomes, symptoms (e.g. WOMAC) and tissue degradation status are expected to be much more subtle in the present case. At this early stage of disease, we do not expect that WOMAC will reflect tissue degradation. The cartilage used in the present study was classified based on WOMAC score, which is a subjective, patient-reported, measurement and does not provide information about cartilage structure.
There are certain limitations to our study. The processing time for the 10th order polynomial registration model was on the order of several hours per subject. The use of lower-order procedures or other more rapid algorithms should be explored in further studies. Since registration was systematically performed using a tenth order polynomial model, there was substantial computational processing time (several hours per patient). Further, we elected to incorporate data from three representative central slices into our analysis in order to efficiently incorporate OA status into the analysis. The question of whether inclusion of all tissue slices, covering the entire knee and perhaps including non-cartilage regions, would improve classification accuracy, may be addressed in further studies. However, in this study we restricted our analysis to the three slices that cover the central weight-bearing region of the femoral condyle and excluded the peripheral slices that are likely subject to partial-volume effects. In addition, our registration method is dependent on the target image. In effect, the accuracy of this approach relies on how well the algorithm can register an image to its corresponding target. We excluded from classification any registered image with interpolation errors, which were occasionally observed in the femur region of patients with substantially smaller bone size than the target. The development of a universal knee atlas has the potential to limit this source of potential bias and would be of particular benefit in the analysis of larger cohorts of patients from the OAI.
Although we analyzed fewer patients than a previous study6 that used similar inclusion criteria, the authors of that study note that in order to maximize the size of the dataset (n = 168), they included patients with a baseline KL grade of two in the progression to symptomatic OA group. This KL score indicates early degeneration, so that inclusion of these subjects would enhance classification accuracy. We elected to exclude patients with a KL grade of two in our analysis, resulting in a smaller sample size but presenting a more realistic, though more challenging, classification task. In spite of this, we achieved an overall classification accuracy of 75%, comparable to the results shown in Reference 6.
Important extensions of this study will be to incorporate additional cartilage slices into the analysis, as noted above, and to combine T2 maps with additional MR imaging contrast modalities. Indeed, it has been shown that incorporation of different MR modalities greatly strengthens classification of early and late cartilage degeneration39-43, so that this may be a useful approach in the context of WND-CHRM analysis as well. Finally, use of cartilage morphology and incorporation of other joint structures, such as periarticular bone, has the potential to greatly improve classification for progression to symptomatic OA. The promise of this has approach is indicated by the fact that WND-CHRM has already shown success in classifying X-ray images based on KL grade32.
In conclusion, we have demonstrated that image features derived from appropriately pre-processed T2 maps of cartilage from the weight-bearing region of the medial femoral condyle may be useful for the detection of OA progression.
Acknowledgments
This work was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. KU acknowledges support through NIH KL2 TR000146.
The OAI is a public-private partnership comprised of five contracts (N01-AR-2-2258; N01-AR-2-2259; N01-AR-2-2260; N01-AR-2-2261; N01-AR-2-2262) funded by the National Institutes of Health, a branch of the Department of Health and Human Services, and conducted by the OAI Study Investigators. Private funding partners include Merck Research Laboratories; Novartis Pharmaceuticals Corporation, GlaxoSmithKline; and Pfizer, Inc. Private sector funding for the OAI is managed by the Foundation for the National Institutes of Health. This manuscript was prepared using an OAI public use data set and does not necessarily reflect the opinions or views of the OAI investigators, the NIH, or the private funding partners.
Footnotes
Author contributions: Substantial contributions to research design, acquisition, analysis and interpretation of data: BGA, MB, CEC, IGG, RGS; drafting the paper and critical revision: all authors; approval of the submitted and final version: all authors.
Conflict of interest: The authors have no conflicts of interest to declare. The study sponsors had no involvement in the study design, collection and interpretation of data, writing of the manuscript or manuscript submission.
References
- 1.Swedberg JA, Steinbauer JR. Osteoarthritis. Am Fam Physician. 1992;45:557–568. [PubMed] [Google Scholar]
- 2.Davis MA. Epidemiology of osteoarthritis. Clin Geriatr Med. 1988;4:241–255. [PubMed] [Google Scholar]
- 3.Braun HJ, Gold GE. Advanced MRI of articular cartilage. Imaging Med. 2011;3:541–555. doi: 10.2217/iim.11.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun HJ, Gold GE. Diagnosis of osteoarthritis: imaging. Bone. 2012;51:278–288. doi: 10.1016/j.bone.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashinsky B, Bouhrara M, Urish KL, Coletta CE, Goldberg IG, Spencer RG. The Orthopaedic Research Society. Las Vegas, NV: 2015. Predicting Early Osteoarthritis in the Human Knee: Pattern Recognition and Machine Learning Classification of Magnetic Resonance Images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urish KL, Keffalas MG, Durkin JR, Miller DJ, Chu CR, Mosher TJ. T2 texture index of cartilage can predict early symptomatic OA progression: data from the osteoarthritis initiative. Osteoarthritis Cartilage. 2013;21:1550–1557. doi: 10.1016/j.joca.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kauffmann C, Gravel P, Godbout B, Gravel A, Beaudoin G, Raynauld JP, et al. Computer-aided method for quantification of cartilage thickness and volume changes using MRI: validation study using a synthetic model. IEEE Trans Biomed Eng. 2003;50:978–988. doi: 10.1109/TBME.2003.814539. [DOI] [PubMed] [Google Scholar]
- 8.Ashinsky BG, Coletta CE, Bouhrara MB, Lukas VA, Boyle JM, Reiter DA, et al. Machine learning classification of OARSI-scored human articular cartilage using magnetic resonance imaging. Osteoarthritis and Cartilage. 2015 doi: 10.1016/j.joca.2015.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shamir L, Orlov N, Eckley DM, Macura T, Johnston J, Goldberg IG. Wndchrm - an open source utility for biological image analysis. Source Code Biol Med. 2008;3:13. doi: 10.1186/1751-0473-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pritzker KP, Gay S, Jimenez SA, Ostergaard K, Pelletier JP, Revell PA, et al. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthritis Cartilage. 2006;14:13–29. doi: 10.1016/j.joca.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Peterfy CG, Schneider E, Nevitt M. The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthritis Cartilage. 2008;16:1433–1441. doi: 10.1016/j.joca.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellamy N, Buchanan WW, Goldsmith CH, Campbell J, Stitt LW. Validation study of WOMAC: a health status instrument for measuring clinically important patient relevant outcomes to antirheumatic drug therapy in patients with osteoarthritis of the hip or knee. J Rheumatol. 1988;15:1833–1840. [PubMed] [Google Scholar]
- 13.Folkesson J, Dam EB, Olsen OF, Pettersen PC, Christiansen C. Segmenting articular cartilage automatically using a voxel classification approach. IEEE Trans Med Imaging. 2007;26:106–115. doi: 10.1109/TMI.2006.886808. [DOI] [PubMed] [Google Scholar]
- 14.Fripp J, Crozier S, Warfield SK, Ourselin S. Automatic segmentation and quantitative analysis of the articular cartilages from magnetic resonance images of the knee. IEEE Trans Med Imaging. 2010;29:55–64. doi: 10.1109/TMI.2009.2024743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glocker B, Komodakis N, Paragios N, Glaser C, Tziritas G, Navab N. Primal/dual linear programming and statistical atlases for cartilage segmentation. Med Image Comput Comput Assist Interv. 2007;10:536–543. doi: 10.1007/978-3-540-75759-7_65. [DOI] [PubMed] [Google Scholar]
- 16.Schneider E, Nevitt M, McCulloch C, Cicuttini FM, Duryea J, Eckstein F, et al. Equivalence and precision of knee cartilage morphometry between different segmentation teams, cartilage regions, and MR acquisitions. Osteoarthritis Cartilage. 2012;20:869–879. doi: 10.1016/j.joca.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kellgren JH, Lawrence JS. Radiological assessment of osteo-arthrosis. Ann Rheum Dis. 1957;16:494–502. doi: 10.1136/ard.16.4.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouhrara M, Bonny JM, Ashinsky BA, Maring MC, Spencer RGS. Noise estimation and reduction in magnetic resonance imaging using a new multispectral nonlocal maximum-likelihood filter. IEEE Trans Med Imaging. 2017;36:181–193. doi: 10.1109/TMI.2016.2601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bearer EL, Zhang X, Janvelyan D, Boulat B, Jacobs RE. Reward circuitry is perturbed in the absence of the serotonin transporter. Neuroimage. 2009;46:1091–1104. doi: 10.1016/j.neuroimage.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan KC, Fu QL, Hui ES, So KF, Wu EX. Evaluation of the retina and optic nerve in a rat model of chronic glaucoma using in vivo manganese-enhanced magnetic resonance imaging. Neuroimage. 2008;40:1166–1174. doi: 10.1016/j.neuroimage.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Chuang KH, Lee JH, Silva AC, Belluscio L, Koretsky AP. Manganese enhanced MRI reveals functional circuitry in response to odorant stimuli. Neuroimage. 2009;44:363–372. doi: 10.1016/j.neuroimage.2008.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tucciarone J, Chuang KH, Dodd SJ, Silva A, Pelled G, Koretsky AP. Layer specific tracing of corticocortical and thalamocortical connectivity in the rodent using manganese enhanced MRI. Neuroimage. 2009;44:923–931. doi: 10.1016/j.neuroimage.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC. Automated image registration: I. General methods and intrasubject, intramodality validation. J Comput Assist Tomogr. 1998;22:139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- 24.Woods RP, Grafton ST, Watson JD, Sicotte NL, Mazziotta JC. Automated image registration: II. Intersubject validation of linear and nonlinear models. J Comput Assist Tomogr. 1998;22:153–165. doi: 10.1097/00004728-199801000-00028. [DOI] [PubMed] [Google Scholar]
- 25.Bouhrara M, Reiter DA, Celik H, Bonny JM, Lukas V, Fishbein KW, et al. Incorporation of rician noise in the analysis of biexponential transverse relaxation in cartilage using a multiple gradient echo sequence at 3 and 7 tesla. Magn Reson Med. 2014 doi: 10.1002/mrm.25111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raya JG, Dietrich O, Horng A, Weber J, Reiser MF, Glaser C. T2 measurement in articular cartilage: impact of the fitting method on accuracy and precision at low SNR. Magn Reson Med. 2010;63:181–193. doi: 10.1002/mrm.22178. [DOI] [PubMed] [Google Scholar]
- 27.Hennig J. Echoes—how to generate, recognize, use or avoid them in MR-imaging sequences. Concepts Magn Reson. 1991;3:125–142. [Google Scholar]
- 28.Prasloski T, Madler B, Xiang QS, MacKay A, Jones C. Applications of stimulated echo correction to multicomponent T2 analysis. Magn Reson Med. 2012;67:1803–1814. doi: 10.1002/mrm.23157. [DOI] [PubMed] [Google Scholar]
- 29.Bouhrara M, Bonny JM. B(1) mapping with selective pulses. Magn Reson Med. 2012;68:1472–1480. doi: 10.1002/mrm.24146. [DOI] [PubMed] [Google Scholar]
- 30.Simmons A, Tofts PS, Barker GJ, Arridge SR. Sources of intensity nonuniformity in spin echo images at 1.5 T. Magn Reson Med. 1994;32:121–128. doi: 10.1002/mrm.1910320117. [DOI] [PubMed] [Google Scholar]
- 31.Orlov N, Shamir L, Macura T, Johnston J, Eckley DM, Goldberg IG. WND-CHARM: Multi-purpose image classification using compound image transforms. Pattern Recognit Lett. 2008;29:1684–1693. doi: 10.1016/j.patrec.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shamir L, Ling SM, Scott WW, Jr, Bos A, Orlov N, Macura TJ, et al. Knee x-ray image analysis method for automated detection of osteoarthritis. IEEE Trans Biomed Eng. 2009;56:407–415. doi: 10.1109/TBME.2008.2006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelletier JP, Raynauld JP, Berthiaume MJ, Abram F, Choquette D, Haraoui B, et al. Risk factors associated with the loss of cartilage volume on weight-bearing areas in knee osteoarthritis patients assessed by quantitative magnetic resonance imaging: a longitudinal study. Arthritis Res Ther. 2007;9:R74. doi: 10.1186/ar2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eckstein F, Maschek S, Wirth W, Hudelmaier M, Hitzl W, Wyman B, et al. One year change of knee cartilage morphology in the first release of participants from the Osteoarthritis Initiative progression subcohort: association with sex, body mass index, symptoms and radiographic osteoarthritis status. Ann Rheum Dis. 2009;68:674–679. doi: 10.1136/ard.2008.089904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matzat SJ, van Tiel J, Gold GE, Oei EH. Quantitative MRI techniques of cartilage composition. Quant Imaging Med Surg. 2013;3:162–174. doi: 10.3978/j.issn.2223-4292.2013.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi JA, Gold GE. MR imaging of articular cartilage physiology. Magn Reson Imaging Clin N Am. 2011;19:249–282. doi: 10.1016/j.mric.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.David-Vaudey E, Ghosh S, Ries M, Majumdar S. T2 relaxation time measurements in osteoarthritis. Magn Reson Imaging. 2004;22:673–682. doi: 10.1016/j.mri.2004.01.071. [DOI] [PubMed] [Google Scholar]
- 38.Boniatis I, Cavouras D, Costaridou L, Kalatzis I, Panagiotopoulos E, Panayiotakis G. Computer-aided grading and quantification of hip osteoarthritis severity employing shape descriptors of radiographic hip joint space. Comput Biol Med. 2007;37:1786–1795. doi: 10.1016/j.compbiomed.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Lin PC, Irrechukwu O, Roque R, Hancock B, Fishbein KW, Spencer RG. Multivariate analysis of cartilage degradation using the support vector machine algorithm. Magn Reson Med. 2012;67:1815–1826. doi: 10.1002/mrm.23189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin PC, Reiter DA, Spencer RG. Classification of degraded cartilage through multiparametric MRI analysis. J Magn Reson. 2009;201:61–71. doi: 10.1016/j.jmr.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Nissi MJ, Toth F, Shaver J, Johnson CP, Zhang J, et al. Multiparametric MRI of Epiphyseal Cartilage Necrosis (Osteochondrosis) with Histological Validation in a Goat Model. PLoS One. 2015;10:e0140400. doi: 10.1371/journal.pone.0140400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rautiainen J, Nissi MJ, Salo EN, Tiitu V, Finnila MA, Aho OM, et al. Multiparametric MRI assessment of human articular cartilage degeneration: Correlation with quantitative histology and mechanical properties. Magn Reson Med. 2014 doi: 10.1002/mrm.25401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griebel AJ, Trippel SB, Emery NC, Neu CP. Noninvasive assessment of osteoarthritis severity in human explants by multicontrast MRI. Magn Reson Med. 2014;71:807–814. doi: 10.1002/mrm.24725. [DOI] [PMC free article] [PubMed] [Google Scholar]


