Abstract
Cell-type specific genes were recognized by interrogating microarrays carrying Dictyostelium gene fragments with probes prepared from fractions enriched in prestalk and prespore cells. Cell-type specific accumulation of mRNA from 17 newly identified genes was confirmed by Northern analyses. DNA microarrays carrying 690 targets were used to determine expression profiles during development. The profiles were fit to a biologically based kinetic equation to extract the times of transcription onset and cessation. Although the majority of the genes that were cell-type enriched at the slug stage were first expressed as the prespore and prestalk cells sorted out in aggregates, some were found to be expressed earlier before the cells had even aggregated. These early genes may have been initially expressed in all cells and then preferentially turned over in one or the other cell type. Alternatively, cell type divergence may start soon after the initiation of development.
INTRODUCTION
Differential gene expression in one cell-type or another can adapt their physiological functions to specialized roles. Although most genes are responsible for processes common to the growth and maintenance of all cells in multicellular organisms, specialized genes are needed for cell-type divergence and differentiation. Recognizing cell-type–specific genes can open the way to a better understanding of development by providing molecular markers to follow the pathways leading to different cell types and can uncover the underlying physiological functions in these processes. The mechanisms that lead to divergence of the initial cell types and subsequent tissue proportioning are more easily approached in Dictyostelium discoideum than in complex metazoans because only two major cell types are formed during the developmental cycle and they are sufficiently different that they can be physically separated in biochemical amounts (Loomis, 1982; Ratner and Borth, 1983). Moreover, when development is initiated by the removal of exogenous nutrients, growth stops and there is no further replication of the chromosomes (Shaulsky and Loomis, 1995). Therefore, studies on the accumulation of proteins and mRNAs do not need to take into account differential growth of specific cell types.
Several hours after the initiation of development in homogenous populations of exponentially growing Dictyostelium amoebae, a small number of cells accumulates certain mRNAs that are later found exclusively in either prespore or prestalk cells (Escalante and Loomis, 1995; Firtel, 1995; Loomis, 1996). The number of cells expressing these cell-type–specific markers increases as aggregates containing up to 105 cells are formed. Prestalk cells then sort out to the top of the aggregate where they form finger-like tips. When the fingers come to lie horizontally, the resulting slug-shaped organisms can migrate away with prestalk cells leading. Prespore cells are found exclusively at the base of tipped mounds and, later, in the posterior three-quarters of slugs. During culmination prespore cells rise up the elongating stalks and encapsulate to form terminally differentiated spores. As the prestalk cells enter the stalk tube, they vacuolize, expand, and become surrounded by a cellulose-containing cell wall from which they never escape.
The 34-Mb Dictyostelium genome is in the process of being sequenced but closure is not expected for several years (Loomis, 1998). The genome is predicted to carry about 8000 genes, many of which are represented among the cDNAs characterized by the Japanese consortium (Morio et al., 1998). Moreover, the predicted protein products of >850 Dictyostelium genes are presently available at NCBI (National Center for Biotechnology Information: www.ncbi.nlm.nih.gov). Genes that are preferentially expressed in either prespore or prestalk cells have been found by analyzing RNA isolated from density-separated cell types (Mehdy et al., 1983; Firtel, 1995; Loomis, 1996). mRNA from most of these marker genes accumulates during the mound stage in one or the other of the cell types but some are expressed earlier and only become cell-type specific after the tipped aggregate stage (Jermyn and Williams, 1995). Strains carrying constructs that express β-galactosidase driven by the regulatory regions of these genes have been used to follow the appearance and progress of the different cell types (Jermyn et al., 1989; Early et al., 1993, Fosnaugh and Loomis, 1993; Nicol et al., 1999). Stained cells first appear during the aggregation stage in a spatially random manner, giving a “salt and pepper” pattern. They then sort out to either the prespore or prestalk domains and are subsequently found either in the sorus where the spores are held at the top of the fruiting body or in the cellular stalk.
The most commonly used cell-type–specific genes encode major structural components such as the extracellular sheath that surrounds slugs (ecmA), the stalk (ecmB), or the spore coat (cotA, cotB, cotC, cotD). Although these genes provide excellent markers for the divergence and regulation of the cell types, they do not provide insights into the underlying mechanisms because deletion or disruption of these genes has no observable effects on cell-type differentiations. In an effort to recognize other cell-type–specific genes, we characterized the expression patterns of a large set of newly identified genes that has recently become available from mapping projects, the Japanese EST project, and saturation mutagenesis screens (Kuspa and Loomis, 1996; Morio et al., 1998; Loomis, unpublished data). Thirty-four cell-type–specific genes were found, only half of which had been previously known. The time courses of accumulation of these genes were determined by probing microarrayed DNA. Initiation of expression was observed at various stages throughout development, although a large number of prespore genes appeared to be coordinately induced at 10 h of development, just after aggregation was completed.
MATERIALS AND METHODS
Growth, Development, and Preparation of RNA
Wild-type strain NC4 was grown for 2 d in association with Klebsiella aerogenes on SM plates (Sussman, 1987). Cells were collected, washed, and deposited on nitrocellulose filters supported on buffer-saturated pads and allowed to develop at 22°C. For cell-type separations, cells were collected from filters after 19 h of development (the “Mexican hat” stage) in 20 mM sodium potassium phosphate, 20 mM EDTA, pH 7.0 (“20/20” buffer) and dissociated by triturating in the presence of 1 mg/ml pronase and 12.5 mM 2,3-dimercaptopropanol (Ratner and Borth, 1983). The dissociated cells were washed, resuspended at 4 × 108 cells/ml and 250-μl aliquots applied to the top of preformed cold 12-ml 45% Percoll gradients and centrifuged at 17,000 × g for 3 min (Ratner and Borth, 1983). Approximately 25% of the cells banded at a lighter density than the remainder of the cells. The separated fractions were washed free of Percoll, pelleted, and dissolved in Trizol (Life Technologies, Grand Island, NY) and RNA processed according to manufacturer's instructions. For time course experiments, the cells were scraped off the filters at 2-h intervals into deionized water, pelleted, dissolved in Trizol reagent, and RNA prepared. PolyA+ RNA was isolated from total RNA preparations with the use of Promega PolyATtract System 1000 (Promega, Madison, WI).
Preparation of Microarrays
Sequences were amplified by polymerase chain reaction (PCR) from 330 Dictyostelium genes chosen from those deposited in GenBank and 360 selected cDNAs. Supplemental material containing a list of all the genes and the size of the arrayed fragments are available at http://www.biology.ucsd.edu/loomis-cgi/microarray/index.html. When amplifying from genomic DNA, primers were designed to bracket 300-1000-bp regions near the 3′ ends such that trinucleotide repeats commonly found in Dictyostelium genes were avoided whenever possible. Total genomic DNA (40 ng) was deposited in each well of a 96-well plate and used as template for 2 U of Taq DNA polymerase (Hot Start Taq; Qiagen, Santa Clarita, CA). Primers were added to 1 μM and the reaction run for 40 cycles with annealing and extension temperatures of 52 and 72°C. Products were visualized by ethidium bromide staining of electrophoretically separated DNA and purified with the use of Millipore 96-well purification plates according to the manufacturer's instructions.
cDNA clones were chosen from the set of Dictyostelium-expressed sequence tags on the basis of sequence homology with named genes (Morio et al., 1998). Plasmids were kindly provided by T. Morio and Y. Tanaka (University of Tsukuba) and used as PCR templates with primers to flanking vector sequences. Reactions were carried out as described above for genomic DNA. All but 30 of the plasmids gave products of the expected size. All cDNAs discussed in the text were confirmed by resequencing.
PCR-amplified DNA targets were printed in duplicate on MD type V glass slides with the use of a Molecular Dynamics GenIII microarray robot (DeRisi et al., 1997; Spellman et al., 1998). Microarrayed slides were stored in the dark under vacuum.
Expression Analyses
Fluorescently labeled DNA probes were prepared from 2 μg of polyA+ RNA with the use of 5 μg of oligo-dT primers with 2 μl of Superscript II DNA polymerase (Life Technologies). Time averaged reference probe was prepared with 3 μl of Cy-5-conjugated dCTP (Amersham Pharmacia Biotech, St. Louis, MO) with the use of polyA+ RNA pooled from samples taken throughout development. Probes from specific time points were prepared with 3 μl of Cy-3 conjugated dCTP (Amersham Pharmacia Biotech). After incubation at 42°C for 3 h, unincorporated dyes were removed with the use of microcon-30 columns (Millipore, Burlington, MA) with three washes with 450 μl of TE buffer before drying and resuspending in 15 μl of 5× standard saline citrate (SSC), 0.3% SDS, 50% formamide.
Labeled probes were mixed together with 1 μg of yeast tRNA and 1 μg of 15-mer oligo-dA, deposited on the microarrays and covered with 22-mm2 coverslips (Amersham Pharmacia Biotech). Hybridization was carried out in sealed chambers (GeneMachines, San Carlos, CA) at 42°C for 10–16 h. The slides were then washed at 42°C in the dark with 2× SSC, 0.2% SDS; 0.4× SSC; and 0.2× SSC followed by a 2-min wash in distilled water at 65°C before drying. Protocols are available with supplemental material at the Web site at http://www.biology.ucsd.edu/loomis-cgi/microarray/index.html.
Probed microarrays were scanned in a Molecular Dynamics GenIII ArrayScanner. Images were quantitated with ScanAlyze available at http://rana.lbl.gov/EisenSoftware.htm. Total Cy3 signal was normalized to total Cy5 signal after background subtraction to allow independent slides to be compared. The ratios of Cy3/Cy5 for individual genes were then calculated.
Statistical Analyses
To extract the times of onset and cessation of transcription, t1 and t2, the microarray signals for cell-type–specific mRNAs (A) were fit to the first order differential equation dA/dt = St − D∗A where the rate of synthesis (S) was taken as constant between t1 and t2 and otherwise zero (Sasik, Iranfar, Hwa, and Loomis, unpublished data). The rate of degradation (D) was adjusted to give mRNA half-lives between 0.5 and 12 h for individual genes. The goodness of fit was measured by the following statistic:
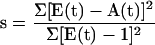 |
where E is the ratio of background-adjusted fluorescence of the time sample (Cy3) and the reference sample (Cy5) normalized so that the mean expression level of each gene throughout development is unity (Sasik, Iranfar, Hwa, and Loomis, unpublished data). Because the denominator of the statistic is a measure of temporal variation found experimentally, s is a measure of how much of that variation can be accounted for by the simple kinetic equation. Statistical significance (p value) of the s statistic for each gene was calculated by permuting the expression pattern 1000 times and calculating the fraction of permutations whose s statistic is less than that of the experimental pattern. These p values were not adjusted for multiple testing.
Northern Analyses
Gel-separated RNA was transferred to nylon membranes (MagnaGraph; MCI Separations, Westborough, MA) and Northern blots were probed as described by Shaulsky and Loomis (1995). 32P-Labeled probes were generated by either random hexamer labeling of DNA fragments or transcription of RNA probes from linearized plasmids.
RESULTS
New Prespore and Prestalk Genes
Microarrays were prepared with a total of 690 target DNAs from Dictyostelium genes. They included cDNAs from 360 newly recognized but uncharacterized genes that show significant homology to established genes as well as DNA that were PCR amplified from 81 genes that were recently recovered from mutant strains. We also included PCR-amplified DNA from 249 Dictyostelium genes with sequences available in GenBank. Expression patterns for many of these have appeared in the literature and can be compared with the patterns observed from probing the chips. These 690 target DNAs were robotically arrayed in duplicate on a set of silanized glass slides with the use of a Molecular Dynamics GenIII robot.
Wild-type cells of strain NC4 were developed to the slug stage before being dissociated to single cells by treatment with pronase and EDTA. Prespore and prestalk cells were then separated on preformed Percoll density gradients (Ratner and Borth, 1983). Prestalk cells are found predominantly in a band at a lower density than the prespore band. However, the low-density band is contaminated with ∼10% prespore cells as judged by their expression of a cotB::lacZ marker (Fosnaugh and Loomis, 1993; Shaulsky et al., 1995). Likewise, the higher density band is contaminated with ∼10% prestalk cells as judged by their expression of a ecmA::lacZ marker. PolyA+ RNA was prepared from prestalk and prespore cells and used to generate fluorescent probes by incorporating Cy3 or Cy5 derivatized nucleotides into DNA copies. The prestalk and prespore probes were mixed and hybridized to the chips to determine the relative levels of expression of the 690 genes. Four independent separations were analyzed. The complete results can be seen with supplemental material at our Web site at http://www.biology.ucsd.edu/loomis-cgi/microarray/index.html.
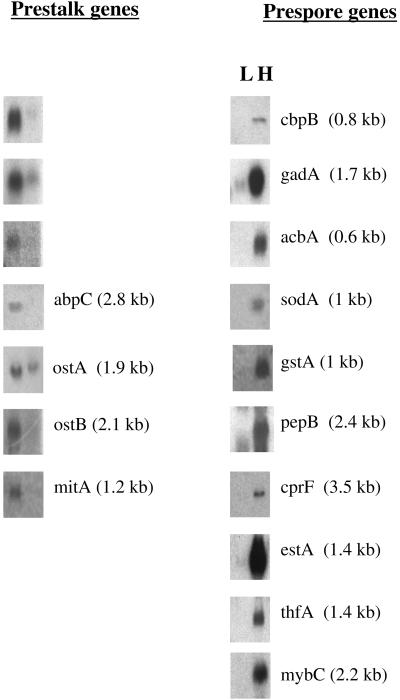
The prestalk signal was found to be more than 2.5-fold that of the prespore signal for 14 genes, whereas the prespore signal was found to be more than 2.5-fold that of the prestalk signal for 21 genes (Table 1). Seven of the prestalk genes had been previously recognized to be enriched in prestalk cells, whereas 11 of the prespore genes had been previously recognized as prespore specific. To confirm the cell-type specificity of these putative prestalk and prespore genes, RNA samples isolated from density-separated slug cells were size fractionated in gels and blots were probed with DNA from the candidate genes. Results from these Northern blots confirmed that mRNAs from the prestalk genes were at least threefold enriched in the low-density fractions, whereas mRNAs from the prespore genes were more than 10-fold enriched in the higher density fractions (Figure 1). In all cases, the sizes of the cell-type–specific mRNAs conformed to those predicted from the open reading frames of the individual genes. Together, these Northern analyses confirmed the cell-type specificity of each of the genes that the microarray analyses indicated were expressed in a cell-type–specific manner.
Table 1.
Cell-type–specific genes of Dictyostelium
| Locus | Gene product | Accession no. | Commentsa |
|---|---|---|---|
| Prestalk genes | |||
| cprA | CP1 | X02407 | Established prestalk gene |
| cprB | CP2 | X03344 | Established prestalk gene |
| lagC | gp150 | U09478 | Established prestalk gene |
| rasD | RAS | Z11804 | Established prestalk gene |
| ampA | D11 | M11012 | Established prestalk gene |
| ecmA | ST430 | X74046 | Established prestalk gene |
| ecmB | ST310 | M73676 | Established prestalk gene |
| cbpD | CBP4 | C24669 | Dorywalska et al., 2000 |
| tpsA | Trehalose-6P synthetase | C92240 | 42% to trehalose phosphate synthetase |
| C89887 | Unknown | C89887 | 66% to short region in laminins |
| abpC | ABP120 | X15430 | Noegel et al., 1989 |
| ostA | P450 1A1 | C84055 | 32% to P450s |
| ostB | P450 2D4 | C92981 | 32% to P450s |
| mitA | Transporter | C92960 | 29% to mitochondrial transporter |
| Prespore genes | |||
| pspA | SP29 | M20909 | Established prespore gene |
| cotA | Spore coat protein SP96 | X16491 | Established prespore gene |
| cotB | Spore coat protein SP70 | M26238 | Established prespore gene |
| cotC | Spore coat protein SP60 | X51892 | Established prespore gene |
| cotD | Spore coat protein SP75 | D13973 | Established prespore gene |
| pspD | Spore coat protein SP87 | U25144 | Established prespore gene |
| wacA | MIP (major intrinsic protein) | U68246 | Established prespore gene |
| aqpA | Intrinsic protein A | C83907 | Established prespore gene |
| pspG | Protein 3F | X16124 | Established prespore gene |
| catB | Catalase B | C23764 | Established prespore gene |
| spiA | Spore protein SpiA | X54452 | Established prespore gene |
| cbpB | Ca++ binding protein | U55379 | Andre et al., 1996 |
| gadA | Glutamate decarboxylase | C84710 | 49% to glutamate decarboxylase |
| acbA | Acyl CoA binding | C90029 | 49% to acyl CoA binding protein |
| sodA | Superoxide dismutase | C23658 | 68% to Cu-Zn SOD |
| gstA | Glutathione S transferase | C83884 | 32% to glutathione S transferase |
| pepB | Protease | C92322 | 38% to pepstatin insensitive protease |
| cprF | Cathepsin O | C84197 | 30% to Dictyostelium CprA |
| estA | Esterase/lipase | AU037382 | 33% to esterases/lipases |
| thfA | Tetrahydrofolate dehydrog | C84726 | 51% to tetrahydrofolate dehydrog |
| mybC | MybC transcription factor | AF098507 | Guo et al., 1999 |
Genes that gave signals at least 2.5-fold stronger with either prestalk or prespore probes in at least six independent microarray measurements were considered potential cell-type specific. Although there was considerable variation between spots, prestalk genes were enriched 4.7-fold on average, whereas prespore genes were enriched 8.7-fold on average. Quantitative values for the individual spots are available with supplementary data at http://www.biology.ucsd.edu/loomis-cgi/microarray/index.html.
References for established cell-type–specific genes can be found in Firtel (1995) and Loomis (1996).
Figure 1.
Northern analyses of cell-type–specific genes. After 18 h of development, cells were dissociated from slugs and the cell types fractionated on Percoll density gradients. Under these conditions, the lighter band (L) is enriched in prestalk cells, whereas the heavier band (H) is predominantly prespore cells (Ratner and Borth, 1983). RNA was extracted from the cell types, 15 μg of total RNA was size fractionated by gel electrophoresis, transferred to nylon membranes, and hybridized to probes from each of the potential cell-type–specific genes. Because the signal for cbpB was at the limit of detection, 1.8 μg of polyA+ RNA from each cell type was analyzed on the Northern. Sizes of the mRNA recognized by different probes are given in parentheses.
We were surprised that the microarray data clearly indicated that mybC mRNA was enriched in prespore rather than prestalk cells because it had been reported that lacZ driven by the mybC upstream region stained only prestalk cells (Guo et al., 1999). However, the results from Northern analyses clearly confirmed the microarray data (Figure 1). It is possible that the region Guo et al. (1999) used to drive lacZ did not result in cell-type regulation representative of the endogenous mybC gene.
Developmental Time Courses
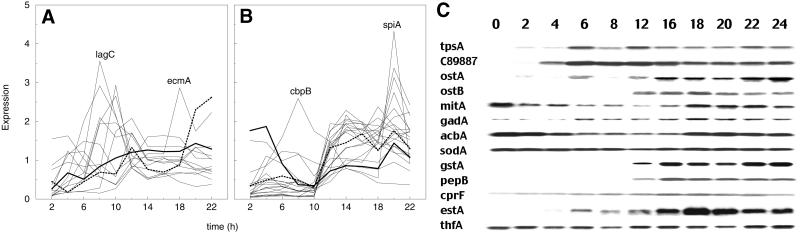
To determine the stages at which each of the cell-type–specific genes are expressed, polyA+ RNA was prepared from cells collected at 2-h intervals throughout development and used to generate Cy3 probes. Cy5 reference probe was prepared from pooled polyA+ RNA prepared from cells that had developed for either 0, 5, 10, 15, and 20 h. Probes were mixed and hybridized to the microarrays. The ratio of the signals at the different time points showed whether specific mRNAs increased or decreased relative to their average levels during development. Average values from three or more determinations for each gene in two independent time courses of development were analyzed. The mean expression level of the cell-type–specific genes throughout development was scaled to unity so as to focus on the temporal aspects of the expression patterns (Figure 2). The previously established prestalk genes, lagC and ecmA, reach maximum expression at 8 and 18 h, respectively, consistent with their expression patterns determined by Northern analyses (Dynes et al., 1994; Jermyn and Williams, 1995). Likewise, the established prespore specific gene spiA reaches maximum expression at 20 h as reported on the basis of Northern results by Richardson et al. (1991). A considerable number of mRNAs from prestalk genes peaked between 8 and 10 h of development and then decreased significantly (Figure 2A). Only one prespore-specific mRNA, cbpB, showed this pattern (Figure 2B). Most prespore-specific genes were not expressed until 10 h of development and continued to be expressed thereafter.
Figure 2.
Time course of gene expression. Microarrays were interrogated with probes prepared from polyA+ RNA isolated at 2-h intervals during development. The relative intensity of the signals compared with that from time-averaged reference probe was normalized to unit mean. The average of two independent time course analyses is given. (A) Prestalk-specific genes. Peak accumulation of lagC and ecmA are indicated. The time course for accumulation of rasD mRNA is shown in bold and that for ecmB is shown as a dotted line. (B) Prespore-specific genes. Peak accumulation of cbpB and spiA are indicated. The time course for accumulation of acbA mRNA is shown in bold and that for gstA is shown as a dotted line. Difficulty in extracting RNA from encapsulated spores and vacuolized stalk cells resulted in slightly less mRNA in the samples from cells developed for 22 h than from cells at earlier times in development. (C) Developmental Northerns. Total RNA (15 μg), from samples collected throughout development at the times shown in hours, was electrophoretically separated and blotted to nylon membranes before being probed for each of the newly recognized cell-type genes. The sizes of the mRNAs shown are those given in Figure 1 for each gene.
Northern Confirmation of Developmental Time Courses
Four of the genes that we found to be enriched in one or the other cell type have been previously shown to be developmentally regulated but were not known to be cell-type enriched. With the use of Northern blots of RNA isolated at various stages during development, the prestalk-enriched mRNAs from cbpD and abpC were shown to accumulate between 6 and 9 h of development and decrease thereafter (Andre et al., 1988; Dorywalska et al., 2000). The prespore-enriched mRNA cbpB could be seen to accumulate after 3 h of development, reach a peak at 12 h, and decrease dramatically thereafter (Andre et al., 1996). Guo et al. (1999) found that mybC mRNA starts to accumulate between 12 and 16 h, reaching a peak at 20 h. The published temporal patterns match those found by probing the microarrays.
To confirm the temporal patterns determined with the use of the microarrays, Northern blots of RNA isolated throughout the development time course were probed for mRNAs from the newly recognized cell-type–specific genes (Figure 2C). The prestalk genes tpsA, C89887, ostA, and ostB were expressed during aggregation and reached a peak at 12–16 h. mRNA from mitA was present in vegetative cells but decreased rapidly after the initiation of development. After 12 h it accumulated once again, apparently in prestalk cells.
mRNA from the prespore gene gadA started to accumulate shortly after the initiation of development and reached a peak at 18 h (Figure 2C). acbA mRNA dropped for the first 10 h and then reaccumulated preferentially in prespore cells. The level of sodA mRNA changed little during development, suggesting that its accumulation in prespore cells is balanced by turnover in prestalk cells. mRNAs from gstA and pepB started to accumulate at 12 h and then remained high. cprF mRNA accumulated during aggregation and reached a peak at 16 h. estA mRNA started to accumulate at 6 h and reached a peak at 18 h. There was a slight drop in mRNA from thfA at 8 h of development followed by reaccumulation, preferentially in prespore cells. The results from these Northern experiments confirmed the microarray data concerning the temporal patterns of newly recognized cell-type–specific genes, which can be seen with supplemental material at our Web site at http://www.biology.ucsd.edu/loomis-cgi/microarray/index.html.
Times of Transcriptional Onset and Cessation
Accumulation of mRNA is determined by the relative rates of synthesis and degradation. When the transcription rate of a gene abruptly increases at a specific stage in development, its mRNA will accumulate until a steady state is reached where the rate of synthesis is equal to the rate of degradation. If transcription abruptly stops, the level of its mRNA will fall exponentially because degradation is a first order process dependent on the level of the specific mRNA (Ai). The expression profiles of the cell-type–enriched mRNAs were fit to the equation describing these events:
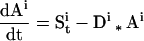 |
where Sit is the mRNA-specific rate of synthesis at time t and Di is the mRNA-specific rate of degradation (Sasik, Iranfar, Hwa, and Loomis, unpublished data). The half-life of the mRNA is ln(2)/Di. We assume that the rate of synthesis is negligible before a specific time (t1) and remains constant until a later time (t2) when it vanishes. Although the mRNA-specific half-life could change during development, in the absence of data to the contrary, we take it to be constant. Half-lives for individual mRNAs were allowed to vary between 0.5 and 12 h to achieve best fit.
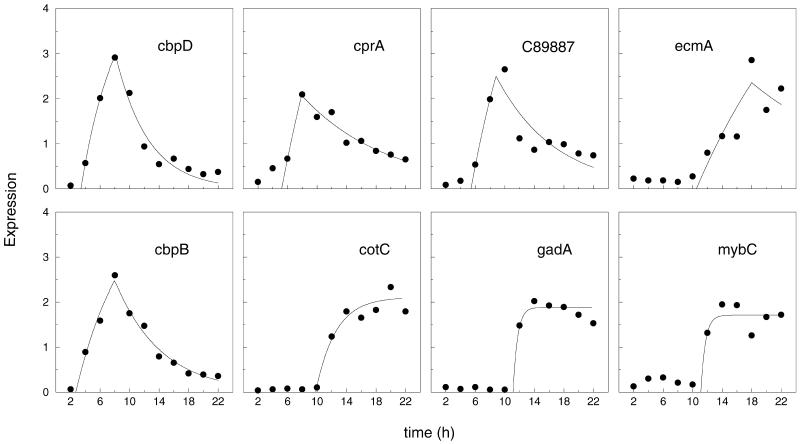
Examples of the best fit for typical cell-type–specific genes are shown in Figure 3. The median half-lives of these genes was calculated to be 4.4 h in good agreement with the 4 h average half-life estimated by Chung et al. (1981). These genes have a single sharp increase in transcription and the time of initiation is well defined. In some cases, the data were nearly equally well fit by an earlier time of cessation (t2) and a longer half-life, hence the determination of the time of cessation is less definitive than the time of onset of transcription. We find that 23 of the mRNAs that are enriched in prespore or prestalk cells at the slug stage fit the first order equation very well (p < 0.02), whereas the remaining mRNAs fit to some extent but appear to be subject to more complex transcriptional regulation.
Figure 3.
Fit of experiment points to simple kinetic equation. Values for individual genes (filled circles) were extracted from the results shown in Figure 2 and fit to the simple first order equation (solid line). The identities of the genes are shown in each panel. The top panels show prestalk-enriched genes, whereas the bottom panels show prespore-enriched genes.
The expression pattern of rasD is shown in bold in Figure 2A. The peak at 4 h followed by a decrease before the accumulation that starts at 6-h precluded a good fit with an equation with the use of a single time of initiation. However, this gene is known to be expressed in all cells early in development and then expressed exclusively in prestalk cells (Jermyn and Williams, 1995). Because prestalk cells are the minority class their expression pattern does not predominate for genes initially expressed in all cells.
The time course of accumulation of ecmB is shown as a dotted line in Figure 2A. It agrees well with the complex expression pattern of this gene where it is initially expressed at a high rate in a cohort of cells that sort to the base of the aggregate before being lost and is subsequently expressed in prestalk and stalk cells late in development (Jermyn et al., 1989; Early et al., 1993). Not surprisingly, this pattern cannot be fit well with an equation that assumes a single time of initial expression.
The expression pattern of acbA is shown in bold in Figure 2B. Its mRNA is present at the start of development and then drops precipitously over the next 10 h before reaccumulating to reach a peak at 20 h when it is strongly enriched in prespore cells. Northern blot analyses of the 0.6-kb mRNA confirmed this expression pattern (Figure 2C). To fit this pattern we would have to use an equation that included two distinct times of expression. It appears that acbA is expressed in growing cells, degraded during aggregation, and then preferentially expressed in prespore cells.
The dotted line in Figure 2B represents the expression pattern of the prespore gene gstA. Its mRNA appears to accumulate at 6 h and then slightly decrease before dramatically increasing after 10 h. The difficulty in fitting this pattern to the simple first order equation was due to a spuriously high value at 6 h that was not observed in the Northern blot analysis (Figure 2C).
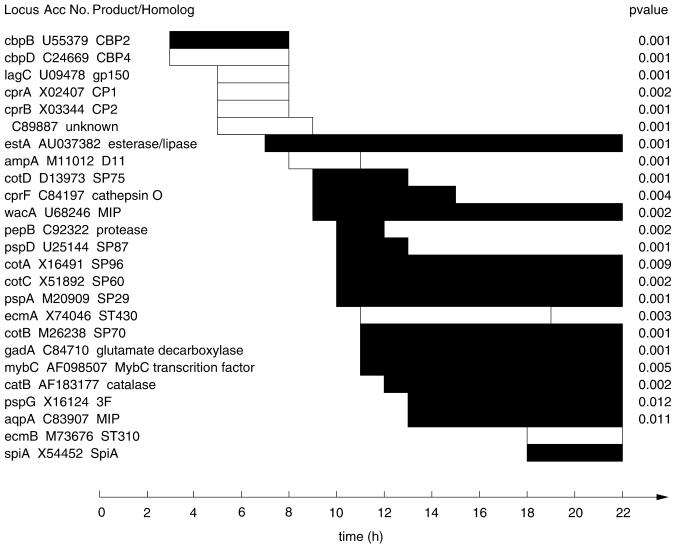
For those cell-type–specific genes whose expression fit well the simple first order equation we could accurately determine the time of onset and cessation (Figure 4). Although the majority of the cell-type–specific genes start to be transcribed between 9 and 13 h when the cell types are sorting out in the mounds, others are transcribed earlier (Figure 4). It is possible that these early genes are expressed in all cells during aggregation and then preferentially turned over in one cell type or the other such that by the slug stage they are enriched in either prespore or prestalk cells. Evidence has been reported for preferential turnover of rasD and cprB in prespore cells leading to prestalk enrichment at the slug stage (Jermyn and Williams, 1995). Although this may be the case for other early genes that become cell-type enriched, it is possible that cell-type divergence starts earlier than had been previously considered.
Figure 4.
Expression patterns of cell-type–specific genes. The periods of expression are given as open bars for the prestalk-enriched genes and filled bars for the prespore-enriched genes. Only expression patterns that could be fit well to the simple first order equation (p < 0.02) and thus gave statistically significant times of initial expression (t1) and cessation of expression (t2) are indicated. Development was subdivided into 1-h windows and t1 and t2 positioned at the nearest border. Genes that are still expressed at 22 h had no apparent time of cessation of transcription during the period of the experiment. Meaningful statistics (p values) could not be calculated for ecmB or spiA because they are expressed so late that their mRNAs accumulate significantly at only two time points.
Transcription of two of the newly recognized cell-type–specific genes, cbpB and cbpD, starts soon after the initiation of development and ceases 5 h later. Both genes had been cloned and characterized previously but were not known to become cell-type enriched by the slug stage (Coukell et al., 1995; Andre et al., 1996). cbpB and cbpD encode small proteins with four consensus EF-hands. That both bind calcium suggests that calcium may be involved in determination of cell fate because cbpB is prespore specific and cbpD is prestalk specific at the slug stage.
The previously established prestalk genes cprA and cprB encode cysteine proteases and are first expressed at 5 h together with lagC and a gene (C89887) encoding a protein that has no significant homologs except for a 20 amino acid domain where there is 66% identity to a portion of the extracellular matrix protein laminin. Its 3.2-kb mRNA is strongly enriched in prestalk cells. A gene encoding a homolog of tetrahydrofoloate dehydrogenase, thfA, is expressed at the same time and its 1.4-kb mRNA becomes enriched in prespore cells. Transcription of these genes ceases a few hours later.
The established prestalk gene ampA (D11) is expressed for a few hours starting at 8 h of development. Its mRNA decreases during the slug stage. Transcription of the prespore-specific gene estA, which encodes a protein related to esterases, also starts at about the same time and continues throughout the remainder of development.
Between 9 and 13 h of development transcription of 15 prespore-specific genes and a prestalk-specific gene, ecmA, is initiated and continues during the slug stage. Many of these genes had been previously characterized and used extensively as cell-type markers (Early et al., 1993; Escalante and Loomis, 1995; Firtel, 1995).
Five hours later mRNA from the spore-specific gene spiA and the stalk-specific gene ecmB accumulate (Figure 3). We did not have sufficient time points during the period of expression of these late genes to warrant statistical analysis but their expression patterns on the microarrays conform well to previous results derived from Northern analyses (Richardson et al., 1991; Early et al., 1993).
Because the times of transcriptional onset are internally comparative for the genes on the microarrays, they clearly demonstrate that both prespore and prestalk genes are regulated at distinct stages throughout early development. A similar analysis that included noncell-type–specific genes defined five statistically significant clusters of genes based on the time of initiation of transcription (Sasik, Iranfar, Hwa, and Loomis, unpublished data). Cell-type–specific genes fall into each of these clusters.
DISCUSSION
These studies have recognized more than a dozen new genes that can serve as useful markers for characterization of the cell types. By separating slug stage cells on Percoll density gradients we were able to recover samples that were 10-fold enriched in either prestalk or prespore cells. RNA prepared from these samples was used to generate DNA probes labeled with either Cy3 or Cy5. Hybridization of these probes to microarrayed targets from 690 Dictyostelium genes indicated that 14 were preferentially expressed in prestalk cells and that 21 were prespore specific. Although the cell-type specificity of seven of the prestalk genes and 11 of the prespore genes had been previously established, these studies uncovered seven new prestalk-enriched genes and 10 new prespore genes, thereby significantly increasing the number of genes known to be cell-type specific. The cell-type specificity of the newly discovered genes was confirmed with the use of Northern blots of prespore and prestalk extracts. The size of the cell-type–specific mRNAs on the Northern blots agreed well with that expected from the sequence, adding further confidence to the characterization.
The relative abundances of the products of 91 of the genes on our microarrays were found to increase threefold or more during development. Eleven genes increased more than 20-fold and all but two of these were found to be cell-type specific. Actinomycin D is known to inhibit all RNA synthesis within an hour of being added to developing cells (Sussman et al., 1967). As expected, addition of actinomycin D after either 4 or 10 h of development blocked the accumulation of all those genes that accumulated threefold or more throughout development, demonstrating that accumulation of these mRNAs was dependent on de novo RNA synthesis (unpublished data).
Two of the cell-type–specific genes, ostA and sodA, increased <3-fold throughout development. Their mRNAs may have turned over more rapidly in one or the other of the cell types, leading to cell-type specificity at the slug stage. The abundance of acbA and mitA mRNA dropped fourfold during the first 10 h and then reaccumulated to reach a peak at 20 h. Apparently these genes are active during growth, turned off after the initiation of development, and then expressed in one or the other cell type after aggregation. Their expression patterns could not be fit well by the simple kinetic equation.
Individual genes showed highly similar expression profiles in the two independent developmental time courses. Although the relative intensity values for specific mRNAs varied somewhat at specific time points, the initial time of accumulation and the time at which maximal accumulation was observed varied by less than an hour between experiments. The temporal patterns determined from microarray data were confirmed by Northern analyses (Figure 2C). Thus, their relative order of appearance is a good indication of progression through the developmental cycle.
Prestalk Genes
The temporal patterns of expression of prestalk genes seen on our microarrays indicate that there are at least five different stages in development when these genes are induced. Four prestalk-enriched genes are expressed 3 h after the initiation of the development, four others are expressed after 5 h, one after 8 h, four after 11 h, and one (ecmB) after 18 h of development. Clearly, individual prestalk genes are not all coordinately induced.
The tpsA gene encodes a protein with 42% identity to trehalose phosphate synthetase of the mustard plant Arabidopsis. It is expressed shortly after the initiation of development and its mRNA accumulates in prestalk cells to levels that are 2.5-fold higher than in prespore cells. Northern analyses confirmed the prestalk enrichment of tpsA mRNA but its mRNA could also be seen in prespore cells (Figure 1). The activity of trehalose phosphate synthetase increases during early development to levels that are somewhat higher in prestalk cells than in prespore cells (Roth and Sussman, 1968; Jefferson and Rutherford, 1976). Thus, the developmental time course and cell-type specificity are consistent with this gene encoding trehalose phosphate synthetase of Dictyostelium. The product generated by this enzyme, trehalose, accumulates in both prestalk and prespore cells and then is rapidly metabolized during terminal differentiation of stalk cells while being stored in spores until germination.
Another of the prestalk enriched genes, abpC, encodes one of the F-actin binding proteins, ABP120 (Noegel et al., 1989). It controls the rate and extent of pseudopod extension by cross-linking F-actin in the cortical cytoskeleton (Cox et al., 1992). It is expressed in all cells during aggregation but is predominantly expressed in prestalk cells during the slug stage.
After 11 h of development the established prestalk gene ecmA is expressed coordinately with two genes encoding members of the P450 family of heme binding proteins, ostA and ostB. Both of these genes encode products that are similar to vertebrate mono-oxidases. A role for such enzymes has been suggested in the degradation of Differentiation Inducing Factor (DIF), a hexaphenone that induces differentiation of a subclass of prestalk cells, because a specific step in its oxidation is sensitive to the P450 inhibitor ancymidol (Morandini et al., 1995). Other enzymes in this pathway are known to be prestalk specific (Traynor and Kay, 1991) and it is possible that one or both of these P450 genes function in this pathway.
Prespore Genes
One of the genes expressed at 11 h of development, gadA, encodes a protein with a high degree of similarity to glutamate decarboxylase. This enzyme catalyzes the conversion of glutamate to γ-aminobutyric acid (GABA). Another prespore gene that is expressed at this stage, acbA, encodes a protein with a high degree of similarity to mammalian proteins referred to as either diazepam binding inhibitor (DBI) or acyl-CoA binding protein (Webb et al., 1987; Chen et al., 1988). DBI has been shown to bind to the γ subunit of the GABA-A receptors in the brain where it reduces binding of diazepam drugs, including valium. However, DBI also affects a variety of peripheral tissues where it binds acyl-CoA (Chen et al., 1988). Peptides derived from this protein affect chemotaxis and phagocytosis of neutrophils apparently by increasing cytosolic calcium levels and also inhibits insulin secretion from pancreatic cells (De Stefanis et al., 1995). Although GABA is clearly not a neurotransmitter in Dictyostelium, it would be interesting if it were used as an intercellular signal. Thus, there are many leads that can be followed up by molecular genetic analyses of gadA and acbA in Dictyostelium.
A gene encoding catalase, catB, was recently shown by Northern analyses to be preferentially expressed in prespore cells (Garcia et al., 2000) and independently confirmed by our microarray data. Two other genes that may be involved in protection from oxidative stress, sodA and gstA, were also found to be prespore specific. sodA encodes a homolog of the copper-zinc superoxide dismutase that generates peroxide from superoxide so that it can be converted to water by catalase with the release of oxygen. The gstA gene encodes a protein related to glutathione S-transferase, an enzyme involved in coupling reactive oxides to reduced glutathione for export. It appears that there may be a selective advantage to prespore cells that can protect themselves from oxidative damage.
Prespore cells also preferentially express genes encoding homologs for two proteases and an esterase/lipase. pepB encodes a protein with similarity to mammalian pepstatin-insensitive protease, whereas cprF encodes a member of the cathepsin O family of proteases that also includes the prestalk-specific genes cprA and cprB. Together with the putative esterase/lipase gene estA, these genes may be involved in protein or lipid modifications specific to prespore cells.
MybC is a nuclear localized protein of the Myb family of transcriptional regulators (Guo et al., 1999). Mutants in which mybC is disrupted remain at the slug stage and express the prespore genes cotA and cotB only at very reduced levels. Overexpression of the catalytic subunit of the cAMP-dependent protein kinase PKA or activation of PKA by addition of 8-Br-cAMP to mybC− cells bypassed the block and permitted efficient sporulation. Moreover, when mybC− cells were developed in chimeric mixtures with wild-type cells, they formed both spores and stalks in the resulting fruiting bodies. This nonautonomous phenotype led Guo et al. (1999) to suggest that MybC is essential for the production or release of intercellular signals that trigger terminal differentiation by activating PKA. Two distinct peptides, SDF-1 and SDF-2, which are released during culmination have been shown to stimulate encapsulation (Anjard et al., 1997, 1998). Neither of these peptides is produced in mybC− mutant cells, which may account for their defects in culmination (Guo et al., 1999).
MybC was thought to be prestalk specific on the basis of the staining pattern in a strain transformed with a construct in which 1.85 kb of upstream sequence and the first 16 amino acids encoded by mybC was ligated to the reporter gene lacZ. Only prestalk cells were stained by X-gal in this strain (Guo et al., 1999). However, in six independent assessments of the cell-type specificity of mybC mRNA with the use of microarrays, we found that the signal was more than 2.5-fold stronger with probes prepared from density-separated prespore cells than from probes from density-separated prestalk cells. Northern analyses showed prespore specificity of mybC comparable with that of established or newly recognized prespore-specific genes (Figure 1). Reassessment of the cell-type specificity of mybC affects the interpretation of its role in terminal differentiation. If MybC acts as a transcription factor necessary for production of SDF-1 and SDF-2, then production of these peptides or their precursors may also be prespore specific. Although SDF-2 stimulates prespore cells to rapidly encapsulate, its activity is dependent on the action of TagC, a prestalk-specific protein (Anjard et al., 1998). Thus, we can envision a process at culmination in which prespore cells provide a precursor to prestalk cells that modify it before it is active on prespore cells. Such a process could closely integrate terminal differentiation of the two cell types.
ACKNOWLEDGMENTS
Roman Sasik is partially supported by a La Jolla Interfaces in Science postdoctoral fellowship from the Burroughs-Wellcome Fund. This work was supported by grants from the National Science Foundation to W.F.L. (0083704) and T.H. (9728463). T.H. further acknowledges the support of an “innovation awards in functional genomics” from the Burroughs-Wellcome Fund.
REFERENCES
- Andre E, Lottspeich F, Schleicher M, Noegel A. Severin, gelsolin, and villin share a homologous sequence in regions presumed to contain F-actin severing domains. J Biol Chem. 1988;263:722–727. [PubMed] [Google Scholar]
- Andre B, Noegel AA, Schleicher M. Dictyostelium discoideum contains a family of calmodulin-related EF-hand proteins that are developmentally regulated. FEBS Lett. 1996;382:198–202. doi: 10.1016/0014-5793(96)00176-7. [DOI] [PubMed] [Google Scholar]
- Anjard C, van Bemmelen M, Veron M, Reymond CD. A new spore differentiation factor (SDF) secreted by Dictyostelium cells is phosphorylated by the cAMP dependent protein kinase. Differentiation. 1997;62:43–49. doi: 10.1046/j.1432-0436.1997.6210043.x. [DOI] [PubMed] [Google Scholar]
- Anjard C, Zeng C, Loomis WF, Nellen W. Signal transduction pathways leading to spore differentiation in Dictyostelium discoideum. Dev Biol. 1998;193:146–155. doi: 10.1006/dbio.1997.8804. [DOI] [PubMed] [Google Scholar]
- Chen ZW, Agerberth B, Gell K, Andersson M, Mutt V, Ostenson CG, Efendic S, Barros-Soderling J, Persson B, Jornvall H. Isolation and characterization of porcine diazepam-binding inhibitor, a polypeptide not only of cerebral occurrence but also common in intestinal tissues and with effects on regulation of insulin release. Eur J Biochem. 1988;174:239–245. doi: 10.1111/j.1432-1033.1988.tb14088.x. [DOI] [PubMed] [Google Scholar]
- Chung S, Landfear SM, Blumberg DD, Cohen NS, Lodish HF. Synthesis and stability of developmentally regulated Dictyostelium mRNAs are affected by cell-cell contact and cAMP. Cell. 1981;24:785–797. doi: 10.1016/0092-8674(81)90104-5. [DOI] [PubMed] [Google Scholar]
- Coukell B, Moniakis J, Grinberg A. Cloning and expression in Escherichia coli of a cDNA encoding a developmentally regulated Ca2+-binding protein from Dictyostelium discoideum. FEBS Lett. 1995;362:342–346. doi: 10.1016/0014-5793(95)00272-b. [DOI] [PubMed] [Google Scholar]
- Cox D, Condeelis J, Wessels D, Soll D, Kern H, Knecht DA. Targeted disruption of the ABP-120 gene leads to cells with altered motility. J Cell Biol. 1992;116:943–955. doi: 10.1083/jcb.116.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stefanis P, Impagnatiello F, Berkovich A, Guidotti A. Inhibitory effect of ODN, a naturally occurring processing product of diazepam binding inhibitor, on secretagogues-induced insulin secretion. Regul Pept. 1995;56:153–165. doi: 10.1016/0167-0115(95)00002-s. [DOI] [PubMed] [Google Scholar]
- DeRisi J, Iyer V, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Dorywalska M, Coukell B, Dharamsi A. Characterization and heterologous expression of cDNAs encoding two novel closely related Ca(2+)-binding proteins in Dictyostelium discoideum. Biochim Biophys Acta. 2000;1496:356–3661. doi: 10.1016/s0167-4889(00)00024-0. [DOI] [PubMed] [Google Scholar]
- Dynes JL, Clark AM, Shaulsky G, Kuspa A, Loomis WF, Firtel RA. LagC is required for cell-cell interactions that are essential for cell-type differentiation in Dictyostelium. Genes Dev. 1994;8:948–958. doi: 10.1101/gad.8.8.948. [DOI] [PubMed] [Google Scholar]
- Early AE, Gaskell MJ, Traynor D, Williams JG. Two distinct populations of prestalk cells within the tip of the migratory Dictyostelium slug with differing fates at culmination. Development. 1993;118:353–362. doi: 10.1242/dev.118.2.353. [DOI] [PubMed] [Google Scholar]
- Escalante R, Loomis WF. Whole-mount in situ hybridization of cell-type-specific mRNAs in Dictyostelium. Dev Biol. 1995;171:262–266. doi: 10.1006/dbio.1995.1278. [DOI] [PubMed] [Google Scholar]
- Firtel RA. Integration of signaling information in controlling cell- fate decisions in Dictyostelium. Genes Dev. 1995;9:1427–1444. doi: 10.1101/gad.9.12.1427. [DOI] [PubMed] [Google Scholar]
- Fosnaugh KL, Loomis WF. Enhancer regions responsible for temporal and cell-type-specific expression of a spore coat gene in Dictyostelium. Dev Biol. 1993;157:38–48. doi: 10.1006/dbio.1993.1110. [DOI] [PubMed] [Google Scholar]
- Garcia MX, Foote C, van Es S, Devreotes PN, Alexander S, Alexander H. Differential developmental expression and cell type specificity of Dictyostelium catalases and their response to oxidative stress and UV-light. Biochim Biophys Acta. 2000;1492:295–310. doi: 10.1016/s0167-4781(00)00063-4. [DOI] [PubMed] [Google Scholar]
- Guo K, Anjard C, Harwood A, Kim HJ, Newell PC, Gross JD. A myb-related protein required for culmination in Dictyostelium. Development. 1999;126:2813–2822. doi: 10.1242/dev.126.12.2813. [DOI] [PubMed] [Google Scholar]
- Jefferson BL, Rutherford CL. Cell specific activity of trehalose-6-phosphate synthetase during differentiation of Dictyostelium discoideum. Cell Differ. 1976;5:189–198. doi: 10.1016/0045-6039(76)90020-8. [DOI] [PubMed] [Google Scholar]
- Jermyn KA, Duffy KT, Williams JG. A new anatomy of the prestalk zone in Dictyostelium. Nature. 1989;340:144–146. doi: 10.1038/340144a0. [DOI] [PubMed] [Google Scholar]
- Jermyn K, Williams J. Comparison of the Dictyostelium rasD and ecmA genes reveals two distinct mechanisms whereby an mRNA may become enriched in prestalk cells. Differentiation. 1995;58:261–267. doi: 10.1046/j.1432-0436.1995.5840261.x. [DOI] [PubMed] [Google Scholar]
- Kuspa A, Loomis WF. Ordered yeast artificial chromosome clones representing the Dictyostelium discoideum genome. Proc Natl Acad Sci USA. 1996;93:5562–5566. doi: 10.1073/pnas.93.11.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis WF. The Development of Dictyostelium. New York: Academic Press; 1982. [Google Scholar]
- Loomis WF. Genetic networks that regulate development in Dictyostelium cells. Microbiol Rev. 1996;60:135. doi: 10.1128/mr.60.1.135-150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis WF. The Dictyostelium genome sequencing project. Protist. 1998;149:209–212. doi: 10.1016/S1434-4610(98)70028-8. [DOI] [PubMed] [Google Scholar]
- Mehdy MC, Ratner D, Firtel RA. Induction and modulation of cell-type specific gene expression in Dictyostelium. Cell. 1983;32:763–771. doi: 10.1016/0092-8674(83)90062-4. [DOI] [PubMed] [Google Scholar]
- Morandini P, Offer J, Traynor D, Nayler O, Neuhaus D, Taylor GW, Kay RR. The proximal pathway of metabolism of the chlorinated signal molecule differentiation-inducing factor-1 (DIF-1) in the cellular slime mold Dictyostelium. Biochem J. 1995;306:735–743. doi: 10.1042/bj3060735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morio T, Urushihara H, Saito T, Ugawa Y, Mizuno H, Yoshida M, Yoshino R, Mitra BN, Pi M, Sato T, Takemoto K, Yasukawa H, Williams J, Maeda M, Takeuchi I, Ochiai H, Tanaka Y. The Dictyostelium developmental cDNA project: generation and analysis of expressed sequence tags from the first-finger stage of development. DNA Res. 1998;5:335–340. doi: 10.1093/dnares/5.6.335. [DOI] [PubMed] [Google Scholar]
- Nicol A, Rappel WJ, Levine H, Loomis WF. Cell-sorting in aggregates of Dictyostelium discoideum. J Cell Sci. 1999;112:3923–3929. doi: 10.1242/jcs.112.22.3923. [DOI] [PubMed] [Google Scholar]
- Noegel AA, Rapp S, Lottspeich F, Schleicher M, Stewart M. The Dictyostelium gelation factor shares a putative actin binding site with alpha-actinin and dystrophin and also has a rod domain containing six 100-residue motifs that appear to have a cross-beta conformation. J Cell Biol. 1989;109:607–618. doi: 10.1083/jcb.109.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner D, Borth W. Comparison of differentiating Dictyostelium discoideum cell types separated by an improved method of density gradient centrifugation. Exp Cell Res. 1983;143:1–13. doi: 10.1016/0014-4827(83)90103-9. [DOI] [PubMed] [Google Scholar]
- Richardson DL, Hong CB, Loomis WF. A prespore gene, Dd31, expressed during culmination of Dictyostelium discoideum. Dev Biol. 1991;144:269–280. doi: 10.1016/0012-1606(91)90421-x. [DOI] [PubMed] [Google Scholar]
- Roth R, Sussman M. Trehalose 6-phosphate synthetase (uridine diphosphate glucose: D-glucose 6-phosphate 1-glucosyltransferase) and its regulation during slime mold development. J Biol Chem. 1968;243:5081–5087. [PubMed] [Google Scholar]
- Shaulsky G, Kuspa A, Loomis WF. A multidrug resistance transporter serine protease gene is required for prestalk specialization in Dictyostelium. Genes Dev. 1995;9:1111–1122. doi: 10.1101/gad.9.9.1111. [DOI] [PubMed] [Google Scholar]
- Shaulsky G, Loomis WF. Mitochondrial DNA replication but no nuclear DNA replication during development of Dictyostelium. Proc Natl Acad Sci USA. 1995;92:5660–5663. doi: 10.1073/pnas.92.12.5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. In: Spudich JA, editor. Methods in Cell Biology. Orlando, FL: Academic Press; 1987. pp. 9–29. [DOI] [PubMed] [Google Scholar]
- Sussman R, Loomis W, Ashworth J, Sussman M. The effect of actinomycin D on cellular slime mold morphogenesis. Biochem Biophys Res Commun. 1967;26:353–359. doi: 10.1016/0006-291x(67)90131-3. [DOI] [PubMed] [Google Scholar]
- Traynor D, Kay RR. The DIF-1 signaling system in Dictyostelium - metabolism of the signal. J Biol Chem. 1991;266:5291–5297. [PubMed] [Google Scholar]
- Webb NR, Rose TM, Malik N, Marquardt H, Shoyab M, Todaro GJ, Lee DC. Bovine and human cDNA sequences encoding a putative benzodiazepine receptor ligand. DNA. 1987;6:71–79. doi: 10.1089/dna.1987.6.71. [DOI] [PubMed] [Google Scholar]