Abstract
Background
Meniscus tears are classified as traumatic or degenerative based on the tear pattern. There is little evidence demonstrating biological differences between the two tear types.
Hypothesis
Gene expression signatures in the injured meniscus are different between traumatic (vertical) and degenerative (complex, horizontal or flap) tears.
Methods
Samples of torn meniscus from the white-white zone were removed at the time of clinically indicated partial meniscectomy from 48 patients (37 with degenerative tears and 11 with traumatic tears). The mRNA expression in the injured menisci was measured by quantitative real-time polymerase chain reaction for selected molecular markers of osteoarthritis, inflammation and cartilage homeostasis (e.g. cytokines/chemokines, aggrecanases/metalloproteinases, transcription factors, cartilage matrix genes and adipokines). The tear pattern (traumatic or degenerative) and location (medial or lateral) were recorded for each patient. Gene expression differences between degenerative and traumatic tears were computed after adjusting for patients’ age, sex and body mass index and for location of the resected meniscus (medial/lateral).
Results
Gene expression in meniscus tears varied by pattern. Chemokines [IL8 (p<0.001) and CXCL6 (p<0.001)] and matrix metalloproteinases [MMP1 (p=0.011) and MMP3 (p=0.016)] were expressed at a significantly higher level in traumatic tears compared to degenerative tears. In contrast, COL1A1 was expressed at a lower level in traumatic tears compared to degenerative tears (p=0.058). None of the genes tested demonstrated significant differences between medial and lateral meniscus tears.
Conclusions
Traumatic meniscus tears overall exhibited higher inflammatory/catabolic response as evidenced by higher levels of chemokines and matrix metalloproteinases expression than degenerative tears. These findings suggest that there is a (molecular) biological distinction between traumatic and degenerative tears.
Clinical relevance
The catabolic/inflammatory differences between traumatic and degenerative tears may be relevant to treatment decisions regarding the meniscus as well as advance our understanding of how meniscus tears relate to the development of knee osteoarthritis.
Level of evidence
Diagnostic Level III.
Keywords: meniscus tear, osteoarthritis, chemokines, metalloproteinases, degenerative tear, traumatic tear
Introduction
Meniscus tears are a common injury in the knee. They are classified based on their pattern into either traumatic tears, defined as longitudinal tears, or degenerative tears, including complex, horizontal and flap tears8. This classification is considered important with regards to clinical decision-making about the surgical treatment of meniscus tears. While most meniscus tears are treated with partial meniscectomy, with an estimate 690,000 partial meniscectomies performed in the U.S. alone each year7, some tears are treated with repair. Healing rates after meniscal repair are no better than 80–85%16, and may be as low as 55%29. While it is generally accepted that degenerative tears are not good candidates for repair in comparison to traumatic tears, there is a need for patient-related factors that might help identify tears at higher risk for repair failure14. The molecular profile of injured tissues from patients undergoing meniscus surgery could help identify such factors.
It has been estimated that 50% of the patients with meniscus tears go on to develop knee osteoarthritis (OA) within 10–20 years9, 10, 13, 21. Tear pattern may also be relevant to this risk as Englund et al.11 reported that degenerative tears are associated with worse outcomes and more OA following meniscectomy. However, very little is known about any biological differences between traumatic and degenerative tears. If there is a distinction in the mechanism and pathophysiology of traumatic and degenerative tears, there may be differences in their biology. Studying the gene expression of tissue resected from these patients at the time of partial meniscectomy is an ideal approach to compare the biology of these tear patterns.
Therefore, the current study was developed to assess whether tear pattern of the injured meniscus relates to gene expression in the injured tissue. The hypothesis of the study is that the expression profile of inflammatory and OA related genes differs between traumatic and degenerative meniscus tears. If such differences exist, they could be relevant to the relative risk for developing OA, as well as the potential for a meniscus tear to heal after attempted repair.
Materials and Methods
Meniscal surgery and grading of chondrosis
The study protocol was approved by the study institution’s human subjects Institutional Review Board. Patients diagnosed with symptomatic isolated medial or lateral meniscus tears without concomitant cartilage or ligament injury treated by a single academic sports medicine surgeon were eligible to be recruited for this study. All patients had preoperative radiographs and MRIs with no evidence for OA. Informed consent was obtained from all subjects prior to knee arthroscopy. Patients were included in this study if they had isolated meniscus tears appropriately treated with partial meniscectomy and no other pathology in the knee. If there was any grade II or greater chondrosis in the knee based on standard diagnostic arthroscopy of all compartments, patients were excluded from the study. There is some overlap of these patients with the cohort in a previously published study looking at the relationship of gene expression in the meniscus to patient body mass index which did not evaluate the association of tear pattern with gene expression19.
The arthroscopic findings were recorded with regard to the tear pattern based on a standard diagnostic arthroscopy performed by the as part of each surgery. Each tear was classified as degenerative (complex, horizontal or flap tear pattern) (Figure 1A) or traumatic (longitudinal tear pattern) (Figure 1B)8. Patients with a meniscus injury involving a radial tear or other patterns that did not specifically meet the definitions above were not included in this study. In all cases, the specimen was collected from the white-white zone of the meniscus as part of the clinically indicated resection of the tear by the treating surgeon. For traumatic tears, time from initial injury to surgery was calculated based on the clinical record.
Figure 1.
Representative images from degenerative (A) and traumatic (B) meniscus tears are shown. Images were taken at the time of arthroscopic partial meniscectomy.
Tissue processing and RNA isolation
The specimens were collected at the time of surgery. They were handled using a previously published technique3. Briefly, the anonymous specimens were transported to the laboratory on ice from the operating room in sterile screw cap containers containing phosphate-buffered saline without calcium and magnesium (PBS, Thermo Fisher Scientific, Rockford, IL, USA). The tissues were weighed and washed twice with PBS to get rid of any blood and debris and to avoid contamination with any other cell types. The blot-dried tissues were put in 50 ml Falcon tubes and 1 ml of TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was added for each 50–100 mg of the tissue wet weight and stored at −80°C until used for total RNA extraction19.
Quantification of gene expression in meniscus
A total of 150–200 ng of isolated RNA was first treated with DNase I to remove traces of contaminating DNA (Invitrogen, Carlsbad, CA, USA). The DNase I treated RNA was then reverse-transcribed to synthesize complementary DNA (Invitrogen, Carlsbad, CA, USA) as per the manufacturer’s instructions. Custom-designed primers (Table 1) for molecular markers of OA including pro-inflammatory cytokines (IL-1α, IL-1β, IL-6, TNFα), aggrecanases (ADAMTS-4, -5), chemokines (IL-8, CCL3, CCL3L1, CXCL1, CXCL3, CXCL6, CCL20), matrix components (BMP-2, Col1a1, Col2a1, aggrecan), metalloproteinases (MMP-1, -3, -9, -13), and transcription factors (NFκB2, NFκBIA, IκBA) were obtained from Invitrogen (Carlsbad, CA, USA). The expression of multiple genes were quantified by quantitative PCR on a 7500 Fast Real Time PCR System (Applied Biosystems, Foster City, CA, USA) using SYBR Green PCR master mix (Applied Biosystems, Foster City, CA, USA) according to manufacturer’s protocols. Samples were amplified with an initial activation step at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 seconds and annealing at 60°C for one minute. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) acted as an endogenous reference gene for normalization of fluorescence thresholds (Ct) values for target genes as this gene showed consistent expression across all samples.
Table 1.
Primers for quantitative real-time polymerase chain reaction
| Gene symbol | Gene name | Forward | Reverse |
|---|---|---|---|
| IL1A | Interleukin 1 alpha | 5′-TGCCTGAGATACCCAAAACC-3′ | 5′-AACAAGTTTGGATGGGCAAC-3′ |
| IL1B | Interleukin 1 beta | 5′-TCCAGGAGAATGACCTGAGC-3′ | 5′-GTGATCGTACAGGTGCATCG-3′ |
| IL6 | Interleukin 6 | 5′-ATGAAGGAGAACTTGCCTGGTG-3′ | 5′-TGCAGGAACTGGATCAGGAC-3′ |
| TNFA | Tumor necrosis factor alpha | 5′-AACCTCCTCTCTGCCATCCA-3′ | 5′-CCAAAGTAGACCTGCCCAGA-3′ |
| ADAMTS4 | A disintegrin and metalloproteinase with thrombospondin motifs 4 | 5′-GAAGGTGCAGTTTTGCCAAG-3′ | 5′-TGTGGTCCACTCTCAATCACTC-3′ |
| ADAMTS5 | A disintegrin and metalloproteinase with thrombospondin motifs 5 | 5′-GCAACCAGTTCTGCATCA-3′ | 5′-TGGCTGCTCGTCTCAAAGTA-3′ |
| BMP2 | Bone morphogenetic protein 2 | 5′-GTCCTCTCTGCACCACTTGC-3′ | 5′-GGAAGATGACACTGGGCTTG-3′ |
| MMP1 | Matrix metalloproteinase 1 | 5′-GGGAATTCACCCCAAGAAC-3′ | 5′-GATGCAGGATTGAGGCAAG-3′ |
| MMP3 | Matrix metalloproteinase 3 | 5′-ACCGAAGTCATAGCCACACTC-3′ | 5′-GGTGCCTCCCCTTGTTCAGTA-3′ |
| MMP9 | Matrix metalloproteinase 9 | 5′-GTTTACGCGTTACGCTGAGAG-3′ | 5′-ACTTCCACCTTGGAGCACTG-3′ |
| MMP13 | Matrix metalloproteinase 13 | 5′-TTTATTGTGGGCTTCACACG-3′ | 5′-GATTTGCGCACACAGACAAC-3′ |
| IL8 | Interleukin 8 | 5′-GGCTAAAGCGCTACCTGCTA-3′ | 5′-GAGTCACACCACCAAGCTGACA-3′ |
| CCL3 | Chemokine (C-C motif) ligand 3 | 5′-TACTTGGCCTCTCCCATGAC-3′ | 5′-CTGTGATGGTGGCTGAAGTG-3′ |
| CCL3L1 | Chemokine (C-C motif) ligand 3-like 1 | 5′-AGGTCTCTGAGGGTCAAGCA-3′ | 5′-CTGGTTGAAAGCATGAGCA-3′ |
| CXCL1 | Chemokine (C-X-C motif) ligand 1 | 5′-TGCTTTGTCCTTTGATGCTG-3′ | 5′-GGAAGAGATGGCCAAAATGA-3′ |
| CXCL3 | Chemokine (C-X-C motif) ligand 3 | 5′-CATCGTCATCCAGTTTGGTG-3′ | 5′-AGGGACCACAACTCGTCATC-3′ |
| CXCL6 | Chemokine (C-X-C motif) ligand 6 | 5′-TGGTCCAGGAGATGAAGACC-3′ | 5′-TCCTCGGAGACTGGTAATGG-3′ |
| CCL20 | Chemokine (C-C motif) ligand 20 | 5′-CTTCTAGCGTTGCTGCTTCC-3′ | 5′-CAACTCGAACTCGCTCAGGA-3′ |
| COL1A1 | Collagen type 1, A1 | 5′-GTGCTAAAGGTGCCAATGGT-3′ | 5′-ACCAGGTTCACCGCTGTTAC-3′ |
| COL2A1 | Collagen type 2, A1 | 5′-CCCAGAGGTGACAAAGGAGA-3′ | 5′-CACCTTGGTCTCCAGAAGGA-3′ |
| ACAN | Aggrecan | 5′-GGCACTAGTCAACCCTTTGG-3′ | 5′-CTGAACCCTGGTAACCCTGA-3′ |
| NFkB1A | Nuclear factor kappa B1 A | 5′-GAACAGCCTTGCATCTAGCC-3′ | 5′-TTTTCAGCATGGATGTCAGC-3′ |
| NFkB2 | Nuclear factor kappa B2 | 5′-TACACCTTGCCTGTGAGCAG-3′ | 5′-TAGCCTTCAGGATGGAGTGG-3′ |
| IkBA | Inhibitory kappa B A | 5′-GATCCGCCAGGTGAAGGG-3′ | 5′-GCAATTTCTGGCTGGTTGG-3′ |
| ADIPOQ | Adiponectin | 5′-GTGATGGCAGAGATGGCAC-3′ | 5′-CGATGTCTCCCTTAGGACCAA-3′ |
| APLN | Apelin | 5′-CAGGGAGGTCGGAGGAAAT-3′ | 5′-ACCAATCTATGGAGGAGACATAACC-3′ |
| LEP | Leptin | 5′-GGATTCTTGTGGCTTTGGC-3′ | 5′-CTTTCTGTTTGGAGGAGACTGACT-3′ |
| RETN | Resistin | 5′-TGGAAGAAGCCATCAATGAGAGG-3′ | 5′-CGCACTGGCAGTGACATGTG-3′ |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | 5′-ACCAGAAGACTGTGGATGG-3′ | 5′-GAGGCAGGGATGATGATGTTCTG-3′ |
Statistical analysis
The data were analyzed using SPSS (SPSS Inc., Chicago, IL). We used a multivariate analysis with genes as independent variables and age, body mass index (BMI), sex, side (location) and tear pattern as fixed factor. All fixed factors included in the model were categorical. Age was categorized as under or over 40 based on a previous study demonstrating differences in gene expression of meniscus tears between these patient populations3. Similarly, BMI was categorized under or over 25 Kg/m2 based on a previous study demonstrating the most significant differences in gene expression of meniscus tears between these populations18. We computed mean and fold change for each tear type. All result were rendered statistically significant at P value of ≤ 0.05. Power analysis indicated that a sample size of 12 per group was necessary to detect an effect size of 1.2 with 80% power and α = 0.05.
Results
Over an 18 month period, 48 patients undergoing clinically indicated arthroscopic partial meniscectomy met the inclusion and exclusion criteria and consented to participate in the study (Table 2). The majority (77%) of tears were degenerative and occurred in the medial meniscus (85%). No patients had proliferative synovitis at the time of surgery.
Table 2.
Characteristics of study patients
| Patient ID | Tear type | Side | Sex | Age (years) | Age category | BMI (Kg/m2) | BMI category | Months from Injury |
|---|---|---|---|---|---|---|---|---|
| P1-Z26F | Degenerative | Lateral | Female | 15 | <40 | 24.32 | <30 | na |
| P1-003 | Degenerative | Medial | Male | 53 | >40 | 22.80 | <30 | na |
| P1-004 | Degenerative | Medial | Female | 51 | >40 | 29.18 | <30 | na |
| P1-005 | Degenerative | Medial | Male | 60 | >40 | 36.26 | >30 | na |
| P1-007 | Degenerative | Lateral | Male | 34 | <40 | 26.32 | <30 | na |
| P1-009 | Degenerative | Medial | Male | 58 | >40 | 26.87 | <30 | na |
| P1-00C | Degenerative | Medial | Male | 59 | >40 | 30.85 | >30 | na |
| P1-015 | Degenerative | Medial | Female | 49 | >40 | 19.48 | <30 | na |
| P1-018 | Degenerative | Medial | Female | 35 | <40 | 22.49 | <30 | na |
| P1-019 | Degenerative | Medial | Male | 36 | <40 | 24.39 | <30 | na |
| P1-Z14F | Degenerative | Lateral | Female | 14 | <40 | 28.57 | <30 | na |
| P1-Z26F | Degenerative | Medial | Female | 26 | <40 | 21.92 | <30 | na |
| P1-Z54M | Degenerative | Medial | Male | 54 | >40 | 25.10 | <30 | na |
| P2-105 | Degenerative | Medial | Female | 59 | >40 | 23.04 | <30 | na |
| P2-107 | Degenerative | Medial | Male | 33 | <40 | 25.36 | <30 | na |
| P2-112 | Degenerative | Medial | Male | 63 | >40 | 29.68 | <30 | na |
| P2-115 | Degenerative | Medial | Male | 55 | >40 | 26.50 | <30 | na |
| P2-119 | Degenerative | Medial | Male | 58 | >40 | 19.93 | <30 | na |
| P2-120 | Degenerative | Medial | Male | 28 | <40 | 36.61 | >30 | na |
| P2-121 | Degenerative | Medial | Female | 50 | >40 | 26.62 | <30 | na |
| P2-123 | Degenerative | Medial | Male | 53 | >40 | 25.11 | <30 | na |
| P2-124 | Degenerative | Medial | Male | 47 | >40 | 23.87 | <30 | na |
| P2-127 | Degenerative | Lateral | Male | 15 | <40 | 19.52 | <30 | na |
| P2-128 | Degenerative | Lateral | Male | 44 | >40 | 33.48 | >30 | na |
| P2-129 | Degenerative | Medial | Male | 48 | >40 | 25.82 | <30 | na |
| P2-130 | Degenerative | Medial | Female | 51 | >40 | 27.98 | <30 | na |
| P2-132 | Degenerative | Medial | Male | 40 | <40 | 41.61 | >30 | na |
| P2-134 | Degenerative | Medial | Male | 60 | >40 | 36.26 | >30 | na |
| P2-135 | Degenerative | Medial | Male | 38 | <40 | 32.49 | >30 | na |
| P2-137 | Degenerative | Medial | Male | 48 | >40 | 21.81 | <30 | na |
| P2-138 | Degenerative | Medial | Female | 67 | >40 | 30.21 | >30 | na |
| P2-142 | Degenerative | Medial | Male | 40 | <40 | 26.44 | <30 | na |
| P2-144 | Degenerative | Medial | Male | 43 | >40 | 24.41 | <30 | na |
| P2-146 | Degenerative | Medial | Male | 53 | >40 | 29.05 | <30 | na |
| P2-148 | Degenerative | Medial | Male | 59 | >40 | 40.62 | >30 | na |
| P2-151 | Degenerative | Medial | Male | 50 | >40 | 33.74 | >30 | na |
| P2-153 | Degenerative | Medial | Male | 48 | >40 | 28.36 | <30 | na |
| Summary≫ | n = 37 | 5 Lat, 32 Med | 10 F, 27 M | 45.78 | 12 <40, 25 >40 | 27.76 | 27 <30, 10 >30 | na |
| P1-011 | Traumatic | Lateral | Female | 14 | <40 | 23.03 | <30 | 0.7 |
| P1-021 | Traumatic | Medial | Male | 30 | <40 | 28.12 | <30 | 1.2 |
| P1-Z49M | Traumatic | Medial | Female | 55 | >40 | 36.02 | >30 | 2.3 |
| P2-104 | Traumatic | Medial | Male | 49 | >40 | 23.75 | <30 | 6.5 |
| P2-106 | Traumatic | Medial | Male | 16 | <40 | 23.05 | <30 | 0.1 |
| P2-108 | Traumatic | Lateral | Male | 41 | >40 | 32.09 | >30 | 3.6 |
| P2-114 | Traumatic | Medial | Female | 16 | <40 | 20.36 | <30 | 1.1 |
| P2-139 | Traumatic | Medial | Female | 56 | >40 | 29.23 | <30 | 6.4 |
| P2-143 | Traumatic | Medial | Male | 36 | <40 | 25.85 | <30 | 2.8 |
| P2-149 | Traumatic | Medial | Male | 46 | >40 | 28.19 | <30 | 8.3 |
| P2-150 | Traumatic | Medial | Female | 43 | >40 | 34.33 | >30 | 0.9 |
| Summary≫ | n = 11 | 2 Lat, 9 Med | 5 F, 6 M | 36.54 | 5 <40, 6 >40 | 27.64 | 8 <30, 3 >30 | 0.1 – 8.3 |
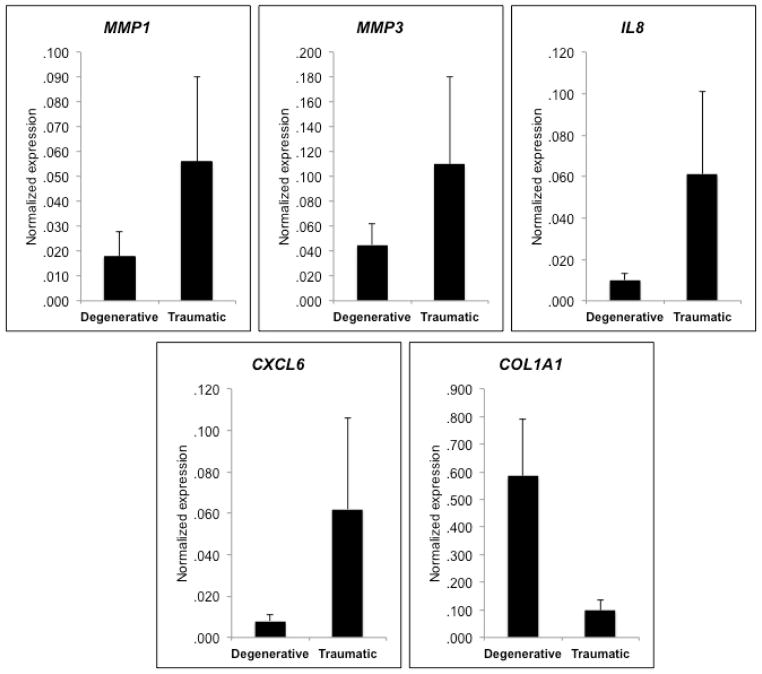
Gene expression differed between traumatic and degenerative meniscus tears, with a number of chemokines and matrix metalloproteinases expressed at higher levels in traumatic tears compared to degenerative tears (Table 3). Two chemokines, IL8 (6.0-fold; P < 0.001) and CXCL6 (8.16-fold; P < 0.001), and two matrix metalloproteinases, MMP1 (3.16-fold; P = 0.011) and MMP3 (2.48-fold; P = 0.016), were expressed at significantly higher levels in traumatic tears compared to degenerative tears (Fig. 2). Most of other chemokines tested in this study were found to be down-regulated in degenerative tears but did not reach a formal statistical significance. COL1A1, a major extracellular matrix gene in meniscus tissue, was expressed at a higher level in degenerative tears compared to traumatic tears (5.98-fold; P = 0.058) (Fig. 2). None of the gene transcripts were different between the medial and lateral meniscus.
Table 3.
Genes differentially expressed between degenerative and traumatic tears
| Gene symbol | Gene name | Class | Degenerative tear (mean±S.E.M.) | Traumatic tear (mean±S.E.M.) | Fold change | Description | P value |
|---|---|---|---|---|---|---|---|
| IL1A | Interleukin 1 alpha | Cytokine | 0.021±0.011 | 0.22±0.016 | 10.48 | Down in degenerative tears | 0.965 |
| IL1B | Interleukin 1 beta | Cytokine | 0.004±0.001 | 0.019±0.012 | 4.75 | Down in degenerative tears | 0.361 |
| IL6 | Interleukin 6 | Cytokine | 0.016±0.007 | 0.023±0.020 | 1.44 | Down in degenerative tears | 0.677 |
| TNF | Tumor necrosis factor alpha | Cytokine | 0.003±0.001 | 0.004±0.003 | 1.33 | Down in degenerative tears | 0.700 |
| ADAMTS4 | A disintegrin and metalloproteinase with thrombospondin motifs 4 | Metalloproteinase | 0.022±0.010 | 0.017±0.010 | 1.26 | Up in degenerative tears | 0.924 |
| ADAMTS5 | A disintegrin and metalloproteinase with thrombospondin motifs 5 | Metalloproteinase | 0.025±0.012 | 0.017±0.010 | 1.48 | Up in degenerative tears | 0.886 |
| BMP2 | Bone morphogenetic protein 2 | Receptor ligand | 0.006±0.002 | 0.025±0.023 | 4.17 | Down in degenerative tears | 0.538 |
| MMP1 | Matrix metalloproteinase 1 | Metalloproteinase | 0.018±0.010 | 0.056±0.034 | 3.11 | Down in degenerative tears | 0.011 |
| MMP3 | Matrix metalloproteinase 3 | Metalloproteinase | 0.045±0.017 | 0.110±0.070 | 2.44 | Down in degenerative tears | 0.016 |
| MMP9 | Matrix metalloproteinase 9 | Metalloproteinase | 0.010±0.005 | 0.021±0.017 | 2.10 | Down in degenerative tears | 0.838 |
| MMP13 | Matrix metalloproteinase 13 | Metalloproteinase | 0.023±0.010 | 0.013±0.006 | 1.79 | Up in degenerative tears | 0.914 |
| IL8 | Interleukin 8 | Chemokine | 0.010±0.003 | 0.061±0.040 | 6.10 | Down in degenerative tears | <0.001 |
| CCL3 | Chemokine (C-C motif) ligand 3 | Chemokine | 0.005±0.002 | 0.028±0.021 | 5.60 | Down in degenerative tears | 0.384 |
| CCL3L1 | Chemokine (C-C motif) ligand 3-like 1 | Chemokine | 0.002±0.001 | 0.016±0.011 | 8.00 | Down in degenerative tears | 0.282 |
| CXCL1 | Chemokine (C-X-C motif) ligand 1 | Chemokine | 0.003±0.001 | 0.003±0.001 | 1.03 | Up in degenerative tears | 0.750 |
| CXCL3 | Chemokine (C-X-C motif) ligand 3 | Chemokine | 0.003±0.001 | 0.016±0.011 | 5.33 | Down in degenerative tears | 0.322 |
| CXCL6 | Chemokine (C-X-C motif) ligand 6 | Chemokine | 0.008±0.003 | 0.062±0.044 | 7.75 | Down in degenerative tears | <0.001 |
| CCL20 | Chemokine (C-C motif) ligand 20 | Chemokine | 0.005±0.002 | 0.017±0.011 | 3.40 | Down in degenerative tears | 0.228 |
| COL1A1 | Collagen type 1, A1 | Extracellular matrix | 0.585±0.205 | 0.098±0.038 | 5.98 | Up in degenerative tears | 0.058 |
| COL2A1 | Collagen type 2, A1 | Extracellular matrix | 0.024±0.009 | 0.026±0.009 | 1.08 | Down in degenerative tears | 0.504 |
| ACAN | Aggrecan | Extracellular matrix | 0.042±0.012 | 0.009±0.006 | 4.39 | Up in degenerative tears | 0.351 |
| NFkB1A | Nuclear factor kappa B1 A | Transcription factor | 0.019±0.007 | 0.023±0.017 | 1.21 | Down in degenerative tears | 0.622 |
| NFkB2 | Nuclear factor kappa B2 | Transcription factor | 0.012±0.008 | 0.016±0.011 | 1.33 | Down in degenerative tears | 0.510 |
| IkBA | Inhibitory kappa B A | Transcription factor | 0.012±0.003 | 0.021±0.012 | 1.75 | Down in degenerative tears | 0.189 |
| ADIPOQ | Adiponectin | Adipokine | 0.012±0.007 | 0.007±0.005 | 1.73 | Up in degenerative tears | 0.755 |
| APLN | Apelin | Adipokine | 0.293±0.269 | 0.158±0.100 | 1.85 | Up in degenerative tears | 0.821 |
| LEP | Leptin | Adipokine | 0.005±0.003 | 0.002±0.001 | 3.18 | Up in degenerative tears | 0.224 |
| RETN | Resistin | Adipokine | 0.014±0.009 | 0.005±0.003 | 2.91 | Up in degenerative tears | 0.210 |
P values in boldface are statistically significant at 5% level. S.E.M. = standard error of the mean
Figure 2.
Gene expression differences between degenerative and traumatic meniscus tears. Normalized mRNA expression of genes significantly up-regulated in traumatic tears (MMP1, MMP3, IL8 and CXCL6) and a gene borderline significantly up-regulated in degenerative tears (COL1A1) are shown. P ≤ 0.05 for MMP1, MMP3, IL8 and CXCL6) and P = 0.058 for COL1A1.
Discussion
Gene expression in the injured meniscus does vary by tear pattern with traumatic tears expressing higher levels of chemokines and matrix metalloproteinases compared to degenerative tears. This is the first study to demonstrate a biologic difference between traumatic and degenerative meniscus tears. These differences could contribute to varying potential for healing following meniscus repair as well as advance our understanding of how meniscus tears contribute to the development of OA in the knee.
Differences in gene expression profile may be important information when deciding whether to resect or repair a meniscus tear. Traditionally, orthopaedic surgeons have shied away from repairing degenerative tears because of concerns about the structural integrity of the longitudinal fibers, even if the repair healed, as well as the lack of a blood supply to the inner meniscus1, 22, 29. While tears in the well-vascularized periphery are associated with good healing rates1, tears in the inner less-vascularized portion of the meniscus are associated with worse healing22, 29. Recently, there has been a more aggressive approach to repairing tears, both due to concerns about the effect of resection on the health of the joint as well as advances in the technical ability to complete a repair. However, biology may be just as important as other factors in terms of determining whether a tear is likely to heal. More research is needed in this area to better understand if and how the variance in gene expression between the 2 tear types effects meniscal healing of an attempted repair. For example, the elevated catabolic environment in traumatic tears raises concerns about their potential to heal after repair, which may partly explain the slow and imperfect healing observed clinically16, 29.
Second, these findings may be important in terms of the overall health of the knee joint and its relative risk for future degeneration into OA. While it has been proposed that meniscal degeneration precedes cartilage degeneration2, 11, a previous study found only limited evidence for a relationship between early degenerative changes in the articular cartilage based on arthroscopy and gene expression in the injured meniscus17. A global survey of gene expression reported that 49 genes were differentially regulated in the meniscus from knees with chondrosis compared to meniscus from knees without chondrosis17. When chondrosis was present in the knee, genes representing cell catabolism (cAMP catabolic process), and tissue and endothelial cell development were repressed while those involved in T cell differentiation and apoptosis were elevated. Another study has reported up-regulation of genes involved in inflammation and cytokine production and down-regulation of genes related to DNA repair processes in meniscal cells from knees with OA compared to meniscal cells from knees without OA27. How our current findings relate to that study is difficult to assess because the knees in the current study did not have OA, only meniscus tears which can be a precursor to OA.
CXCL6, also known as Granulocyte Chemotactic Protein 2 (GCP2), is a ligand shared by two receptors that mediate neutrophil recruitment to inflammatory sites4. While there are no published studies to date reporting on its function in the knee joint, CXCL6 was shown to correlate with the severity of periodontal disease, acting as a functional adjunct to IL8, a functionally and structurally related chemokine12. The elevation of both of these markers in traumatic meniscus tears suggest they may play a significant role in the inflammatory response of the meniscus to injury.
IL8 has been shown to be induced from articular cartilage in response to mechanical, inflammatory and metabolic stresses5, with the authors concluding that this cytokine is likely to play a role in OA. Osteoblasts derived from osteophytes produce IL8, and the production increased under conditions of nonphysiologic load23. IL8 was recently shown to be significantly higher in the synovial fluid of patients undergoing TKA compared to controls and the level of IL-8 was strongly correlated with the radiographic severity of OA15. Meniscus cells from normal and OA knees have been shown to increase production of IL-8 in response to pro-inflammatory stimulation25. Since this chemokine appears to play an important role in the meniscus, as well as the articular cartilage, bone and synovial fluid from the knee, the elevated expression of IL-8 in traumatic meniscus tears may play a key role in how the meniscus tear affects the rest of the joint.
Static compression of the meniscus has been shown to induce the expression of MMP1 in the meniscus28. The expression of MMP1 and MMP3 has also been shown to be elevated in the synovial fluid of patients with OA24. A recent animal study demonstrated elevated expression of MMP1 and MMP3 from the menisci of knees with OA compared to menisci from knee without OA26. However, MMP3 expression was lower in the meniscus of human patients with OA compared to controls20, which is congruent with our findings that traumatic meniscus tears have elevated MMP-3 compared to degenerative meniscus tears. Furthermore, MMP3 has been shown to be elevated in synovial fluid from knees undergoing arthroscopy and directly correlated to preoperative Visual Analogue Scores (VAS)6. More research is needed to assess how levels of MMP-3 in the injured meniscus relate to levels of MMP3 in the synovial fluid as well as clinical symptomatology.
There are a number of limitations to the current study that can be overcome by additional research in this area to better understand if and how our findings relate to the potential for meniscus healing, as the relevance of molecular markers for OA to the meniscus is not well studied. First, an unbiased transcriptome analysis would provide additional information. Quantifying the gene expression signatures in articular cartilage and synovial fluid as well as meniscus from the same patients would provide a better comparison of molecular changes in these tissues and the overall effect on joint health. There is no in situ analysis to confirm the origin of the mRNA or protein validation. While the skewed distribution towards degenerative tears is a potential source of bias, the large fold, highly significant differences between tear patterns makes any bias, if present, unlikely to alter the findings. Biomarkers from blood and serum were not analyzed for systemic signs of inflammation or OA. We did not analyze the association of time from injury with gene expression in traumatic tears, as similar data is lacking for degenerative tears. While there is no comparison to gene expression in normal menisci, the purpose of the study was to find differences between the 2 types of injury, not compare them to normal menisci. Finally, a prospective study over 2 to 5 years could investigate the relationship of variance in meniscal gene expression by tear pattern to future degeneration in the joint.
Despite these limitations, this study demonstrates that gene expression in the injured meniscus does relate to the tear pattern. The elevated expression of chemokines and matrix metalloproteinases in traumatic tears may be associated with healing potential following attempted meniscal repair as well as the future risk of OA in the knee. In general, traumatic meniscus tears appear to have a more active biologic response to injury compared to degenerative tears. Future mechanistic studies would better characterize these differences to understand how they can be used to guide clinical decision making in the management of meniscus tears.
What is known about this subject
The classification of meniscus tears into degenerative and traumatic is considered important. While it is known that meniscus tears are associated with the development of osteoarthritis, the biologic basis for this association is not well elucidated.
What this study adds to existing knowledge
This study reports the first comparison of the gene expression profile of degenerative and traumatic tears, demonstrating a more inflammatory/catabolic profile in traumatic tears.
Footnotes
Study was performed at the Department of Orthopaedic Surgery, Washington University School of Medicine, St. Louis, MO
References
- 1.Asahina S, Muneta T, Yamamoto H. Arthroscopic meniscal repair in conjunction with anterior cruciate ligament reconstruction: factors affecting the healing rate. Arthroscopy. 1996;12(5):541–545. doi: 10.1016/s0749-8063(96)90191-7. [DOI] [PubMed] [Google Scholar]
- 2.Berthiaume MJ, Raynauld JP, Martel-Pelletier J, et al. Meniscal tear and extrusion are strongly associated with progression of symptomatic knee osteoarthritis as assessed by quantitative magnetic resonance imaging. Ann Rheum Dis. 2005;64(4):556–563. doi: 10.1136/ard.2004.023796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brophy RH, Rai MF, Zhang Z, Torgomyan A, Sandell LJ. Molecular analysis of age and sex-related gene expression in meniscal tears with and without a concomitant anterior cruciate ligament tear. J Bone Joint Surg Am. 2012;94(5):385–393. doi: 10.2106/JBJS.K.00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catusse J, Liotard A, Loillier B, Pruneau D, Paquet JL. Characterization of the molecular interactions of interleukin-8 (CXCL8), growth related oncogen alpha (CXCL1) and a non-peptide antagonist (SB 225002) with the human CXCR2. Biochem Pharmacol. 2003;65(5):813–821. doi: 10.1016/s0006-2952(02)01619-2. [DOI] [PubMed] [Google Scholar]
- 5.Chauffier K, Laiguillon MC, Bougault C, et al. Induction of the chemokine IL-8/Kc by the articular cartilage: possible influence on osteoarthritis. Joint Bone Spine. 2012;79(6):604–609. doi: 10.1016/j.jbspin.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Cuellar VG, Cuellar JM, Kirsch T, Strauss EJ. Correlation of Synovial Fluid Biomarkers With Cartilage Pathology and Associated Outcomes in Knee Arthroscopy. Arthroscopy. 2015 doi: 10.1016/j.arthro.2015.08.033. [DOI] [PubMed] [Google Scholar]
- 7.Cullen KA, Hall MJ, Golosinskiy A. Ambulatory surgery in the United States, 2006. Natl Health Stat Report. 2009;(11):1–25. [PubMed] [Google Scholar]
- 8.Englund M, Guermazi A, Lohmander SL. The role of the meniscus in knee osteoarthritis: a cause or consequence? Radiol Clin North Am. 2009;47(4):703–712. doi: 10.1016/j.rcl.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Englund M, Guermazi A, Roemer FW, et al. Meniscal tear in knees without surgery and the development of radiographic osteoarthritis among middle-aged and elderly persons: The Multicenter Osteoarthritis Study. Arthritis Rheum. 2009;60(3):831–839. doi: 10.1002/art.24383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Englund M, Roemer FW, Hayashi D, Crema MD, Guermazi A. Meniscus pathology, osteoarthritis and the treatment controversy. Nat Rev Rheumatol. 2012;8(7):412–419. doi: 10.1038/nrrheum.2012.69. [DOI] [PubMed] [Google Scholar]
- 11.Englund M, Roos EM, Lohmander LS. Impact of type of meniscal tear on radiographic and symptomatic knee osteoarthritis: a sixteen-year followup of meniscectomy with matched controls. Arthritis Rheum. 2003;48(8):2178–2187. doi: 10.1002/art.11088. [DOI] [PubMed] [Google Scholar]
- 12.Kebschull M, Demmer R, Behle JH, et al. Granulocyte chemotactic protein 2 (gcp-2/cxcl6) complements interleukin-8 in periodontal disease. J Periodontal Res. 2009;44(4):465–471. doi: 10.1111/j.1600-0765.2008.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDermott ID, Amis AA. The consequences of meniscectomy. J Bone Joint Surg Br. 2006;88(12):1549–1556. doi: 10.1302/0301-620X.88B12.18140. [DOI] [PubMed] [Google Scholar]
- 14.Mesiha M, Zurakowski D, Soriano J, Nielson JH, Zarins B, Murray MM. Pathologic characteristics of the torn human meniscus. Am J Sports Med. 2007;35(1):103–112. doi: 10.1177/0363546506293700. [DOI] [PubMed] [Google Scholar]
- 15.Monibi F, Roller BL, Stoker A, Garner B, Bal S, Cook JL. Identification of Synovial Fluid Biomarkers for Knee Osteoarthritis and Correlation with Radiographic Assessment. J Knee Surg. 2015 doi: 10.1055/s-0035-1549022. [DOI] [PubMed] [Google Scholar]
- 16.Paxton ES, Stock MV, Brophy RH. Meniscal repair versus partial meniscectomy: a systematic review comparing reoperation rates and clinical outcomes. Arthroscopy. 2011;27(9):1275–1288. doi: 10.1016/j.arthro.2011.03.088. [DOI] [PubMed] [Google Scholar]
- 17.Rai MF, Patra D, Sandell LJ, Brophy RH. Transcriptome analysis of injured human meniscus reveals a distinct phenotype of meniscus degeneration with aging. Arthritis Rheum. 2013;65(8):2090–2101. doi: 10.1002/art.37984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rai MF, Patra D, Sandell LJ, Brophy RH. Relationship of gene expression in the injured human meniscus to body mass index: a biologic connection between obesity and osteoarthritis. Arthritis Rheumatol. 2014;66(8):2152–2164. doi: 10.1002/art.38643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rai MF, Sandell LJ, Cheverud JM, Brophy RH. Relationship of age and body mass index to the expression of obesity and osteoarthritis-related genes in human meniscus. Int J Obes (Lond) 2013;37(9):1238–1246. doi: 10.1038/ijo.2012.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roller BL, Monibi FA, Stoker AM, Kuroki K, Bal BS, Cook JL. Characterization of knee meniscal pathology: correlation of gross, histologic, biochemical, molecular, and radiographic measures of disease. J Knee Surg. 2015;28(2):175–182. doi: 10.1055/s-0034-1376333. [DOI] [PubMed] [Google Scholar]
- 21.Roos H, Adalberth T, Dahlberg L, Lohmander LS. Osteoarthritis of the knee after injury to the anterior cruciate ligament or meniscus: the influence of time and age. Osteoarthritis Cartilage. 1995;3(4):261–267. doi: 10.1016/s1063-4584(05)80017-2. [DOI] [PubMed] [Google Scholar]
- 22.Rubman MH, Noyes FR, Barber-Westin SD. Arthroscopic repair of meniscal tears that extend into the avascular zone. A review of 198 single and complex tears. Am J Sports Med. 1998;26(1):87–95. doi: 10.1177/03635465980260013301. [DOI] [PubMed] [Google Scholar]
- 23.Sakao K, Takahashi KA, Arai Y, et al. Osteoblasts derived from osteophytes produce interleukin-6, interleukin-8, and matrix metalloproteinase-13 in osteoarthritis. J Bone Miner Metab. 2009;27(4):412–423. doi: 10.1007/s00774-009-0058-6. [DOI] [PubMed] [Google Scholar]
- 24.Sauerschnig M, Stolberg-Stolberg J, Schulze A, Salzmann GM, Perka C, Dynybil CJ. Diverse expression of selected cytokines and proteinases in synovial fluid obtained from osteoarthritic and healthy human knee joints. Eur J Med Res. 2014;19:65. doi: 10.1186/s40001-014-0065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stone AV, Loeser RF, Vanderman KS, Long DL, Clark SC, Ferguson CM. Pro-inflammatory stimulation of meniscus cells increases production of matrix metalloproteinases and additional catabolic factors involved in osteoarthritis pathogenesis. Osteoarthritis Cartilage. 2014;22(2):264–274. doi: 10.1016/j.joca.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stone AV, Vanderman KS, Willey JS, et al. Osteoarthritic changes in vervet monkey knees correlate with meniscus degradation and increased matrix metalloproteinase and cytokine secretion. Osteoarthritis Cartilage. 2015;23(10):1780–1789. doi: 10.1016/j.joca.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y, Mauerhan DR, Honeycutt PR, et al. Analysis of meniscal degeneration and meniscal gene expression. BMC Musculoskelet Disord. 2010;11:19. doi: 10.1186/1471-2474-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Upton ML, Chen J, Guilak F, Setton LA. Differential effects of static and dynamic compression on meniscal cell gene expression. J Orthop Res. 2003;21(6):963–969. doi: 10.1016/S0736-0266(03)00063-9. [DOI] [PubMed] [Google Scholar]
- 29.van Trommel MF, Simonian PT, Potter HG, Wickiewicz TL. Arthroscopic meniscal repair with fibrin clot of complete radial tears of the lateral meniscus in the avascular zone. Arthroscopy. 1998;14(4):360–365. doi: 10.1016/s0749-8063(98)70002-7. [DOI] [PubMed] [Google Scholar]