Abstract
Mycoplasma genitalium (MG) is a sexually transmitted pathogen for which there is no FDA approved diagnostic test available in the U.S. A modified real-time PCR assay for detecting MG and simultaneously identifying macrolide resistance mutations from clinical specimens was evaluated and proved to be sensitive and accurate for diagnostic purposes.
Keywords: Mycoplasma genitalium, macrolide, resistance, real-time PCR
Mycoplasma genitalium (MG) is a sexually transmitted pathogen associated with urethritis in men and several inflammatory syndromes in women. The clinical significance of MG infection has been recognized by the Centers for Disease Control and Prevention, listing MG under “Emerging Issues” in the 2015 Sexually Transmitted Diseases Treatment Guidelines (Workowski et al., 2015). There is no FDA approved test for MG in the US, therefore treatment is usually empiric. The macrolide azithromycin is the first line recommended antibiotic for treating MG infections (Workowski et al., 2015). However, MG resistance to macrolides is increasing, occurring in up to 100% in some populations (Gesink et al., 2012). Macrolide treatment failure is mainly associated with mutations in domain V of the 23S rRNA gene that affect affinity of the drug. Methods for detecting macrolide resistance mutations in MG have been published and commercial assays are available in some countries (Gosse et al., 2016, Kristiansen et al., 2016, Tabrizi et al., 2016, Tan et al., 2017, Touati et al., 2014, Twin et al., 2012, Wold et al., 2015). We adapted Touati’s assay for directly detecting MG and macrolide resistance mutations for research and clinical diagnostic purposes (Touati et al., 2014).
Initial evaluation of the Touati method on a Roche LightCycler® 480 II (Roche Diagnostics, Indianapolis, IN) showed a very weak signal while the amplification was robust as indicated by the agarose gel. Examination of the probes revealed that the Tm of the sensor probe was significantly lower than that of the donor probe (57.8°C vs 66.4°C). We then modified the sensor probe (LC-Red 705-AACGGGACGGAAAGACCCCG-phosphate, Tm=66.1°C), optimized the PCR conditions, and named the assay Mycoplasma genitalium macrolide resistant (MRMR) PCR. The 20 μL of reaction mixture contained 0.375 μM of forward primer, 0.5 μM of reverse primer, 0.1 μM of each probe, 0.4 U of LightCycler® Uracil-DNA Glycosylase (Roche Diagnostics, Indianapolis, IN), 10 μL of LightCycler® 480 Probes Master (2X, Roche Diagnostics, Indianapolis, IN) and 2 μL of template DNA. A touchdown program was used: after 10 min of pre-incubation at 40°C and 95°C, respectively, there were 10 cycles of pre-amplification with 95°C, 10s; 60°C, 15s (ramp rate 1.1°C/s); and 72°C, 15s, followed by 45 cycles amplification with 95°C, 10s; 60°C to 55°C, 1°C/step, ramp rate 2.2°C/s, 10s; and 72°C, 15s. Then a melting curve analysis was performed: 95°C, 10s; 50°C, 15s; and a slow increase to 75°C with continuous requisition with a ramp rate of 0.11°C/s. Equipment was finally cooled down by 40°C, 30s. Data were analyzed using LightCycler 480 Software 1.5. “Abs quant/2nd Derivative Max” and “Tm calling” analyses were performed.
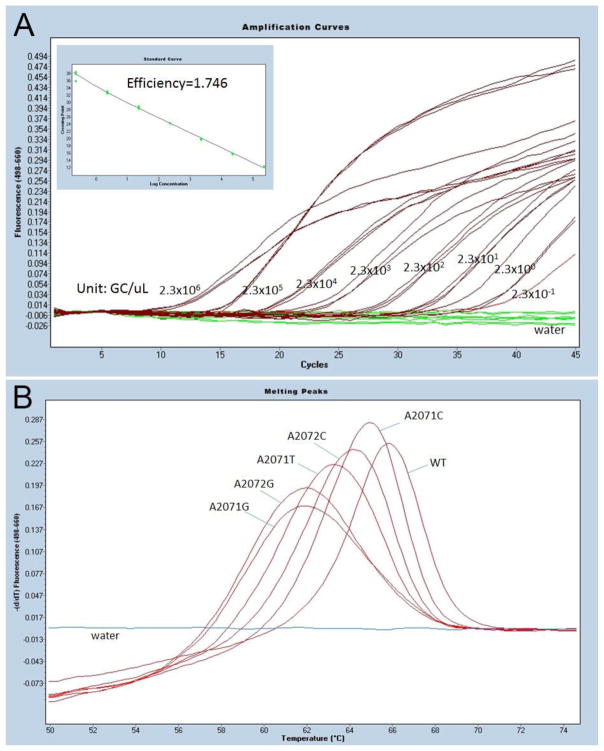
The limit of detection (LOD) was determined using 1:10 serial dilutions of genomic DNA of strain G37 (ATCC 33530). DNA was purified by DNeasy® Blood & Tissue Kit (Qiagen, Gaithersburg, MD) and concentration was determined by Nanodrop 1000 (Thermo Fisher, Canoga Park, CA). Each diluent was tested in triplicates. The serial dilutions were tested 17 times on 7 different days to ensure the PCR reproducibility. The LOD of the MGMR PCR was 2.3 genome copy (GC)/μL in the PCR template (~5 GC/test) (Figure 1A). The lowest concentration detected was 0.23 GC/μL with a cycle number of 40.
Figure 1. MGMR PCR limit of detection (LOT) and melting peak analysis.
A) The limit of detection was determined by testing the 1:10 serial dilution of MG 37 (ATCC 33530) genomic DNA. Each diluent was tested in triplicates. The starting concentration was 2.3 x 106 genome copy (GC)/μL in PCR template. The LOD was 2.3 GC/μL in PCR template, or ~5 GC/test. The PCR efficiency was 1.746. B) The representative melting peaks of 5 known mutations associated with MG macrolide resistance.
The assay specificity was validated using DNA from 98 reference organisms, including human, Mycoplasma and Ureaplasma species infecting humans and other organisms sharing the same physiological niche with MG (Table S1). Most were from ATCC; some were clinical isolates. No cross-reactions were observed. Five known mutations at position 2071 and 2072 (E. coli numbering 2058 and 2059; A2071G, A2072G, A2071C, A2072C, and A2071T) associated with macrolide resistance were cloned into plasmid pMiniT (New England BioLabs, Ipswich, MA) and transformed into NEB 10-beta competent E. coli (New England BioLabs, Ipswich, MA). All mutations were successfully detected by Tm calling analysis (Figure 1B). Four different melting curves were identified. A2071G and A2072G had a similar Tm (61.9±0.8°C). Tm of wild type (65.8±0.9°C) was about 4°C higher than that of A2071G and A2072G, while Tms of other mutations (A2071C, A2071T, and 2072C) were between that of wild type and A2071G/A2072G.
To evaluate the clinical performance of the MGMR PCR, an in-house validated real-time PCR assay targeting MG glyceraldehyde-3-phosphate dehydrogenase gene (MGGAP PCR) (Svenstrup et al., 2005, Waites et al., 2012) was used as a reference method for comparison. We tested 530 specimens (99 urine samples, 96 oral swabs, and 79 rectal swabs from men; 89 vaginal swabs, 88 oral swabs, and 79 rectal swabs from women) from 188 patients attending a sexually transmitted diseases clinic in Birmingham, AL between February 2015 and March 2017. DNA was purified using a cobas 4800 system (Roche Diagnostics, Indianapolis, IN) and stored at −80°C. Positive MGMR PCR amplicons from clinical specimens were cleaned by ExoSAP-IT (Thermo Fisher, Canoga Park, CA) and sequenced to verify the genotypes. Among 530 clinical specimens, 20 (from 20 patients) were positive by MGGAP PCR and 27 (from 24 patients) by MGMR PCR (Table 1). Compared to MGGAP PCR, the percent positive agreement and percent negative agreement were 95.0% (95% CI: 75.1–99.9) and 98.4% (95% CI: 96.9–99.3), respectively. The overall agreement of the two methods was very good for urine and vaginal samples (Kappa= 0.939 and 0.816, respectively), while MGMR PCR detected more MG positive rectal samples (6 vs 1, p=0.063, McNemar’s test, kappa=0.278) than MGGAP PCR. Sequencing of the 27 positive MGMR PCR products revealed 100% agreement in genotype detection: 15 were wild type and 12 of 27 (44%) were macrolide resistant mutants (8 were A2071G and 4 were A2072G).
Table 1.
Comparison of MGMR PCR versus MGGAP PCR for detection of MG
| MGGAP PCR | % positive agreement (95% CI) | % negative agreement (95% CI) | |||
|---|---|---|---|---|---|
| MGMR PCR | Positive | Negative | Total | ||
| Positive | 19 | 8 | 27 | 95.0 (75.1–99.9) | 98.4 (96.9–99.3) |
| Negative | 1a | 502 | 503 | ||
| Total | 20 | 510 | 530 | ||
Note:
The only sample that was positive by MGGAP PCR and negative by MGMR PCR had a very low bacterial load. In addition, MGGAP PCR was performed earlier on a fresher sample while MGMR PCR was performed several weeks later and thus DNA degradation could have contributed to the discordant result between these tests.
In a summary, we modified the sensor probe and cycling conditions for the MGMR PCR. The extended sensor probe matched the higher Tm of the donor probe and the touchdown program improved sensitivity and specificity (Korbie and Mattick, 2008). The LOD of MGMR PCR is about 20 times lower than the original method (50 genome copy/μL) (Touati et al., 2014). Compared to the MGGAP PCR, MGMR PCR showed a superior performance in MG detection from clinical specimens, especially from rectal samples. MGMR PCR also accurately detected wild type and macrolide-resistant mutants from plasmid and clinical specimens, compared to a previous high resolution melt analysis method (Twin et al., 2012) which detected ≤10 copies per reaction but failed to differentiate wild type and one mutant form.
This study was limited by the low number of positive samples, which could have limited power to accurately estimate performance of the assays.
Overall, MGMR PCR is a sensitive and accurate assay to simultaneously detect MG and macrolide resistance from clinical specimens. This assay could serve as a diagnostic tool for clinicians and help guide treatment decisions.
Highlights.
A modified assay to detect Mycoplasma genitalium and identify macrolide resistance mutations.
Modified probes and program improve assay sensitivity and specificity.
Validated for research and clinical diagnostic purposes.
Acknowledgments
We thank Dr. Patricia Totten at the University of Washington for providing DNA from a macrolide-resistant mutant strain of MG for initial method development.
Funding: This work was supported by a National Institute of Allergy and Infectious Diseases Sexually Transmitted Infection Cooperative Research Centers Developmental Research Project Award (U19AI113212).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Gesink DC, Mulvad G, Montgomery-Andersen R, Poppel U, Montgomery-Andersen S, Binzer A, et al. Mycoplasma genitalium presence, resistance and epidemiology in Greenland. Int J Circumpolar Health. 2012;71(1):18203. doi: 10.3402/ijch.v71i0.18203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosse M, Lysvand H, Pukstad B, Nordbo SA. A Novel SimpleProbe PCR Assay for Detection of Mutations in the 23S rRNA Gene Associated with Macrolide Resistance in Mycoplasma genitalium in Clinical Samples. J Clin Microbiol. 2016;54(10):2563–7. doi: 10.1128/JCM.01233-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbie DJ, Mattick JS. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat Protoc. 2008;3(9):1452–6. doi: 10.1038/nprot.2008.133. [DOI] [PubMed] [Google Scholar]
- Kristiansen GQ, Lisby JG, Schonning K. A 5′ Nuclease Genotyping Assay for Identification of Macrolide-Resistant Mycoplasma genitalium in Clinical Specimens. J Clin Microbiol. 2016;54(6):1593–7. doi: 10.1128/JCM.00012-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenstrup HF, Jensen JS, Bjornelius E, Lidbrink P, Birkelund S, Christiansen G. Development of a quantitative real-time PCR assay for detection of Mycoplasma genitalium. J Clin Microbiol. 2005;43(7):3121–8. doi: 10.1128/JCM.43.7.3121-3128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrizi SN, Tan LY, Walker S, Twin J, Poljak M, Bradshaw CS, et al. Multiplex Assay for Simultaneous Detection of Mycoplasma genitalium and Macrolide Resistance Using PlexZyme and PlexPrime Technology. PLoS One. 2016;11(6):e0156740. doi: 10.1371/journal.pone.0156740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan LY, Walker SM, Lonergan T, Lima NE, Todd AV, Mokany E. Superior Multiplexing Capacity of PlexPrimers Enables Sensitive and Specific Detection of SNPs and Clustered Mutations in qPCR. PLoS One. 2017;12(1):e0170087. doi: 10.1371/journal.pone.0170087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touati A, Peuchant O, Jensen JS, Bebear C, Pereyre S. Direct detection of macrolide resistance in Mycoplasma genitalium isolates from clinical specimens from France by use of real-time PCR and melting curve analysis. J Clin Microbiol. 2014;52(5):1549–55. doi: 10.1128/JCM.03318-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twin J, Jensen JS, Bradshaw CS, Garland SM, Fairley CK, Min LY, et al. Transmission and selection of macrolide resistant Mycoplasma genitalium infections detected by rapid high resolution melt analysis. PLoS One. 2012;7(4):e35593. doi: 10.1371/journal.pone.0035593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites KB, Xiao L, Paralanov V, Viscardi RM, Glass JI. Molecular methods for the detection of Mycoplasma and ureaplasma infections in humans: a paper from the 2011 William Beaumont Hospital Symposium on molecular pathology. J Mol Diagn. 2012;14(5):437–50. doi: 10.1016/j.jmoldx.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold C, Sorthe J, Hartgill U, Olsen AO, Moghaddam A, Reinton N. Identification of macrolide-resistant Mycoplasma genitalium using real-time PCR. J Eur Acad Dermatol Venereol. 2015;29(8):1616–20. doi: 10.1111/jdv.12963. [DOI] [PubMed] [Google Scholar]
- Workowski KA, Bolan GA Centers for Disease C, Prevention. Sexually transmitted diseases treatment guidelines, 2015. MMWR Recomm Rep. 2015;64(RR-03):1–137. [PMC free article] [PubMed] [Google Scholar]