Abstract
The related transcriptional co-factors YAP (Yes-associated protein) and TAZ (transcriptional co-activator with PDZ-binding motif) have been proposed to either promote or inhibit osteoblast differentiation. Here we investigated the skeletal consequences of deleting YAP and TAZ at different stages of the osteoblast lineage using Prx1-Cre, Osx1-Cre, and Dmp1-Cre transgenic mice. Prx1-Cre-mediated deletion resulted in embryonic lethality. Mice lacking both copies of TAZ and one copy of YAP in cells targeted by Prx1-Cre were viable and displayed elevated bone mass associated increased bone formation. Deletion of YAP and TAZ using Osx1-Cre mice led to perinatal lethality. Suppression of Osx1-Cre activity until 21 days of age permitted postnatal deletion of YAP and TAZ, which resulted in increased osteoblast number at 12 weeks of age but no change in bone mass. Mechanistic studies revealed that YAP and TAZ suppress canonical Wnt signaling and Runx2 activity in osteoblast progenitors. Consistent with this, deletion of YAP and TAZ from osteoprogenitor cells increased osteoblast differentiation in vitro. Deletion of YAP and TAZ from mature osteoblasts and osteocytes using Dmp1-Cre mice led to reduced osteoblast number and bone formation, as well as increased osteoclast number, but no changes in known regulators of bone turnover such as RANKL, OPG, and Sost. Together these results suggest that YAP and TAZ in osteoblast progenitors oppose differentiation towards the osteoblast lineage but in mature osteoblasts and osteocytes, they promote bone formation and inhibit bone resorption.
Keywords: osteoblast, osteocyte, yes-associated protein (YAP), transcriptional co-activator with PDZ-binding motif (TAZ), Wnt signaling
1. Introduction
Bone is constantly remodeled via a process in which osteoclasts resorb bone that is subsequently replaced by osteoblasts [1]. Excessive bone resorption or insufficient bone formation results in bone loss and increases fracture risk. Osteoblast differentiation from mesenchymal progenitors requires the transcription factors Runx2 and Osterix (Osx), as well as canonical Wnt signaling [2-7]. The molecular mechanisms by which these factors interact to control osteoblastogenesis and bone formation are not fully understood.
YAP (Yes-associated protein), encoded by the Yap1 gene, and TAZ (transcriptional co-activator with PDZ-binding motif), encoded by the Wwtr1 gene, are transcriptional cofactors that have been implicated as regulators of osteoblast differentiation. These proteins have similar structures and redundant functions in several cell types [8-11]. They stimulate gene expression by binding to members of the TEAD family of transcription factors and promote expression of genes that stimulate cell proliferation [12]. Recent studies have shown that YAP and TAZ also interact with the Wnt/β-catenin pathway. Specifically, cytoplasmic YAP and TAZ bind Axin and facilitate sequestering of β-catenin in the destruction complex thereby promoting its proteosomal degradation [13]. Cytoplasmic TAZ also inhibits phosphorylation of Dishevelled (DVL), which inhibits release of β-catenin from the destruction complex [14]. These studies suggest that cytoplasmic YAP and TAZ may inhibit osteoblast differentiation by antagonizing Wnt/β-catenin signaling. Consistent with this, inhibition of YAP in murine mesenchymal progenitors increased osteoblast differentiation in vitro [15].
In vivo suppression of TAZ expression in zebrafish abolished skeletal development as well as osteoblast differentiation [16]. In contrast, germline deletion of TAZ in mice did not result in any obvious defects in skeletal development [17, 18]. Mice lacking YAP die during embryogenesis [19]. Thus, the role of YAP and TAZ in osteoblast formation in mammals remains unclear. To elucidate the role of YAP and TAZ in osteoblast lineage cells in vivo, we deleted YAP and TAZ from cells at different stages of osteoblast differentiation using Prx1-Cre, Osx1-Cre, and Dmp1-Cre transgenic mice.
2. Material and methods
2.1. Mice
Mice harboring both YAP and TAZ conditional alleles, kindly provided by Eric N. Olson (UT Southwestern Medical Center, Texas), were described previously [8, 20]. The generation of transgenic mice expressing the Cre recombinase in different cell populations have been described previously: Prx1-Cre [21], Osx1-Cre [22], and Dmp1-Cre [23]. Prx1-Cre; YAPf/+,TAZf/f mice and littermates were obtained by mating YAPf/f,TAZf/f mice (mixture of 129/Sv and C57BL/6) and Prx1-Cre mice (crossed into C57BL/6 for more than 10 generations). Osx1-Cre; YAPf/f,TAZf/f mice and littermates were obtained by mating YAPf/f,TAZf/f mice (mixture of 129/Sv and C57BL/6) with Osx1-Cre mice (mixed background crossed into C57BL/6 for 6 generations). DMP1-Cre; YAPf/f,TAZf/f mice and littermates were obtained by mating YAPf/f,TAZf/f (mixture of 129/Sv and C57BL/6) with DMP1-Cre (crossed into C57BL/6 for more than 10 generations).
The experimental animals used in most of the studies were obtained by a two-step breeding strategy. Hemizygous Cre transgenic mice were crossed with homozygous YAPf/f,TAZf/f mice to generate heterozygous YAPf/+,TAZf/+ offspring with and without a Cre allele. The YAPf/+,TAZf/+ mice with a Cre allele were then backcrossed with the YAPf/f,TAZf/f mice to obtain the experimental YAPf/f,TAZf/f mice with and without a Cre allele. For the Prx1-Cre model, we analyzed Prx1-Cre;YAPf/+,TAZf/f and YAPf/+,TAZf/f mice using YAPf/+,TAZf/f mice as the control since the conditional double knockout mice were embryonic lethal. For the Dmp1-Cre model, YAPf/f,TAZf/f mice were analyzed as the control. To generate mice with postnatal YAP ad TAZ deletion using Osx1-Cre transgenic mice, adult Osx1-Cre; YAPf/+,TAZf/+ and YAPf/f,TAZf/f mice were fed a diet containing doxycycline (Bio-Serv) beginning 1 week before breeding to suppress expression of the Cre transgene in the resulting offspring. After birth, the offspring were maintained on the doxycycline-containing diet until 3 weeks of age, after which they were switched to regular chow for 9 weeks or 21 weeks. To generate the Osx1-Cre control mice in a mixed background similar to that of the conditional knockout mice, YAPf/+,TAZf/+ mice were bred with Osx1-Cre mice and the resulting progeny carrying YAP and TAZ wild-type alleles were intercrossed to generate control mice. The control mice were fed the doxycycline-containing diet in the same way as Osx1-Cre;YAPf/f,TAZf/f mice.
To quantify bone formation, mice were injected with calcein (20 mg/kg body weight) 7 and 3 days before harvesting. All mice were housed in the animal facility of the University of Arkansas for Medical Sciences. The Institutional Animal Care and Use Committees of the University of Arkansas for Medical Sciences approved protocols involving these mice.
2.2. Skeletal analysis
BMD of the lumbar spine and femur were measured by dual-energy X-ray absorptiometry using a PIXImus Densitometer (GE-Lunar Corp.) and the manufacturer's software as previously described [24, 25]. Three dimensional bone volume and architecture of L4 vertebra and femurs were measured using μCT (model μCT40, Scanco Medical) as previously described [25, 26].
2.3. Histology
Femurs and L1-L3 vertebra were fixed in 10% Millonig's formalin for 24 hours and were gradually transferred to 100% ethanol, and embedded undecalcified in methyl methacrylate. Histomorphometric analysis was performed on 5μm longitudinal sections using the OsteoMeasure Analysis System (OsteoMetrics Inc.). Static and dynamic histomorphometry measurements of the cancellous bone were restricted to the secondary spongiosa. Terminology used were recommended by the Histomorphometry Nomenclature Committee of the American Society for Bone and Mineral Research [27]. Growth plate analysis was also performed using the OsteoMeasure Analysis System (OsteoMetrics Inc.) on 5μm longitudinal femoral sections.
2.4. Quantitative PCR
Bones, from which soft tissues were removed, were harvested from animals and stored immediately in liquid nitrogen. Cortical bone was prepared by removing the ends of femurs and tibias and then flushing the bone marrow with PBS. The bones were then scraped with a scalpel to remove cells on the periosteal surface and frozen in liquid nitrogen for later RNA isolation, or decalcified for genomic DNA isolation. Total RNA was isolated using Trizol (Thermo Fisher Scientific, MA), according to the manufacturer's instructions and cDNA was prepared as previously described [28]. The following Taqman assays from Applied Biosystems were used for quantitative RT-PCR: Yap1 (Mm01143263_m1), wwtr1 (Mm01289583_m1), Axin2 (Mm00443610_m1), Sp7 (Mm00504574_m1), Col1a1 (Mm00801666_g1), Tnfsf11b (Mm0041908_m1), Tnfrsf11b (Mm00435452_m1), SOST (Mm00470479_m1), Dkk1 (Mm00438422_m1), csf1 (Mm00432686_m1), Bglap2 (for, 5′-GCTGCGCTCTGTCTCTCTGA-3′, rev, 5′-TGCTTGGACATGAAGGCTTTG-3′, probe, 5′-AAGCCCAGCGGCC-3′), and ribosomal protein S2 (Mrps2) (for, 5′-CCCAGGATGGCGACGAT-3′, rev, 5′-CCGAATGCTGTAATGGCGTAT-3′, probe, 5′-FAM-TCCAGAGCAGGATCC-NFQ-3′). The relative mRNA amounts were calculated using the ΔCt method [29]. Genomic DNA was isolated from decalcified bone fragments after digestion with proteinase K and phenol/chloroform extraction. Two custom Taqman assays from Applied Biosystems were used for quantifying the YAP and TAZ gene deletion efficiency: one specific for sequences between the loxP sites and the other specific for sequences downstream from the 3′ loxP site. The sequence of primers are as follows: Yap1 (for, 5′- CAGACAACAACATGGCAGGAC-3′, rev, 5′-CCGCTGGGCTGGCA-3′, probe, 5′-FAM-CTTTCGCAACTGAACGTT-NFQ-3′) and wwtr1 (for, 5′-CCCCAGGAAGGTGATGAATCAG-3′, rev, 5′-GCACCGAGGTGGAAGTGAT-3′, probe, 5′-FAM-ACGGGTGGAGGTTCAC-NFQ-3′).
2.5. Plasmid construction, viral production, and cell transduction
The 7TFP Wnt/β-catenin luciferase reporter construct was obtained from Addgene (Plasmid #24308) [30]. The 7×OSE Runx2 luciferase reporter was constructed by replacing the 7×TCF promoter with a 7×OSE promoter within the PstI and NheI sites of the 7TFP lentiviral vector. Single OSE was built by annealing two oligos AGCTGCAATCACCAACCACAGCA and CTTGCTGTGGTTG-GTGATTGCAG (Integrated DNA Technologies) and the product was ligated to generate series multi copies of OSEs. The ligation products were then separated in an agarose gel and 7×OSE was recovered according to the size of the DNA using Zymoclean Gel DNA Recovery kit (Zymo Research, Irvine, CA). 7×OSE was then cloned into PstI and NheI sites of the 7TFP lentiviral vector and the sequence was confirmed by DNA sequencing. Lentiviral vectors used in this study are second generation vectors. For virus production, HEK293T cells were cultured in a 6-well culture plate and co-transfected with a total 3 μg of lentiviral reporter vector, pMD2G (Addgene plasmid #12259, a gift from Didier Trono), and psPAX2 (Addgene plasmid # 12260, a gift from Didier Trono) at the ratio of 2:0.9:0.4 using TransIT-LT1 transfection reagent (Mirus, Madison, WI). The media was changed 12 hours after transfection. The viral supernatant was collected 48 h after media change, passed through a 0.45 μm filter, and used fresh. Bone marrow stromal cells were transduced with 1ml of viral supernatants containing 8 μg/ml polybrene (Sigma-Aldrich, St. Louis, MO) for 24 hours. Cells were then selected in α-MEM medium containing 2 μg/ml puromycin (Sigma-Aldrich) for 3 days. After selection, cells were replated in 24-well plates at 5×104 and cultured for 24 hours. Luciferase activity was measured using Dual-Luciferase Reporter Assay System (E1910, Promega, Madison, WI) and normalized to total protein which is measured using Pierce BCA Protein Assay Kit (23225, Thermo Fisher Scientific).
2.6. Cell culture
Bone marrow stromal cells were harvested from long bones as previously described [31]. Osteoblast differentiation of bone marrow precursors was evaluated by plating bone marrow cells in 12-well plates at 5×106 cells/well and culturing in α-MEM containing 10% fetal bovine serum, 1% penicillin/streptomycin/glutamine, 1% ascorbic acid, and 10 mM β-glycerolphosphate. Culture medium was changed every 3 days. After 21 days, the cultures were fixed with 10% Millonig's modified phosphate buffered formalin and then stained with an aqueous solution of 40 mM alizarin red. Osteoblast-specific gene expression was evaluated in parallel cultures lacking β-glycerolphosphate and harvested after 12 days. Calvaria cells were isolated from 5-day-old YAPf/f,TAZf/f mice as described previously [32] and were transduced with adenovirus encoding Cre recombinase (Ad-Cre) or GFP (Ad-GFP) (Vector Biolabs, Malvern, PA) at an MOI of 30 for 6 hours and allowed to recover for 72 hours. The cells were trypsinized and replated in 6-well plates for RNA analysis or in 10-cm dishes for protein analysis.
2.7. Western blot
The nuclear and cytoplasmic fractions were isolated from the total cell lysates using the Nuclear Extraction Kit (Active Motif, Carlsbad, CA) according to the instructions of the manufacturer. Proteins were then resolved in 10% SDS-polyacrylamide gels and electroblotted onto polyvinylidene difluoride membranes. Membranes were subsequently blocked with 5% nonfat dry milk in TBS and were then incubated with primary antibodies and an appropriate horseradish peroxidase-linked secondary antibody. The following antibodies were used: β-catenin (#610154; BD Biosciences, San Jose, CA), lamin A (sc-20680; Santa Cruz Biotechnology Inc., Dallas, Texas), α-tubulin (T5168, Sigma, St. Louis, MO), and YAP (4912, Cell Signaling Technology, Danvers, MA). The immunoblot was quantified using ImageJ (NIH).
2.8. ELISA
The blood was collected by retro-orbital bleeding. The blood samples were left at room temperature for 30 minutes for clotting. Serum was then collected by centrifuging the blood sample at 500g for 5 minutes. Serum CTX was measured using mouse CTX ELISA kit (Novatein Biosciences, Woburn, MA) according to the manufacturer's instruction.
2.9. Statistics
Student's t-test was used to detect statistically significant treatment effects, after determining that the data were normally distributed and exhibited equivalent variances. All t-tests were two-sided. P-values less than 0.05 were considered as significant. Values in all graphs represent means ± s.d..
3. Results
3.1. Haploinsufficiency of YAP and complete loss of TAZ in Prx1-Cre-targeted cells increases bone mass
To determine whether YAP and TAZ have an essential role in osteoblast-lineage cells, we deleted the genes for YAP and TAZ using the Prx1-Cre transgene [21], which leads to DNA recombination in all mesenchyme-derived cells in the limbs. Complete loss of YAP and TAZ in mesenchymal progenitors caused embryonic lethality with severe hemorrhage and edema during embryo development (Fig. 1A). However, mice with haploinsufficiency of YAP and complete loss of TAZ in mesenchymal progenitors, referred to here as Prx1-Cre;YAPf/+,TAZf/f mice, developed normally. We analyzed the skeletal phenotype of these mice at 5 weeks of age, and the analysis was focused on the femur since the Prx1-Cre transgene is active in the limbs but not the spine [21]. Deletion of the YAP and TAZ genes was detected by quantitative real time PCR using genomic DNA isolated from cortical bone (Fig. 1B). The body weight of Prx1-Cre;YAPf/+,TAZf/f male mice at 5 weeks of age was indistinguishable from control YAPf/+, TAZf/f littermates (Fig. 1C).
Figure 1. Haploinsufficiency of YAP and complete deletion of TAZ in Prx1-Cre-targetted cells increases bone mass.
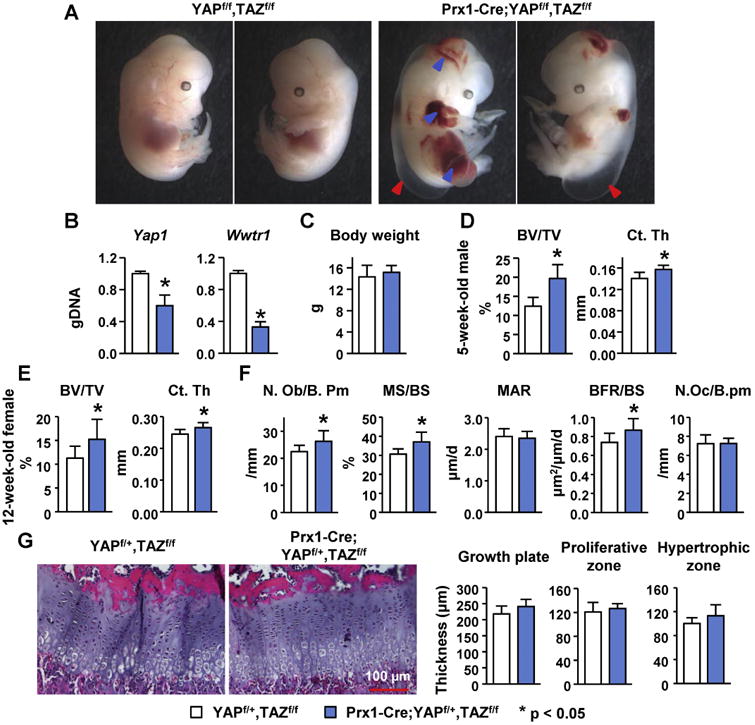
(A) Hemorrhage and edema in YAP/TAZ mutant embryos at E13.5. Blue arrowheads indicate hemorrhage and red arrowheads indicate edema. (B) Quantitative PCR of loxP-flanked genomic DNA isolated from cortical bone of male YAPf/+,TAZf/f (n = 6) and Prx1-Cre;YAPf/+,TAZf/f (n = 7) littermates. (C) Body weight of 5-week-old male YAPf/+,TAZf/f (n = 6) and Prx1-Cre;YAPf/+,TAZf/f (n = 7) littermates. (D) Cancellous bone volume (BV/TV) and cortical thickness in the distal femur of 5-week-old YAPf/+,TAZf/f (n = 6) and Prx1-Cre;YAPf/+,TAZf/f (n = 7) littermates. (E) Cancellous bone volume (BV/TV) and cortical thickness (Ct.Th) in the femur of 12-week-old female YAPf/+,TAZf/f (n = 11) and Prx1-Cre;YAPf/+,TAZf/f (n = 10) littermates. (F) Histomorphometric analysis of femoral cancellous bone in 12-week-old female YAPf/+,TAZf/f (n = 11) and Prx1-Cre;YAPf/+,TAZf/f (n = 10) littermates.(G) Representative images and histomorphometric analysis of the growth plate (in the femur of 5-week-old male YAPf/+,TAZf/f (n = 6) and Prx1-Cre;YAPf/+,TAZf/f (n = 7) mice. Values are the mean ± s.d.. *P < 0.05 versus YAPf/+,TAZf/f using Student's t-test.
Micro-CT analysis of the femur of 5-week-old male mice revealed increased cancellous bone volume with increased trabecular number and decreased trabecular separation in Prx1-Cre;YAPf/+,TAZf/f mice compared to control littermates (Fig. 1D and supplementary Fig. 1). Moreover, cortical thickness was also increased in these mice (Fig. 1D). The increases in femoral cancellous bone volume and cortical thickness were also present in 12-week-old female Prx1-Cre;YAPf/+,TAZf/f mice (Fig. 1E). Histomorphometric analysis of femoral cancellous bone of 12-week-old female mice revealed increased osteoblast number, mineralizing surface, and bone formation rate in Prx1-Cre;YAPf/+,TAZf/f mice (Fig. 1F). However, the osteoclast number did not change (Fig. 1F). Histological analysis of the growth plate in the distal femur revealed no obvious changes (Fig. 1G). Quantification of the growth plate of 5-week-old male mice indicated that there was no significant difference in overall thickness, width of proliferative zone, and width of hypertrophic zone (Fig. 1G). The overall growth plate thickness in 12-week-old female mice also did not change (supplementary Fig. 1), suggesting that chondrocyte biology was not notably altered in 5- and 12-week old Prx1-Cre;YAPf/+,TAZf/f mice. These results demonstrate that reduced expression of YAP and TAZ in the entire mesenchymal lineage, which includes all stages of the osteoblast lineage, increases bone mass.
3.2. Postnatal deletion of YAP and TAZ from the osteoblast lineage promotes osteoblast formation
To determine the role of YAP and TAZ in cells committed to the osteoblast lineage, we next deleted the YAP and TAZ genes using Osx1-Cre transgenic mice, which express the Cre recombinase at the earliest stages of the osteoblast lineage [22]. Mice lacking YAP and TAZ in cells targeted by the Osx1-Cre transgene displayed perinatal lethality (not shown). By taking advantage of temporal regulation of the Osx1-Cre transgene by doxycycline, we deleted the YAP and TAZ genes from osteoblast progenitors postnatally. To do this, breeders were kept on a doxycycline-containing diet to turn off the Osx1-Cre transgene, which enabled us to obtain live Osx1-Cre;YAPf/f,TAZf/f offspring. These mice were kept on a doxycycline diet until 3 weeks of age, then switched to normal diet, and harvested at 12 or 24 weeks of age to analyze the skeletal phenotype. Mice that harbor the Osx1-Cre transgene alone were used as controls since previous reports have demonstrated a negative impact of the Osx1-Cre transgene on skeleton [33-36]. Deletion of the YAP and TAZ genes was detected by quantitative PCR of genomic DNA isolated from cortical bone and showed a more profound deletion in the 24 week old animals (Fig. 2A and supplementary Fig. 2). Micro-CT analysis of 12-week-old female mice did not reveal a significant difference in cancellous bone volume in either the distal femur or the L4 vertebra of Osx1-Cre;YAPf/f,TAZf/f mice compared to control Osx1-Cre mice (Fig. 2B and C). Micro-CT analysis of 24-week-old male and female mice also did not reveal a significant difference in cancellous bone volume in either the distal femur or the L4 vertebra (supplementary Fig. 2). Cortical thickness was also unaffected in these mice at both ages (Fig. 2B and supplementary Fig. 2). Nonetheless, osteoblast number and mineralizing surface were significantly increased in vertebral cancellous bone of 12-week-old female Osx1-Cre;YAPf/f,TAZf/f mice (Fig. 2D). In contrast to the increased mineralizing surface, the mineral appositional rate was reduced in Osx1-Cre;YAPf/f,TAZf/f mice, resulting in a bone formation rate that was not different from control mice (Fig. 2D). Osteoclast number was unaffected by postnatal deletion of YAP and TAZ using the Osx1-Cre transgene (Fig. 2E).
Figure 2. Postnatal deletion of YAP and TAZ from osteoblast progenitors promotes osteoblast formation in vivo.
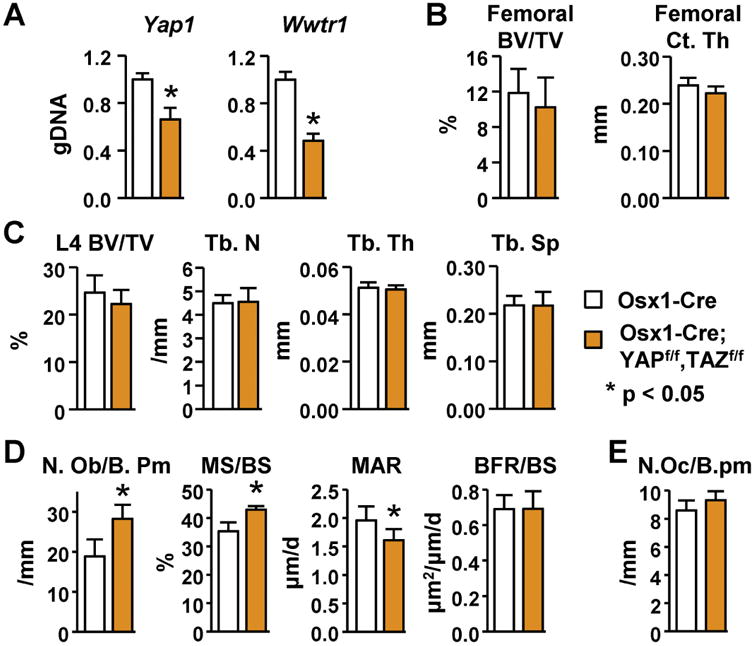
(A) Quantitative PCR of loxP-flanked genomic DNA isolated from tibial cortical bone of Osx1-Cre;YAPf/f,TAZf/f (n = 7) and Osx1-Cre (n = 7) mice. (B) Femoral cancellous bone volume (BV/TV) and cortical thickness (Ct.Th) of 12-week-old female Osx1-Cre;YAPf/f,TAZf/f (n = 7) and Osx1-Cre (n = 7) mice. (C) Cancellous bone volume (BV/TV), trabecular number (Tb.N), trabecular thickness (Tb.Th), and trabecular spacing (Tb.Sp.) in the L4 vertebra of 12-week-old female Osx1-Cre;YAPf/f,TAZf/f (n = 7) and Osx1-Cre (n = 7) mice. (D) Histomorphometric analysis of osteoblast number per bone perimeter (N.Ob/B.Pm), mineralizing surface per bone surface (MS/BS), mineral apposition rate (MAR), and bone formation rate per bone surface (BFR/BS) in cancellous bone in the L1-L3 lumbar vertebra of 12-week-old female Osx1-Cre;YAPf/f,TAZf/f (n = 7) and Osx1-Cre (n = 7) mice. (E) Histomorphometric analysis of osteoclast number per bone perimeter (N.Oc/B.Pm) of mice described in (D). *P < 0.05 using Student's t-test. All mice were exposed to doxycycline in utero and maintained on a doxycycline-containing diet until 3 weeks of age.
3.3. Deletion of YAP and TAZ in osteoblast progenitors increases Wnt signaling and Runx2 activity
To determine whether osteoblast differentiation was affected by deletion of YAP and TAZ in osteoblast progenitors, we examined the osteoblastogenic potential of bone marrow progenitors isolated from conditional knockout mice. Ex vivo culture of bone marrow stromal cells revealed increased osteoblastogenesis in cultures from Prx1-Cre;YAPf/+,TAZf/f mice, as indicated by Alizarin-red staining (Fig. 3A). As expected, YAP mRNA was reduced by approximately 50% and TAZ mRNA by more than 80% in these cultures (Fig. 3B). This was associated with increased expression of osteoblast marker genes, including osterix-1 (Sp7), osteocalcin (Bglap2), and collagen 1a1 (Col1a1) (Fig. 3B).
Figure 3. Deletion of YAP and TAZ from mesenchymal progenitors increases osteoblast formation associated with increased Wnt signaling and Runx2 activity.
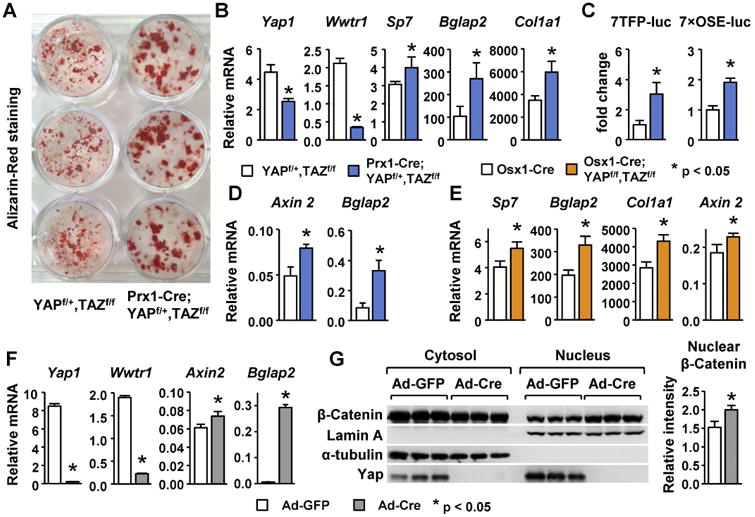
(A) Alizarin Red staining of bone marrow stromal cells isolated from 5-week-old YAPf/+,TAZf/f (n = 5) and Prx1-Cre;YAPf/+,TAZf/f (n = 5) mice and cultured for 21 days in osteoblast differentiation medium (n = 3 wells/group). (B) Quantitative RT-PCR of YAP (Yap1), TAZ (Wwtr1), osterix-1 (Sp7), osteocalcin (Bglap2), and collagen1a1 (Col1a1) in 12-day primary bone marrow cells cultured in osteoblast differentiation medium (n = 3 wells/group). (C) Relative luciferase activity measured in bone marrow stromal cells isolated from YAPf/+,TAZf/f (n = 5) and Prx1-Cre;YAPf/+,TAZf/f mice (n = 5) and transduced with lentivirus harboring 7TFP (Wnt) or 7×OSE (Runx2) luciferase reporters. (n = 3 wells/group). (D) Axin2 and Bglap2 mRNA levels in bone marrow stromal cells isolated from YAPf/+,TAZf/f (n = 5) and Prx1-Cre;YAPf/+,TAZf/f mice (n = 5) and cultured for 5 days. (n = 3 wells/group). (E) Quantitative RT-PCR of Sp7, Bglap2, Col1a1, and Axin2 mRNAs in bone marrow stromal cells isolated from Osx1-Cre;YAPf/f,TAZf/f (n = 5) and Osx1-Cre (n = 5) mice and cultured for 12 days in osteoblast differentiation medium (n = 3 wells/group). (F) Quantitative RT-PCR of Yap1, Wwtr1, Bglap2, and Axin2 mRNAs in YAPf/f,TAZf/f calvaria cells transduced with adenovirus harboring GFP or Cre. (n = 3 wells/group). (G) Immunoblot of protein extracted from the cytoplasm and nucleus of calvaria cells described in (F) and the quantification of nuclear β-catenin normalized to Lamin A. *p < 0.05 using Student's t-test.
We next sought molecular explanations for the increased osteoblast differentiation in the conditional knockout mice. YAP has been reported to suppress Runx2 activity in vitro [37]. Moreover, YAP and TAZ have been shown to interact with β-catenin and promote its degradation [13, 14]. Given the essential role of Runx2 and β-catenin in osteoblastogenesis, the interaction of YAP and TAZ with these proteins could potentially explain the skeletal phenotype we observed in Prx1-Cre;YAPf/+,TAZf/f and Osx1-Cre;YAPf/f,TAZf/f mice. Therefore, we examined whether deletion of YAP and TAZ affected Wnt signaling or Runx2 activity in osteoblast progenitor cells. To assess the transcriptional activity of β-catenin and Runx2, lentiviruses harboring luciferase reporters for TCF activity (7TFP-luc) or Runx2 activity (7×OSE-luc) were transduced into cultured bone marrow stromal cells from Prx1-Cre;YAPf/+,TAZf/f and control mice. Reduced levels of YAP and TAZ increased the activity of both reporters (Fig. 3C). Consistent with these results, mRNA levels of Axin2, a Wnt target gene, and Bglap2, a Runx2 target gene, were higher in bone marrow stromal cells isolated from Prx1-Cre;YAPf/+,TAZf/f mice compared to cells from control mice (Fig. 3D). Ex vivo culture of bone marrow mesenchymal cells from Osx1-Cre;YAPf/f,TAZf/f mice also showed increased expression of osteoblast specific genes Sp7, Bglap2, and Col1a1, as well as the Wnt target gene Axin2, compared with control cultures (Fig. 3E).
To achieve more complete deletion of YAP and TAZ, calvaria cells isolated from YAPf/f,TAZf/f mice were transduced with an adenovirus expressing the Cre recombinase. This led to a potent reduction in the mRNAs of both genes and a concomitant increase in the expression of Axin2 and Bglap2 (Fig. 3F). In addition, immunoblot analysis revealed increased accumulation of β-catenin in the nucleus of knockout calvaria cells (Fig. 3G). Taken together, these results suggest that YAP and TAZ suppress Wnt signaling and Runx2 activity in osteoblast progenitors and this may account for the increase in osteoblast number we observed in mice with reduced YAP and TAZ in the osteoblast lineage.
3.4. Deletion of YAP and TAZ in mature osteoblasts and osteocytes decreases bone mass
The phenotypes we observed by deleting YAP and TAZ with Prx1-Cre or Osx1-Cre could be due to loss of YAP and TAZ activity at any stage of the osteoblast lineage. Therefore, we sought to determine whether these genes have an essential function in mature osteoblasts and osteocytes. To do this, we deleted them using the Dmp1-Cre transgene, which causes DNA recombination in osteocytes as well as mature osteoblasts [26, 38].
Mice lacking both the YAP and TAZ genes in Dmp1-Cre expressing cells, hereafter referred to as Dmp1-Cre; YAPf/f,TAZf/f mice, were born at the expected Mendelian ratio and their body weight was indistinguishable from control YAPf/f,TAZf/f littermates (Supplementary Fig. 3). Deletion of the YAP and TAZ genes was detected by quantitative real time PCR using genomic DNA isolated from cortical bone (Fig. 4A). Dmp1-Cre;YAPf/f,TAZf/f mice exhibited decreased bone mineral density (BMD) at 5 and 12 weeks of age as measured by dual energy x-ray absorptiometry (DXA) (Supplementary Fig. 3). Quantification of the femoral cancellous bone compartment revealed decreased bone volume, decreased trabecular number, and increased trabecular separation in 12-week-old female and male Dmp1-Cre;YAPf/f,TAZf/f mice compared to control littermates (Fig. 4B, C, and Supplementary Fig. 3). Cortical thickness was also decreased in Dmp1-Cre;YAPf/f,TAZf/f mice (Fig. 4B and Supplementary Fig. 3). Similar results were observed in the cancellous bone of lumbar vertebra of Dmp1-Cre;YAPf/f,TAZf/f mice in both sexes (Fig. 4D and Supplementary Fig. 3).
Figure 4. Deletion of YAP and TAZ in mature osteoblasts and osteocytes decreases bone mass and osteoblast number.
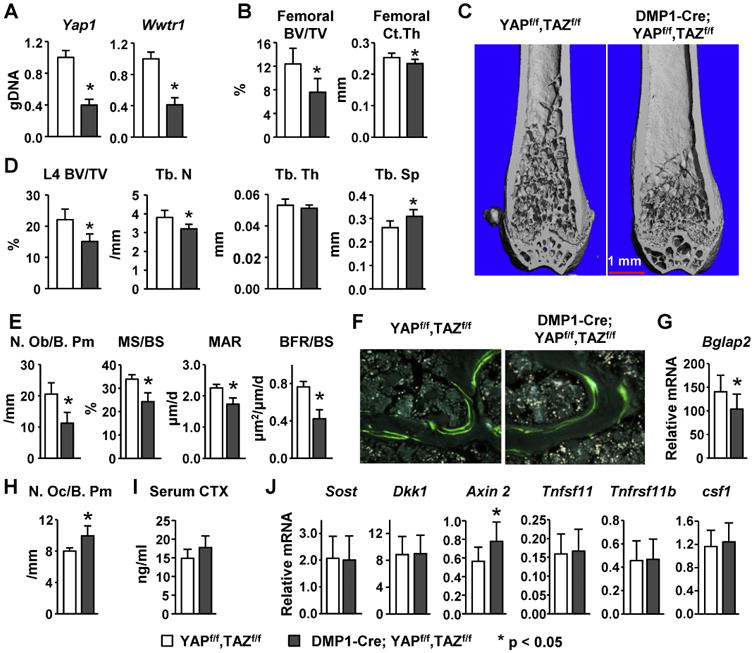
(A) Quantitative PCR of loxP-flanked genomic DNA isolated from tibial cortical bone of YAPf/f,TAZf/f (n = 10) and Dmp1-Cre;YAPf/f,TAZf/f (n = 11) mice. (B) Femoral cancellous bone volume (BV/TV) and cortical thickness (Ct.Th.) of 12-week-old female YAPf/f,TAZf/f (n = 10) and Dmp1-Cre;YAPf/f,TAZf/f (n = 11) mice. (C) Representative micro-CT images of the distal femur of 12-week-old female mice. Scale bar, 1 mm. (D) Vertebral cancellous bone volume (BV/TV), trabecular number (Tb.N), trabecular thickness (Tb.Th), and trabecular spacing (Tb.Sp.) measured in the L4 vertebra of 12-week-old female YAPf/f,TAZf/f (n = 10) and Dmp1-Cre;YAPf/f,TAZf/f (n = 11) mice. (E) Osteoblast number per bone perimeter (N.Ob/B.Pm), mineralizing surface per bone surface (MS/BS), mineral apposition rate (MAR), and bone formation rate per bone surface (BFR/BS) in cancellous bone of L1-L3 lumbar vertebra of 12-week-old female YAPf/f,TAZf/f (n = 10) and Dmp1-Cre;YAPf/f,TAZf/f (n = 11) mice. (F) Photomicrographs of calcein-labeled surfaces in vertebral cancellous bone (20×). (G) Osteocalcin (Bglap2) mRNA abundance in L5 vertebra from YAPf/f,TAZf/f and Dmp1-Cre;YAPf/f,TAZf/f mice. (H) Osteoclast number per bone perimeter (N.Oc/B.Pm) of the same animals described in E. (I) Serum CTX measured by ELISA in 12-week-old female YAPf/f,TAZf/f (n = 8) and Dmp1-Cre;YAPf/f,TAZf/f (n = 9) mice. (J) Quantitative RT-PCR of SOST, Dkk1, Axin2, RANKL (Tnfsf11), OPG (Tnfrsf11b), and m-csf (Csf1) mRNA in tibia shafts from 12-week-old female YAPf/f,TAZf/f (n = 10) and Dmp1-Cre;YAPf/f,TAZf/f (n = 11) mice. *P < 0.05 versus YAPf/f,TAZf/f using Student's t-test.
Consistent with the changes in bone mass and architecture, histomorphometric analysis of female vertebral cancellous bone revealed that osteoblast number was significantly decreased in 12-week-old Dmp1-Cre;YAPf/f,TAZf/f mice (Fig. 4E). Moreover, the bone formation rate was reduced in Dmp1-Cre;YAPf/f,TAZf/f mice with decreased mineralizing surface and mineral appositional rate (Fig. 4E and F). Consistent with the decrease of osteoblast number in the vertebra cancellous bone, expression of osteocalcin in the 5th lumbar vertebra was also significantly decreased in Dmp1-Cre;YAPf/f,TAZf/f mice (Fig. 4G). On the other hand, osteoclast number was increased in 12-week-old female Dmp1-Cre;YAPf/f,TAZf/f mice (Fig. 4H). However, the bone resorption marker CTX in the circulation was not significantly increased in Dmp1-Cre;YAPf/f,TAZf/f mice (Fig. 4I). We then sought molecular explanations for the reduced osteoblast number and bone mass in Dmp1-Cre;YAPf/f,TAZf/f mice. We measured the expression of genes that are known to regulate osteoblast an osteoclast formation including Sost, Dkk1, Axin2, RANKL (Tnfsf11), OPG (Tnfrsf11b), and M-csf (Csf1). The measurements were focused on cortical bone, which is enriched in osteocytes and osteoblasts. However, measurement of these mRNAs in cortical bone did not reveal any changes in their expression in Dmp1-Cre;YAPf/f,TAZf/f mice except for Axin2 which was increased in conditional knockout mice (Fig. 4J). Taken together, these results demonstrate that, in contrast to deletion of YAP and TAZ using the Prx1-Cre or Osx1-Cre transgene, deletion of YAP and TAZ gene using the Dmp1-Cre transgene decreases osteoblast number and bone mass.
4. Discussion
The goal of our study was to determine whether YAP and TAZ play an essential role in osteoblast differentiation. We found that deletion of YAP and TAZ from osteoblast progenitors increased osteoblast formation and this was associated with increased Wnt signaling and Runx2 activity. In contrast, deletion of YAP and TAZ from differentiated osteoblasts and osteocytes decreased osteoblast number and bone formation. These results suggest that YAP and TAZ perform distinct functions in mesenchymal progenitors versus mature osteoblasts and osteocytes.
Previous studies have not provided a clear consensus on the role of YAP and TAZ in osteoblast differentiation. Specifically, some studies suggest that these factors promote osteoblast differentiation while others conclude the opposite. For example, suppression of TAZ in zebrafish using morpholinos abolished osteoblast differentiation and skeletal development [39]. In addition, knockdown of YAP and TAZ in human mesenchymal progenitors completely inhibited expression of osteoblastic markers in vitro [40]. Similarly, haploinsufficiency of YAP and TAZ almost completed abolished osteoblast differentiation of murine osteoblast progenitors in vitro [41]. More recent studies suggest that TAZ promotes osteoblast progenitor proliferation and differentiation by binding to TEAD and Runx2, respectively [42, 43]. In contrast to these studies, knockdown of YAP in murine bone marrow-derived mesenchymal cells stimulated osteoblast differentiation in vitro [15]. In addition, deletion of YAP in osteochondroprogenitors using a Col2-Cre transgene increases bone mass in mice [44]. These studies are consistent with early work suggesting that YAP inhibits Runx2 transcriptional activity [45]. Also, contrary to the morpholino experiment in zebrafish, mice with germline deletion of TAZ undergo apparently normal skeletal development [17, 18].
We have examined the roles of YAP and TAZ at several stages of osteoblast differentiation and found that they play distinct roles at these different stages. To interpret the phenotypes observed in our study, it may be helpful to consider only the models in which both copies of YAP and TAZ were deleted. The low bone formation and bone mass resulting from deletion with Dmp1-Cre clearly demonstrates that YAP and TAZ function in mature osteoblasts and osteocytes to promote bone formation. Since the Osx1-Cre transgene deletes in mature osteoblasts and osteocytes, in addition to osteoblast progenitors, one might have expected low bone mass similar to that obtained with Dmp1-Cre. However, since this was not the case, the loss of positive actions in mature cells must have been counterbalanced by loss of negative actions in progenitors in the Osx1-Cre-targeted mice.
Our mechanistic studies suggest that canonical Wnt signaling and Runx2 activity were elevated in osteoblast progenitors with reduced YAP and TAZ. These changes were associated with robust increases in markers of commitment to the osteoblast lineage and osteoblast differentiation. Why this translated into elevated bone mass in the mice targeted with Prx1-Cre but not Osx1-Cre is unclear but may be related to the remaining YAP allele, deletion at an earlier stage of the mesenchymal lineage, or deletion at an earlier stage of development in the Prx1-Cre-targeted mice.
In mature osteoblasts and osteocytes, deletion of YAP and TAZ decreased osteoblast number, increased osteoclast number, and lowered bone mass. However, the mechanisms underlying these changes remain unknown. We measured expression of several factors known to regulate osteoblast and osteoclast formation such as Sost, RANKL, OPG, and M-CSF. However, we did not observe any changes in expression of these genes. Numerous studies have demonstrated that YAP and TAZ participate in the response of cells and tissues to changes in mechanical loading [40, 46-48]. Moreover, loss of mechanical load in bone causes a decline in osteoblast number and an increase in osteoclast number, similar to what we observed in the conditional knockout mice. Therefore, it is possible that deletion of these genes from osteocytes impaired the ability of the conditional knockout mice to respond to normal physiological loading.
While preparing the revision of this manuscript, Kegelman et al. published a study in which YAP and TAZ were deleted from osteoblast progenitors using Osx1-Cre transgenic mice [49]. In contrast to our findings, they reported that deletion of YAP and TAZ from Osx1-Cre expressing cells decreases bone mass and that this is associated with reduced osteoblast number and increased osteoclast number. Similar to our study, these authors found that the bone formation rate did not change in the conditional knockout mice. Differences in results between the two studies may be explained by several important differences in approach. For example, we compared our conditional knockout mice with Osx1-Cre transgenic mice, which have previously been shown to display low bone mass and even fractures at some skeletal sites shortly after birth [33-35]. In contrast, Kegelman et al. used YAPf/f,TAZf/f mice as controls. Secondly, in our study the deletion was initiated at 3 weeks of age, while the other study deleted YAP and TAZ from embryonic development onward.
In conclusion, the results presented here provide evidence that YAP and TAZ in osteoblast progenitors play a negative role in regulating osteoblast differentiation possibly through interfering with Wnt signaling and Runx2 activity. In contrast, YAP and TAZ in mature osteoblasts and osteocytes play a positive role in osteoblast formation and function, potentially by transducing mechanical signals in osteocytes. Further studies will be required to address this latter possibility. Taken together, our results suggest that YAP and TAZ have opposing effects at different stages of osteoblast differentiation.
Supplementary Material
YAP and TAZ expression in mesenchymal progenitors inhibits their differentiation towards osteoblasts
TAZ expression in mesenchymal progenitors is completely dispensable for osteoblastogenesis in mice
YAP and TAZ expressed in mature osteoblasts and osteocytes promote osteoblast number and bone formation
Acknowledgments
The authors would like to thank I. B. Gubrij, P.E. Cazer, and J.J. Goellner for technical support and E. N. Olson for supplying the YAPf/f,TAZf/f mice. We also thank the staff of the UAMS Department of Laboratory Animal Medicine for animal care. This work was supported by the National Institutes of Health (AR049794 and GM125503-01 to C.A.O.), the Central Arkansas Veteran's Healthcare System (Merit Review 1I01BX000294 to C.A.O.), and UAMS (Bone and Joint Initiative to J.X.). Additional support was provided by UAMS Translational Research Institute (UL1 RR029884) and UAMS tobacco settlement funds.
Footnotes
Conflict of interest: C. O'Brien owns shares in Radius Health, Inc.
Author contributions: J.X. and C.A.O. designed the experiments. J.X. performed the experiments. J.X., M.A., and C.A.O. analyzed the results. J.X. and C.A.O. wrote the first draft of the manuscript. All authors edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Manolagas SC. Birth and death of bone cells: basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr Rev. 2000;21:115–37. doi: 10.1210/edrv.21.2.0395. [DOI] [PubMed] [Google Scholar]
- 2.Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2011;13:27–38. doi: 10.1038/nrm3254. [DOI] [PubMed] [Google Scholar]
- 3.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–50. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8:727–38. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–64. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 6.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–71. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 7.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 8.Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci U S A. 2013;110:13839–44. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poitelon Y, Lopez-Anido C, Catignas K, Berti C, Palmisano M, Williamson C, et al. YAP and TAZ control peripheral myelination and the expression of laminin receptors in Schwann cells. Nat Neurosci. 2016;19:879–87. doi: 10.1038/nn.4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reginensi A, Hoshi M, Boualia SK, Bouchard M, Jain S, McNeill H. Yap and Taz are required for Ret-dependent urinary tract morphogenesis. Development. 2015;142:2696–703. doi: 10.1242/dev.122044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piccolo S, Dupont S, Cordenonsi M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev. 2014;94:1287–312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 12.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–74. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azzolin L, Zanconato F, Bresolin S, Forcato M, Basso G, Bicciato S, et al. Role of TAZ as mediator of Wnt signaling. Cell. 2012;151:1443–56. doi: 10.1016/j.cell.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 14.Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell. 2010;18:579–91. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Seo E, Basu-Roy U, Gunaratne PH, Coarfa C, Lim DS, Basilico C, et al. SOX2 regulates YAP1 to maintain stemness and determine cell fate in the osteo-adipo lineage. Cell Rep. 2013;3:2075–87. doi: 10.1016/j.celrep.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–8. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 17.Hossain Z, Ali SM, Ko HL, Xu J, Ng CP, Guo K, et al. Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc Natl Acad Sci U S A. 2007;104:1631–6. doi: 10.1073/pnas.0605266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian Y, Kolb R, Hong JH, Carroll J, Li D, You J, et al. TAZ promotes PC2 degradation through a SCFbeta-Trcp E3 ligase complex. Mol Cell Biol. 2007;27:6383–95. doi: 10.1128/MCB.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, et al. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol. 2006;26:77–87. doi: 10.1128/MCB.26.1.77-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, et al. Regulation of insulinlike growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4:ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 22.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–44. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 23.Lu Y, Xie Y, Zhang S, Dusevich V, Bonewald LF, Feng JQ. DMP1-targeted Cre expression in odontoblasts and osteocytes. J Dent Res. 2007;86:320–5. doi: 10.1177/154405910708600404. [DOI] [PubMed] [Google Scholar]
- 24.O'Brien CA, Jilka RL, Fu Q, Stewart S, Weinstein RS, Manolagas SC. IL-6 is not required for parathyroid hormone stimulation of RANKL expression, osteoclast formation, and bone loss in mice. Am J Physiol Endocrinol Metab. 2005;289:E784–E793. doi: 10.1152/ajpendo.00029.2005. [DOI] [PubMed] [Google Scholar]
- 25.Xiong J, Piemontese M, Thostenson JD, Weinstein RS, Manolagas SC, O'Brien CA. Osteocyte-derived RANKL is a critical mediator of the increased bone resorption caused by dietary calcium deficiency. Bone. 2014;66:146–54. doi: 10.1016/j.bone.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong J, Piemontese M, Onal M, Campbell J, Goellner JJ, Dusevich V, et al. Osteocytes, not Osteoblasts or Lining Cells, are the Main Source of the RANKL Required for Osteoclast Formation in Remodeling Bone. PLoS One. 2015;10:e0138189. doi: 10.1371/journal.pone.0138189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, et al. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 1987;2:595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
- 28.Galli C, Fu Q, Wang W, Olsen BR, Manolagas SC, Jilka RL, et al. Commitment to the osteoblast lineage is not required for RANKL gene expression. J Biol Chem. 2009;284:12654–62. doi: 10.1074/jbc.M806628200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Fuerer C, Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One. 2010;5:e9370. doi: 10.1371/journal.pone.0009370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Gregorio GB, Yamamoto M, Ali AA, Abe E, Roberson P, Manolagas SC, et al. Attenuation of the self-renewal of transit-amplifying osteoblast progenitors in the murine bone marrow by 17 beta-estradiol. J Clin Invest. 2001;107:803–12. doi: 10.1172/JCI11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jilka RL, Weinstein RS, Bellido T, Parfitt AM, Manolagas SC. Osteoblast programmed cell death (apoptosis): modulation by growth factors and cytokines. J Bone Miner Res. 1998;13:793–802. doi: 10.1359/jbmr.1998.13.5.793. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Mishina Y, Liu F. Osterix-Cre transgene causes craniofacial bone development defect. Calcif Tissue Int. 2015;96:129–37. doi: 10.1007/s00223-014-9945-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang W, Olsen BR. Skeletal defects in Osterix-Cre transgenic mice. Transgenic Res. 2015;24:167–72. doi: 10.1007/s11248-014-9828-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davey RA, Clarke MV, Sastra S, Skinner JP, Chiang C, Anderson PH, et al. Decreased body weight in young Osterix-Cre transgenic mice results in delayed cortical bone expansion and accrual. Transgenic Res. 2012;21:885–93. doi: 10.1007/s11248-011-9581-z. [DOI] [PubMed] [Google Scholar]
- 36.Piemontese M, Onal M, Xiong J, Han L, Thostenson JD, Almeida M, et al. Low bone mass and changes in the osteocyte network in mice lacking autophagy in the osteoblast lineage. Sci Rep. 2016;6:24262. doi: 10.1038/srep24262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaidi SK, Sullivan AJ, Medina R, Ito Y, van Wijnen AJ, Stein JL, et al. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 2004;23:790–9. doi: 10.1038/sj.emboj.7600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong J, Onal M, Jilka RL, Weinstein RS, Manolagas SC, O'Brien CA. Matrix-embedded cells control osteoclast formation. Nat Med. 2011;17:1235–41. doi: 10.1038/nm.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–8. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 40.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–83. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 41.Tang Y, Feinberg T, Keller ET, Li XY, Weiss SJ. Snail/Slug binding interactions with YAP/TAZ control skeletal stem cell self-renewal and differentiation. Nat Cell Biol. 2016;18:917–29. doi: 10.1038/ncb3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsumoto Y, La RJ, Lim M, Adissu HA, Law N, Mao X, et al. Ubiquitin ligase RNF146 coordinates bone dynamics and energy metabolism. J Clin Invest. 2017;127:2612–25. doi: 10.1172/JCI92233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsumoto Y, La RJ, Kent OA, Wagner MJ, Narimatsu M, Levy AD, et al. Reciprocal stabilization of ABL and TAZ regulates osteoblastogenesis through transcription factor RUNX2. J Clin Invest. 2016;126:4482–96. doi: 10.1172/JCI87802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng Y, Wu A, Li P, Li G, Qin L, Song H, et al. Yap1 Regulates Multiple Steps of Chondrocyte Differentiation during Skeletal Development and Bone Repair. Cell Rep. 2016;14:2224–37. doi: 10.1016/j.celrep.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 45.Zaidi SK, Sullivan AJ, Medina R, Ito Y, van Wijnen AJ, Stein JL, et al. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 2004;23:790–9. doi: 10.1038/sj.emboj.7600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang KC, Yeh YT, Nguyen P, Limqueco E, Lopez J, Thorossian S, et al. Flow-dependent YAP/TAZ activities regulate endothelial phenotypes and atherosclerosis. Proc Natl Acad Sci U S A. 2016;113:11525–30. doi: 10.1073/pnas.1613121113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertero T, Oldham WM, Cottrill KA, Pisano S, Vanderpool RR, Yu Q, et al. Vascular stiffness mechanoactivates YAP/TAZ-dependent glutaminolysis to drive pulmonary hypertension. J Clin Invest. 2016;126:3313–35. doi: 10.1172/JCI86387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–59. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 49.Kegelman CD, Mason DE, Dawahare JH, Horan DJ, Vigil GD, Howard SS, et al. Skeletal cell YAP and TAZ combinatorially promote bone development. FASEB J. 2018:fj201700872R. doi: 10.1096/fj.201700872R. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


