Abstract
Objective
Infantile hemangiomas (IH) are the most common benign vascular neoplasms of infancy, characterized by a rapid growth phase followed by a spontaneous involution, or triggered by propranolol treatment by poorly understood mechanisms. LIN28/let-7 axis plays a central role in the regulation of stem cell self-renewal and tumorigenesis. However, the role of LIN28B/let-7 signaling in IH pathogenesis has not yet been elucidated.
Approach and Results
LIN28B is highly expressed in proliferative IH and is less expressed in involuted and in propranolol treated IH samples as measured by immunofluorescence staining and qRT-PCR. Small RNA sequencing analysis of IH samples revealed a decrease in microRNAs that target LIN28B, including let-7, and an increase in microRNAs in the mir-498(46) cluster. Overexpression of LIN28B in HEK293 cells induced the expression of miR-516b in the mir-498(46) cluster. Propranolol treatment of induced pluripotent stem cells (iPSCs), which express mir-498(46) endogenously, reduced the expression of both LIN28B and mir-498(46) and increased the expression of let-7. Furthermore, propranolol treatment reduced the proliferation of iPSCs and induced epithelial-to-mesenchymal transition.
Conclusions
This work uncovers the role of the LIN28B/let-7 switch in IH pathogenesis and provides a novel mechanism by which propranolol induces IH involution. Furthermore, it provides therapeutic implications for cancers in which the LIN28/let-7 pathway is imbalanced.
Keywords: Induced pluripotent stem cells, Differentiation, Epigenetics, Methylation, microRNA, Epithelial Mesenchymal Transition, C19MC, mir-498(46) cistron
Subject codes: Cell Signaling/Signal Transduction, Developmental Biology, Vascular Biology
Introduction
Infantile hemangiomas (IH) are highly vascularized benign tumors diagnosed in 3–10% of children before they are one year old1. IH lesions have a unique pattern of growth in which the initial phase of rapid proliferation is followed by slow spontaneous involution that leaves behind a fibro-fatty residuum2. During the proliferative phase, immature endothelial cells, positive for glucose transporter-1 (GLUT1), form aberrant blood vessels, rich in α-smooth muscle actin-positive pericytes and mast cells3. This endothelial GLUT1 expression distinguishes IH from other types of vascular tumors and vascular malformations4, 5. Stem cell reprograming factors OCT4, SOX2, NANOG and MYC are also highly expressed in IH6.
The chromosome 19 microRNA (miRNA) cluster (C19MC), referred to as miRNA cistron mir-498(46) or mir-498(46), was recently reported to be highly expressed in IH7. Mir-498(46) is unique to primates and is the largest known human miRNA gene cluster that spans over 100 kb and contains 46 miRNA genes flanked by Alu elements8. Expression of miRNA genes in this cluster is restricted to the placenta, embryonic stem cell (ESC) and certain tumors8–11.
While some studies have shown considerable similarity between IH and placental transcriptomes12, which suggests a placental origin for proliferating cells in IH13, other studies propose that IH originate from hemangioma stem cells14. In support of the latter notion, human CD133+ hemangioma stem cells (HemSCs) implanted subcutaneously into immunodeficient mice induce GLUT1-positive microvessels, which are gradually replaced by adipocytes, thereby reproducing the IH involution phase. Importantly, when grown in culture, GLUT1-positive/mir-498(46)-expressing endothelial cells, isolated from proliferative IH, lose the expression of mir-498(46) and GLUT1 and undergo an endothelial-mesenchymal transition (EndMT), a process similar to epithelial-mesenchymal transition (EMT)7, 15.
Serendipitously, it was discovered by Léauté-Labrèze that the non-selective beta-adrenergic receptor blocker, propranolol, triggers early involution of IH16. Consequently, propranolol has become the first line therapy for IH17. Previous work has shown that propranolol-treated IH tumors contain significantly lower levels of mir-498(46) than proliferative IH7. Moreover, levels of circulating mir-498(46) fall during the involution phase and correlate with the clinical response to propranolol treatment7, suggesting the potential involvement of mir-498(46) in the pathogenesis of IH.
The self-renewal capacity of ESCs is regulated in part by a set of interactive gene products, LIN28 and the let-7 family of miRNAs. Two human LIN28 paralogs, LIN28A and LIN28B, are highly expressed in ESCs. They inhibit the post-transcriptional maturation of let-7 miRNAs and influence mRNA translation18, 19. Conversely, let-7 negatively regulates the expression of LIN28 by interacting with the 3′ untranslated regions of both LIN28A and LIN28B mRNAs20. This interaction creates a double negative feedback loop, which is highly conserved. Ectopic expression of LIN28A or LIN28B alongside the reprogramming factors Oct4, Sox2, and Nanog21–23 has been successfully used to generate induced pluripotent stem cells (iPSCs) from fibroblasts. In addition, LIN28B overexpression has been successfully used to generate iPSCs from fibroblasts24.
EMT is the biological process through which normally polarized epithelial cells lose their apical-basal polarity and acquire a mesenchymal cell phenotype characterized by enhanced migration and invasiveness, resistance to apoptosis and increased deposition of extracellular matrix components24. This trans-differentiation is mediated by key transcription factors such as SNAIL, SLUG, TWIST1 and ZEB that regulate downstream epithelial markers, which among other effects, leads to decreased E-cadherin levels and increased expression of mesenchymal cell markers, such as N-cadherin and vimentin25. iPSC generation from fibroblasts requires suppression of EMT and activation of mesenchymal-epithelial transition (MET) signals26.
IH represents a unique model to study postnatal vasculogenesis and vessel regression. Despite the prevalence of these tumors, the complex pathogenesis of IH is poorly understood. Given the role of LIN28/let-7 axis in governing stem cell self-renewal and cell differentiation, we hypothesized that LIN28/let-7 signaling is dysregulated in IH. In this study, we demonstrate that LIN28B is highly expressed in proliferative IH samples and is downregulated both in involuted and propranolol-treated IH samples. Consistent with those observations, in vitro treatment of iPSCs with propranolol caused a decrease in LIN28B, mir-498(46) expression, decreased cell proliferation and increased the expression of let-7 family of miRNAs and EMT genes. Our results highlight the role of LIN28 and let-7 in the pathogenesis of IH and offers a new mechanism by which propranolol induces involution of IH.
Material and methods
The authors declare that all supporting data are available within the article and its online supplementary files.
Tissue specimens
Institutional review board approval for collection of resected human hemangiomas was obtained from Columbia University College of Physicians and Surgeons (IRB #AAAA9976). The clinical characteristics of the patients used for this study are shown in Supplementary Table SI. Term placental tissues from normotensive patients who were delivered by cesarean section were obtained from Tampa General Hospital/University of South Florida (IRB # 00015578). Written informed consent was obtained from all patients. De-identified foreskin was used as normal infant skin (NS) and obtained from Tampa General Hospital, Tampa Florida.
Cell Culture
The human iPSC line SCVI274 was a gift from Dr Joseph C. Wu at Stanford Cardiovascular Institute, Stanford University School of Medicine. iPSCs were cultured on matrigel coated 6-well plates and maintained in Essential 8 medium (A1517001, Life Technologies) and passaged every fourth day. HEK293 cells (Life Technologies) were maintained in DMEM supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich). Propranolol (Sigma-Aldrich, P0884) was reconstituted in DMSO and used at 50 μM concentration for all experiments. The HemSC line H42 was a gift from Dr. June Wu at the Department of Surgery, Columbia University College of Physicians and Surgeons. Briefly, freshly dissected IH tissues were digested in 0.2% collagenase A (Roche Diagnostics, Indianapolis, IN). Suspension of single cells were selected using anti-CD133–coated magnetic beads (Miltenyi Biotec) and cultured on fibronectin-coated (1 μg/cm2) plates with endothelial growth media-2 SingleQuot (EBM-2, CC-3156; Cambrex) supplemented with 20% FBS 27, 28. HemSCs were maintained in EBM-2 SingleQuot media supplemented with 20% FBS.
Transient transfection
For LIN28B over-expression experiments, 105 HEK293 cells per ml were seeded in 12-well plates (Greiner Bio). The following day, the cells were transfected with pcDNA3-FLAG-Lin28B, a gift from Narry Kim (Addgene plasmid # 51373) using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. The culture medium was changed 24 hours after transfection and total RNA was harvested at 72 hours.
Cell proliferation
iPSCs were grown in 6 well plates and split 1:10 at 3-day intervals. Cells were seeded at subconfluency on a matrigel-coated 96-well plate in Essential 8 media then treated with 50 μM propranolol or vehicle control for 72 hours. WST1 proliferation assays were performed as previously described29.
Cell viability
The viability of iPSCs after propranolol treatment was analyzed by staining of live and dead cells with calcein AM and Propidium iodide (PI), respectively, 72 hours after treatment. Cells were washed with PBS before the addition of 2 μM calcein AM and 4 μM PI solution. Fluorescence microscopy images were captured after 15 minutes.
RNA isolation, RT–PCR and quantitative PCR (qPCR)
Total RNA was isolated from human tissues and cultured cells using an RNeasy Mini Kit (Qiagen) and stored at −80°C in RNAse-free water. For RT–PCR analysis, 1 μg total RNA was reverse transcribed using random hexamer or oligodT primers and M-MuLV reverse transcriptase (New England Biolabs) according to the manufacturer’s specifications.
For transcription assessments, cDNA products were amplified for 25 PCR cycles using previously described P0, P1, P5, P8 and P9 primer sets30 and Q5 High Fidelity DNA polymerase (New England Biolabs). PCR products were run on 1% agarose gels and imaged with a BioDoc-It imaging system (UVP). All primers used in this study were obtained from Sigma Aldrich. To assess relative mRNA and miRNA expression levels, quantitative PCR (qRT-PCR) of cDNA products was performed using the following ThermoFisher TaqMan qRT-PCR probes: LIN28A (Hs00702808_s1), LIN28B (Hs01013729_m1), SERPINE1 (Hs00167155_m1), TWIST1 (Hs01675818_s1), GAPDH (Hs02786624_g1) miR-515-5p (001112), miR-516a-5p (002416), miR-516b (001150), miR-517a (002402), miR-518c (002401), miR-519d (002403), let-7a (000377), let-7c (000379), U18 (001204). Taqman probes were used according to manufacturer’s instructions with TaqMan Fast Advanced Master Mix and QuantStudio 3 instrument (Life Technologies) as previously described31. Data were analyzed by the ΔΔCt method: target Ct values and were normalized to GAPDH Ct values.
DNA isolation and HpaII sensitivity assay
DNA was isolated from cultured cells using the Blood and Tissue Kit (Qiagen) and stored at −20°C in RNAse-free water. 300ng of DNA was digested with 2 μl of 10,000 units/ml HpaII (NEB) restriction enzyme or water (mock) for 12 hours. qPCR was performed using 60 ng of DNA from mock or HpaII reactions with primers designed to amplify 340bp region containing 6 HpaII restriction sites in the C19MC CpG island (F: 5′-GCGCCGGCTGCACGTCCCTTAGGAG and R:5′-CCCGCTGCCTGGAAGTATCGCCACC) and SYBR Green (SYBR Green PCR Master mix, Bio-Rad) in a QuantStudio 3 instrument (Applied Biosystems). HpaII sensitivity was calculated using the formula [1 − 2Ct(mock) − Ct(HpaII)] × 100% as described32.
Immunofluorescence
Paraffin embedded infantile hemangioma and placenta sections (10 μm) were immunostained with primary antibodies against LIN28B 40 ug/ml (Abcam Cat# ab71415, RRID:AB_2135050) and GLUT1 3.4 ug/ml (Abcam Cat# ab40084, RRID:AB_2190927) overnight at 4°C. Sections were washed 3 times in PBS and incubated with 4 ug/ml secondary antibody Alexa Fluor 488 (Life Technologies Cat# a21202, RRID:AB_141607) for GLUT1 and alexa fluor cy3 (Life Technologies Cat# a10520, RRID:AB_2534029) for LIN28B for 30 minutes at room temperature. The sections were then washed with PBS and mounted with ProLong Diamond Antifade Mountant with DAPI, (Invitogen). Confocal images were obtained using an Olympus FV 1200 instrument. Immunofluorescence intensities were measured using ImageJ software as described33. Fluorescence intensities were normalized to background intensities of secondary antibody only controls.
Immunoblotting
Immunoblot analyses were performed as previously described34 using 24 ng/ml antibodies against GAPDH (Cell Signaling Technology Cat# 2118L RRID:AB_561053), 0.2 μg/μl antibodies against LIN28B (Abcam Cat# ab71415 RRID:AB_2135050) followed by IRDye 680 donkey anti-rabbit IGG secondary antibodies(0.2 μg/ml, LI-COR, 926-68073 RRID:AB_10954442). Immunoblots were imaged using the Odyssey Infrared Imaging System (LI-COR) and quantified using Image Studio software (LI-COR).
miRNA- and mRNA-sequencing
Two μg of total RNA were converted into a small-RNA cDNA library according to the previously published protocol35. Briefly, the RNA input for each sample was ligated to a 3′ adaptor barcoded sequence, pooled, size selected and gel purified, followed by 5′ adapter ligation and then subjected to size selection and gel purification. The cDNA library preparation was completed by second strand synthesis using SuperScript III, alkaline RNA hydrolysis, and PCR amplification for 10 cycles. mRNA libraries were prepared by utilizing the Illumina TruSeq Stranded mRNA LT protocol using 500 ng total RNA and NEB’s Protoscript II reverse transcriptase for the first-strand cDNA synthesis according to the manufacturer’s protocol. Individual RNAseq libraries were quality controlled on an Agilent TapeStation with a High Sensitivity D1000 ScreenTape. Indexed samples were quantified using the Qubit dsDNA HS assay and pooled at equimolar concentration (10 nM). The libraries were sequenced on an Illumina NextSeq 500 sequencer 75-bp paired-end in mid-output mode in the Genomics Core Facility of The Rockefeller University.
Bioinformatics analysis
The miRNA read annotation for sRNAseq experiments was performed as previously described36, 37 using an slightly updated, manually curated miRNA reference based on previous work 38. For nomenclature of single miRNAs and definition and nomenclature of miRNA cistrons (or “pre-cursor clusters”) see Figure S1 of Akat et al. 39. The RNAseq data was aligned to the Human Genome Build 38 using the STAR aligner40 (version 2.0.4j) allowing for two mismatches. Expression values (count matrices) were generated using featureCounts against gene definitions from Ensembl release 88 (GTF file) using fractional counting of multi-mapping reads.
Statistical analysis
sRNAseq and RNAseq data analyses were performed using the R statistical language. Differential analysis was performed using the Bioconductor package edgeR. All RT-PCR and immunoblotting data are reported as mean ± standard error of the mean (SEM). All data were tested for normality. Comparisons between two groups of normally distributed data were made by two-tailed Student’s t-tests with correction for unequal variance, while comparisons of data that was not normally distributed were done using Mann-Whitney U tests. Comparisons with more than two groups were subject to one-way ANOVA with Dunnett’s post hoc tests against controls. Statistical testing was performed with IBM SPSS 24. P < 0.05 was considered statistically significant or as specified in the relevant tables and figure legends.
Results
LIN28B is highly expressed in proliferative IH and is reduced upon involution
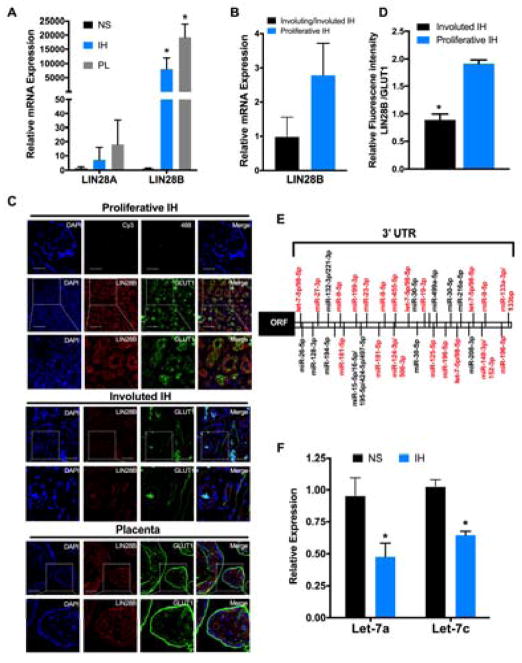
IH are derived from dysregulated stem cells41 and represent a unique tumor model characterized by a proliferative phase followed by spontaneous involution, regulated by yet unknown mechanisms. Given the key role of the LIN28/let-7 axis in governing stem cell self-renewal and cell differentiation, the expression levels of LIN28A and LIN28B were first assessed in IH samples by qRT-PCR and compared to normal infant skin (NS) and samples of normal human placentas at term. IH samples exhibited a 7- and 7500-fold increase (p < 0.05) in LIN28A and LIN28B, respectively, compared to NS (Figure 1A). The expression levels of LIN28A and LIN28B in IH were comparable to their expression levels in the placenta (Figure 1A).
Figure 1. LIN28B is highly expressed in proliferative IH.
(A and B) qRT-PCR analysis of LIN28A and LIN28B (A) or LIN28B (B) expression normalized to GAPDH. IH (n=4), NS (n=4), normal term placenta (PL, n=4), proliferative IH (n=2) and involuting/involuted IH (n=2). (C) Representative immunostaining for LIN28B and GLUT1 of proliferative IH (n=4), involuting/involuted IH (n=5) and term placenta (n=3) samples. Nuclei were counterstained with DAPI. No positive immunostaining was observed in the negative control sections (Alexa Fluor 488 or cy3). Scale bars: 50 μm; original magnification, ×60 and ×120 (insets). (D) Quantification of LIN28B/GLUT1 fluorescence signal ratio in proliferative and involuted IF samples. (E) Predicted miRNAs target sites in LIN28B 3′UTR (TargetScan Human Release 7.1). In red, downregulated miRNA in sRNAseq analysis of IH vs. NS. (F) qRT-PCR analysis of let-7a and let-7c expression in IH (n=4) and NS (n=4). Graphs represent means ± SEM. * p < 0.05 vs. NS (A and F), vs proliferative IH (D) by one-way ANOVA with Dunnett’s post hoc test (A) or two-tailed Student’s t-test (D and F).
Given the dramatic increase of LIN28B expression in IH, all subsequent experiments were focused on LIN28B. To determine if LIN28B is differentially expressed in proliferative IH compared to involuting/involuted IH, we assessed LIN28B expression by qRT-PCR and found that it was 2.7-fold higher (p < 0.33) in proliferative compared to involuting/involuted IH samples (Figure 1B).
To determine LIN28B cellular localization, dual immunofluoresence staining for LIN28B and the IH marker, GLUT1, was performed in paraffin embedded IH sections obtained from proliferative and involuted IH specimens. Term placenta samples were used as positive controls. In proliferative IH samples, LIN28B was highly expressed and co-localized with the GLUT1 positive endothelial cells as well as perivascular non-endothelial cells, whereas involuted IH samples showed weak staining for both LIN28B and GLUT1 (Figure 1C). As expected, in term placentas LIN28B and GLUT1 were co-localized in trophoblast and endothelium of chorionic villi (Figure 1C). Quantifications of LIN28B to GLUT1 immunofluorescence signal intensities showed ~2-fold increase in proliferative IH compared to involuted IH samples (Figure 1D). These data confirm the increased expression of LIN28B in proliferative IH and its localization in GLUT1-positive endothelial cells and perivascular non-endothelial cells that may display properties of facultative stem cells15.
The let-7 miRNA family and numerous other miRNAs negatively regulate LIN28B by binding to its 3′UTR (Figure 1E). To determine whether the increase in LIN28B in IH samples is associated with decreased expression of these miRNAs, small RNA sequencing (sRNAseq) analysis was performed on eight IH and five NS samples. Looking at the most abundant, or highly expressed, miRNAs (i.e. consuming the top 90% of sequencing reads) in IH compared to NS samples showed 32 significantly changed miRNA cistrons (FDR 0.25), of which 21 were upregulated in IH and 11 downregulated in IH compared to NS (Table 1). Of the 21 upregulated miRNA cistrons, mir-498(46) was most extensively upregulated with a 492-fold increase (Table 1). The downregulated cistrons include miRNAs that are predicted to bind to LIN28B 3′UTR, namely let-7a, let-7c, let-7f, let-7g, let-7i, miR-19b(2), miR-506(1), miR-196a/b, miR-455-5p(1), miR-9(3), miR-124(3), miR-27a(1), miR-27b(1), miR-23b(1), miR-125b(2), miR-199b-5p(1), miR-19a(1) and miR-148a(1), which were 2- to 45.6-fold downregulated (Supplementary Table SII). Importantly, let-7, miR-196 and miR-9 bind to multiple target sites on LIN28B 3′ UTR (Figure 1E). These data indicate that the upregulation of LIN28B in IH may be due, at least in part, to its decreased post-transcriptional regulation by miRNAs. Additionally, LIN28B is an established repressor of miRNA biogenesis42, among which let-7 is the most extensively studied43. Consequently, sRNAseq analysis of IH and NS revealed that let-7g(1), let-7c(1), and miR-200c(1) were 2.9-, 3.6- and 135-fold downregulated, respectively, in IH compared to NS (Supplementary Table SII). To determine whether additional members of the let-7 family were affected, yet did not pass the sRNAseq analysis filters, qRT-PCR analysis was performed for randomly selected members let-7a and let-7c. As expected, both were downregulated (p <0.05) in IH compared to NS (Figure 1F).
Table 1. Expression differences of highly expressed miRNA cistrons in IH vs. NS by sRNAseq.
Highly expressed cistrons were defined as being within the top 90% RNAseq reads in any of the 8 IH and 5 NS samples. Shown are cistrons with a false discovery rate of less than 25% in the differential analysis.
| Cistron (mir-) | Normalized frequency (%) | Fold change | P Value | FDR | |
|---|---|---|---|---|---|
|
| |||||
| Infantile hemangioma | Normal skin | ||||
| 498(46) | 8.888 | 0.018 | 492.7 | 3.99E-12 | 1.27E-09 |
| 195(2) | 2.312 | 0.543 | 4.3 | 3.65E-05 | 4.29E-04 |
| 378(1) | 3.760 | 0.899 | 4.2 | 1.25E-05 | 1.80E-04 |
| 140(1) | 1.079 | 0.279 | 3.9 | 1.76E-04 | 1.66E-03 |
| 30a(4) | 5.075 | 1.473 | 3.4 | 9.02E-05 | 9.22E-04 |
| 10b(1) | 2.428 | 0.724 | 3.4 | 5.04E-02 | 1.49E-01 |
| 15a(4) | 1.576 | 0.523 | 3.0 | 7.01E-03 | 3.27E-02 |
| 142(1) | 0.536 | 0.195 | 2.8 | 9.35E-03 | 4.11E-02 |
| 29a(4) | 1.902 | 0.698 | 2.7 | 3.17E-02 | 1.06E-01 |
| 181a-1(4) | 1.570 | 0.596 | 2.6 | 7.35E-04 | 5.98E-03 |
| 424(2) | 3.495 | 1.437 | 2.4 | 3.05E-03 | 1.90E-02 |
| 26a-1(2) | 9.431 | 3.934 | 2.4 | 1.94E-02 | 7.49E-02 |
| 22(1) | 3.401 | 1.420 | 2.4 | 1.30E-02 | 5.43E-02 |
| 144(2) | 3.972 | 1.679 | 2.4 | 6.92E-02 | 1.91E-01 |
| 143(2) | 18.461 | 9.717 | 1.9 | 8.68E-02 | 2.24E-01 |
| 21(1) | 4.601 | 2.758 | 1.7 | 3.72E-02 | 1.22E-01 |
| 26b(1) | 1.467 | 0.901 | 1.6 | 7.79E-02 | 2.06E-01 |
| 191(2) | 0.675 | 0.438 | 1.5 | 1.17E-02 | 4.99E-02 |
| 126(1) | 1.724 | 1.103 | 1.6 | 8.90E-02 | 2.27E-01 |
| 30b(2) | 0.970 | 0.640 | 1.5 | 6.42E-02 | 1.80E-01 |
| 25(3) | 0.988 | 0.706 | 1.4 | 8.19E-02 | 2.13E-01 |
| 135a-1(3) | 0.764 | 1.097 | −1.4 | 7.29E-02 | 1.96E-01 |
| 17(12) | 1.145 | 1.754 | −1.5 | 4.18E-02 | 1.32E-01 |
| 130a(1) | 0.299 | 0.501 | −1.7 | 3.72E-02 | 1.22E-01 |
| 199b(1) | 0.785 | 1.437 | −1.8 | 2.66E-02 | 9.59E-02 |
| 148a(1) | 0.993 | 4.427 | −4.5 | 1.39E-06 | 2.45E-05 |
| 221(2) | 0.226 | 1.130 | −5.0 | 2.08E-09 | 1.65E-07 |
| 210(1) | 0.025 | 0.376 | −14.9 | 1.40E-08 | 6.34E-07 |
| 141(2) | 0.049 | 1.947 | −40.8 | 6.17E-06 | 9.31E-05 |
| 200a(3) | 0.010 | 0.553 | −58.5 | 6.34E-08 | 1.78E-06 |
| 203(1) | 0.014 | 6.200 | −461.0 | 1.69E-10 | 7.56E-09 |
| 205(1) | 0.013 | 1.730 | −150.3 | 1.14E-07 | 2.59E-06 |
LIN28B activates mir-498(46) miRNAs independently of the upstream CpG-island
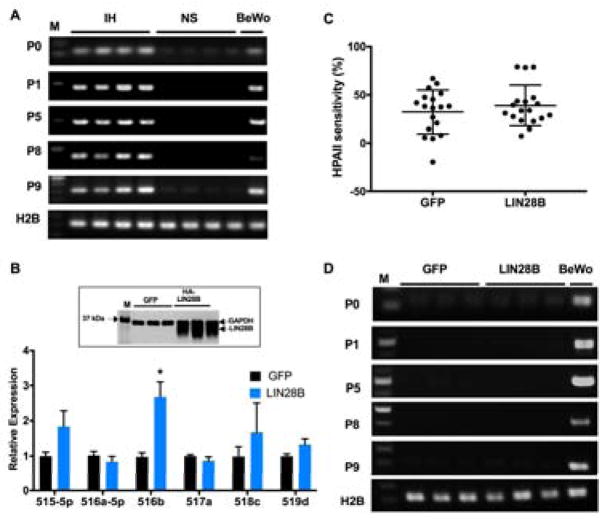
The expression of mir-498(46) is controlled by methylation of an upstream CpG-rich promoter region that includes a transcription start site located ~17 kb upstream of the first miRNA gene30. To test whether the increase in mir-498(46) expression found in IH samples is due to transcriptional activation of the upstream CpG-rich island, we performed qRT-PCR using primer sets designed to amplify regions downstream of the CpG-island, as previously described30. The trophoblast-derived choriocarcinoma BeWo cell line, which endogenously expresses mir-498(46), was used as a positive control. IH samples and BeWo cells displayed active transcription starting at the ~17kb upstream mir-498(46) CpG-related promoter region, whereas no transcripts were found in NS samples (Figure 2A). This indicates that the promoter region upstream of mir-498(46) is transcriptionally active in IH.
Figure 2. LIN28B increases the expression of miRNAs of the mir-498(46) cistron.
(A) RT–PCR analysis using P0, P1, P5, P8 and P9 primer sets reveal the presence of transcripts generated upstream of mir-498(46) in IH (n=4) but not in NS. BeWo cells were used as positive control and H2B as reference gene. (B–D) HEK293 cells were transfected with FLAG-LIN28B or GFP control vector for 72 hours followed by (B) qRT-PCR analysis of six randomly selected miRNAs of the mir-498(46) cistron normalized to U18. Insert, representative immunoblot of LIN28B and GAPDH; (C) HPAII sensitivity assay of the mir-498(46) cistron CpG-island promoter region and (D) RT–PCR analysis as described in A. Data represents means ± SEM of 3 independent experiments. * p < 0.05 vs. GFP transfected cells by Mann-Whitney U test.
Given that LIN28B expression is 7000-fold higher in IH than NS, we tested for a possible positive regulation of mir-498(46) miRNAs by LIN28B. To that end, HEK293 cells were transiently transfected with a LIN28B-coding plasmid, and the expression levels of LIN28B and of six randomly selected miRNAs of mir-498(46) miRNAs were measured after 72 h by immunoblotting and qRT-PCR, respectively. Interestingly, overexpression of LIN28B enhanced the expression of miR-516b by more than 2.6-fold (p < 0.05), but did not significantly alter the expression of miR-515-5p, miR-516a-5p, miR-517a, miR-518c and miR-519d (Figure 2B).
To test whether LIN28B affects the methylation of the ~17kb CpG- island upstream of mir-498(46) which activates its transcription, we performed an HpaII-sensitivity assay followed by qPCR. Overexpression of LIN28B did not induce a significant increase in HpaII sensitivity of the CpG island (Figure 2C) or induce transcriptional activation downstream of the CpG island (Figure 2D). These data indicate that LIN28B induces the expression of mir-498(46) independently of the ~17kb upstream promoter region.
Propranolol inhibits LIN28B in IH and iPSC
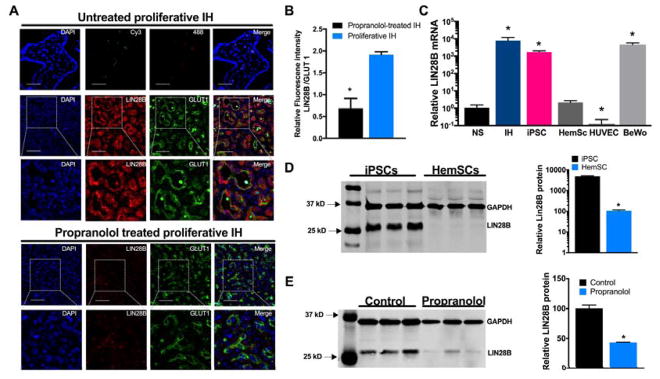
The standard of care propranolol treatment of patients with IH induces rapid IH involution. To determine whether propranolol affects the expression of LIN28B in IH, we performed dual immunofluorescence staining of LIN28B and the IH marker, GLUT1, in paraffin embedded IH sections obtained from propranolol-treated proliferative IH and compared them to untreated proliferative IH samples. Both LIN28B and GLUT1 were markedly decreased in propranolol-treated proliferative IH compared to untreated proliferative IH (Figure 3A). In addition, quantification of LIN28B to GLUT1 immunofluorescence signal intensities showed ~2-fold increase in proliferative IH compared to propranolol treated IH samples (Figure 3B).
Figure 3. Propranolol decreases the expression of LIN28B in IH treated patients and iPSCs.
(A) Representative immunostaining for LIN28B and GLUT1 of proliferative IH (n=4) and propranolol treated proliferating IH (n=4) samples. Nuclei were counterstained with DAPI. No positive staining was observed in the negative control sections (Alexa Fluor 488 or Cy3). Scale bars: 50 μm; original magnification, ×60 and ×120 (insets).(B) Quantification of LIN28B/GLUT1 fluorescence signal ratio in proliferative and propranolol-treated samples. (C) qRT-PCR analysis of LIN28B expression normalized to GAPDH in IH, iPSC, HemSC (line H42, from 3 independent passages), HUVEC and BeWo cells compared to NS. (D and E) Representative immunoblot for LIN28B and GAPDH expression in (D) iPSC and HemSC line H42, from 3 independent passages and (E) iPSCs treated with 50uM propranolol or vehicle control for 72 hours accompanied by densitometric quantification of LIN28B normalized to GAPDH. Data represent the mean ± SEM of at least 3 independent experiments. * p < 0.05 vs. proliferative IH (B), vs. NS (C), vs. iPSCs (D), or vs. vehicle control (E) by Student’s t-test (B, D and E) or by one-way ANOVA with Dunnett’s post hoc test (C).
To further investigate the effects of propranolol on the expression of LIN28B and let-7 in IH, we used the CD133+ HemSCs, which form hemangioma-like tumors when injected subcutaneously in immunodeficient mice14. First, we tested whether HemSCs express LIN28B and the miR-498(46) cluster in NS and IH. We also tested their expression in BeWo cells and iPSCs, which are known to express high levels of both. As negative controls, we used HUVECs that express neither LIN28B nor miR-498(46). Surprisingly, qRT-PCR revealed that compared to NS, LIN28B showed only a 2-fold increase in HemSCs, whereas in IH, iPSCs and BeWo cells LIN28B was >1000-fold (p< 0.05) higher (Figure 3C). As expected, negligible LIN28B expression was found in HUVECs (Figure 3C). Moreover, sRNAseq analysis revealed that miR-498(46) expression levels were 3.1-, 4.1-, 28- and 492.7- fold higher in HemSCs, HUVECs, iPSCs and IH, respectively, compared to NS (Supplementary Table SII). Lastly, the expression of LIN28B protein in HemSC and iPSCs was also assessed by Western blot analysis, which showed negligible levels of LIN28B in HemSCs (Figure 3D). Based on these findings, we proceeded to test the effect of propranolol in iPSCs, that express LIN28B, miR498(46) and beta-adrenergic receptors. After propranolol treatment for 72 h, iPSCs showed a 2.3-fold decrease (p<0.05) in LIN28B protein levels (Figure 3E), but no significant differences were found at the mRNA levels (data not shown).
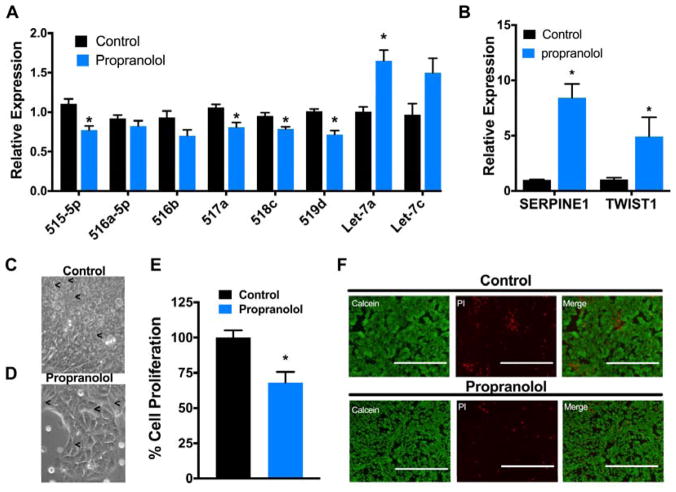
Propranolol induces mir-98(13) and suppresses mir-498(46) in iPSCs
To further investigate the effects of propranolol on the expression of mir-498(46) and let-7, iPSCs were treated with propranolol for 72 h and sRNAseq analysis was performed. Compared to vehicle control, propranolol treated-iPSCs showed a 3.2-fold increase in the expression of cistron mir-98(13), which encodes 9 of 12 let-7 family miRNA genes, and a 1.6-fold downregulation of mir-498(46) (Table 2). The results were confirmed by qRT-PCR, which revealed significant reduction of mir-498(46) members miR-515-5p, miR-517a, miR-518C and miR-519d and >1. 65-fold increase in let-7a expression in iPSCs treated with propranolol for 72 hr (Figure 4A).
Table 2. Propranolol treatment decreases miR-498(46) expression and increases let-7 miRNA expression in iPSCs.
Shown are all cistrons with and FDR < 25%. In bold mir-498(46) and mir-98(13).
| Cistron (mir-) | Normalized frequency (%) | Fold change | P Value | FDR | |
|---|---|---|---|---|---|
|
| |||||
| Propranolol | Control | ||||
| 1226(1) | 0 | 0.0029 | −579.2 | 0.0004 | 0.0994 |
| 33b(1) | 0.002 | 0.0087 | −4.3 | 0.0094 | 0.2276 |
| 488(1) | 0.0022 | 0.0082 | −3.8 | 0.0024 | 0.1167 |
| 498(46) | 0.4075 | 0.6333 | −1.6 | 0.0021 | 0.1167 |
| 21(1) | 10.5844 | 6.864 | 1.5 | 0.002 | 0.1167 |
| 188(8) | 0.1459 | 0.0842 | 1.7 | 0.0063 | 0.2276 |
| 98(13) | 0.2313 | 0.0836 | 2.8 | 0.0074 | 0.2276 |
| 10b(1) | 0.0176 | 0.0024 | 7.5 | 0.0086 | 0.2276 |
| 10a(1) | 0.0168 | 0.0001 | 120.8 | 0.0018 | 0.1167 |
| 3155(1) | 0.0014 | 0 | 284.3 | 0.0086 | 0.2276 |
Figure 4. Propranolol decreases the expression of mir-498(46) cistron and induces let-7 and EMT genes in iPSCs.
iPSCs were treated with 50uM propranolol or vehicle control for 72 hours. (A and B) qRT-PCR analysis of indicated miRNAs or genes normalized to U18 or GAPDH, respectively. (C and D) Representative 40x bright field images of control and propranolol-treated iPSCs. (E and F) Proliferation assay and representative fluorescence microscopy images of Calcein and propidium iodide (PI) staining for life and dead cells respectively (Scale bars: 200 μm; 20x magnification). Data represent the mean ± SEM of at least 3 independent experiments. * p < 0.05 vs. vehicle control by Student’s t-test (A, B and E).
Propranolol induces EMT markers in iPSC
Propranolol has been shown to induce early involution of IH, during which immature vascular endothelial cells undergo EndMT, transitioning to a mesenchymal phenotype with subsequent differentiation into adipocytes15. To test whether propranolol induces the expression of mesenchymal markers, we performed mRNA sequencing (RNAseq) on the same propranolol and vehicle treated-iPSC samples that were used for sRNAseq. Gene set enrichment analysis performed on RNAseq data showed an enrichment of genes defining EMT (FDR 0.00046) in propranolol treated iPS cells compared to control (Supplementary Table S III). Gene set analysis also showed upregulation of the pathways for adipogenesis and TGF-β signaling (FDR 0.09 and 0.06 respectively, Supplementary Table SIII). Among the most highly upregulated genes, two mesenchymal cell markers, SERPINE1 and TWIST1, showed a 12- and 2.9-fold increase (FDR 1.06 × 10−6 and 0.004), respectively (Supplementary Table SIII). Upregulation of SERPINE1 and TWIST1 was further confirmed by qRT-PCR, which showed 8.4- and 5-fold increased expression (p<0.05), respectively (Figure 4B). Furthermore, gene set analysis of the RNAseq data also showed downregulation of E2F targets, G2M checkpoint and mitotic spindle pathways (FDR<0.25, Supplementary Table SIII). We also examined propranolol-treated iPSCs for EMT associated morphological changes and observed that propranolol treated iPSCs exhibited a more elongated cellular shape compared to control (Figure 4C and D). Next, we tested the effect of propranolol on iPSC proliferation using the WST1 proliferation assay. Propranolol treatment induced a 33% decrease in cell proliferation without affecting cell viability (Figure 4E and F). Furthermore, gene set enrichment analysis also showed that E2F target genes and genes involved in G2/M cell cycle progression are downregulated by propranolol treatment in iPSCs (FDR 0.006 and 0.017 respectively, Supplementary Table III) indicating that propranolol strongly inhibits proliferation.
Discussion
IH is a unique tumor model, characterized by a rapid proliferative phase followed by a spontaneous or propranolol-induced involution phase. The mechanisms that trigger spontaneous involution and the action of propranolol are still under investigation. The present study shows that the reprogramming factor LIN28B is highly expressed in the proliferative IH phase but is significantly decreased in involuted IH and in IH tissues from propranolol-treated patients. The high LIN28B expression in proliferative IH correlates with the expression of the ESC-enriched mir-498(46)44–46 and is inversely correlated with the expression of let-7 miRNAs. Treatment of iPSCs with propranolol reduced the expression of LIN28B and mir-498(46) and induced the expression of let-7 family of miRNAs and EMT genes. Moreover, propranolol treatment reduced iPSC cell proliferation.
The increase in LIN28B and reduction in let-7 reported in this study highlight the role of stem cells in the pathology of IH. Although HemSCs, which are 0.2% of the proliferative hemangioma cell population, are believed to be the cellular origin of IH, our data show that these cells do not express LIN28B and miR-498(46) in tissue culture 14, 15. However, our in vivo data show that LIN28B is localized in GLUT1 positive endothelial cells that were previously characterized as stem cell-like15. These cells formed colonies that could be induced to re-differentiate into endothelial cells, pericytes/smooth muscle cells or adipocytes. Interestingly, when these GLUT1 positive endothelial cells were cultured for 3 weeks they converted to a mesenchymal phenotype and lost GLUT1 and mir-498(46) expression 7, 15. It would be of interest to test whether freshly isolated HemSCs express LIN28B and mir-498(46) or implantation of HemSC into immunodeficient mice restores LIN28B and mir-498(46) expression. Nevertheless, increased expression of stem cell transcription factors OCT4, SOX2, NANOG and MYC have been reported in IH 6. In addition, our in vivo data also show that in proliferative IH LIN28 was not limited to GLUT1 positive cells but was also expressed in perivascular non-endothelial cells, which are essential for the maintenance of IH vessels stability47.
The increase in LIN28B in IH reported here may be due to transcriptional and/or post transcriptional activation. The reprogramming transcription factors such as MYC has been shown to transactivate LIN28B48, 49. Although little is known about the transcriptional activation of LIN28B, posttranscriptional regulation by miRNAs such as let-7 and other miRNAs has been extensively studied23, 50. Furthermore, LIN28B was reported to bind its own mRNA, increasing its stability and protein abundance51. Thus, our sRNAseq results are in line with previous reports and highlight the central role of the LIN28B/let-7 switch in governing stem cell self-renewal in the proliferative phase of IH.
We also showed that the ESC-enriched miRNA cluster mir-498(46) is highly expressed in IH. This cluster of miRNAs is normally imprinted, with only the paternally inherited allele expressed in the placenta30. The overexpression of mir-498(46) seen in IH could be regulated by methylation of the upstream CpG island or by chromosomal rearrangements such as amplifications of the corresponding chromosome 19 region, as found in embryonal tumor with multilayered rosettes (ETMRs) 16, primitive neuroectodermal tumors (PNETs) and in thyroid adenomas 11, 52, 53. Given the ability of IH to undergo spontaneous involution, the elevated expression of mir-498(46) in proliferative IH is unlikely to be due to DNA amplifications or translocations, but rather, due to epigenetic modification. We therefore proceeded to investigate the previously identified promoter region and transcription start site that overlap an annotated CpG island located ~17 kb upstream of the first miRNA gene of mir-498(46). Although our findings show that, unlike NS, IH samples exhibited active transcription starting from the upstream CpG island, similar to that seen in the placenta30, they do not preclude the existence of additional active promoter regions within mir-498(46). In fact, mir-498(46) carries numerous CpG dinucleotide islands extensively interspersed with Alu-rich repeats, which account for over 50% of the 100-kb sequence. Numerous studies showed that Alu methylation is highly dynamic and Alu-rich regions can function as independent promoters for RNA polymerase II and RNA polymerase III in both mesenchymal stem cells and cancer stem cells44, 54, 55.
Although LIN28 is a well-established inhibitor of miRNA maturation18, 42, 51, 56, the present study revealed that overexpression of LIN28B increased the expression of miR-516b of mir-498(46). This is especially noteworthy given that LIN28B can be used along with other reprogramming factors to generate pluripotent stem cells enriched with mir-498(46). In fact, a recent study showed that LIN28A activates gene expression by binding directly to a consensus DNA sequence at promoter regions and recruiting the CpG demethylase TET157. Previous study showed that both mesenchymal stem and cancer stem cells exhibit disperse expression patterns of miRNAs of the mir-498(46) cluster rather than a bloc expression regulated by the upstream promoter 44. Accordingly, our data show that overexpression of LIN28B did not affect the methylation of the upstream CpG-island or increase the transcription in that region indicating the existence of downstream promoter regions. LIN28B may bind to the Alu repeats within mir-498(46), which may function as independent RNA polymerase II promoters and activate transcription by recruiting TET1. In fact, analyses of previously published PAR-CLIP data show that LIN28B binds numerous Alu repeats located in the mir-498(46)51. This unexpected result points to a potential binding of LIN28B to the CpG-rich Alu repeats to activate transcription. Moreover, LIN28 is a potent RNA-binding protein that regulates splicing factor abundance58, and therefore may induce processing of the transcript of mir-498(46) cistron.
In the last decade, propranolol has become the preferred treatment for morbid proliferating IH. To date, more than 500 published articles describe the observed propranolol-induced involution of IH and the various hypotheses regarding its mechanisms of action. Here we propose a new model through which propranolol triggers rapid IH involution. We demonstrate that propranolol induces a shift in LIN28B/let-7 balance to favor cell differentiation and senescence. This model is based on the data presented here, which show that propranolol reduced the expression of LIN28B and increased the expression of let-7 in IH samples in vivo and in iPSCs in vitro. Furthermore, we show that propranolol reduces the proliferation of iPSCs and initiates the conversion to mesenchymal phenotype as evidenced by the increase in SERPINE1 and TWIST1 expression, while reducing the expression of miRNAs of the mir-498(46) cistron. RNAseq and gene set analyses confirmed the downregulation in E2F targets, G2M checkpoint and mitotic spindle pathways and showed upregulation in EMT, adipogenesis, and TGF-β signaling pathways. To our knowledge, this is the first report to show that propranolol reduces the proliferation of iPSCs and induces EMT and adipogenesis. These profound effects of propranolol may be due in part to the reduction in LIN28B that binds and regulates mRNAs of cell cycle regulators51 and/or as a consequence of the increase in let-7 miRNAs, which regulate Rb1/E2F genes59. The induction of EMT genes by propranolol is in agreement with a previous study that showed that activation of adenylyl cyclase and elevation in cAMP and EPAC signaling replaced the need for OCT4 for iPSC generation. This cAMP-dependent reprograming increased cellular division rate and induced genes involved in MET60. Therefore, by reducing cAMP levels, propranolol may be inducing EMT and thus accelerating IH involution. Of note, propranolol has been shown to induce adipogenesis in hemangioma stem cells61, 62. Lastly, the reduction in miRNAs of the mir-498(46) cistron in propranolol treated iPSCs reported here is in agreement with previous work that showed that propranolol treated IH tissues contained significantly lower levels of mir-498(46) miRNAs than did proliferative, untreated IH7.
The LIN28/let-7 axis is implicated not only in pluripotency but also in tumorigenesis, especially in cancer stem cells, which are resistant to chemotherapies and promote metastasis50, 63. In fact, numerous pre-clinical and clinical studies provide evidence that propranolol increases chemosensitivity and reduces the metastatic rates in multiple cancer types64. The present work highlights the role of the LIN28B/let-7 switch in IH pathogenesis and propranolol induced IH involution. This study may also have therapeutic implications for IH and for cancers in which the LIN28/let-7 pathway is imbalanced.
Supplementary Material
Highlights.
LIN28B and miR-498(46) miRNA cluster are highly expressed in proliferative infantile hemangioma.
Propranolol significantly decreases LIN28B expression in infantile hemangioma patients.
Propranolol decreased LIN28B and increase let-7 expression in iPSCs.
Propranolol reduced iPSC proliferation and induced epithelial to mesenchymal transition
Propranolol decreases the expression of the miR-498(46) miRNA cluster in iPSCs.
Acknowledgments
We thank Dr. B. Jake Cha at the Lisa Muma Weitz Laboratory for Advanced Microscopy and Cell Imaging (Morsani College of Medicine, University of South Florida) for help with image analyses.
Sources of Funding
JC is supported by the American Heart Association 15PRE25850019 and HTJ is supported by the NIH, National Heart, Lung, and Blood Institute R00HL109133 and R01HL128411.
Non-standard Abbreviations and Acronyms
- C19MC
Chromosome 19 miRNA cluster
- EMT
Epithelial to mesenchymal transition
- EndMT
Endothelial to mesenchymal transition
- ESC
Embryonic stem cells
- HemSC
Hemangioma stem cell
- IH
Infantile hemangioma
- iPSC
Induced Pluripotent Stem Cells
- MET
Mesenchymal to epithelial transition
- NS
Normal skin
Footnotes
Disclosures
The authors have declared that no conflict of interests exist.
Author contributions
EFM and JV conducted all experimentation and analyzed data. KMA and TT performed all RNAseq experiments and their bioinformatics analysis. JL conducted bioinformatics analysis. JC conducted the immunostaining. AM conducted methylation experiments. HTJ conceived and directed the study and analyzed the data. JW and JCMT provided the specimens and technical support; EFM, KMA and HTJ wrote the manuscript.
References
- 1.Smith CJ, Friedlander SF, Guma M, Kavanaugh A, Chambers CD. Infantile hemangiomas: An updated review on risk factors, pathogenesis, and treatment. Birth Defects Res. 2017;109:809–815. doi: 10.1002/bdr2.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darrow DH, Greene AK, Mancini AJ, Nopper AJ, Neck S Section On Plastic S, Section On Dermatology SOO-H. Diagnosis and management of infantile hemangioma: Executive summary. Pediatrics. 2015;136:786–791. doi: 10.1542/peds.2015-2482. [DOI] [PubMed] [Google Scholar]
- 3.Greenberger S, Bischoff J. Pathogenesis of infantile haemangioma. Br J Dermatol. 2013;169:12–19. doi: 10.1111/bjd.12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.North PE, Waner M, Mizeracki A, Mihm MC., Jr Glut1: A newly discovered immunohistochemical marker for juvenile hemangiomas. Hum Pathol. 2000;31:11–22. doi: 10.1016/s0046-8177(00)80192-6. [DOI] [PubMed] [Google Scholar]
- 5.Mulliken JB, Glowacki J. Hemangiomas and vascular malformations in infants and children: A classification based on endothelial characteristics. Plast Reconstr Surg. 1982;69:412–422. doi: 10.1097/00006534-198203000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Amaya CN, Bryan BA. Enrichment of the embryonic stem cell reprogramming factors oct4, nanog, myc, and sox2 in benign and malignant vascular tumors. BMC Clin Pathol. 2015;15:18. doi: 10.1186/s12907-015-0018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strub GM, Kirsh AL, Whipple ME, Kuo WP, Keller RB, Kapur RP, Majesky MW, Perkins JA. Endothelial and circulating c19mc micrornas are biomarkers of infantile hemangioma. JCI Insight. 2016;1:e88856. doi: 10.1172/jci.insight.88856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human micrornas. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 9.Kleinman CL, Gerges N, Papillon-Cavanagh S, et al. Fusion of ttyh1 with the c19mc microrna cluster drives expression of a brain-specific dnmt3b isoform in the embryonal brain tumor etmr. Nat Genet. 2014;46:39–44. doi: 10.1038/ng.2849. [DOI] [PubMed] [Google Scholar]
- 10.Morales-Prieto DM, Ospina-Prieto S, Chaiwangyen W, Schoenleben M, Markert UR. Pregnancy-associated mirna-clusters. J Reprod Immunol. 2013;97:51–61. doi: 10.1016/j.jri.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Li M, Lee KF, Lu Y, Clarke I, et al. Frequent amplification of a chr19q13.41 microrna polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell. 2009;16:533–546. doi: 10.1016/j.ccr.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnes CM, Huang S, Kaipainen A, Sanoudou D, Chen EJ, Eichler GS, Guo Y, Yu Y, Ingber DE, Mulliken JB, Beggs AH, Folkman J, Fishman SJ. Evidence by molecular profiling for a placental origin of infantile hemangioma. Proc Natl Acad Sci U S A. 2005;102:19097–19102. doi: 10.1073/pnas.0509579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.North PE, Waner M, Mizeracki A, Mrak RE, Nicholas R, Kincannon J, Suen JY, Mihm MC., Jr A unique microvascular phenotype shared by juvenile hemangiomas and human placenta. Arch Dermatol. 2001;137:559–570. [PubMed] [Google Scholar]
- 14.Khan ZA, Boscolo E, Picard A, Psutka S, Melero-Martin JM, Bartch TC, Mulliken JB, Bischoff J. Multipotential stem cells recapitulate human infantile hemangioma in immunodeficient mice. J Clin Invest. 2008;118:2592–2599. doi: 10.1172/JCI33493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang L, Nakayama H, Klagsbrun M, Mulliken JB, Bischoff J. Glucose transporter 1-positive endothelial cells in infantile hemangioma exhibit features of facultative stem cells. Stem Cells. 2015;33:133–145. doi: 10.1002/stem.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leaute-Labreze C, Dumas de la Roque E, Hubiche T, Boralevi F, Thambo JB, Taieb A. Propranolol for severe hemangiomas of infancy. N Engl J Med. 2008;358:2649–2651. doi: 10.1056/NEJMc0708819. [DOI] [PubMed] [Google Scholar]
- 17.Leaute-Labreze C, Hoeger P, Mazereeuw-Hautier J, et al. A randomized, controlled trial of oral propranolol in infantile hemangioma. N Engl J Med. 2015;372:735–746. doi: 10.1056/NEJMoa1404710. [DOI] [PubMed] [Google Scholar]
- 18.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor microrna. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microrna processing by lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 rna regulates developmental timing in caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 22.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Ratanasirintrawoot S, Chandrasekaran S, et al. Lin28 regulates stem cell metabolism and conversion to primed pluripotency. Cell Stem Cell. 2016;19:66–80. doi: 10.1016/j.stem.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 24.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Science Signaling. 2014:7. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li RH, Liang JL, Ni S, Zhou T, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 27.Munabi NC, England RW, Edwards AK, Kitajewski AA, Tan QK, Weinstein A, Kung JE, Wilcox M, Kitajewski JK, Shawber CJ, Wu JK. Propranolol targets hemangioma stem cells via camp and mitogen-activated protein kinase regulation. Stem Cells Transl Med. 2016;5:45–55. doi: 10.5966/sctm.2015-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan ZA, Melero-Martin JM, Wu X, Paruchuri S, Boscolo E, Mulliken JB, Bischoff J. Endothelial progenitor cells from infantile hemangioma and umbilical cord blood display unique cellular responses to endostatin. Blood. 2006;108:915–921. doi: 10.1182/blood-2006-03-006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santulli G, Wronska A, Uryu K, Diacovo TG, Gao M, Marx SO, Kitajewski J, Chilton JM, Akat KM, Tuschl T, Marks AR, Totary-Jain H. A selective microrna-based strategy inhibits restenosis while preserving endothelial function. J Clin Invest. 2014;124:4102–4114. doi: 10.1172/JCI76069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefevre A, Coullin P, Moore GE, Cavaille J. The primate-specific microrna gene cluster (c19mc) is imprinted in the placenta. Hum Mol Genet. 2010;19:3566–3582. doi: 10.1093/hmg/ddq272. [DOI] [PubMed] [Google Scholar]
- 31.Totary-Jain H, Sanoudou D, Ben-Dov IZ, Dautriche CN, Guarnieri P, Marx SO, Tuschl T, Marks AR. Reprogramming of the microrna transcriptome mediates resistance to rapamycin. J Biol Chem. 2013;288:6034–6044. doi: 10.1074/jbc.M112.416446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by tet1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen EC. Quantitative analysis of histological staining and fluorescence using imagej. Anatomical Record-Advances in Integrative Anatomy and Evolutionary Biology. 2013;296:378–381. doi: 10.1002/ar.22641. [DOI] [PubMed] [Google Scholar]
- 34.Totary-Jain H, Sanoudou D, Dautriche CN, Schneller H, Zambrana L, Marks AR. Rapamycin resistance is linked to defective regulation of skp2. Cancer Res. 2012;72:1836–1843. doi: 10.1158/0008-5472.CAN-11-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hafner M, Renwick N, Farazi TA, Mihailovic A, Pena JT, Tuschl T. Barcoded cdna library preparation for small rna profiling by next-generation sequencing. Methods. 2012;58:164–170. doi: 10.1016/j.ymeth.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farazi TA, Brown M, Morozov P, Ten Hoeve JJ, Ben-Dov IZ, Hovestadt V, Hafner M, Renwick N, Mihailovic A, Wessels LF, Tuschl T. Bioinformatic analysis of barcoded cdna libraries for small rna profiling by next-generation sequencing. Methods. 2012;58:171–187. doi: 10.1016/j.ymeth.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown M, Suryawanshi H, Hafner M, Farazi TA, Tuschl T. Mammalian mirna curation through next-generation sequencing. Front Genet. 2013;4:145. doi: 10.3389/fgene.2013.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farazi TA, Horlings HM, Ten Hoeve JJ, et al. Microrna sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71:4443–4453. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akat KM, Moore-McGriff D, Morozova P, Browna M, Gogakos T, Da Rosa JC, Mihailovic A, Sauer M, Ji RP, Ramarathnam A, Totary-Jain H, Williams Z, Tuschl T, Schulze PC. Comparative rna-sequencing analysis of myocardial and circulating small rnas in human heart failure and their utility as biomarkers. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11151–11156. doi: 10.1073/pnas.1401724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. Star: Ultrafast universal rna-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harbi S, Wang R, Gregory M, Hanson N, Kobylarz K, Ryan K, Deng Y, Lopez P, Chiriboga L, Mignatti P. Infantile hemangioma originates from a dysregulated but not fully transformed multipotent stem cell. Sci Rep. 2016;6:35811. doi: 10.1038/srep35811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. Tut4 in concert with lin28 suppresses microrna biogenesis through pre-microrna uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen PN, Huang CJ, Sugii S, Cheong SK, Choo KB. Selective activation of mirnas of the primate-specific chromosome 19 mirna cluster (c19mc) in cancer and stem cells and possible contribution to regulation of apoptosis. J Biomed Sci. 2017;24:20. doi: 10.1186/s12929-017-0326-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laurent LC, Chen J, Ulitsky I, Mueller FJ, Lu C, Shamir R, Fan JB, Loring JF. Comprehensive microrna profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem cells. 2008;26:1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- 46.Bar M, Wyman SK, Fritz BR, et al. Microrna discovery and profiling in human embryonic stem cells by deep sequencing of small rna libraries. Stem cells. 2008;26:2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edwards AK, Glithero K, Grzesik P, Kitajewski AA, Munabi NC, Hardy K, Tan QK, Schonning M, Kangsamaksin T, Kitajewski JK, Shawber CJ, Wu JK. Notch3 regulates stem-to-mural cell differentiation in infantile hemangioma. JCI Insight. 2017:2. doi: 10.1172/jci.insight.93764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cotterman R, Knoepfler PS. N-myc regulates expression of pluripotency genes in neuroblastoma including lif, klf2, klf4, and lin28b. PLoS One. 2009;4:e5799. doi: 10.1371/journal.pone.0005799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beckers A, Van Peer G, Carter DR, et al. Mycn-driven regulatory mechanisms controlling lin28b in neuroblastoma. Cancer Lett. 2015;366:123–132. doi: 10.1016/j.canlet.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shyh-Chang N, Daley GQ. Lin28: Primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12:395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hafner M, Max KE, Bandaru P, Morozov P, Gerstberger S, Brown M, Molina H, Tuschl T. Identification of mrnas bound and regulated by human lin28 proteins and molecular requirements for rna recognition. RNA. 2013;19:613–626. doi: 10.1261/rna.036491.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 53.Rippe V, Dittberner L, Lorenz VN, Drieschner N, Nimzyk R, Sendt W, Junker K, Belge G, Bullerdiek J. The two stem cell microrna gene clusters c19mc and mir-371-3 are activated by specific chromosomal rearrangements in a subgroup of thyroid adenomas. PLoS One. 2010;5:e9485. doi: 10.1371/journal.pone.0009485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borchert GM, Lanier W, Davidson BL. Rna polymerase iii transcribes human micrornas. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 55.Saito Y, Suzuki H, Tsugawa H, Nakagawa I, Matsuzaki J, Kanai Y, Hibi T. Chromatin remodeling at alu repeats by epigenetic treatment activates silenced microrna-512–5p with downregulation of mcl-1 in human gastric cancer cells. Oncogene. 2009;28:2738–2744. doi: 10.1038/onc.2009.140. [DOI] [PubMed] [Google Scholar]
- 56.Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28a and lin28b inhibit let-7 microrna biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng Y, Yao B, Shin J, et al. Lin28a binds active promoters and recruits tet1 to regulate gene expression. Mol Cell. 2016;61:153–160. doi: 10.1016/j.molcel.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilbert ML, Huelga SC, Kapeli K, et al. Lin28 binds messenger rnas at ggaga motifs and regulates splicing factor abundance. Mol Cell. 2012;48:195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. Senescence is an endogenous trigger for microrna-directed transcriptional gene silencing in human cells. Nat Cell Biol. 2012;14:266–275. doi: 10.1038/ncb2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fritz AL, Adil MM, Mao SR, Schaffer DV. Camp and epac signaling functionally replace oct4 during induced pluripotent stem cell reprogramming. Mol Ther. 2015;23:952–963. doi: 10.1038/mt.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.England RW, Hardy KL, Kitajewski AM, Wong A, Kitajewski JK, Shawber CJ, Wu JK. Propranolol promotes accelerated and dysregulated adipogenesis in hemangioma stem cells. Ann Plast Surg. 2014;73(Suppl 1):S119–124. doi: 10.1097/SAP.0000000000000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma X, Zhao T, Ouyang T, Xin S, Ma Y, Chang M. Propranolol enhanced adipogenesis instead of induction of apoptosis of hemangiomas stem cells. Int J Clin Exp Pathol. 2014;7:3809–3817. [PMC free article] [PubMed] [Google Scholar]
- 63.Balzeau J, Menezes MR, Cao S, Hagan JP. The lin28/let-7 pathway in cancer. Front Genet. 2017;8:31. doi: 10.3389/fgene.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pantziarka P, Bouche G, Sukhatme V, Meheus L, Rooman I, Sukhatme VP. Repurposing drugs in oncology (redo)-propranolol as an anti-cancer agent. Ecancermedicalscience. 2016;10:680. doi: 10.3332/ecancer.2016.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.