Abstract
ADY1 is identified in a genetic screen for genes on chromosome VIII of Saccharomyces cerevisiae that are required for sporulation. ADY1 is not required for meiotic recombination or meiotic chromosome segregation, but it is required for the formation of four spores inside an ascus. In the absence of ADY1, prospore formation is restricted to mainly one or two spindle poles per cell. Moreover, the two spores in the dyads of the ady1 mutant are predominantly nonsisters, suggesting that the proficiency to form prospores is not randomly distributed to the four spindle poles in the ady1 mutant. Interestingly, the meiosis-specific spindle pole body component Mpc54p, which is known to be required for prospore membrane formation, is localized predominantly to only one or two spindle poles per cell in the ady1 mutant. A partially functional Myc-Pfs1p is localized to the nucleus of mononucleate meiotic cells but not to the spindle pole body or prospore membrane. These results suggest that Pfs1p is specifically required for prospore formation at selected spindle poles, most likely by ensuring the functionality of all four spindle pole bodies of a cell during meiosis II.
INTRODUCTION
In Saccharomyces cerevisiae, cells undergo sporulation to generate haploid gamete cells. In wild type, most of the asci have four spores and are named tetrads. Less commonly, asci with three, two, or one spore are also produced and are named triads, dyads, or monads, respectively. Spore formation is initiated after meiosis II at the spindle pole body (SPB), a multilayered organelle embedded in the nuclear envelope (Byers, 1981; Winey and Byers, 1993). Besides its main role as the microtubule-organizing center in S. cerevisiae, the SPB is also important for spore formation, because mutants with defective SPBs also have defects in sporulation (Moens and Rapport, 1971; Davidow et al., 1980; Okamoto and Iino, 1982; Uno et al., 1985; Brachat et al., 1998; Knop and Strasser, 2000). Soon after meiosis II, the outer plaque of the SPB begins to expand in both width and thickness. A double-membraned structure called a prospore membrane is then synthesized juxtaposed to the cytoplasmic face of the outer plaque (Moens and Rapport, 1971; Byers, 1981). It has been shown that the prospore membrane results from the fusion of vesicles derived from the late Golgi complex (Neiman, 1998). Eventually, the prospore membrane encapsulates a haploid nuclear lobe and a portion of the cytoplasm to form a prospore (Moens and Rapport, 1971; Byers, 1981). The prospores, however, are not visible under the light microscope. Subsequently, spore wall materials are deposited into the lumen of the prospore membrane (Lynn and Magee, 1970), which later matures into the spore wall and the spores become distinct by light microscopy.
Despite this well accepted paradigm of spore formation, its molecular mechanism remains poorly understood. In recent years the characterization of several proteins has added to our understanding of this process. First, the molecular nature of the morphologically unique SPB outer plaques at meiosis II has for the first time begun to be unraveled. Mpc54p and Mpc70p are two meiotic proteins that are expressed during a short period around meiosis II, and they are localized to the expanded outer plaques of the SPB (Knop and Strasser, 2000). In the absence of either protein, the meiotic SPB outer plaque is much less prominent and no prospore membrane is formed. These results suggest that Mpc54p and Mpc70p are specific components of the expanded outer plaques of the SPB during meiosis II, and confirm that this morphologically unique structure is required for prospore membrane formation (Knop and Strasser, 2000). The subsequent synthesis of the prospore membrane involves a number of gene products, including the septin gene SPR3 (Fares et al., 1996) and SPO14. SPO14 encodes a phospholipase D enzyme that is important for meiosis (Rose et al., 1995). The spo14 mutant cells can enter meiosis, and many of them can finish meiosis I and II, but no spores are formed. Electron microscopic (EM) studies have shown that no prospore membrane can be detected in the mutant (Rudge et al., 1998). The fusion protein of SPO14 and the green fluorescent protein (GFP), when overexpressed, is first seen in the cytoplasm and then relocalized to the outer plaques of SPBs and the growing prospore membrane (Rudge et al., 1998). Don1p has a similar localization pattern as Spo14p and is thought to localize to precursors of the prospore membrane, although the deletion of DON1 does not cause obvious defects in prospore membrane formation (Knop and Strasser, 2000).
In a screen for genes required for sporulation, we identified a novel gene ADY1 (accumulates dyads), corresponding to the open reading frame (ORF) YHR185C. The ady1 mutant can finish both meiosis I and II but rarely forms tetrads. Here we present evidence that suggests that ADY1 is required for the proper localization of the meiosis-specific SPB component Mpc54p to all four spindle poles during meiosis II, and that it is required for prospore membrane formation at selected spindle poles.
MATERIALS AND METHODS
Yeast Strains and Media
Yeast strains were derivatives of S288C and are listed in Table 1. Deletion strains were provided frozen in 96-well microtiter dishes. YEPD was 1% yeast extract B, 2% peptone, 2% dextrose (and 2% agar in plates). Sporulation medium was 1% potassium acetate, 0.5 mg/ml dextrose, and supplemented with the necessary amino acids at a final concentration of 20 μg/ml. YPEG medium is the same as YEPD except that 5% (vol/vol) of glycerol replaces the 2% dextrose.
Table 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| WSY33 | MATα his3Δ200 leu2-3,112 ura3-52 lys2-801 |
| WSY1079 | MATα ade1 his3-Δ leu2 can1R ady1∷GFP:HIS3 |
| WSY1086 | MATa his3Δ200 leu2-3,112 ura3-52 trp1-903 ady1∷GFP:HIS3 |
| WSY1217 | MATα his3Δ200 leu2-3,112 ura3-52 ade2-R8 spo11-Δ3 atr∷HIS3 |
| WSY1218 | MATa his3Δ200 leu2-3,112 ura3-52 ade2-1 trp1Δ spo11-Δ3 atr∷HIS3 |
| WSY1223 | MATα his3Δ200 leu2-3,112 ura3-52 ade2-1 trp1-903 cyh2 ady1∷GFP:HIS3 |
| WSY1224 | MATa his3Δ200 leu2-3,112 ura3-52 lys2-801 ade2-1 cyh2 ady1∷GFP:HIS3 |
| WSY1226 | MATa his3Δ200 leu2-3,112 ura3-52 lys2-801 ade2-1 cyh2 |
| WSY1244 | MATα his3Δ200 leu2-3,112 ura3-52 trp1-Δ ade2-R8 ady1∷GFP:HIS3 |
| WSY1245 | MATα his3Δ200 leu2-3,112 ura3-52 trp1-Δ lys2-801 ade2-R8 |
| WSY1248 | MATa/MATα his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 ura3-52/ura3-52 lys2-801/LYS2 TRP1/trp1-903 cyh2/CYH2 ady1∷GFP:HIS3/ady1∷GFP:HIS3 |
| WSY1250 | MATa/MATα his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 ura3-52/ura3-52 lys2-801/LYS2 TRP1/trp1-903 cyh2/CYH2 |
| WSY1312 | MATα his3Δ200 leu2-3,112 ura3-52 lys2-801 ady1∷GFP:HIS3 |
| WSY1313 | MATa his3Δ200 leu2-3,112 ura3-52 trp1-903 ady1∷GFP:HIS3 |
| WSY1794 | MATa/MATα his3Δ200/his3Δ200 leu2-3,112/LEU2 ade2/ade2 lys2-801/lys2-801 URA3/ura3 can1R/CAN1s TRP1/trp1-903 |
| WSY1797 | MATa/MATα his3Δ200/his3Δ200 LEU2/leu2-3,112 ade2/ade2 lys2-801/LYS2 TRP1/trp1-903 ura3/URA3 can1R/CAN1s ady1∷GFP:HIS3/ady1∷GFP:HIS3 |
| WSY1834 | MATa/MATα his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 ura3/ura3 trp1-903/TRP1 ADE2/ade2-1 ady1∷GFP:HIS3/ady1∷GFP:HIS3 don1∷Don1-eGFP-kanMX/don1∷Don1-eGFP-kanMX |
| WSY1837 | MATa/MATα his3Δ200/his3Δ200 leu2-3,112/leu2-3,112 ura3/ura3 trp1-903/TRP1 ADE2/ade2-1 lys2/LYS2 CYH2/cyh2 don1∷Don1-eGFP-kanMX/don1∷Don1-eGFP-kanMX |
| WSY1846 | MATa/MATα his3/his3 leu2/leu2 ura3/ura3 trp1-903/trp1-903 ADE2/ade2-1 lys2/LYS2 ady1∷GFP:HIS3/ady1∷GFP:HIS3 mpc54∷Mpc54-eGFP-KanMX/MPC54 |
| WSY1849 | MATa/MATα his3/his3 leu2/leu2 ura3/ura3 lys2-801/LYS2 TRP1/trp1-903 cyh2/CYH2 mpc54∷Mpc54-eGFP-KanMX/MPC54 |
All strains were isogenic to S288C. Double colon refers to a deletion of the preceding gene and replacement by the marker gene that follows. A single colon refers to linkage without deletion.
Screening
Two hundred fifteen MATa haploid strains were transformed with the pHO plasmid (Table 2) to enable mating type switching and self-mating (Herskowitz and Jensen, 1991). To induce sporulation, transformants were grown in liquid leucine-free medium (to select for the plasmid) at 30°C overnight to stationary phase (OD600 = >3.0). The cultures were washed once in sterile distilled water and resuspended in sporulation medium. Cells were examined for sporulation defects every 24 h for 72 h. Once a strain was found to be defective for sporulation, the ORF deleted in that strain was identified in Saccharomyces Genome Database (SGD) to determine whether it was previously described. Only strains with deletions of novel genes were studied further. Those mutants were first grown on YEPG plates to eliminate the petites, which do not sporulate because of defects in respiration. Strains that grew on YEPG were then examined for possible defects in diploidization. Two kinds of defects could lead to failure to form diploids in this screen: failure to switch mating type with pHO and failure to mate. We checked mating type switching by testing the mating types of single colonies of the pHO transformants. Briefly, pHO transformants were inoculated to grow on YEPD plates for several generations so that in the wild type more than half of the transformed cells had lost the plasmid. Single colonies on YEPD were then mated to mating type testers by replica plating. Four types of single colonies on YEPD were present in wild type: colonies that mated only with MATa tester, or only with the MATα tester, or with both tester strains, or with neither tester. Deletion mutants that cannot switch mating type have only MATa colonies. Mutants that can switch mating type but cannot mate may include two categories: those that fail to mate with wild type (unilateral mating mutant) and those that fail to mate with a strain bearing the same mutation but can mate with a wild-type strain (bilateral mating mutant). Unilateral mating mutants were excluded by demonstrating an inability to mate with wild type. The rest of the sporulation-defective strains in this screen include either bilateral mating mutants or authentic sporulation mutants. These strains were then backcrossed to wild type. We focused only on the strains that showed a 2:2 segregation of the deleted locus. Bilateral mating mutants were excluded by failure of two His+ spores of opposite mating types to form a diploid. The true sporulation mutants were then tested for the segregation of the sporulation defect by two means. First, all four spores from two tetrads were transformed with the pHO plasmid, and the transformants were examined for sporulation as described above. Second, diploid cells were made by mating either two His+ or two His− spores together and then testing for their ability to sporulate. For a true sporulation mutant caused by the HIS3 replacement, only His− diploid or transformants will sporulate in these tests.
Table 2.
Plasmids
| Plasmid | Relevant plasmid loci | Reference |
|---|---|---|
| pHO | CEN-LEU2-HO | Rudin et al., 1989 |
| pGD55 | CEN-TRP1-ADY1 | This study |
| pGD67 | 2μ-LEU2-GAL1-Myc-ADY1 | This study |
| pRS200 | CEN-TRP1 | Sikorski and Hieter, 1989 |
| pSPO14-GFP | 2μ-LEU2-GFP-SPO14 | Rudge et al., 1998 |
Plasmid Construction
For the subcloning of ADY1, a cosmid clone #70899 was purchased from American Type Culture Collection (Manassas, VA), and a 2.6-kb PstI-PstI fragment was cloned into the PstI site of the yeast shuttle vector pRS200 (Sikorski and Hieter, 1989). This 2.6-kb fragment includes the entire ORF of ADY1 and 1657 bp upstream of the start codon and 229 bp downstream of the stop codon. No other ORF is included in the subcloned fragment.
The myc-tagged Pfs1p was made as described below: the ADY1 coding sequence was polymerase chain reaction amplified with the XhoI sequence inserted on both ends. This fragment was then cloned into the XhoI site of the vector pEsc-LEU2 (Stratagene, La Jolla, CA) to yield the plasmid pGD67, in which the expression of myc-Pfs1 is directed by the GAL1 promoter (Table 2). To test whether pGD67 was functional, wild-type or ady1 mutant cells were transformed with the vector alone or pGD67. The transformants were then grown in leucine-free medium to stationary phase and transferred to sporulation medium. After 12–14 h in the sporulation medium, cells were about to enter meiosis I. Galactose was then added at the final concentration of 1% to induce the expression of Myc-Pfs1. Sporulation was examined 2 d later, and 300 cells were counted for each strain. The frequencies of triads and tetrads was both 3.33% in the ady1 mutant transformed with pGD67, compared with 0.33% triads and 0% tetrads in the same mutant transformed with the vector pEsc-LEU2. In contrast, wild-type cells transformed with pGD67 generated 14.67% triads and 14% tetrads, compared with 11.33% triads and 8.33% tetrads when transformed with the vector. The relatively lower frequency of triads and tetrads in the wild type (compared with Table 3) in this experiment may be due to the addition of galactose to induce the expression of the fusion protein. The nature of the apparent enhancement of sporulation in the wild type transformed with pGD67 compared with the vector is uncertain but was observed in two separate trials.
Table 3.
Sporulation frequency of the ady1 mutant and wild type
| Strain | No spore | Monad | Dyad | Triad | Tetrad |
|---|---|---|---|---|---|
| WT | 48.5 | 2.25 | 6.25 | 20.8 | 22.2 |
| ady1/ady1 | 61.7 | 20.2 | 16.3 | 1.80 | 0 |
The frequency of sporulation was tested in the wild type (WT, WSY1250) or homozygous ady1 mutant (WSY1248) after 3 d in sporulation conditions. Cells (400) were counted in each case.
Microscopic Analysis
Anti-tubulin indirect immunofluorescence was performed on cells fixed with 3.7% formaldehyde for 2 h, and the monoclonal antibody (mAb) YOL 1/34 was added at 1:50 (Serotec, Oxford, United Kingdom), and goat anti-rat rhodamine-conjugated secondary antibody added at 1:100 (Jackson Immunoresearch, West Grove, PA). DNA-specific fluorescent dye 4, 6-diamidino-2-phenylindole (DAPI; Sigma, St Louis, MO) was added at 1 μg/ml to identify the nucleus. To detect Myc-Pfs1, cells were fixed in 3.7% CH2O for 15 min, and the primary antibody was anti-Myc mAb at 1:150, and the secondary antibody was Cy3-conjugated goat anti-mouse at 1:1000. To detect Spc98, cells were fixed for 20 min in 3.7% CH2O, and the mouse anti-90 antibody (gift of John Kilmartin, MRC, Cambridge, United Kingdom) was added without dilution, and the secondary antibody was Cy3-conjugated goat anti-mouse at 1:1000. For the double labeling experiment, cells were fixed for 15 min, and antibodies were added in the following sequence: anti-Myc, anti-mouse Cy3, anti-tubulin, and anti-rat fluorescein isothiocyanate.
Thin-section electron microscopy of spore walls was done as described (Krisak et al., 1994), with the use of cells cultured in sporulation medium for 72 h
Commitment to Meiotic Recombination
Intragenic recombination was tested as described (Kassir and Simchen, 1991).
Intergenic Recombination
Intergenic recombination can be calculated by the following equations, assuming that triple or more crossover events are rare (Perkins, 1949):
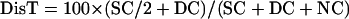 |
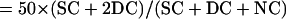 |
where DisT, SC, DC, and NC stand for the distance in centimorgan (cM), single crossover, double crossover, and noncrossover, respectively. In a tetrad analysis, the numbers of tetratype (T), nonparental ditype (NPD), and parental ditype (PD) are known, and therefore can be used to derive the values of SC and DC. SC = T − 2 × NPD, and DC = 4 × NPD. Therefore, DisT can be calculated by the following equation:
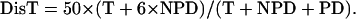 |
In dyads composed exclusively of nonsister haploid spores, the
theoretical distribution of the recombinants is shown
below.
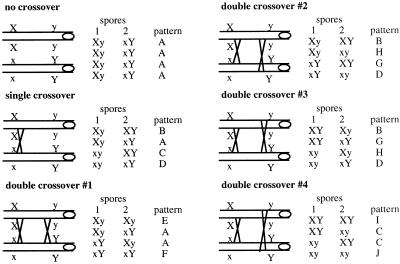
A–J represent the different patterns of spore phenotype. The value of DC can be derived in the following equations:
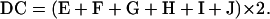 |
The value of B, C, and D contributed by double crossover is as follows:
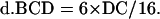 |
The value of B, C, and D from single crossover is as follows:
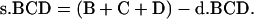 |
The value of pattern A from single crossover is as follows:
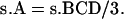 |
The value of pattern A contributed by double crossover is as follows:
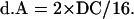 |
The value of no crossover is as follows:
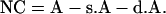 |
The final measurement of map distance can be derived with the use of the first equation and with the values of SC, DC, and NC as calculated in the above-mentioned equations.
Dissection of Individual Dyads
Dissection of individual dyads was done as described (Davidow et al., 1980).
UV-induced Mutagenesis
The ploidy of the spores was determined based on the frequencies of canavanine-resistant colonies after UV treatment. Spores or control strains were grown in YEPD overnight and about equal number of cells (8 μl at OD600 = 6.0) were inoculated onto predetermined spots on a YEPD plate and a canavanine plate. After the liquid was absorbed into agar, the canavanine and YEPD plates were exposed to UV light to induce mutagenesis. The UV light was delivered at sub-LD values from a UV cross-linker (FB-UVXL-1000; Fisher Scientific, Fair Lawn, NJ) with 9000 μj/cm2 energy. The UV-treated cells were then incubated at 30°C for 3 d to allow canavanine-resistant colonies to grow.
SDS-Polyacrylamide Gel and Immunoblotting
Protein extracts were prepared, run on SDS polyacrylamide gels, transferred to polyvinylidene difluoride membranes and blotted as described (Saunders et al., 1997). The primary antibody was mAb anti-Myc (Sigma) at 1:100, and the secondary antibody was horseradish peroxidase-conjugated goat anti-mouse IgG (Roche Molecualr Biochemicals, Indianapolis, IN) at 1:2000.
Chi-Square Test
A χ2 test was done to test whether the observed distribution pattern of genetic markers in the dyads was statistically significant. The null hypothesis is that the spores of the dyads package the haploid meiotic nuclei randomly, which gives rise to 4/6 chances dyads of 1:1 nonsister spores, 2/6 chances of dyads of sister spores (1/6 as 2:0, and 1/6 as 0:2). For simplicity, the ADE1 locus was tested and is shown in Table 4 (see Table 6 for genetic data).
Table 4.
The frequency of segregation of the ADE1 locus in ady1/ady1 mutants.
| Distribution | O | E | (O − E)2/E |
|---|---|---|---|
| Nonsister | 69 | 51.3 | 6.1 |
| Sister | 8 | 25.7 | 12.2 |
| Total | 77 | 77 | 18.3 |
Seventy-seven dyads were dissected and tested for growth on −ade plates. The frequency of ade+/ade+ spores (1:1) or ade+/ade− (2:0 or 0:2) spores is shown.
O, observed; E, expected.
Table 6.
Segregation of markers in the dyads of an ady1 mutant
| Genetic markers | Distance to centromere (cM) | 1:1 | 2:0 | 0:2 | Total |
|---|---|---|---|---|---|
| ADE1 | 4 | 69 | 6 | 2 | 77 |
| URA3 | 7 | 66 | 10 | 1 | 77 |
| TRP1 | 1 | 69 | 8 | 0 | 77 |
A ady1 mutant (WSY1079 × WSY1086) heterozygous for the three centromere-linked markers was cultured in sporulation medium. Dyads were dissected individually (see MATERIALS AND METHODS) and the spores were tested on drop-out plates for the genetic markers. 1:1, one spore was wild type for the marker and the other was mutant; 2:0, both spores were wild type for the marker; 0:2, both markers were mutant for the marker. The markers' distances to centromeres were as described in SGD.
Degree of freedom = 1
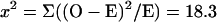 |
Probability <0.001.
Null hypothesis is rejected. Therefore, the high frequency of dyads comprised of nonsister spores was unlikely due to random packaging.
Ether Resistance
Experiments were done in a ventilated hood. Cells sampled at different stages of growth were spread onto YEPD on glass plates at various dilutions. Ether (0.75 ml) was added to a piece of sterilized 3M filter paper, which was then placed on the lids of inverted plates. After various amount of exposure times, the filter was removed and the plates left in the hood with their lids half open for ∼45 min before being returned to incubator for 3 d at 30°C.
RESULTS
ADY1 Is a Novel Gene Required for Sporulation
It has been shown that the expression of ∼500 genes is induced during meiosis and many of them may be functionally required for meiosis (Chu et al., 1998). Previous work has identified >60 genes that are uniquely required for sporulation (Kupiec et al., 1997). The collaborative effort of several groups to delete every ORF in the S. cerevisiae genome (Winzeler et al., 1999) has made it possible to identify the other genes required for sporulation in a systematic way. We have screened 215 strains that collectively represent the deletion of every nonessential ORF on chromosome VIII (strains kindly provided by Drs. Linda Riles and Mark Johnston, Washington University, St. Louis, MO; Niedenthal et al., 1999). To begin the screen, the deletion strains were first transformed with a plasmid harboring the HO gene to enable mating type switching and self-mating to form diploid cells (see MATERIALS AND METHODS; pHO plasmid courtesy of Dr. Jim Haber, Brandeis University, Waltham, MA; Strathern and Herskowitz, 1979). The transformants were then grown in rich medium to stationary phase, transferred into sporulation medium, and examined by the differential contrast microscopy at various time points for up to 3 d. In the wild-type transformants, typically ∼50% of the cells formed triads and tetrads by 24 h in sporulation condition. One of the deletion mutants, believed to be missing the ORF YHR185C, named as ADY1 in this study, formed asci at a reduced frequency. Moreover, tetrads were rarely seen in this mutant, and most of the asci were comprised of monads and dyads (Table 3). The sporulation defect in the ady1 mutant was not due to a defect in diploidization with pHO (see MATERIALS AND METHODS). Standard linkage analysis on 15 tetrads confirmed that the defect in the mutant phenotype was caused by a single gene mutation, which was tightly linked with the genetic marker HIS3 that replaces the ORF YHR185C. A plasmid pGD55 harboring the full-length ADY1 gene but no other ORF (see MATERIALS AND METHODS) was constructed to test whether the ADY1 gene complements the sporulation defect of the ady1 mutant. As shown in Figure 1, when transformed with the vector alone, a homozygous ady1 diploid mutant sporulated only poorly, with mostly monads and dyads; however, when the same strain was transformed with the plasmid pGD55 it sporulated with >50% triads and tetrads. We therefore conclude that a novel gene ADY1 at YHR185C is required for the formation of tetrads in sporulation.
Figure 1.
ADY1 rescues the sporulation phenotype of the ady1 mutant. (Left) MATa/MATα ady1/ady1 strain (WSY1248) transformed with the vector pRS200 showed a low level of sporulation after 3 d of culture in sporulation medium, with mostly monads and dyads. (Middle) Same strain showed sporulation comparable with wild type when transformed with a plasmid harboring the ADY1 gene. (Right) Sporulation in a wild-type MATa/MATα diploid strain (WSY1250).
The ADY1 gene encodes a hypothetical protein of 27.9 kDa. There is a middle gene sporulation element motif (Hepworth et al., 1995; Ozsarac et al., 1997) starting at −18 bp of its 5′-flanking sequence. Middle gene sporulation element is found upstream of many middle meiotic genes whose expression is mediated by the meiotic transcription activator NDT80 (Chu and Herskowitz, 1998). Consistent with the presence of an MSE motif, ADY1 was found to be expressed midway in the sporulation program (Chu et al. 1998; our unpublished results). ADY1 has no other recognizable structural motifs and shares no strong sequence similarity to other known proteins.
ADY1 Is Not Required for Meiotic Chromosomal Segregation
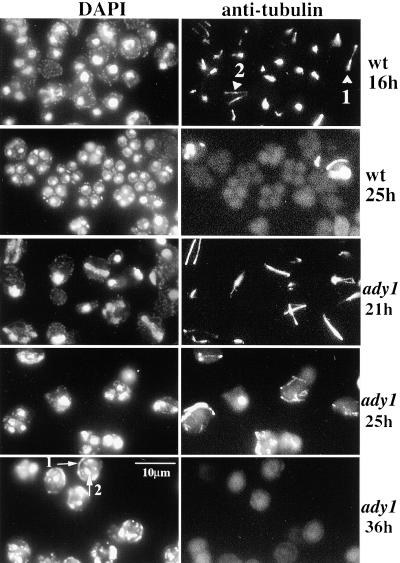
The reduced number of spores in the ady1 mutant could be explained by a reduction in the number of meiotic divisions (Klapholtz and Esposito, 1980; Shuster and Byers, 1989). To understand what causes the phenotype of rare tetrads and relatively abundant dyads in the ady1 mutant, we examined the progress of meiosis in the mutant. A homozygous ady1 mutant or wild-type strain was cultured for sporulation as described above. Aliquots of cells were taken at different time points during sporulation, fixed with 3.7% formaldehyde, and stained with anti-tubulin for spindles and DAPI for DNA (see MATERIALS AND METHODS). Cells were then examined by epifluorescence microscopy. In the wild-type cells, meiosis I began after ∼12 h in sporulation condition (our unpublished results); after 16 h, meiosis II had started in many of the cells (Figure 2). Refractive spores were present in >50% of the cells after 25 h in sporulation conditions, with one nucleus in each spore. In the ady1 mutant cells, meiosis I was not started until after ∼16 h in sporulation conditions. However, by 25 h in sporulation conditions, >40% of the mutant cells were in meiosis II, with four well separated nuclei and normal-appearing meiosis II spindles. By 36 h in the sporulation medium, refractive spores were visible, although predominantly in monads and dyads. At this stage the frequency of cells with four nuclei often became difficult to determine in the mutant, because in many cells one mass of DAPI staining was seen in each spore of the monads or dyads, whereas more DAPI staining was also seen at the rim of the spores (Figure 2). These results indicate that ADY1 is not essential for meiotic chromosomal segregation.
Figure 2.
Examination of meiosis by immunofluorescence microscopy. Samples were taken out of a MATa/MATα ady1/ady1 strain (ady1: WSY1248) and a MATa/MATα ADY1/ADY1 strain (WT: WSY1250) at different time points in sporulation medium. After fixation, cells were stained with anti-tubulin antibodies for meiotic spindles and counterstained with DAPI for DNA. Arrowheads 1 and 2, meiotic I and II spindles, respectively. Arrow 1, chromosomal masses outside a spore in a monad of the ady1 mutant; arrow 2, a nucleus inside a spore in a monad of the ady1 mutant.
ADY1 Is Not Required for Spore Wall Maturation
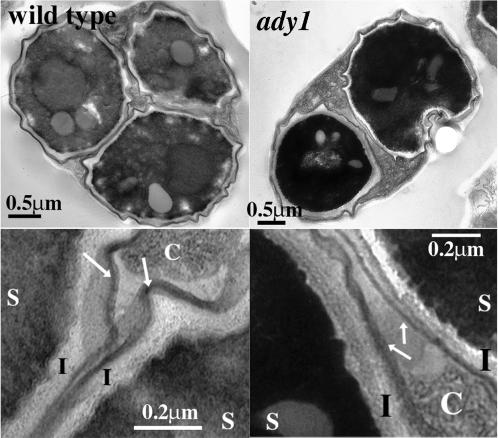
Because meiosis nuclear divisions are normal in the ady1 mutant, two scenarios could explain the rare tetrads. A packaging defect may cause fewer than four of the haploid meiotic nuclear lobes to be encapsulated to form prospores. Alternatively, four prospores may be produced, but only some of them then undergo normal maturation to form the refractive spores, whereas the others are not developed well enough to be seen by the light microscope. Because immature spores are less resistant to environmental stress, we tested maturation of the spores by their resistance to ether. Wild type and a ady1 mutant were cultured for sporulation and aliquots of cells were taken at 0 and 72 h and plated out on YEPD (see MATERIALS AND METHODS). The cells were then exposed to ether vapors for 30 min and the surviving colonies were counted after 3 d of culture. As shown in Table 5, vegetative cells were very sensitive to ether treatment, with a survival rate of <1% for both wild type and the ady1 mutant. Sporulating ady1 cells were >20-fold more resistant than vegetative cultures, although they were only 20% as resistant as the sporulating wild-type cells. This decrease of ether resistance in the sporulating culture of the ady1 mutant could be due to immature spores but could also be due to a lower percentage of total spores (Table 3). To investigate further whether ADY1 is required for spore wall maturation, we examined the spore wall structure in both the ady1 mutant and wild type. Cells were cultured in the sporulation medium for 3 d and then fixed with gluteraldehyde and osmium tetroxide, and stained with uranyl acetate (see MATERIALS AND METHODS). Thin sections were cut and examined by EM. As shown in Figure 3, usually three spores were seen on one section in the wild type, but rarely more than two in the ady1 mutant. The wild-type spore walls have been shown to include four layers (Kreger-van Rij, 1978). As shown in Figure 3, the outer layers were thin and osmophilic, representing the dityrosine and chitosan layers. The inner two layers, namely, glucan and mannan, appeared as one single electron-lucent layer. In the ady1 mutant, the outer layers were osmophilic, compact, and structurally indistinguishable from the wild type, and the inner layers also appeared normal (Figure 3). The spore walls in the ady1 mutant appeared to function normally in excluding cytoplasmic materials from the spores. These results suggest that when spores do form in the ady1 mutant they are structurally and functionally normal. ADY1 is therefore not required for spore wall maturation.
Table 5.
Resistance to ether
| Vegetative ady1/ady1 | Sporulating ady1/ady1 | Vegetative WT | Sporulating WT | |
|---|---|---|---|---|
| 0 min in ether | 100 | 100 | 100 | 100 |
| 30 min in ether | 0.61 | 14.2 | 0.45 | 68 |
Wild type (WT, WSY1250) and the homozygous ady1 mutant (WSY1248) in vegetative growth or after 3 d in sporulation condition were tested for their resistance to ether. The percentage of survival was shown under different situations.
Figure 3.
Spore wall structures of wild type (WSY1250) and the ady1 mutant (WSY1248). (Top) Low-magnification images of the spore walls in the wild type (left) and ady1 mutant (right). (Bottom) High-magnification images of the cells in the corresponding top panel showing spores (S), the inner (I) and outer (arrows) layers of the spore wall, and the excluded cytoplasm (C). The nuclear staining in the ady1 mutant was not consistently darker than wild type. Arrows point to the outer layers of the spore wall.
ADY1 Is Required for Formation of Spores at Both Poles of a Spindle
The normal meiotic nuclear divisions and normal spore maturation observed in the ady1 mutant suggest that the rare tetrads and relatively common monads and dyads most likely result from a packaging defect. However, it is also possible that the monads and dyads represent a minor subset of the mutant cells that only undergo a single meiotic division. To distinguish between these possibilities, it was necessary to determine what meiotic products were included in the spores of the ady1 mutant. Because it was difficult to distinguish monads from cells that did not enter meiosis under the dissection microscope, we examined only dyads for genetic analysis. Wild-type or homozygous ady1 mutant strains were created that were heterozygous for three centromere-linked markers, namely, ADE1, URA3, and TRP1, and cultured for sporulation as described above. Dyads were identified and dissected individually (see MATERIALS AND METHODS) to monitor the segregation of the genetic markers to the two spores (Davidow et al., 1980). The segregation was considered 1:1 if one spore is prototrophic and the other is auxotrophic, and 2:0 if both are prototrophic, and 0:2 if both are auxotrophic.
A single meiotic division leads to formation of spores that are diploid. If the dyads are due to a single reductional division, the centromere-linked markers are expected to segregate in the 1:1 pattern with slightly more than half of the dyads containing two nonmating spores. If the dyads are due to a single equational division, the centromere-linked markers are expected to segregate in the 2:0 pattern, and slightly less than half of the dyads will contain two nonmating spores. On the other hand, if the dyads are due to the inclusion of only two of the four meiotic products after meiosis II, both spores in a dyad will be capable of mating, because they are haploid.
As shown in Table 6, the centromere-linked markers showed predominantly the 1:1 segregation pattern, suggesting a single reductional division might have caused the dyads in the mutant. However, all the spores from the dyads of the ady1 mutant were maters, arguing against a single meiotic division (Table 7). Alternatively, a single reductional division combined with a very low recombination at the MAT locus could explain the formation of dyads. However, the high frequency of dyads with both spores of the same mating type (Table 7) suggests that recombination at the MAT locus is normal (see Table 8 and Figure 8 for recombination at other loci). The data are most consistent with the conclusion that the dyads were generated by a packaging defect, giving rise to haploid spores that are all maters.
Table 7.
Summary of marker segregation
| 3(1:1) | 2(1:1) 1(2:0) | 1(1:1) 2(2:0) | 3(2:0) | 3(0:2) | Total |
MAT
locus
|
Total | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| a:α* | a:a* | α:α* | N:N* | ||||||||
| Observed | 65 | 5 | 0 | 5 | 0 | 75** | 54 | 10 | 12 | 1 | 77 |
| Expected | |||||||||||
| Reductional | 59.72 | 16.11 | 1.16 | <0.01 | 0 | 77 | 30.8 | 0 | 0 | 46.2 | 77 |
| Equational | <0.01 | 1.16 | 16.11 | 59.72 | 0 | 77 | 46.2 | 0 | 0 | 30.8 | 77 |
MAT locus, 30 cM to centromere. Expected values were calculated as described (Shuster and Byers, 1989).
a:α, one spore capable of mating with MATα tester, the other with MATa; a:a, both spores capable of mating with MATα tester; α:α, both spore capable of mating with MATa tester; N:N, neither spore capable of mating with either tester.
Total value was not equal to 77 because two dyads had the pattern of 2 (2:0) 1 (0:2), which was not expected in either case of single meiotic division and was therefore not shown in this table to avoid confusion.
Table 8.
Intergenic recombination in wild type at two loci
| Interval | T | NPD | PD | Total | Distance calculated (cM) | Distance reported (cM) |
|---|---|---|---|---|---|---|
| CAN1-URA3 | 28 | 4 | 37 | 69 | 37.7 | 45 |
| LEU2-MAT | 39 | 3 | 27 | 69 | 41.3 | 30.6 |
A wild-type diploid heterozygous for the CAN1, URA3, and LEU2 (WSY1794) was constructed and cultured to sporulate. Tetrads were dissected and the numbers of tetrads of tetratype (T), nonparental ditype (NPD), and parental ditype (PD) were counted to calculate genetic recombination at the specified intervals (see MATERIALS AND METHODS). The expected distances at the two intervals were cited from SGD.
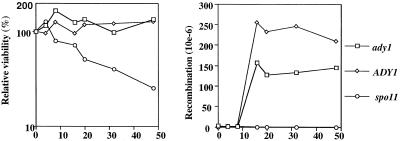
Figure 8.
Test of viability and meiotic recombination. A MATa/MATα ade2-1/ade2-R8 ady1/ady1 mutant (ady1) (WSY1224 × WSY1244), MATa/MATα ade2-1/ade2-R8 spo11/spo11 (spo11) (WSY1217 × WSY1218), or MATa/MATα ade2-1/ade2-R8 strain (WT) (WSY1226 × WSY1245) was tested for commitment to meiotic recombination. Recombination frequency was calculated as the frequency of adenine prototrophic colonies relative to total colonies on YEPD plates. (Left) Viability of the ady1, spo11, and wild-type cells at different time points during sporulation. (Right) Recombination frequency of the ady1, spo11, and wild-type cells at different time points in the sporulation medium.
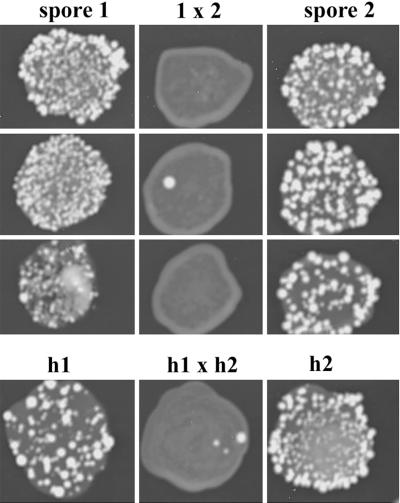
To confirm that the spores in the dyads of the ady1 mutant were haploid, the ploidy was tested by examining the frequency of UV-induced mutation at the CAN1 locus of the spores (see MATERIALS AND METHODS). The wild-type CAN1 allele is dominant, and the recessive can1 allele encodes the resistance to the drug canavanine. Because a diploid with two CAN1 genes is much less likely to undergo mutation of both genes than a haploid will undergo mutation of the single CAN1 gene, the frequency of UV-induced canavanine resistant colonies in a diploid strain is much lower than in a haploid strain. As shown in Figure 4, six spores were tested and all had a frequency of canavanine-resistant colonies comparable with the haploid controls. In contrast, strains made from mating two mutant spores together had a much lower frequency of canavanine resistant colonies, comparable with the diploid control containing two copies of the wild-type CAN1 allele. The result confirms that haploid spores are generated in the dyads of the ady1 mutant. Together, the above-mentioned experiments show that the dyads of the ady1 mutant contain predominantly two nonsister spores. If two of the four meiotic nuclear lobes were randomly packaged into prospores, 66.7% of the dyads are expected to contain two nonsister spores. In the ady1 mutant, >86% of the dyads were composed of spores segregating 1:1 for the each of the three centromere-linked genetic markers (Table 6). A Chi-square test (see MATERIALS AND METHODS) confirms that the occurrence of nonsister spores in the ady1 mutant is not due to chance (p < 0.001), suggesting that in the case of dyads, most often spores are generated from one pole of each spindle at meiosis II.
Figure 4.
UV-induced mutagenesis at the CAN1 locus in the spores of the ady1 mutant (WSY1079 × WSY1086). (Top) Strains derived from the spores (side columns) of the ady1 mutant and from mating two spores (middle) of the ady1 mutant were exposed to UV light. The colonies growing up on the canavanine plate represented mutagenesis at the wild-type CAN1 locus in these strains. (Bottom, middle column) Diploid control (h1 × h2) that resulted from mating the two haploid controls (h1, WSY33; h2, WSY1086) were induced by UV for canavanine-resistant colonies.
ADY1 Is Required for Localization of SPO14p and Don1p to All Four Prospore Membranes
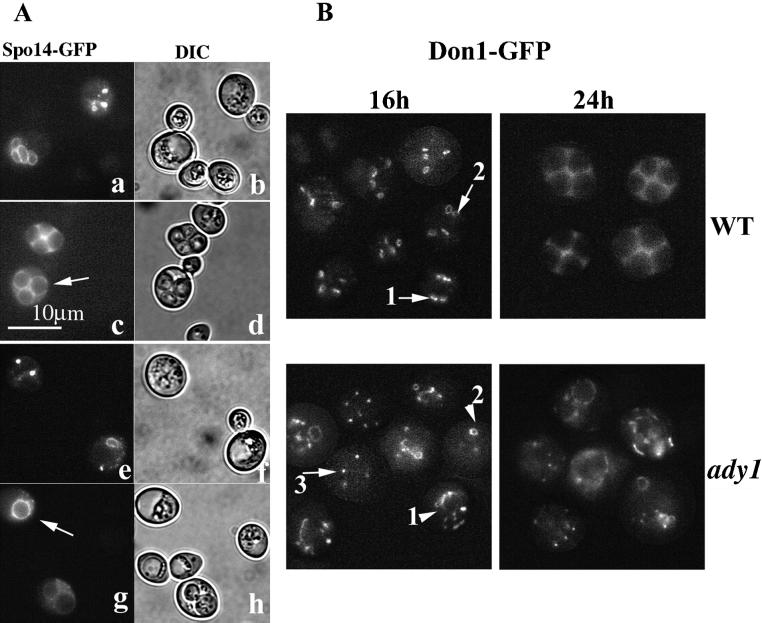
The above-mentioned results strongly suggest that ADY1 is involved in prospore formation but not in spore maturation. Because SPO14 has been shown to be required for prospore membrane formation and is localized to the prospore membrane (Rudge et al., 1998), we decided to use it as a probe to examine prospore membrane formation in the ady1 mutant. Wild-type or ady1 cells transformed with the plasmid pSPO14-GFP (plasmid courtesy of Dr. JoAnne Engebrecht, SUNY at Stony Brook, NY) were cultured for sporulation as described above. In the wild type, many cells were in meiosis II by 16 h in sporulation medium. At this stage, no spores were visible by differential contrast microscopy. However, four small dots or small circles of SPO14-GFP signal can be seen in some cells, most likely representing SPO14 near the SPBs (Figure 5A; Rudge et al., 1998). Later in sporulation, larger circles of SPO14-GFP signal were seen in some cells. The circles of GFP signal decorated the spore walls that were visible at this stage, consistent with a previous report (Rudge et al., 1998). In the ady1 mutant, small dots of SPO14-GFP signal were also seen at early stages of meiosis (20 h). However, there were usually only one or two sites of the SPO14-GFP fluorescence per cell in the mutant. At later stages one or two larger circles of GFP signal were seen to colocalize with the spore wall. These results suggest that SPO14-GFP can only be localized to a subset of the four SPBs in the ady1 mutant; however, once it is localized to a SPB, the change of SPO14-GFP localization and therefore prospore membrane development can proceed normally in the mutant.
Figure 5.
Fluorescence of Spo14-GFP and Don1-GFP. (A) Spo14-GFP signal in live wild-type (WSY1250) (a–d) and the ady1 mutant (WSY1248) (e–h) cells transformed with the SPO14-GFP plasmid. Arrows point to a cell with three rings of Spo14-GFP signal in the wild type, or a cell with only one ring of Spo14-GFP signal, respectively. (B) Don1-GFP signal in live wild-type (WSY1837) or ady1 mutant cells (WSY1834). Cells were cultured for sporulation and sampled for microscopic examination and imaging at specified time points. The images were a montage of cells with GFP stainings from different fields and represented the typical views. Wild-type cells almost always showed four small bars (arrow 1) or circles (arrow 2) of fluorescence. The ady1 mutant often showed aberrant stainings of Don1-GFP, either in the form of multiple signals of various shape (arrowhead 1), or only one or two rings or dots (arrowhead 2). Occasionally, cells with normal looking Don1-GFP were seen (arrow 3).
To confirm whether ADY1 is required to initiate prospore membrane formation at all four spindle poles, we examined the localization of Don1p, a precursor of prospore membrane, during meiosis. In the wild type, the first specific staining of Don1-GFP was in most cases (>75%) as four small bar-like or small circular structures at the four SPBs during meiosis (Don1-GFP strain is a gift from Dr. Michael Knop, Max-Planck-Institut fur Biochemie, Martinsreid, Germany; Knop and Strasser, 2000; Figure 5B). Don1-GFP later became larger circles that colocalized with the spores (Knop and Strasser, 2000; Figure 5B). In the ady1 mutant, cells with four bar- or ring-like stainings of Don1-GFP were rarely seen (<4%). Instead, various types of aberrant localization of Don1-GFP were seen: some with only one or two short bar- or dot-like stainings, and many with multiple sites of stainings of different brightness, size, and shape in the cytoplasm (Figure 5B). Even though the mutant cells were cultured for prolonged time in the sporulation medium, cells with four regularly shaped short bar or circular Don1-GFP stainings were rarely seen (Figure 5B; our unpublished results), suggesting that these aberrant stainings of Don1-GFP in the ady1 mutant is not merely a delay in the correct localization of the protein to the SPBs. The abnormal localization of Don1p is consistent with the abnormal prospore membrane formation in the ady1 mutant.
Meiosis-specific SPB Component Mpc54p Is Mislocalized in ady1 Mutant
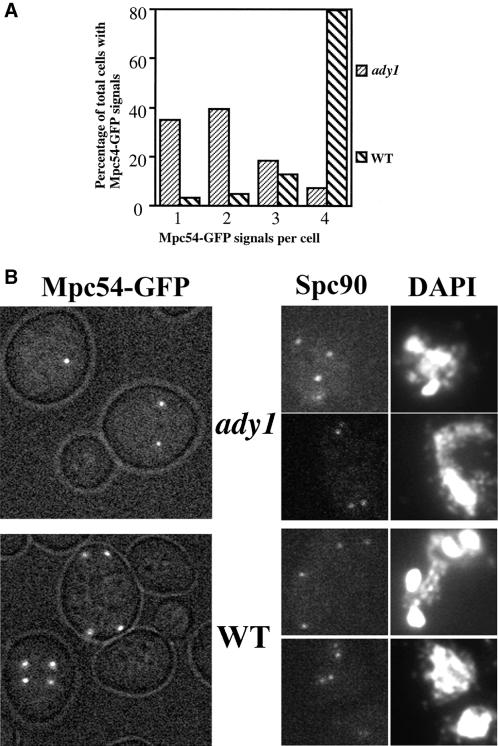
Because the SPB is known to be required for prospore membrane formation, it is possible that a structural defect in the SPB causes the block of prospore formation in the ady1 mutant, as suggested by the abnormal localization patterns of Spo14p and Don1p. We next tested a very early step in prospore membrane formation, namely, the morphological modification of the SPB outer plaques (Moens and Rapport, 1971; Byers, 1981). Because Mpc54p has been shown to localize to the expanded outer plaque of the SPB, and to be required for prospore membrane formation (Knop and Strasser, 2000), we examined the expansion of the outer plaques with the use of Mpc54p as a specific probe. As shown in Figure 6, A and B, Mpc54-GFP showed four discrete spots in most cases in the wild type. In contrast, only a small percentage of the ady1 mutant cells showed four dots of Mpc54-GFP staining; In most cases, only one or two spots of Mpc54-GFP staining were observed. These results suggest that the meiosis-specific SPB component Mpc54p is not present in every spindle pole of the ady1 mutant. Therefore, we propose that the ady1 mutant cells are blocked at the very earliest steps of prospore membrane formation at the affected SPBs.
Figure 6.
Localization of Mpc54p and Spc90p. (A and B) Mpc54-GFP signals in live cells. Wild type (WSY1849) or ady1 mutant (WSY1846) was cultured for sporulation as before. Samples were taken at 14–16 h for live cell imaging. (A) Quantitation of cells with one, two, three, or four sites of Mpc54-GFP signals. More than 60 cells with Mpc54-GFP signals were counted in the wild type or the mutant. (B) Typical microscopic view of Mpc54-GFP in wild-type or ady1 mutant cells. (C) Spc90 staining in fixed cells. Wild type (WSY1250) or ady1 mutant (WSY1248) was cultured for sporulation for 20–22 h and fixed for immunofluorescence microscopic analysis. Cells were stained with the anti-Spc90 antibody for the SPB and counterstained with DAPI for DNA.
The lack of Mpc54p at a subset of the SPBs may be because these SPBs have a global structural defect, in which case other, if not all, SPB components may also be missing. Spc98p has been shown to be a SPB marker in mitotic cells (Wigge et al., 1998). At meiosis II, both the wild-type and the ady1 mutant cells showed four dots when stained by the anti-Spc90 antibody (Figure 6C). We therefore conclude that the ady1 mutants have intact SPBs during meiosis, but one or more meiosis-specific components are missing from a subset of the SPBs.
Pfs1p Is Localized to Nucleus in Meiosis
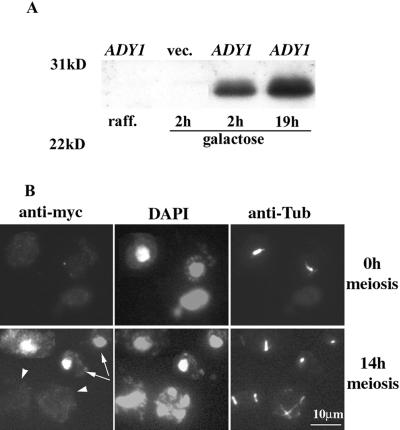
Because ADY1 is required for the localization of Mpc54p and spore formation at every spindle pole, it may act on the SPB to anchor Mpc54p and possibly other meiosis-specific SPB components that are required for prospore membrane formation. Alternatively, ADY1 may play a regulatory role, and therefore may not necessarily localize to the SPB. To address these questions, we decided to examine the localization pattern of ADY1. A Myc-Pfs1 fusion protein was expressed by the GAL1 promoter (Figure 7A) and was found to be partially functional, because the ady1 mutant transformed with the hybrid gene produced triads and tetrads at a level of 23% of that of the wild type transformed with the same gene (see MATERIALS AND METHODS). In the mitotic cells, the fusion protein most often accumulated in the cytoplasm, but sometimes also in the nucleus (our unpublished results). In contrast, it was observed only in the nucleus in the sporulating cells. Furthermore, the high accumulation was only observed in mononucleate cells that showed no meiotic spindles, but not in cells with two or four nuclear lobes and meiotic spindles (Figure 7B).
Figure 7.
Expression and localization of the fusion protein Myc-Pfs1. (A) Fusion protein was induced by galactose. Wild-type cells (WSY1250) transformed with the hybrid gene Myc-ADY1 (Pfs1) or the vector (vec.) were grown to log phase in the vegetative medium with raffinose (raff.) as the only sugar. Galactose was then added to the final concentration of 1%. Cells were sampled after 2 h or 19 h. Protein extracts were run on SDS-PAGE and transferred to the polyvinylidene difluoride membrane, which was probed with the anti-Myc antibody. (B) Cells transformed with the Myc-ADY1 hybrid gene or vector was either grown to log phase in the vegetative medium (veg.) or cultured in the sporulation medium (spo.) for 11 h, at which point cells were about to enter meiosis I. Galactose was then added to both cultures to the final concentration of 1%. After 2–4 h, cells were fixed and probed with the anti-Myc and anti-tubulin antibodies (see MATERIALS AND METHODS). Arrows indicate cells that have not entered meiosis and arrowheads indicate cells in meiosis.
ADY1 Is Not Required for Meiotic Recombination
To further characterize ADY1 during meiosis, we tested its involvement in meiotic recombination. First, the commitment of ady1 to meiotic recombination was examined with the use of a return-to-growth protocol (Kassir and Simchen, 1991) because it was difficult to carry out tetrad analysis with the rare tetrads in the ady1 mutant. For this purpose, homozygous ady1 or wild-type diploid strains were created that had one copy of the ade2-1 allele and one copy of the ade2-R8 allele (ade2 mutant strains courtesy of Dr. Giora Simchen, The Hebrew University, Jerusalem, Israel). These two strains normally do not grow on adenine-free plates. However, Ade+ colonies can form in these strains at a very low frequency, usually in the range of 1–3 Ade+ colonies per million viable colonies, either due to mitotic recombination or spontaneous reversion. In the wild type, there was a dramatic increase of Ade+ colonies after 16 h in sporulation conditions due to meiotic recombination. In the ady1 mutant, the Ade+ colonies also increased substantially after 16 h in sporulation conditions. The recombination level of the ady1 mutant is ∼50–80% that of the wild type (Figure 8). In contrast, the frequency of Ade+ colonies in the spo11 mutant (mutant strain a gift of Dr. Rochelle Esposito, University of Chicago, Chicago, IL) did not increase during sporulation. These results suggest that ADY1 may not play a major role in meiotic recombination.
To further address the above-mentioned possibility, we decided to test intergenic recombination of the ady1 mutant during sporulation. The recombination at two intervals was tested. Those were MAT-LEU2 on chromosome III and URA3-CAN1 on chromosome V. In the wild type, the occurrence of no crossover, single crossover, or double crossover was indicated by tetrads of parental ditype, tetratype, and nonparental ditype. As shown in Table 8, the calculated results (see MATERIALS AND METHODS) were close to reported genetic distances between the tested loci. Because the mutant was known to form predominantly nonsister haploid spores (Figure 4 and Table 6), it was possible to deduce the occurrence of no crossover, single crossover, or double crossover from the frequency of different phenotypic patterns of the two spores in the dyads (see MATERIALS AND METHODS; Tables 9 and 10). The recombination frequencies at the two intervals of the ady1 mutant were either higher than or very close to the wild type (Tables 8 and 10). These results demonstrate that ADY1 is not required for meiotic recombination.
Table 9.
Patterns of segregation in the ady1 mutant following recombination
| Interval | A | B | C | D | E | F | G | H | I | J | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| URA3-CAN1 | 72 | 15 | 26 | 21 | 8 | 8 | 1 | 1 | 0 | 0 | 152 |
| MAT-LEU2 | 84 | 19 | 15 | 19 | 2 | 1 | 6 | 5 | 1 | 0 | 152 |
A ady1 mutant diploid heterozygous for the CAN1, URA3, and LEU2 (WSY1797) was constructed and cultured to sporulate. Dyads were dissected individually and the numbers of dyads demonstrating different patterns of segregation (A–J) of the recombinant chromosomes were counted (see MATERIALS AND METHODS).
Table 10.
Intergenic recombination in the ady1 dyads at two loci
| Interval | Double crossover | Single crossover | No crossover | Total | Distance calculated (cM) | Distance reported (cM) |
|---|---|---|---|---|---|---|
| URA3-CAN1 | 36 | 64.7 | 51.3 | 152 | 44.9 | 45 |
| MAT-LEU2 | 30 | 55.7 | 66.3 | 152 | 38.1 | 30.6 |
Calculation of distance follows the description in MATERIALS AND METHODS, with the values of double crossover, single crossover, and no crossover derived from the values of pattern A–J in Table 6B (see MATERIALS AND METHODS).
DISCUSSION
The function of a novel gene ADY1 is characterized in this study. Pfs1p is not required for the meiotic nuclear divisions or meiotic recombination but is required to form four spores per cell. In the absence of Pfs1p, monads and dyads, but not tetrads, are generated. The production of monads and dyads is not because of an arrest during spore wall maturation, because few immature spores are seen by EM. Moreover, the spores that are detected have normal spore wall structures and most likely normal resistance to ether vapors. The lack of four spores per cell in the ady1 mutant therefore is due to a defect that occurs between the normal meiotic nuclear division and spore maturation. This period can be arbitrarily divided into two stages: initiation of prospore membrane formation, and growth of the prospore membrane and encapsulation of the haploid meiotic products. The mislocalization of Spo14p, Don1p, and Mpc54p in the ady1 mutant suggests that the defect is in the early initiation of prospore membrane synthesis.
Spo14p is a phospholipase D enzyme and is localized to and required for prospore membrane formation (Rose et al., 1995; Rudge et al., 1998). In the wild type, Spo14-GFP is first seen as four dots, presumably at the SPBs, and later as four rings that are colocalized with the spore walls. In the ady1 mutant, Spo14-GFP is seen as one or two dots at the early onset of prospore membrane formation, and later as one or two rings. These results suggest that the prospore membrane in the ady1 mutant can develop normally, but only at one or two SPBs.
The inability to initiate four prospore membranes in the ady1 mutant cell may be because the outer plaques of some SPBs are unable to undergo proper morphological expansion known to be required to start prospore membrane assembly. Mpc54p is a component of the expanded SPB outer plaque (Knop and Strasser, 2000). In the wild type, Mpc54-GFP is almost always present at four sites, most likely the four expanded outer plaques of the SPBs. In the mutant, however, Mpc54-GFP is most often seen at only one or two sites. We believe it is the lack of Mpc54p and the resulting lack of properly constructed outer plaques at some SPBs that cause the inability of Spo14p to localize to these SPBs and start the assembly of the prospore membrane.
The inability to form the prospore membrane in the absence of ADY1 does not seem to be randomly distributed among the four spindle poles of an ady1 mutant cell. In the dyads of the ady1 mutant, the two spores are predominantly nonsisters, suggesting that Pfs1p may be more critical for one SPB of a spindle to initiate prospore membrane formation than for the other. In the ady1 mutant, it is very likely that Mpc54p and Spo14p are localized most often to nonsister SPBs, which then are able to initiate prospore membrane synthesis. As a result, predominantly nonsister spores are included in the dyads of the ady1 mutant.
Cells can be induced to produce dyads composed of predominantly nonsister spores under several other conditions. One example is the hfd1-1 mutant, where sporulating cells have been shown to contain two SPBs with the expanded outer plaques and two others without the expanded outer plaques (Okamoto and Iino, 1982). The identity of the hfd1-1 locus is unknown. As another example, when wild-type cells subjected to a reversible thermal arrest were allowed to sporulate, the majority of the sporulated cells were dyads, with two haploid nonsister spores (Davidow et al., 1980). When fresh medium was supplemented after the thermal arrest, the frequency of tetrads increased substantially. It was therefore proposed that an unknown metabolic “substance” might become limiting under certain conditions. The hypothetical substance is sequestered preferentially to the outer plaques of the parental or daughter SPBs to determine the morphogenesis of the outer plaques and the formation of spores. This limiting factor could be regenerated with the addition of fresh medium (Davidow et al., 1980). In the ady1 mutant, the addition of fresh medium did not improve either the general sporulation frequency or the frequency of tetrads among sporulated cells (our unpublished results). The mechanism that leads to formation of monads and dyads in the ady1 mutant may not be a renewable limiting factor that is preferentially sequestered by some SPBs.
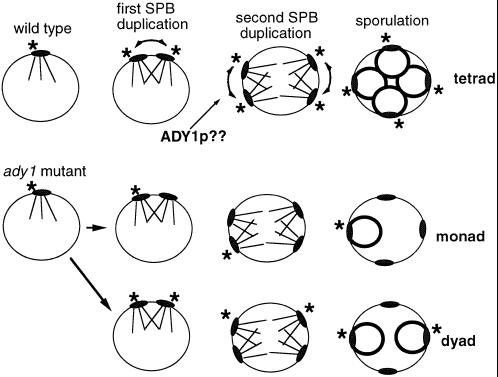
The mechanism by which ADY1 promotes the initiation of prospore membrane formation is presently unknown. It may directly help to recruit or anchor Mpc54p and possibly other proteins to the SPB outer plaque, which only then is able to undergo the morphological expansion in preparation for the prospore membrane assembly. This model does not explain why predominantly nonsister spores are included in the dyads of the ady1 mutant. Moreover, the partially functionally Myc-Pfs1p is not localized to the SPBs, a pattern strongly suggested by this model. Alternatively, Pfs1p may not be involved in prospore membrane formation per se but instead required for completing SPB duplication during meiosis. In this model, SPBs in the ady1 mutant duplicate incorrectly so that typically only the original SPB is capable of recruiting Mpc54p and initiating prospore membrane formation, leading to formation of monads. Less commonly, the first SPB duplication occurs correctly but the second duplication fails, giving rise to two nonsister spores in a dyad. Only rarely does the second duplication proceed correctly, giving rise to the infrequent tetrads (Figure 9).
Figure 9.
Model for Pfs1p function. (Top) Pfs1p is required for all four SPBs in a cell at meiosis II to acquire the ability to initiate prospore membrane formation. (Bottom) In absence of Pfs1p, one or both of the parental SPBs at meiosis II are capable of prospore membrane formation, whereas the two daughter SPBs are not. As a result, predominantly monads or dyads containing two nonsister spores are produced in the ady1 mutant. ∗, SPBs that are capable of initiating prospore membrane formation.
ACKNOWLEDGMENTS
We thank the following individuals for useful comments or reagents: Drs. Linda Riles and Mark Johnston (Washington University) for providing the chromosome VIII deletion strains; Dr. JoAnne Engebrecht (SUNY at Stony Brook) for the SPO14-GFP plasmid; Dr. Michael Knop (Max-Planck-Institut fur Biochemie, Germany) for Mpc54-GFP and Don1-GFP strains; Dr. Jim Haber (Brandeis University) for the pHO plasmid; Dr. John Kilmartin (MRC, United Kingdom) for the anti-Spc90 antibody; Dr. Giora Simchen (The Hebrew University, Israel) for the ade2 mutants; Dr. Rochelle Esposito (University of Chicago, Chicago, IL) for the spo11 mutant; Dr. Jonathan Warner (Albert Einstein College of Medicine, New York, NY) for the anti-L3 antibody; and Drs. Jason Kahana (Ludwig Institute for Cancer Research, San Diego, CA) and Pamela Silver (Harvard University, Boston, MA) for the anti-GFP antibody. This study was funded through American Cancer Society award CB-171, and Grant FY00-139 from the March of Dime Foundation.
REFERENCES
- Brachat A, Kilmartin JV, Wach A, Philippsen P. Saccharomyces cerevisiae cells with defective spindle pole body outer plaques accomplish nuclear migration via half-bridge-organized microtubules. Mol Biol Cell. 1998;9:977–991. doi: 10.1091/mbc.9.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B. Cytology of the yeast life cycle. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces cerevisiae: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1981. pp. 59–96. [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Chu S, Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- Davidow LS, Goetsch L, Byers B. Preferential occurrence of nonsister spores in two-spored asci of Saccharomyces cerevisiae: evidence for regulation of spore-wall formation by the spindle pole body. Genetics. 1980;94:581–595. doi: 10.1093/genetics/94.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H, Goetsch L, Pringle JR. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J Cell Biol. 1996;132:399–411. doi: 10.1083/jcb.132.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Ebisuzaki LK, Segall J. A 15-base-pair element activates the SPS4 gene midway through sporulation in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:3934–3944. doi: 10.1128/mcb.15.7.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I, Jensen RE. Putting the HO gene to work: practical use for mating type switching. In: Guthrie C, Fink GR, editors. Guide to Yeast Genetics and Molecular Biology. New York: Academic Press; 1991. [DOI] [PubMed] [Google Scholar]
- Kassir Y, Simchen G. Monitoring meiosis and sporulation in Saccharomyces cerevisiae. In: Guthrie C, Fink GR, editors. Guide to Yeast Genetics and Molecular Biology. New York: Academic Press; 1991. [DOI] [PubMed] [Google Scholar]
- Klapholtz S, Esposito RE. Isolation of spo12-1 and spo13-1 from a natural variant of yeast that undergoes a single meiotic division. Genetics. 1980;96:567–588. doi: 10.1093/genetics/96.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Strasser K. Role of spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 2000;19:3657–3667. doi: 10.1093/emboj/19.14.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreger-van Rij NJ. Electron microscopy of germinating ascospores of Saccharomyces cerevisiae. Arch Microbiol. 1978;117:73–77. doi: 10.1007/BF00689354. [DOI] [PubMed] [Google Scholar]
- Krisak L, Strich R, Winters RS, Hall JP, Mallory MJ, Kreitzer D, Tuan RS, Winter E. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 1994;8:2151–2161. doi: 10.1101/gad.8.18.2151. [DOI] [PubMed] [Google Scholar]
- Kupiec M, Byers B, Esposito RE, Mitchell AP. Meiosis and sporulation in Saccharomyces cerevisiae. In: Pringle JR, Broach JR, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 889–1036. [Google Scholar]
- Lynn RR, Magee PT. Development of the spore wall during ascospore formation in Saccharomyces cerevisiae. J Cell Biol. 1970;44:688–692. doi: 10.1083/jcb.44.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Rapport E. Spindle, spindle plaques and meiosis in the yeast (Hansen) J Cell Biol. 1971;50:344–361. doi: 10.1083/jcb.50.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J Cell Biol. 1998;140:29–37. doi: 10.1083/jcb.140.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenthal R, Riles L, Guldener U, Klein S, Johnston M, Hegemann JH. Systematic analysis of S. cerevisiae chromosome VIII genes. Yeast. 1999;15:1775–1796. doi: 10.1002/(SICI)1097-0061(199912)15:16<1775::AID-YEA496>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Iino T. Genetic block of outer plaque morphogenesis at the second meiotic division in an hfd1-1 mutant of Saccharomyces cerevisiae. J Gen Microbiol. 1982;128:1309–1317. doi: 10.1099/00221287-128-6-1309. [DOI] [PubMed] [Google Scholar]
- Ozsarac N, Straffon MJ, Dalton HE, Dawes IW. Regulation of gene expression during meiosis in Saccharomyces cerevisiae: SPR3 is controlled by both ABFI and a new sporulation control element. Mol Cell Biol. 1997;17:1152–1159. doi: 10.1128/mcb.17.3.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DD. Biochemical mutants in the smut fungus Ustilago maydis. Genetics. 1949;34:607. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose K, Rudge SA, Frohman MA, Morris AJ, Engebrecht J. Phospholipase D signaling is essential for meiosis. Proc Natl Acad Sci USA. 1995;92:12151–12155. doi: 10.1073/pnas.92.26.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin N, Sugarman E, Haber JE. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics. 1989;122:519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge SA, Morris AJ, Engebrecht J. Relocalization of phospholipase D activity mediates membrane formation during meiosis. J Cell Biol. 1998;140:81–90. doi: 10.1083/jcb.140.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W, Hornack D, Lengyel V, Deng C. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster EO, Byers B. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics. 1989;123:29–43. doi: 10.1093/genetics/123.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern JN, Herskowitz I. Asymmetry and directionality in production of new cell types during clonal growth: the switching pattern of homothallic yeast. Cell. 1979;17:371–381. doi: 10.1016/0092-8674(79)90163-6. [DOI] [PubMed] [Google Scholar]
- Uno I, Matsumoto K, Hirata A, Ishikawa T. Outer plaque assembly and spore encapsulation are defective during sporulation of adenylate cyclase-deficient mutants of Saccharomyces cerevisiae. J Cell Biol. 1985;100:1854–1862. doi: 10.1083/jcb.100.6.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Byers B. Assembly and functions of the spindle pole body in budding yeast. Trends Genet. 1993;9:300–304. doi: 10.1016/0168-9525(93)90247-f. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–909. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Zeng X, Kahana JA, Silver PA, Morphew MK, McIntosh JR, Fitch IT, Carbon J. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J Cell Biol. 1999;146:415–425. doi: 10.1083/jcb.146.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]