Abstract
Post-translational modifications (PTMs) are key modulators of protein function. Huntington disease (HD) is a dominantly inherited neurodegenerative disorder caused by an expanded CAG trinucleotide repeat in the huntingtin (HTT) gene. A spectrum of PTMs have been shown to modify the normal functions of HTT, including proteolysis, phosphorylation and lipidation, but the full contribution of these PTMs to the molecular pathogenesis of HD remains unclear. In this study, we examine all commonly occurring missense mutations in HTT to identify potential human modifiers of HTT PTMs relevant to HD biology. We reveal a SNP that modifies post-translational myristoylation of HTT, resulting in downstream alterations to toxic HTT proteolysis in human cells. This is the first SNP shown to functionally modify a PTM in HD and the first validated genetic modifier of post-translational myristoylation. This SNP is a high-priority candidate modifier of HD phenotypes and may illuminate HD biology in human studies.
Introduction
Huntington disease (HD) is a debilitating neurodegenerative disease with no treatment to delay progression of the disease. HD is caused by an expanded CAG repeat in the huntingtin gene (HTT) that translates into a polyglutamine tract at the N-terminus of the huntingtin protein (HTT)1. Wild-type HTT is critical for cell viability2 and has been shown to play a role in a variety of pathways including orientation of the mitotic spindle3, trafficking of autophagosomes in neurons4, and in regulating autophagy5. Expanded polyglutamine in HTT leads to loss of function and a toxic gain of function in neurons of the striatum and cortex, causing behavioral changes, movement deficits and ultimately death. Age of disease onset is inversely proportional to the length of the CAG repeat6. However, age of onset may be accelerated or delayed by genetic modifiers, including a SNP at the transcription factor binding site of Nf-kB in the HTT promoter7, and in genes related to DNA repair8.
The HTT protein is a large monomeric protein whose function is intricately regulated by post-translational modifications including phosphorylation, acetylation, ubiquitination, proteolysis, and fatty acylation9. While some PTMs of HTT have been shown to be protective against toxicity of mutant HTT, such as phosphorylation at S13/16 and S42110, others are crucial for HD pathogenesis or increase mutant HTT toxicity. In particular, caspase-mediated proteolysis of HTT at amino acid D586 has been shown to be necessary for the development of disease phenotypes in HD mouse models11,12. Consequently, modulating PTMs has become a focus of therapeutic strategies for HD. We sought to identify human SNPs that lead to missense mutations that may alter PTMs in HTT and, consequently, modify progression or pathogenic effects of the disease.
Results
To identify SNPs that could alter HTT PTMs and potentially modify HTT function, all common missense mutations (≥0.1% minor allele frequency; MAF) within HTT were curated from Phase 3 of the 1000 Genomes Project (1 KG) and from the Genome Aggregation Database (gnomAD). Nineteen common missense SNPs with ≥0.1% MAF were found in 1 KG, and 19 common missense SNPs with ≥0.1% MAF were found in gnomAD (Table 1). The top 14 most common missense SNPs in 1 KG and in gnomAD were shared in both data sets, highlighting convergent allele discovery by distinct methodologies.
Table 1.
Functional SNPs in gnomAD and 1000 Genomes Phase 3.
| RSID | Chromosome Position (GRCh37.p13) | Reference | Alternate | Amino acid position (NP_002102.4) | Mutation | Allele Frequency (gnomAD) | Allele Frequency (1000 G) | PROVEAN27 PREDICTION | SIFT28 PREDICTION | SNAP29 PREDICTION |
|---|---|---|---|---|---|---|---|---|---|---|
| rs362331 | 3215835 | T | C | 2311 | Y/H | 0.43680 | 0.44129 | Neutral | Tolerated | Neutral |
| rs362272 | 3234980 | G | A | 2788 | V/I | 0.28390 | 0.21026 | Neutral | Tolerated | Neutral |
| rs363125 | 3189547 | C | A | 1722 | T/N | 0.12750 | 0.18910 | Neutral | Tolerated | Neutral |
| rs149109767 | 3230410 | AGAG | A | 2645 | E/– | 0.06067 | 0.02656 | Deleterious | NA | — |
| rs35892913 | 3148570 | G | A | 1066 | V/I | 0.04200 | 0.02356 | Neutral | Tolerated | Neutral |
| rs363075 | 3137674 | G | A | 895 | G/R | 0.04069 | 0.02376 | Deleterious | Damaging | Effect |
| rs1143646 | 3148653 | T | G | 1093 | I/M | 0.03962 | 0.02236 | Neutral | Damaging | Neutral |
| rs3025837 | 3174845 | A | C | 1387 | N/H | 0.01489 | 0.04153 | Neutral | Tolerated | Neutral |
| rs34315806 | 3162034 | C | T | 1262 | T/M | 0.00837 | 0.02756 | Neutral | Damaging | Neutral |
| rs151106561 | 3132038 | A | G | 627 | Q/R | 0.00523 | 0.00280 | Neutral | Tolerated | Neutral |
| rs150003356 | 3189510 | T | C | 1710 | C/R | 0.00281 | 0.00100 | Deleterious | Tolerated | Effect |
| rs3025843 | 3156038 | A | G | 1175 | T/A | 0.00256 | 0.00679 | Neutral | Tolerated | Neutral |
| rs118005095 | 3129240 | G | A | 553 | G/E | 0.00242 | 0.00619 | Neutral | Tolerated | Neutral |
| rs34389685 | 3133113 | G | A | 698 | G/E | 0.00190 | 0.00200 | Deleterious | Damaging | Effect |
| rs186355914 | 3109095 | C | T | 233 | A/V | 0.00170 | 0.00020 | Neutral | Damaging | Neutral |
| rs201702168 | 3221998 | A | T | 2446 | E/D | 0.00154 | 0.00140 | Neutral | Tolerated | Neutral |
| rs375957195 | 3230428 | C | A | 2647 | D/E | 0.00153 | 0.00319 | Neutral | Tolerated | Neutral |
| rs1065746 | 3148624 | G | C | 1084 | D/H | 0.00135 | 0.00020 | Deleterious | Damaging | Effect |
| rs190593027 | 3144528 | C | G | 996 | T/R | 0.00110 | 0.00140 | Neutral | Damaging | Neutral |
| rs201646519 | 3210567 | T | C | 2076 | S/P | 0.00018 | 0.00100 | Neutral | Tolerated | Neutral |
| rs201760820 | 3134411 | G | A | 789 | V/M | 0.00047 | 0.00100 | Neutral | Tolerated | Neutral |
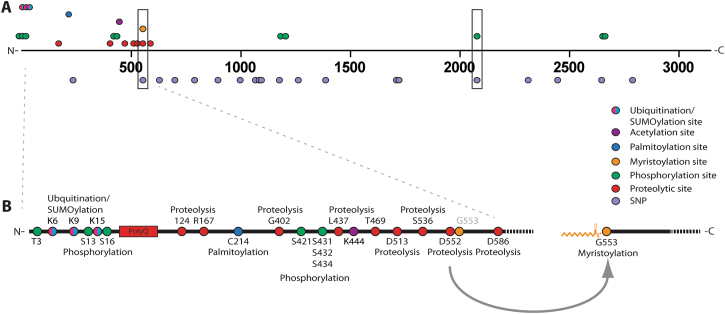
A manually curated list of known HTT PTMs was intersected with all common missense SNPs identified from 1 KG and gnomAD (Fig. 1). Among the 21 common missense SNPs identified from 1 KG and gnomAD, two overlapped directly with known HTT PTMs; rs118005095 (G553E) and rs201646519 (S2076P) (Table 1, boxed; Supplemental Table 1). SNP rs201646519 intersects with the predicted phosphorylated residue S2076, resulting in a proline substitution thereby blocking phosphorylation at this site. Myristoylated G55313,14 overlapped with the missense SNP rs118005095, which leads to a substitution of glycine to glutamic acid (G553E) and would therefore completely abrogate post-translational myristoylation at this site (Fig. 2A). Myristoylation involves the covalent addition of the 14 carbon fatty acid, myristate, to N-terminal glycines either co-translationally on the nascent polypeptide as it is being translated on the ribosome following the removal of the initiator methionine or post-translationally following proteolysis that exposes an N-terminal glycine15. The fatty acid moiety promotes membrane binding of proteins. No other common missense SNPs directly intersected with a known HTT PTM residue. Due to the low frequency of rs201646519, we focused on rs118005095.
Figure 1.
Schematic representing where HTT PTMs and HTT missense SNPs intersect. (A) Linear map of HTT SNPs leading to missense mutations mapped to known HTT PTMs. The two missense mutations that directly intersect with myristoylation at G553 and phosphorylation at S2076, G553E and S2076P, are boxed. (B) PTMs within the first 586 amino acids of HTT are highlighted. Proteolytic caspase sites are indicated on the bottom while non-caspase mediated proteolytic sites are displayed on top. G553 is myristoylated following caspase cleavage at D552.
Figure 2.
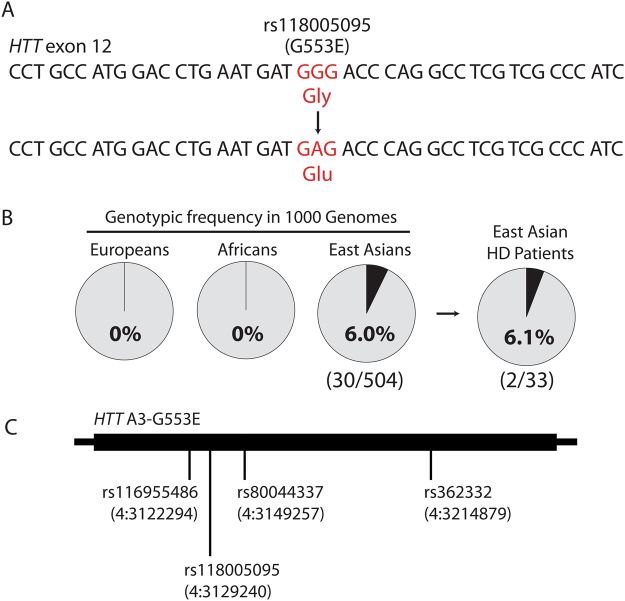
The rs118005095 missense variant is a naturally occurring human SNP that alters the HTT amino acid sequence. (A) rs118005095 results in mutation of the HTT 553 glycine residue to glutamic acid. (B) rs118005095 occurs exclusively in populations of East Asian ancestry. (C) The rs118005095 variant is one of four SNPs defining a gene-spanning HTT haplotype in the East Asian population.
The G553E SNP rs118005095 shows pronounced ethnic differences in frequency, being most common in individuals of East Asian ancestry. In 1 KG, rs118005095 is observed on 31 out of 5008 chromosomes from all populations, of which 30 instances occur in subjects of defined East Asian origin (n = 504 subjects) at a genotypic frequency of 6.0% (30/504) and allelic frequency of 3.0% (30/1008) (Fig. 2B). The one remaining 1 KG chromosome with rs118005095 outside East Asian individuals occurs in a Bengali subject from Bangladesh, close to East Asia. In the gnomAD data, rs118005095 occurs in chromosomes from East Asian subjects at an allelic frequency of 2.914% (552/18942). In contrast, rs118005095 is observed in <0.1% of chromosomes from European, African, South Asian, and Latino subjects, reflecting its absence in similar reference populations from 1 KG. Therefore rs118005095 is expected to occur in approximately 6.0% of individuals from the East Asian general population.
We have previously shown that HTT is characterized by a haplotype block of low recombination and that SNPs within the gene represent specific haplotypes16. HTT haplotype analysis in 1 KG reveals that rs118005095 occurs on a specific A3b haplotype variant in the East Asian population, tagged by three additional haplotype-defining SNPs: rs116955486, rs80044337, and rs362332 (Fig. 2C). Of note, previously published haplotypes of the HD mutation in East Asian patients do not include this haplotype, suggesting that this SNP is not found on mutant HTT chromosomes in the East Asian population. We thus predicted that rs118005095 may occur in wild-type HTT of East Asian HD patients, potentially impacting the normal function of myristoylation in the context of the disease.
We have previously shown that the expanded polyglutamine mutation dramatically reduces myristoylation of HTT at G55314. HD patients with the G553E mutation (rs118005095) would therefore be unable to compensate for deficient myristoylation with a wild-type copy bearing the rs118005095 variant. Genotyping of archived DNA from 33 East Asian HD patients in the UBC HD Biobank confirmed the presence of the SNP in two patients (Fig. 2B). As expected, phasing of rs118005095 to the mutation revealed that this missense variant occurred on wild-type HTT in these patients. However, no clinical information was available to assess the potential modifying effects of the SNP.
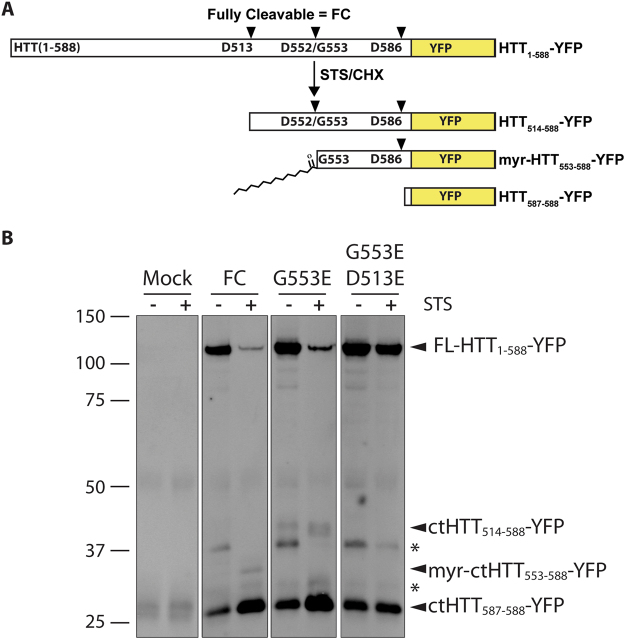
To investigate the predicted cellular effects of the rs118005095 (G553E) variant, we generated the mutation in a biologically relevant form of HTT bearing the first 1–588 amino acids appended to YFP (HTT1–588-YFP, Fig. 3A), previously shown to be post-translationally myristoylated14 and disease-causing in HD mouse models17. Myristoylation at G553 was first validated in WT and mHTT1–588-YFP using an alkyne-myristate analog that can be covalently linked to biotin-azide via Click chemistry for detection (Supplementary Fig. 1)18. Caspase-cleavage at D552 was promoted using staurosporine (STS) and cycloheximide (CHX) in order to increase post-translational myristoylation at G553, as previously described14. Post-translational myristoylation was detected in fully-cleavable (FC) HTT (HTT1–588-YFP with no point mutations added) even in the absence of STS/CHX, likely due to basal levels of caspase activity (Supplementary Fig. 1). Myristoylation was increased in the presence of STS/CHX. A point mutation at the primary site of caspase-cleavage in HTT at position D586 to glutamate was made in order to more easily measure myristoylation at G553 following caspase cleavage at D552 (D586E, Supplementary Fig. 1). This led to a dramatic increase of post-translational myristoylation at G553.
Figure 3.
The G553E substitution blocks post-translational myristoylation and alters proteolysis of wtHTT. (A) Schematic representation of proteolytic cleavage of WT fully cleavable (FC) HTT1–588-YFP. HTT is proteolysed by caspases at D586 and D552 and post-translationally myristoylated at G553. (B) HeLa cells were transfected with WT HTT1–588-YFP bearing the indicated point mutations. Proteolysis was induced with staurosporine (STS) and cycloheximide. In the presence of the G553E mutation, a higher molecular weight band was detected that was blocked when D513 was mutated to E, confirming the D513 caspase cleavage site. *Denotes unknown or non-specific bands. Image is a composite of lanes from the same Western blot.
As predicted, substitution of the essential glycine to alanine or serine (G553A/D586E and G553S/D586E) completely abrogated post-translational myristoylation of HTT at G553 following caspase-induced cleavage. Similarly, blocking caspase cleavage at D552 by substituting the aspartate to glutamate (D552E, Supplementary Fig. 1) completely blocked post-translational myristoylation at G553. Inhibiting caspase-cleavage at D552 appeared to block the generation of both the myristoylated and non-myristoylated bands. Therefore, we determined that myristoylation status at G553 could also be determined by gel mobility. Non-myristoylated C-terminal (ct) HTT553–588-YFP migrates at a slightly higher molecular weight than myristoylated-ctHTT553–588-YFP (Supplementary Fig. 1). This mobility shift was shown previously in a shorter myristoylated fragment14 and mimics other lipidated proteins such as LC319.
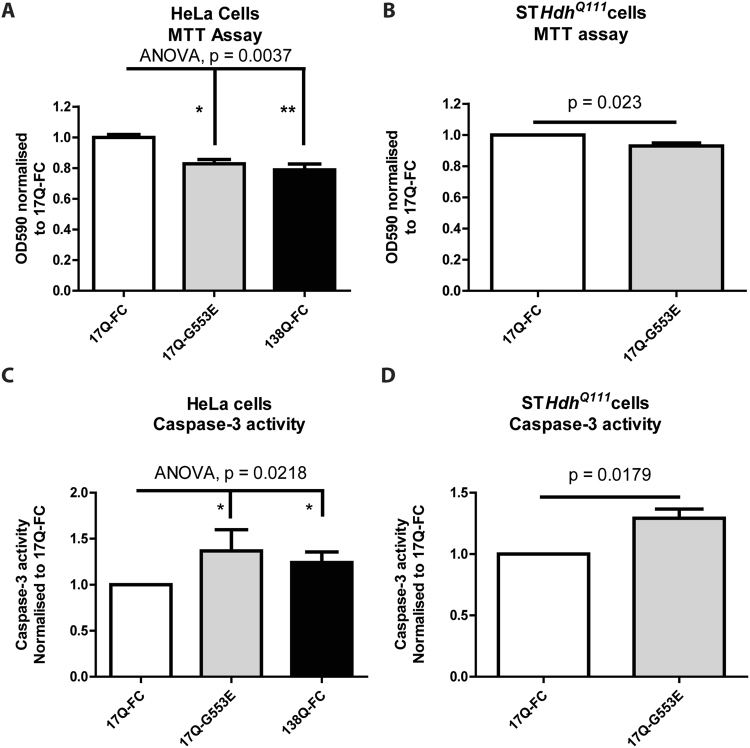
Consequently, the G553E mutation was further characterized in the absence of the myristate analog in WT-HTT1–588-YFP (Fig. 3). As expected, substitution of the essential N-terminal glycine at position 553 completely abrogated post-translational myristoylation of the wild-type form (Fig. 3B). Surprisingly, the G553E mutation also inhibited caspase cleavage of HTT at D552, which was associated with a concomitant increase in caspase cleavage of HTT at D513 (Fig. 3B). Cleavage at D586 did not appear to be affected. Of note, blocking myristoylation conventionally by substituting glycine to alanine did not prevent cleavage at D552 (Supplemental Fig. 1). Proteolysis at D513 was recently shown to be toxic in cells11. Due to the increased cleavage of HTT at the toxic site D513 in the presence of the G553E mutation, we hypothesized that the G553E variant may induce toxicity of wild-type HTT. Indeed, HeLa cell viability, measured by the MTT assay, was significantly reduced, similar to mutant 138Q HTT alone, in the presence of the G553E mutation compared wild-type HTT (Fig. 4A). Toxicity was also evaluated by measuring cleavage of spectrin (Supplemental Fig. 2), a common marker for cell death, particularly in HD studies20–22. Spectrin cleavage was significantly increased (Supplemental Fig. 2) in HeLa cells expressing mHTT, while the G553E mutation had an intermediate effect corresponding to the cell viability MTT assay. Cell viability was also significantly decreased in STHdhQ111 cells in the presence of the G553E mutation compared to wild-type FC-HTT (Fig. 4B). These cells are striatal derived cells from transgenic mice in which human exon 1 has been inserted into the mouse gene and represent a cellular model for HD23. In agreement with the cell viability MTT assay, we detected a significant increase in caspase-3 activity in HeLa cells expressing the 17Q-HTT-G553E-YFP or mHTT (Fig. 4C), suggesting the intiation of apoptosis. Similarly, the G553E substitution significantly increased caspase-3 activity in STHdhQ111 cells (Fig. 4D). Altogether, this suggests that expression of G553E mutation is toxic alone and toxicity is increased in the presence of the disease causing polyQ mutation.
Figure 4.
The G553E mutation leads to decreased cell viability and increased caspase-3 activity in HeLa cells and the HD cell model STHdhQ111 cells. (A,B) The MTT assay was used to measure cell viability in cells expressing the indicated constructs. (A) HeLa cells (ANOVA, p = 0.0037, h = 11.22, n = 6, 2–3 replicates per n). (B) STHdhQ111 cells (paired t-test, p = 0.0234, n = 5, 3 replicates per n). (C,D) Caspase-3 activity was measured in cells expressing the indicated constructs by following cleavage of the fluorescent substrate Ac-DEVD-AFC. Caspase-3 activity is represented as relative fluorescence units normalised to 17Q-FC. (C) HeLa (ANOVA, p = 0.0218, h = 7.652, n = 5, 3 replicates per n). (D) STHdhQ111 cells (paired t-test, p = 0.0179, n = 5, 3 replicates per n). (A) and (C) Represent Kruskal-Wallis non-parametric ANOVA with Dunn’s Multiple comparison test post hoc analysis. *p < 0.05, **p < 0.01). (B) and (D) Represent 2-tailed paired Student’s t-test. Error bars represent SEM.
Discussion
Previously, we have shown that proteolysis at D586 is essential to the pathogenesis of HD in a mouse model12. In addition, we identified post-translational myristoylation as a novel PTM in HTT that is decreased in the presence of the expanded polyQ suggesting it is a protective PTM14. Now, we have identified the first SNP leading to a missense mutation that alters multiple PTMs in HTT. This is also the first demonstration of a naturally occurring missense mutation that blocks post-translational myristoylation. The G553E mutation was shown to completely abrogate post-translational myristoylation of HTT and induce cellular toxicity of the protein in cellulo. Surprisingly, this mutation gives rise to a toxic fragment of wild-type HTT caused by proteolysis at D513.
We have shown the G553E mutation blocks caspase cleavage at D552 leading to increased cleavage of HTT at the toxic caspase site D513. It has recently been shown that proteolysis of either wild-type HTT or mutant HTT at more than one site, particularly caspase-cleavage at D513 and D586, is toxic11. This is mediated by a newly appreciated toxicity associated with the C-terminal HTT that is exacerbated by toxic shorter N-terminal fragments11,24. It is thought that the N- and C-terminal fragments interact in the presence of single site cleavage, which is considered less toxic. However, increased cleavage at multiple sites leads to decreased interaction following proteolysis at more than one site, particularly caspase cleavage at D513 and D586 and calpain cleavage at R16711. This proteolysis is toxic in both wild-type and mutant HTT.
Consequently, increased proteolysis of wild-type HTT at D513 caused by the G553E mutation, in conjunction with cleavage at D586 and loss of protective myristoylation, is predicted to be toxic. Loss of function of the mutant HTT allele would likely exacerbate this toxicity (Fig. 5), which is supported by the increased toxicity when the G553E mutation is expressed in the presence of the HD mutation in STHdhQ111 cells (Fig. 4B and D). Remarkably, blocking cleavage at D586 in an HD mouse model has been shown to completely ameliorate the disease phenotype in mice12. Therefore, we predict that the G553E mutation may be an important disease modifier and will lead to an earlier onset of disease in HD patients. It will be important to screen East Asian HD patients for the G553E mutation to validate the proteolytic consequences and phenotypic impact of the rs118005095 variant.
Figure 5.
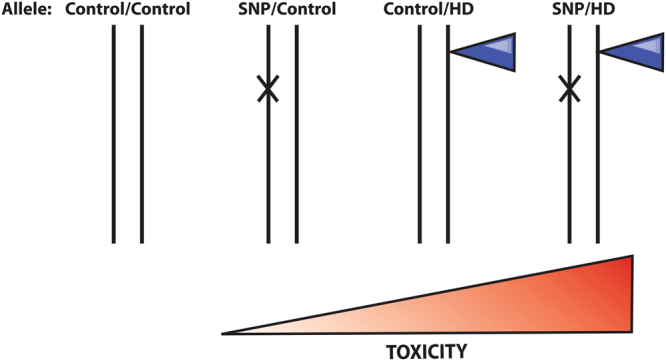
Allelic representation of the compound effect of the G553E mutation with the HD mutation. HD is a monogenic disease caused by a CAG expansion. The SNP encoding the G553E mutation occurs on the control allele causing toxicity. It is predicted that carriers of both mutations with have an earlier onset of disease.
Materials and Methods
Genotyping and Haplotype Analysis
Haplotype analysis of rs118005095 in 1000 Genomes Phase 3 was performed using custom scripts in the R statistical computing environment applied to a variant call file spanning HTT variants (chr4:3034088-3288007) in all 5008 subjects16. SNPs in high pairwise linkage disequilibrium (r2 > 0.90) with rs118005095 were found in similarly high linkage disequilibrium on a common gene-spanning haplotype in subjects of East Asian ancestry among all populations in 1000 Genomes. Direct genotyping of rs118005095 was performed on banked DNA samples from East Asian HD subjects from the UBC HD Biobank. PCR amplicons containing the G553E variant (rs118005095) were generated using rs118005095 forward (GCGGACTCAGTGGATCTGGC) and rs118005095 reverse (GTCTGAAGGGGTAACAGCTGAATC) primers and Sanger sequenced using one or both primers. All subject DNA samples were collected, stored and examined with informed consent and approval of the University of British Columbia, Children’s and Women’s Health Centre of British Columbia and the Vancouver Coastal Health Research Institute’s Research Ethics Boards, (UBC-C&W H06-70467 and H06-70532, VCHRI V09-0129). The study of both previously collected de-identified archived DNA samples from the HD BioBank at UBC as well as patient samples collected from the Centre for HD at UBC was conducted in accordance with the protocols and procedures required by UBC Clinical Research Ethics Board and their affiliated REBs.
Cloning
All HTT-YFP point mutations were generated using gBlocks, as described previously25, except the D513E mutation was generated by Top Gene Technologies. All point mutations were confirmed by DNA sequencing (Eurofins Genomics).
Click chemistry and fragment analysis
Post-translational myristoylation of HTT1–588-YFP was detected as described previously14. Briefly, HeLa cells were transiently transfected using X-tremeGene (Roche) as per the manufacturer’s directions. After 20 h post-transfection, the media was exchanged with DMEM containing charcoal filtered FBS and cells were incubated with alkyne-myristate (Cayman Chemicals) for 30 mins prior to the addition of 1 µM staurosporine (STS) and 5 µg/mL of cycloheximide (CHX) to promote caspase activity and prevent synthesis of new protein, respectively. After 4 h, cells were lysed in modified RIPA buffer. HTT1–588-YFP was immunoprecipitated using goat anti-GFP (Eusera) and subjected to Click chemistry using azido-biotin (Invitrogen). Following SDS-PAGE on large 10% gels, post-translational myristoylation was detected by Western blot analysis using streptavidin Alexa680 (Invitrogen) and Licor. YFP was detected using rabbit anti-GFP (Eusera).
Viability assays
Cell viability was measured using the MTT assay (Sigma) as per the manufacturer’s directions. Briefly, 20 h post-transfection in HeLa cells and 44 h in StHdh(Q111) cells, MTT was added for an additional 4 h. OD readings were normalized to WT fully cleavable HTT. The experiment was performed 5–6 times with 3 technical replicates per experiment for each condition. Alternatively, toxicity of each construct was evaluated by measuring cleavage of spectrin. As above, HeLa cells were transfected with the indicated constructs for 24 h, lysed in RIPA buffer, lysates were separated on 7% SDS-PAGE gels, transferred to PVDF, probed for spectrin (AA6, Enzo Life Sciences) and detected by Licor. Full-length and cleaved spectrin were quantified using Image Studio Lite V.4 and presented as a ratio of cleaved spectrin over total spectrin (n = 4).
Caspase-3 activity was measured as previously described26. Briefly, adherent and floating cells were collected 24 h post-transfection and lysed (50 mM Tris pH 8, 150 mM NaCl, 1% Igepal). Lysates were diluted in 2X caspase assay buffer (100 mM HEPES pH 7.4, 200 mM NaCl, 0.2% CHAPS, 2 mM EDTA and 20% glycerol). Fresh DTT (20 mM) and 200 µM Ac-DEVD-AFC (Enzo Biosciences) was added in a 384-well plate. The relative fluorescence units (RFU)/min were acquired by measuring fluorescence every 2–5 min with excitation at 400 nm and emission at 505 nm.
Data availability
Data described in this study will be available from the corresponding authors upon request.
Electronic supplementary material
Acknowledgements
This work was supported by the CHDI Foundation and the Canadian Institutes of Health Research (CIHR 20R90174). DDOM was supported by postdoctoral fellowships from CIHR, Michael Smith Foundation for Health Research and the Bluma Tischler Fellowship from UBC. CK was supported by a CIHR Doctoral Research Award. The authors would like to thank Dr. Niels Skotte for contributing to illustrations in Figure 1 and Dr. Dagmar Ehrnhoefer for help in setting up the caspase activity assay.
Author Contributions
D.D.O.M., C.K. and M.R.H. conceived of experimental design. D.D.O.M. and C.K. wrote the manuscript. D.D.O.M. performed the molecular biology and biochemical analyses as well as the comparisons between SNP and PTMs. C.K. completed the haplotype and genotype analysis. J.A.C. aided in analyses and phasing of haplotypes, CAG repeats and SNP sequencing data from patient DNA. Y.T.N. aided in the biochemical analyses. R.A.S. performed direct sequencing of the G553E SNP in patient DNA.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25903-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
D. D. O. Martin, Email: dale.martin@uwaterloo.ca
M. R. Hayden, Email: mrh@cmmt.ubc.ca
References
- 1.The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- 2.Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat. Genet. 2000;26:300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 3.Godin JD, et al. Huntingtin Is Required for Mitotic Spindle Orientation and Mammalian Neurogenesis. Neuron. 2010;67:392–406. doi: 10.1016/j.neuron.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 4.Wong YC, Holzbaur ELF. The Regulation of Autophagosome Dynamics by Huntingtin and HAP1 Is Disrupted by Expression of Mutant Huntingtin, Leading to Defective Cargo Degradation. J. Neurosci. 2014;34:1293–1305. doi: 10.1523/JNEUROSCI.1870-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin DDO, Ladha S, Ehrnhoefer DE, Hayden MR. Autophagy in Huntington disease and huntingtin in autophagy. Trends Neurosci. 2015;38:26–35. doi: 10.1016/j.tins.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Langbehn DR, Brinkman RR, Falush D, Paulsen JS, Hayden MR. A new model for prediction of the age of onset and penetrance for Huntington’s disease based on CAG length. Clin. Genet. 2004;65:267–277. doi: 10.1111/j.1399-0004.2004.00241.x. [DOI] [PubMed] [Google Scholar]
- 7.Bečanović K, et al. A SNP in the HTT promoter alters NF-κB binding and is a bidirectional genetic modifier of Huntington disease. Nat. Neurosci. 2015;18:807–816. doi: 10.1038/nn.4014. [DOI] [PubMed] [Google Scholar]
- 8.Lee J-M, et al. Identification of genetic factors that modify clinical onset of Huntington’s disease. Cell. 2015;162:516–526. doi: 10.1016/j.cell.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrnhoefer DE, Sutton L, Hayden MR. SmallChanges, Big Impact: Posttranslational Modifications and Function of Huntingtin in Huntington Disease. Neurosci. 2011;17:475–492. doi: 10.1177/1073858410390378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warby SC, et al. Huntingtin phosphorylation on serine 421 is significantly reduced in the striatum and by polyglutamine expansion in vivo. Hum. Mol. Genet. 2005;14:1569–1577. doi: 10.1093/hmg/ddi165. [DOI] [PubMed] [Google Scholar]
- 11.El-Daher M-T, et al. Huntingtin proteolysis releases non-polyQ fragments that cause toxicity through dynamin 1 dysregulation. EMBO J. 2015;34:2255–71. doi: 10.15252/embj.201490808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham RK, et al. Cleavage at the Caspase-6 Site Is Required for Neuronal Dysfunction and Degeneration Due to Mutant Huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 13.Martin DDO, et al. Tandem reporter assay for myristoylated proteins post-translationally (TRAMPP) identifies novel substrates for post-translational myristoylation: PKCε, a case study. FASEB J. 2012;26:13–28. doi: 10.1096/fj.11-182360. [DOI] [PubMed] [Google Scholar]
- 14.Martin DDO, et al. Identification of a post-translationally myristoylated autophagy-inducing domain released by caspase cleavage of huntingtin. Hum. Mol. Genet. 2014;23:3166–3179. doi: 10.1093/hmg/ddu027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin DDO, Hayden MR. Post-translational myristoylation at the cross roads of cell death, autophagy and neurodegeneration. Biochem. Soc. Trans. 2015;43:229–234. doi: 10.1042/BST20140281. [DOI] [PubMed] [Google Scholar]
- 16.Kay, C. et al. Huntingtin Haplotypes Provide Prioritized Target Panels for Allele-specific Silencing in Huntington Disease Patients of European Ancestry. Molecular Therapy, 10.1038/mt.2015.128 (2015). [DOI] [PMC free article] [PubMed]
- 17.Zuleta A, Vidal RL, Armentano D, Parsons G, Hetz C. AAV-mediated delivery of the transcription factor XBP1s into the striatum reduces mutant Huntingtin aggregation in a mouse model of Huntington’s disease. Biochem. Biophys. Res. Commun. 2012;420:558–563. doi: 10.1016/j.bbrc.2012.03.033. [DOI] [PubMed] [Google Scholar]
- 18.Yap MC, et al. Rapid and selective detection of fatty acylated proteins using omega-alkynyl-fatty acids and click chemistry. J. Lipid Res. 2010;51:1566–80. doi: 10.1194/jlr.D002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). 8627 (2016). [DOI] [PMC free article] [PubMed]
- 20.Skotte, N. H. et al. Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: Providing a therapeutic option for all Huntington disease patients. PLoS One9 (2014). [DOI] [PMC free article] [PubMed]
- 21.Wang KK, et al. Simultaneous degradation of alphaII- and betaII-spectrin by caspase 3 (CPP32) in apoptotic cells. J. Biol. Chem. 1998;273:22490–22497. doi: 10.1074/jbc.273.35.22490. [DOI] [PubMed] [Google Scholar]
- 22.Weber JJ, Ortiz Rios MM, Riess O, Clemens LE, Nguyen HP. The calpain-suppressing effects of olesoxime in Huntington’s disease. Rare Dis. 2016;4:e1153778. doi: 10.1080/21675511.2016.1153778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trettel F. Dominant phenotypes produced by the HD mutation in STHdhQ111 striatal cells. Hum. Mol. Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 24.Ochaba J, et al. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc. Natl. Acad. Sci. 2014;111:16889–16894. doi: 10.1073/pnas.1420103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong BKY, et al. Partial rescue of some features of Huntington Disease in the genetic absence of caspase-6 in YAC128 mice. Neurobiol. Dis. 2015;76:24–36. doi: 10.1016/j.nbd.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 26.Ehrnhoefer DE, et al. A quantitative method for the specific assessment of caspase-6 activity in cell culture. PLoS One. 2011;6:e27680. doi: 10.1371/journal.pone.0027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi, Y., Sims, G. E., Murphy, S., Miller, J. R. & Chan, A. P. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS One7 (2012). [DOI] [PMC free article] [PubMed]
- 28.Sim N-L, et al. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–7. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hecht M, Bromberg Y, Rost B. Better prediction of functional effects for sequence variants. BMC Genomics. 2015;16:S1. doi: 10.1186/1471-2164-16-S8-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data described in this study will be available from the corresponding authors upon request.