Abstract
Structural studies of integral membrane proteins have been limited by the intrinsic conformational flexibility and the need to stabilize the proteins in solution. Stabilization by mutagenesis was very successful for structural biology of G protein-coupled receptors (GPCRs). However, it requires heavy protein engineering and may introduce structural deviations. Here we describe the use of specific calixarenes-based detergents for native GPCR stabilization. Wild type, full length human adenosine A2A receptor was used to exemplify the approach. We could stabilize native, glycosylated, non-aggregated and homogenous A2AR that maintained its ligand binding capacity. The benefit of the preparation for fragment screening, using the Saturation-Transfer Difference nuclear magnetic resonance (STD-NMR) experiment is reported. The binding of the agonist adenosine and the antagonist caffeine were observed and competition experiments with CGS-21680 and ZM241385 were performed, demonstrating the feasibility of the STD-based fragment screening on the native A2A receptor. Interestingly, adenosine was shown to bind a second binding site in the presence of the agonist CGS-21680 which corroborates published results obtained with molecular dynamics simulation. Fragment-like compounds identified using STD-NMR showed antagonistic effects on A2AR in the cAMP cellular assay. Taken together, our study shows that stabilization of native GPCRs represents an attractive approach for STD-based fragment screening and drug design.
Introduction
G protein-coupled receptors (GPCR) represent one of the largest family of integral membrane proteins and constitute highly druggable targets1–5. GPCRs are known to undergo conformational changes upon ligand binding and signal transduction6,7. This conformational flexibility represents a bottleneck in protein production and crystallographic studies. To improve thermostability and conformational homogeneity, mutagenesis and protein fusion (such as T4 lysozyme) approaches have been developed for various GPCRs such as β1 adrenergic or adenosine A2A receptors8–10. Co-expression of mini-protein G has also helped to stabilize A2A receptor11. In addition to acting at the expression level, high-affinity ligands, lipids or lipid-like molecules can be added during membrane preparation, solubilization and/or purification to provide conformational or oligomeric stabilization12–17. Recently the adenosine A1 receptor was crystallized bound to a selective covalent antagonist, revealing therefore basis for subtype selectivity18. Another approach is also possible using antibody fragments (Fab) or nanobodies as crystallization chaperones to stabilize GPCRs and facilitate their crystallization19–26. Recently, in situ reconstitution of the adenosine A2A receptor in spontaneously formed synthetic liposomes was described to allow microscopy visualization and radio-ligand binding27. Other artificial membrane generation tools were also described28,29.
Those technologic and methodologic advancements led to the resolution of 251 GPCR 3D structures deposited in the PDB30–35 and open new routes for drug discovery, including the fragment-based approach. The fragment-based method consists in screening weak-affinity small molecular-weight compounds against protein targets36. The technique is well established for soluble therapeutic targets, while few studies have been described for membrane proteins. Yet fragment screening could be particularly valuable in the case of GPCRs, for the development of allosteric modulators that can overcome the selectivity issue of orthosteric ligands17,37. Fragment screening has been performed using biological and biophysical assays, including SPR38–41 and NMR-based TINS technology41,42. Although these techniques have proved to be valuable, it is of high importance to develop orthogonal methods that enable robust identification and validation of fragment hits. Besides, both SPR and TINS approaches require the immobilisation of the receptors. By comparison to the Carr-Purcell-Meiboom-Gill sequence (CPMG) experiment used in the TINS technology, the so-called Saturation-Transfer Difference (STD) NMR experiment provides structural information through the discrimination of solvent exposed and buried hydrogens of the ligand bound to the receptor43,44. While it is acknowledged that the STD experiment is particularly efficient for fragment screening, this technique has not been successfully applied to purified GPCRs yet.
To allow drug design and fragment screening on wild-type GPCRs using STD experiments, we have developed a strategy using calixarene-based detergent to solubilize and stabilize native, full length and functional GPCRs. We have lately reported on a systematic solubilization method for membrane proteins that allows screening for suitable detergents45–47 and have described the use of novel calixarene-based detergents46,48–50. Here, we report our solubilization strategy using the adenosine A2A receptor (A2AR) as a case study. A2AR belongs to the GPCR class-A family of membrane spanning proteins that is involved in the brain and immune system regulation51. A2AR is of high medical interest particularly in Parkinson disease52, and also in cancer immunology53. A2AR is also responsible for regulating blood flow to the cardiac muscle and is important in the regulation of glutamate and dopamine release54,55. The purified adenosine A2A receptor shows enhanced thermostability, while its behavior in solution shows no sign of aggregation and the presence of homogenous populations of monomers and oligomers. Functionality was assessed by radioligand binding. The feasibility of the STD-based fragment screening is demonstrated with the observation of the binding of characterized agonist and antagonist compounds to A2AR. The STD-NMR screening results illustrate the advantage of the experiment to obtain rapid structural information and gain additional insight into the ligand interaction.
Taken together, this work shows that wild-type GPCRs can be screened to identify fragment hits using STD-NMR experiments, which will bring new information for drug discovery in particular for the identification of allosteric modulators. This work also changes the dogma that GPCRs are by default unstable proteins requiring stabilization by mutagenesis and describes a new strategy for the fragment screening of highly unstable and druggable targets.
Results
Functional expression of WT and full length A2AR
Full length and wild type A2AR was expressed in yeast (Pichia pastoris) and Sf9 (Spodoptera frugiperda) with a His-tag at the amino-terminal. As shown in Fig. S1A, A2AR expression in yeast was clone-dependent. A specific band was observed at ~40 kDa band using a specific A2AR antibody, mainly for clones 2 and 3. A sixty-nine hours induction gives higher yield than 21 hours for A2AR expression. Therefore, 69 hours induction and clone 2 were selected for further expression. Regarding insect cells expression, Sf9 insect cells exhibit better expression 48 and 72 hours post-infection (Fig. S1B). Forty-eight hours post-infection time was used for further expression steps. To evaluate the localization of the expressed protein, we performed cell lysis and membrane fractionation. From both yeast and insect cells, two fractions corresponding to enriched internal membranes (15000 G centrifugation, 15 K) and plasma membranes (100000 G centrifugation, 100 K) were analyzed by Western blot (Fig. S1C–F). Fractionation of Sf9 cells shows that A2AR was expressed in the 100 K and 15 K fractions, similarly (Fig. S1D and S1F). This was not the case for yeast expression since most of A2AR was observed in the 15 K fraction (Fig. S1C and S1E). To verify if A2AR expression was functional, we performed radioligand binding using the well-characterized agonist CGS-21680. Saturation curves show a specific binding of 3H-CGS-21680 to all A2AR containing membranes (Fig. S1C–F and Table S9). Extrapolated Kd was generated for each membrane fraction. Interestingly, similar Kd values of ~0.11 (±0.05) and 0.29 µM (±0.06) were observed for 15 K and 100 K Sf9 membranes fractions, respectively. In contrast, yeast enriched plasma membranes (100 K) showed a lower Kd (0.80 ± 0.26 µM) in comparison to enriched internal membranes (15 K) (0.31 ± 0.13 µM). Thus, A2AR expression was functional.
Solubilization, purification and ligands binding
Since dodecylmaltoside (DDM) was already reported to successfully solubilize A2AR31,33,56 and given that calixarene based detergent (CALX) were recently described to have a positive impact on membrane proteins stability46,47,49,57, we performed co-solubilization with DDM/CHS in combination with CALX-R10 detergent. Figure S3A shows that it was easier to solubilize A2AR from Sf9 than from P. pastoris (compare lane 3 to 2). To assess A2AR N-glycome, total N-glycans attached to A2AR purified from both plasma and internal membranes of yeast and Sf9 were enzymatically released, fluorescently labeled, and analyzed by hydrophilic-interaction ultra-high-performance liquid chromatography with fluorescence detection (HILIC-UHPLC-FLD).
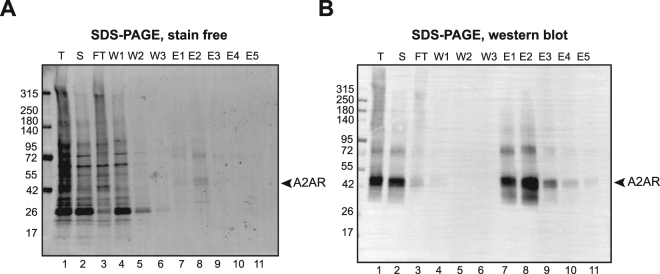
Figure S3 shows that glycosylation profile is preserved regardless of the localization of expressed protein (plasma vs internal membranes). This is true for A2AR expressed in P. pastoris and in Sf9. A2AR from Pichia was difficult to solubilize in comparison to Sf9. In addition to that, solubilization of A2AR from Sf9 offers the possibility to use both internal and plasma membranes for purification since they both have similar glycosylation pattern and similar ligand binding properties. We therefore combined Sf9 membranes (internal and plasma) for larger scale solubilization, purification and A2AR characterization. Good solubilization yields (~90%) were obtained for this detergent mixture as shown in Fig. 1B (compare lane 2 to 1). Most of A2AR could bind to the Talon-His column (Fig. 1B, compare lane 3 and 2) and elute specifically with a good purity (>90%) as shown in Fig. 1A (lane 8). Higher molecular weight gel migration of A2AR at ~80 kDa was observed. This corresponds most probably to SDS-resistant dimers since protein samples were not heated to avoid aggregation. This is commonly observed for membrane proteins. We then assessed radioligand binding of purified A2AR using ZM241385 (antagonist) and CGS-21680 (agonist). Table S9 shows obtained Kd values of 3.6 nM (±1.12) and 0.5 µM (±0.127 µM) for 3H-ZM241385 and 3H -CGS-21680, respectively. As comparative study, we have solubilized and purify A2AR using DDM/CHS and evaluated its ligand binding using 3H -CGS-21680 (Table S9). The obtained Kd was very similar to the one obtained using A2AR solubilized/purified in DDM/CHS/CALX-R10 (Table S9). This was also similar to Sf9 membrane bound forms and different from yeast membranes as shown in Fig. S1D, S1F and Table S9. This data shows that purified A2AR has maintained its ligand binding properties during the expression, solubilization and purification process.
Figure 1.
Purification of native A2AR. Talon affinity purification of A2AR from DDM/CHS/CALX-R10 solubilized total Sf9 membranes and analyzed by stain free SDS-PAGE (A) and western blot using antibody against A2AR (B).T, S, FT, W and E correspond to Total, Soluble, Flow Through, Wash and Elution fractions, respectively.
Behavior in solution and stability of purified native A2AR
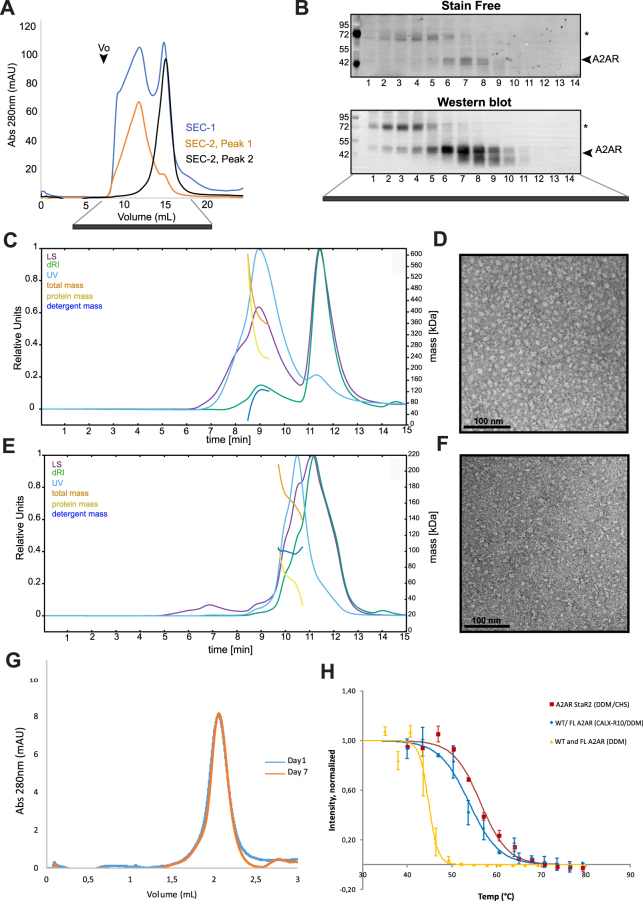
The next step was to assess the behavior of purified A2A R in solution. To this end, we loaded his-tag affinity purified A2AR on a gel filtration column. Fig. 2A shows a typical profile of a non-aggregated protein since no peak was observed at the void volume Vo. Two peaks were noticed on size exclusion chromatography corresponding to two protein assemblies of different sizes. SDS-PAGE (stain free) and western blot analyses (Fig. 2B) show the existence of two populations of A2AR that migrate at ~80 and ~40 kDa and are abundant in peak 1 and peak 2 fractions, respectively. Similar profile in Size exclusion chromatography was also observed for A2AR StaR2 construct (96 aminoacid c-terminal truncation and 8-point mutations). The 80 kDa band corresponds most probably to SDS-resistant dimers. Peak 2 shows a second faster band consistent with a degradation product of A2AR. To investigate masses and thus oligomeric states of the protein, we performed a SEC-MALS experiment on both samples (peak 1 and 2, Fig. 2C, E respectively). The sample corresponding to peak 1 has a clearly defined protein peak at approximately 9.25 minutes, well separated from the free micelles peak at around 11.5 minutes. With dn/dc of the protein component set to 0.185 ml/g and dn/dc of the detergent set to 0.1618 ml/g, the mass of the protein component stabilizes around 240 ± 10 kDa. This corresponds approximatively to the theoretical mass of a A2AR pentamer, which is about 238 kDa. In peak 2, the protein and the free micelles are only partially separated. The protein UV maximum is at around 10.5 minutes and the free micelles dRI maximum at 11.2 minutes. Moreover, the shapes of all three spectra (UV, LS, dRI) suggest the presence of a second, smaller peak of protein with a maximum around 10 minutes. To evaluate the mass of the main peak of the protein, we limited the calculations only to data points between 9.7 and 10.7 min. Setting the dn/dc values as in sample/peak 1, we obtained the mass of the protein component strongly decreasing from 105 to 70 kDa between 9.7 and 10.0 minutes, then slightly stabilizing at 65 ± 5 kDa between 10.0 and 10.4 minutes, and finally strongly decreasing to 30 kDa until 10.7 minutes. The stable part thus displays a value slightly higher than that of a monomer. If we hypothesize that it corresponds to a mixture of monomer and dimer, then the proportions would be 63% monomer and 37% dimer. Figure 2D shows that A2AR particles of a size of ~10 by 10 nm could be observed for SEC peak 1 fractions. Much smaller particles were observed for SEC peak 2 fractions (Fig. 2F). This is consistent with SEC-MALS finding that the first and second peaks correspond to higher and lower-order oligomers, respectively. To evaluate their ligand binding capacity, both peaks were analyzed by radioligand binding. Only A2AR from peak 2 showed convincing ligand binding (Table S9) in contrast to protein from peak 1. This strongly suggests that even if oligomeric A2AR was not obviously “aggregated”, it was not folded correctly enough to allow good ligand binding. We therefore focused on peak 2 for the next studies.
Figure 2.
Behavior in solution and stability of purified native A2AR. (A) Gel filtration profile of A2AR showing two peaks (indicated as 1 and 2). Fractions corresponding to each peak were pooled, concentrated and used to run a second SEC as indicated by the red and black chromatograms. (B) Gel filtration fractions were analyzed by SDS-PAGE revealed by stain free (total protein) or western blot (A2AR only). Full length original gels are presented in Fig. S5. SEC-MALS analysis show the profile of peak 1 (C) and peak 2 (E). Light scattering (LS), differential refractive index (dRI), OD at 280 nm (y1 axes) and calculated masses (y2 axes) were plotted as a function of experiment’s time. OD, LS and dRI were rescaled to range from 0 to 100%, with 100% corresponding to maximum value of the curve. Negative stain image of A2AR fractions from Size exclusion chromatography peak 1 (D) and peak 2 (F). Scale bar correspond to 100 nm. Stability of solubilized/purified A2AR by Analytical Size exclusion chromatography (G). SEC was performed on affinity purified A2AR and gel filtrated protein (pool 2) after incubation 1 and 7 days at room temperature. SEC chromatograms were superimposed. Thermalshift assay (H). The assay was performed as described in methods on wild type and full length A2AR solubilized using two different conditions, CALX and reference corresponding to DDM/CHS/CALX-R10/ZM241385 and DDM/CHS, respectively. For comparison, A2AR StaR2, truncated (96 aminoacid c-terminal deletion) and mutated 8-point mutations solubilized and purified in DDM/CHS as described78 was also analyzed by thermalshift.
To evaluate the stability of the native GPCR, we submitted purified monomeric A2AR (peak 2 of a first gel filtration chromatography) to a second gel filtration run after 1 and 7 days incubation at room temperature. Figure 2G shows no decay of A2AR signal in size exclusion chromatography, arguing for good stability. To confirm A2AR stability we performed a western blot-based thermal shift assay. This assay relies on the assumption that unstable heated proteins will aggregate and after ultracentrifugation and western blot the band intensity corresponding to the protein will decay proportionally to its instability58. The result shown in Fig. 2H indicates that using CALX-R10/DDM condition (in the presence of ZM241385), A2AR exhibits a Tm of ~55 °C. The same A2AR is less stable in DDM with a Tm of ~43 °C as previously reported58 and confirmed in Fig. 2H. As a comparative study, we have expressed A2AR StaR2, solubilized it using DDM/CHS as described8,59 and submitted it to the same thermal shift assay. Figure 2H shows a 4 °C higher stability of A2AR StaR2 in comparison to A2AR wild-type and full length solubilized using CALX-R10/DDM. This is relatively minor considering that StaR2 contains 8 points mutations and a 96 amino acids truncation in the C-terminus. Thus, we could stabilize native, glycosylated, non-aggregated and homogenous A2AR that maintained its ligand binding capacity.
Binding investigation of antagonists and agonists on A2AR using STD-NMR
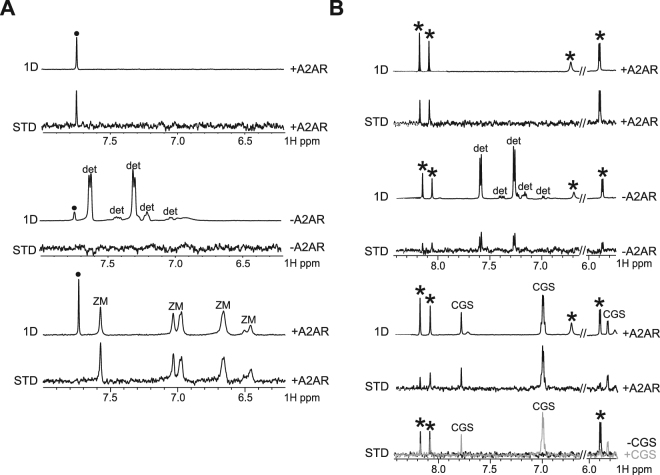
STD experiment, which is a well-established NMR method for fragment screening against soluble therapeutic targets44, has not yet been used against purified GPCRs. Thus, we first wanted to demonstrate the feasibility of the approach through the binding investigation of known antagonists and agonists to A2AR. Fig. 3A shows the STD binding signal of caffeine bound to A2AR. By comparison, in the absence of the protein, the STD signal is considerably weaker, showing that the unspecific binding of caffeine to the micelles is insignificant. Competition experiment was performed by adding the A2AR antagonist ZM241385. As illustrated in Fig. 3A, the binding signal of caffeine disappears, while the binding signal of ZM241385 is observed. The STD experiment indicates that the caffeine binds to the same binding pocket as ZM241385, in agreement with the previously reported X-ray structures30,31,60. One expected advantage of the preparation of native A2AR is the possibility to observe the binding of agonists since the conformational flexibility of the receptor is not constrained in such a preparation. We have therefore investigated the binding of adenosine to A2AR. Figure 3B shows the STD spectrum of adenosine bound to A2AR. As for the caffeine, the STD signals are significantly weaker in the control experiment performed in the absence of the receptor. A competition experiment was achieved by adding the agonist compound CGS-21680. The STD intensities of adenosine decrease in the presence of CGS-21680, showing the competition between adenosine and CGS-21680 that both bind in the same binding pocket. Interestingly, adenosine still exhibits a significant STD signal in the presence of CGS-21680. This suggests that adenosine binds to another binding pocket when CGS-21680 is bound to A2AR. This finding will be further discussed in the discussion part. As illustrated in Fig. 3, the observation of the binding of agonists and antagonists to native A2AR as well as the competition experiments demonstrate that the fragment screening can be achieved using STD-NMR on the A2AR preparation.
Figure 3.
STD-NMR binding of A2AR to antagonists and agonists. 1D and STD NMR spectra of the caffeine antagonist bound to A2AR (A). The 1D and STD NMR spectra are also shown in the absence (middle) of the A2A protein. NMR resonance of caffeine is indicated with a black dot and the aromatic compounds of the detergent buffer are labelled (det). The 1D and STD NMR spectra of caffeine are shown in the presence (bottom) of the ZM241385 compound. NMR resonances of the ZM241385 antagonist compound are labelled with the letters ZM. The STD binding signal of caffeine disappears in the presence of ZM241385. 1D and STD NMR spectra of the adenosine agonist bound to A2AR (B). The 1D and STD NMR spectra are also shown in the absence (middle) of the A2A protein. NMR resonances of adenosine are indicated with a star and the aromatic compounds of the detergent buffer are labelled (det). The 1D and STD NMR spectra of adenosine are shown in the presence of the CGS-21680 agonist compound. NMR resonances of the CGS-21680 compound are labelled with the letters CGS. The STD binding signal of adenosine is weaker in the presence of CGS-21680. The intensities of the STD signals of adenosine in the presence and absence of CGS-21680 are superimposed (bottom) to illustrate the change in the STD intensities, particularly for the ribose resonance at 5.9 ppm.
Fragment screening against A2AR using STD-NMR
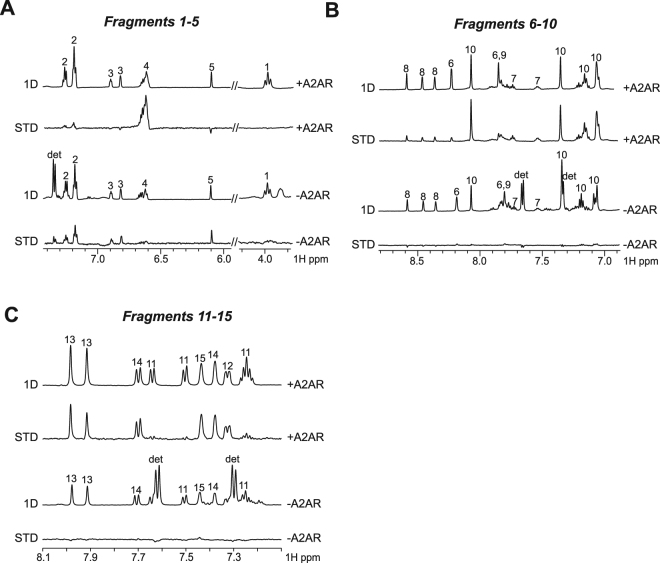
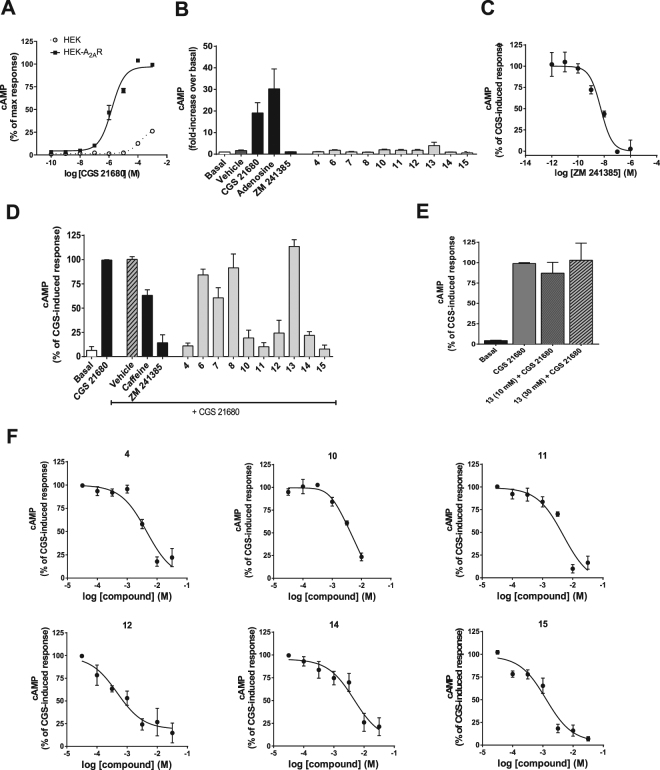
We then performed fragment screening against A2AR using a hundred fragments. The molecules were screened into mixtures of 5 to 10, as typically done with soluble proteins. Fragments were then classified into three groups displaying strong binding, weak binding or no binding, depending on the intensity of the STD signals observed. Nineteen fragments (19%) were shown to exhibit significant STD intensities upon A2AR binding (Fig. S4). To further analyse the fragment screening results, cAMP cell-based assay was performed on ten fragments displaying either strong (fragments 4, 10, 12, 13, 14, 15) or weak binding (fragments 6, 7, 8 and 11) (Figs 4 and S7).
Figure 4.
Fragment screening against native A2AR STD-NMR. The 1D and STD NMR spectra for fragments 1–5 (A), fragments 6–10 (B) and fragments 11 to 15 (C) are shown in the presence (top) and in the absence (bottom) of A2AR. The fragments bound to A2AR display STD signals, while the non-binders have no binding signal observed in the absence of A2AR, showing that the binding observed in the presence of native A2AR is specific.
Functional validation of fragments in the cAMP cell based assay
We then tested the capacity of the fragment binders shown in Fig. 4 to induce cAMP production in HEK293 cells stably expressing A2AR. CGS-21680 titration curves show that cAMP production is A2AR expression dependent (Fig. 5A). Non-transfected HEK293 cells were used as negative control and a small increase in cAMP production was observed associated to high concentrations of CGS-21680, probably due to the known presence of endogenous A2AR61. We then investigated the potential agonistic effect of the fragments displayed in Fig. 4. Stimulation of the A2AR cell line with different concentrations (10 µM, 100 µM, 1 mM and 10 mM) of each fragment for 30 minutes at room temperature had no effect on cAMP production even at the highest concentration of 10 mM as shown in Fig. 5B. CGS-21680 and adenosine served as positive controls and showed robust agonistic effects as expected. Accordingly, the well-established A2AR antagonist ZM241385 did not show any effect in this agonistic assay, while its efficiency in inhibiting CGS-21680-induced increase in cAMP production was confirmed (Fig. 5C). We then investigated the antagonistic effect of the fragments in the cAMP signaling assay. This test was performed by pre-incubating the A2AR cell line with the fragments (15 min, room temperature), followed by addition of the CGS-21680 agonist (30 min, room temperature). Fragments, 4, 10, 11, 12, 14 and 15 behave as full antagonists at 10 mM, whereas fragment 6, 7, 8 and 13 were without effect. Compound 13 remained inactive in this assay at concentrations up to 30 mM (Fig. 5E). To further characterize the observed antagonistic effect, we generated full competition curves for fragments 4, 10, 11, 12, 14 and 15, which confirmed their antagonistic effect (Fig. 5F).
Figure 5.
Functional validation of A2AR compounds on the cAMP signaling pathway. Concentration-response curves of CGS-21680-induced cAMP production in control HEK293 cells and in HEK293 cells stably expressing the A2A receptor (HEK-A2AR) (A). Analysis of agonist effect of compounds on cAMP production (10 mM, 30 min). Vehicle: DMSO (1%); CGS-21680: reference agonist (1 µM); adenosine: reference agonist (1 µM); ZM241385: reference antagonist (1 µM) (B). Concentration-response curve of ZM241385 antagonist (15 min pre-incubation) on CGS-21680-induced (1 µM, 30 min) cAMP production (C). Analysis of antagonist effect of compounds (10 mM, 15 min pre-incubation) on CGS-21680-induced (1 µM, 30 min) cAMP production. Vehicle: DMSO (1%); CGS-21680: reference agonist (1 µM); caffeine: reference antagonist (10 mM); ZM241385: reference antagonist (1 µM) (D). Absence of antagonist effect of compound 13 (at 10 mM and 30 mM) on CGS-21680-induced cAMP production (E). Concentration-response curves of compounds, 4, 10, 11, 12, 14, 15 on CGS-21680-induced cAMP production (F). Data are expressed as mean ± S.E.M. of 3 to 6 independent experiments and normalized to either basal or CGS-21680-induced levels.
This result confirms the value of combining NMR-STD experiments and cell-based assays to discover functionally relevant fragments. Thus, using stabilized native A2AR, we could identify fragments with antagonistic effects on A2AR.
Discussion
While dramatic progress has been achieved for structural and biophysical studies of membrane proteins such as GPCRs37,62, innovative approaches are still needed to discover new drugs targeting GPCRs. Significant efforts for the improvement of GPCRs stability has been made thanks to thermostabilization approaches by truncation, multiple alanine scan mutations and protein fusion. A2AR was significantly thermostabilized by mutating up to 8 residues at once and removing 96 amino-acids at the carboxy-terminal of the receptor8. A similar approach was successfully applied to other GPCR such as β-adrenergic receptor9,63. However, despite these approaches being very successful for structure determination, the modification of the protein sequence may restrict the repertoire of protein conformations existing in the native receptor and introduce a bias that may be misleading or limiting for structure-based drug discovery. Indeed, a recent NMR study demonstrates structural deviations of the fused receptor in the crystal due to the fusion64. In addition to that, even if the deletion of the c-terminus or the replacement of intra-cellular loops of GPCRs does not systematically impair ligand binding, these domains are crucial binding sites for interacting proteins important for receptor function65 and are thus likely to cause conformation deviations or restrictions from the receptor’s native state.
Here we report a stabilization approach for native, non-mutated GPCR. Here, we have used a specific calixarene-based detergent to solubilize and stabilize native, full length and functional A2AR. Native A2AR was stable for at least one week at 25 °C and showed a Tm of ~ 55 °C, corresponding to a significant stabilization shift in comparison to that previously reported for WT A2AR truncated at the C-terminus8,58 where a Tm of ~40 °C was measured. The native A2AR showed no sign of aggregation in SEC or EM in solution. Functionality was assessed by radioligand binding demonstrating binding of well-characterized agonist and antagonist compounds. This illustrates also the absence of conformational constraint since agonist and antagonist compounds were both able to bind to the receptor. This was not the case for StaR preparation that is not able to bind correctly to agonists such as CGS-21680 or NECA in comparison to the wild type protein8,59. The present work describes a natural alternative to systematic mutagenesis/fusion approaches and changes the dogma that GPCRs are unstable proteins requiring systematic stabilization by mutagenesis. The strategy described here is certainly not a time-consuming task in comparison to systematic scanning mutagenesis. This approach may be generalized across GPCRs and other highly challenging and druggable targets such as ion channels and transporters. NMR has been previously used to study the interaction of small molecules to GPCRs66–71. However, only the TINS technology was applied to screen fragments against GPCRs prepared in micelles and immobilized on a resin41,42. Here we aimed to use the STD method, which has the advantage to provide structural information through the discrimination of solvent-exposed hydrogens from buried hydrogens for the ligand bound to the receptor43,44. STD experiments recorded for the antagonist caffeine and the agonist adenosine showed that both types of ligands could be observed as binders with the native A2AR preparation. As shown in Fig. 3B, the STD intensities of adenosine bound to A2AR not only are weaker upon addition of the agonist CGS-21680, but the profile of the STD intensities are also modified. Notably, the STD signal of the proton of the adenosine ribose moiety at 5.9 ppm is considerably smaller when CGS-21680 binds A2AR. This indicates that the adenosine ribose moiety is buried in A2AR in the absence of CGS-21680, while it is solvent-exposed in the presence of CGS-21680 (Fig. 6). This observation corroborates with previous investigation of the binding mechanism of GPCR ligands using molecular dynamics simulation72, showing the presence of transient binding sites also called metastable binding sites or ligand-entry sites as potential allosteric sites37. In particular, a metastable binding site was proposed for adenosine bound to A2AR73. It was shown that adenosine could bind at the entrance of the orthosteric binding site, with the ribose oriented towards the entrance, solvent-exposed, in agreement with the NMR observation. These results show that the benefit of the STD-NMR experiment is to provide structural information for ligands bound to the receptor in the presence or absence of other compounds. This information will likely be of high interest for the discovery of allosteric binders. In the reported study, 19% of the fragments displayed significant binding on A2AR using STD-NMR. Comparison of NMR results with the cAMP cell-based assays achieved for 10 fragments showed that four fragments (6, 7, 8 and 13) displaying STD signals did not exhibit biological activity. It is acknowledged that fragment screening typically requires orthogonal techniques to identify and validate fragment hits, due to the weak affinity of such binders74. Therefore, it is not surprising to observe differences between the STD-based screening and the cAMP cell-based assay. While STD signals for fragments 6, 7 and 8 were classified as weak, fragment 13 displayed large STD signals upon binding to A2AR (Fig. 4C). The binding of compound 13 to A2AR was confirmed by testing the fragment alone (not in mixture) using STD (Fig. S8). In addition, STD-based competition experiment with the agonist CGS-21680 shows that fragment 13 binds in the orthosteric binding site of A2AR (Fig. S8). While the chemical structure of fragment 13 is similar to the adenine of adenosine, no conclusion can be drawn based on the cAMP cell-based assay only. It cannot be excluded that compound 13 may exhibit a pattern of agonism or even antagonism, as it is widely accepted that the classification of ligands in terms of their pharmacological properties is entirely dependent on the functional readout that is assayed75,76. Further investigation will be achieved for fragment 13, which is not the focus of this study. In conclusion, the current study describes the first stabilizing detergent/surfactant-based approach for native GPCR stabilization. Our goal in this study was to provide an alternative to systematic mutagenesis approach. Our preparation of A2AR could bind to well characterized agonists (adenosine and CGS-21680) and antagonists (caffeine and ZM241385). This suggests a full conformational space of the receptor. Also, the benefit of the STD-based fragment screening was discussed, showing that structural information for the binders can be inferred from the screening experiments. The reported approach represents an attractive alternative to the classical large-scale library compounds screening using cell-based assays.
Figure 6.
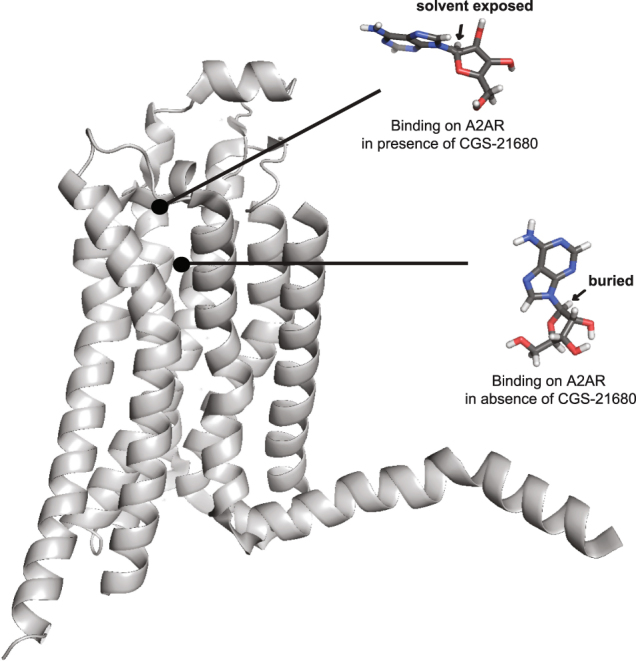
STD-NMR indicates a modification of the exposure to solvent for the ribose moiety of adenosine upon binding to A2AR, in the presence or in the absence of CGS-21680. The structure proposed for the adenosine with the buried ribose proton corresponds to the structure of the adenosine solved in complex with A2AR33 (PDB ID: 2YDO). The structure proposed for the adenosine with the solvent exposed ribose proton is inspired by the study of Sabbadin et al.73.
Methods
Full length and wild type A2AR Expression
For insect cells expression, the full–length human A2AR was cloned into pOET1 transfer plasmid in frame with N-terminal hemagglutinin signal sequence, Strep-tag II and 8xHis tag, and baculovirus was produced according to the manufacturer’s protocol (flashback ULTRA™ system, Oxford Expression Technologies). Sf9 insect cells were infected with baculovirus at a density of 1.5 × 106 cells ml−1, using a MOI of 1, and grown at 28 °C for 64 hours in an orbital shaker. After 64 hours, cell pellets were collected, washed in Hepes buffer pH 7.4, 200 mM NaCl, 1x protease inhibitor cocktail (Sigma), then stored at −80 °C until use. For yeast expression, the full–length human A2AR was cloned into into the pPICα A expression vector (Thermo Fisher Scientific) in frame with the α-factor signal sequence, a Strep-tag II and a 8xHis tag, and linearized using the restriction enzyme DraI. The linearized vector was transformed into the P. pastoris strains KM71 and GS115 by using the Pichia EasyComp™ Transformation Kit (Thermo Fisher Scientific). Clone selection was performed by selecting recombinant His+ clones on MD agar plates (1.34% (w/v) yeast nitrogen base without amino acids, 2% (w/v) dextrose, 0.00004% (w/v) biotin, and 1.5% (w/v) agar). To select for multicopy transformants, His+ clones were grown on Zeocin-YPD agar plates (1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) dextrose, 2% (w/v) agar, and 0.1 or 0.025 mg/ml Zeocin). Representative clones exhibiting resistance to Zeocin were tested for recombinant protein production by Western-blotting. The selected transformants were stored as glycerol stocks at −80 °C. Single P. pastoris colonies from high expressing clones were selected on YPD plates containing 0.1 mg/ml Zeocin. Cells from a single colony were used to inoculate 300 ml of BMGY medium. The culture was grown overnight at 30 °C to an OD600 of 2–6. A total of 1.5 L of BMGY was inoculated with 300 ml of the starter culture and grown for 4 hr to an OD600 of 2. The cells were spun down at 4,000 g for 15 min, the cell pellet was washed with double distilled water, and then the cells were spun down. The cell pellet was resuspended in BMMY to an OD600 of 1. The culture was incubated for 20 hours at 20 °C with shaking at 150 rpm at 28 °C, and cell pellets were collected, washed in Hepes buffer pH 7.4, 200 mM NaCl, 1x protease inhibitor cocktail, then stored at −80 °C until use.
Lysis and Membrane fractionation
Frozen cell pellets were thawed, resuspended in Hepes buffer pH 7.4, 200 mM NaCl, 1x protease inhibitor cocktail, and lysed by mechanical cell lysis. Cell lysis was performed on ice using a BeadBeater homogenizer with 0.1 mm diameter glass beads. Membrane fractionation was then carried out at 4 °C by sequential centrifugations. For both insect or yeast cells expressing A2A, 3 centrifugations were performed: 500 g for 5 min, 15000 g for 30 min, and 100000 g for 45 min. Membrane pellets were washed twice in buffer containing high salt (1 M NaCl) to remove membrane associated proteins. Membrane enriched pellets were resuspended in Hepes buffer pH 7.4, 200 mM NaCl, 1x protease inhibitor cocktail and glycerol 10%, quantified using the Pierce Micro BCA Protein Assay Kit (Thermo Scientific), flash-frozen and stored at −80 °C until use.
Protein solubilization & purification
Protein solubilization
Proteins from internal or plasma membrane fractions were incubated for 2 h at 4 °C at a final concentration of 5 mg/ml in 50 mM Hepes buffer pH 7.4, 200 mM NaCl, 1x protease inhibitor cocktail, and with 0.155% CALX-R10 (10-fold the critical micelle concentration or CMC) in combination with 0.5% DDM and 0.06% CHS (57-fold the CMC). Extraction without detergent and with SDS served as negative and positive controls, respectively. After solubilization samples were centrifuged at 100000 g for 45 min at 4 °C and an aliquot of the total extract, the pellet and the supernatant from each solubilization condition was analyzed by SDS-PAGE and western-blot.
His-tag affinity chromatography
The soluble protein fraction was loaded onto a TALON column equilibrated with 50 mM Hepes buffer pH 7.4, 200 mM NaCl, 0.05% DDM and 0.006% CHS. After 2 h incubation at 4 °C, resin was washed with 12 Column Volumes (CV) of Wash Buffer containing 50 mM Hepes buffer pH 7.4, 200 mM NaCl, 0.05% DDM and 0.006% CHS, 20 mM Imidazole. Target protein was eluted with 4 CV of washing buffer with 150 mM Imidazole. Samples of each fraction T, S, FT, W and E (corresponding to Total, Solubilized, Flow through, Wash and Elution, respectively) were analyzed by SDS-PAGE and western-blot.
Size exclusion chromatography
Affinity purified A2AR was concentrated using Centriprep contractors with a 50 K cut-off and loaded on a superdex 200 Increase 10/300 GL(GE-Healthcare) at 0.3 ml/min. Running buffer was 50 mM Hepes buffer pH 7.4, 200 mM NaCl, 0.05% DDM and 0.006% CHS. Elution was performed with 1.5 CV of running buffer and 150 µl-fractions were collected. Fractions were analyzed by SDS-PAGE and western-blot. To assess stability of A2AR, superdex 200 Increase 5/150 GL (3 ml) was used.
SDS-PAGE and Western-blot
A2AR samples were denatured with 5x Laemmli buffer and incubated for 20 min at RT prior to analysis without heating to avoid aggregates formation. Proteins were separated by SDS-PAGE on a 4–15% acrylamide gel (4–15% Mini-PROTEAN® TGX Stain-Free™ Gel, Bio-Rad) and subsequently immobilized by electro-transfer to PVDF membrane. The immunodetection of A2AR was performed by using the SNAP i.d. system (Millipore) with either a primary A2AR antibody (mAb 7F6-G5-A2), Santa Cruz Biotechnology) or an anti-His HRP antibody. Quantification of the signal was performed using Image Lab 4.1 software from Bio-Rad to evaluate the extraction efficiency. SDS-PAGE were silver stained using Bio-Rad Dodeca Silver Stain Kit following supplier protocol or coomassie stained using the PageBlue™ protein staining solution.
Clear Native-PAGE (CN-PAGE) and Western-blot
Non-denaturated proteins were separated by native-PAGE on a 4–15% acrylamide gel (4–15% Mini-PROTEAN® TGX Stain-Free™ Gel, Bio-Rad) using 25 mM imidazole as anode buffer and 7.5 mM imidazole, 0.05% deoxycholate, 0.01% DDM as cathode buffer). Clear Native PAGE gels ran for 90 min at 200 V and 4 °C. Proteins were then immobilized by electro-transfer to PVDF membrane. The immunodetection of A2AR was performed by using the SNAP i.d. system (Millipore) with A2AR antibody.
Protein quantification
Total protein concentrations in the plasma and the internal membrane fractions were determined with the micro BCA protein assay kit (Pierce) using the bovine serum albumin (BSA) as a standard.
Negative staining electron microscopy
Protein samples at 40 µg/ml were adsorbed on 200 Mesh copper grids coated with formvar-C for 2 min at RT. Then grids with suspension were colored with 1% uranyl acetate for 1 min and observed on a transmission electron microscope (Jeol 1400 JEM, Tokyo, Japan) equipped with a Gatan camera (Orius 600) and Digital Micrograph Software.
N-glycosylation analysis
Prior to deglycosylation membrane samples were desalted using ice-cold methanol (Merck, Darmstadt, Germany). Briefly, dried membrane samples were resuspended in 1 ml of ice-cold methanol and centrifuged for 15 min at 2200 g. The supernatant was carefully removed and the procedure was repeated. The remaining methanol was evaporated by drying down in the vacuum concentrator. Dried samples were dissolved in 30 μL of 1.33% SDS (w/v) and denatured by incubation at 65 °C for 10 minutes. The following steps of N-glycan release and fluorescent labelling were essentially as described previously77. After labelling, the free label and reducing agent were removed from the samples by hydrophilic interaction liquid chromatography solid-phase extraction (HILIC-SPE) using 0.2 μm GHP filter plates and ice-cold 96% acetonitrile. Fluorescently labelled N-glycans were separated by HILIC on a Waters Acquity ultra-performance liquid chromatography (UPLC) system (Milford, MA, USA) as described previously77. Briefly, labelled N-glycans were separated on a Waters BEH Glycan chromatography column, 150 × 2.1 mm i.d., 1.7 μm BEH particles, with 100 mM ammonium formate, pH 4.4, as solvent A and acetonitrile as solvent B. Separation method used linear gradient of 70–53% acetonitrile (v/v) at flow rate of 0.56 ml/min in a 23 minutes’ analytical run. Samples were maintained at 10 °C before injection, and the separation temperature was 25 °C. The identity of N-glycans separated by HILIC-UPLC was determined by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS). Prior to MS analysis, fractions of each N-glycan chromatography peaks were collected, dried down in a vacuum concentrator and resuspended in 10 μL of ultrapure water. Aliquots of 2 μL were spotted onto a MTP AnchorChip 384 BC MALDI target (Bruker Daltronics, Bremen, Germany), mixed on plate with 1 µL of matrix solution (5 mg/ml 2,5-DHB, 1 mM NaOH in 50% acetonitrile) and left to dry by air. Recrystallization was performed by adding 0.2 µL of ethanol to each spot. Analyses were performed in positive-ion reflectron mode on an UltrafleXtreme MALDI-TOF-MS equipped with a Smartbeam-II laser and FlexControl 3.4 software Build 119 (Bruker Daltonics). The instrument was calibrated using a plasma N-glycome standard. A 25-kV acceleration voltage was applied after a 140-ns extraction delay. A mass window of m/z 1000 to 5000 with suppression up to m/z 900 was used for N-glycan samples. For each spectrum, 10 000 laser shots were accumulated at a laser frequency of 2000 Hz, using a complete sample random walk with 200 shots per raster spot.
Ligands binding assay
Radioligand binding
This assay was performed at 4 °C in triplicate using 96 wells plate with U bottom. Protein at a final concentration of 24 µg/ml (1 µg–50 µL per well) was incubated at 4 °C in the presence of 3.6 µM 3H-CGS-21680 (0.6 µM final, 10 µL per well) (Perkin-Elmer NET1021250UC)+/− 1 mM of cold ligand (0.17 mM final) in binding buffer (50 mM Tris-HCl pH 7.4, 10 mM MgCl2, 0.5 mM EDTA) or ZM241385. After 2 h of incubation, 60 µL of 0,1% γ-globulin (prepared in wash buffer) and 120 µl of 25% PEG6000 (prepared in wash buffer) were added per well, mixed and incubated for 15 mins at RT.
Samples were then filtered using PEI-pre-coated GF/B plates (Perkin-Elmer, cat#6005177). Plates were washed 4 times with ice-cold wash buffer (50 mM Tris-HCl pH 7.4) and 25 µl of scintillation reagent was added per well. After 1 h of incubation, CPM detection was done using the Microbeta2 equipment (Perkin Elmer), applying 5 mins counting per well.
NMR binding
NMR experiments were acquired at 293 K on a Bruker AVIII 600 MHz spectrometer equipped with a cryoprobe and a SampleJet auto-sampler. NMR sample containing protein was recorded with 2 µM A2AR in a buffer consisting of 50 mM Hepes at pH 7.5, 200 mM NaCl and 0,05% DDM/0,005% CHS and 10% D2O. NMR experiment in the absence of the protein was recorded in a buffer consisting of 50 mM Hepes at pH 7.5, 200 mM NaCl, 0,05% DDM/0,005% CHS and 0,02% CALX-R10/0,002% CHS (1CMC). Saturation time was 2 secs per experiment. Fragment screening was performed in mixtures of 5 to 10 fragments at 600 µM for each fragment.
Competition experiments: NMR competition experiments were acquired at 293 K in the presence of 10% D2O on a Inova Agilent 600 MHz spectrometer equipped with a cryoprobe and an auto-sampler. NMR sample contained 1 µM of A2AR in a buffer consisting of 50 mM Hepes at pH 7.5, 200 mM NaCl and 0,05% DDM/0,005% CHS. Caffeine and adenosine were used at a final concentration of 600 µM; ZM241385 and CGS-21680 were solubilized in 100% DMSO-d6 and used at a final concentration of 360 µM. Saturation time for the STD was 2 secs.
Thermostability assay
Membranes of A2AR (4 mg/ml total protein) were solubilized in different conditions (see solubilization method above) for 2 hours at 4 °C. Solubilized fractions were obtained after 100,000 g ultracentrifugation for 1 h at 4 °C. Solubilized fraction serves to make 50 μl aliquots to be submitted to one temperature each as part of a gradient of temperature ranging from 25 to 72 °C using PCR thermal cycler (PeqSTAR 2x gradient; Peqlab). Samples were then centrifuged 40 min at 20000 g and supernatants were analyzed by SDS-PAGE and western-blot using anti- A2AR antibody (7F6-G5-A2). The relative intensity of the target protein at each temperature was quantified on Western-blot using Image Lab software 4.1 from Bio-rad. Each condition was performed twice. Intensity was plotted as a function of the temperature, normalized and fit to the Boltzmann equation with the least square method using Solver Adds-in of Excel software. The method is described by58.
SEC-MALS
SEC-MALS experiment were performed with a Phenomenex Yarra Sec. 3000-3, 300 × 4.6 mm column, using a setup of consecutive: Agilent 1260 Infinity UV detector with 1 μL G4212-60008 cartridge, Wyatt Dawn Heleos 18-angles light scattering detector and Wyatt Optilab T-rEX refractometer. Temperature was held constant at 20 °C. Flow rate was set to 0.3 ml/min. OD was measured at 280 nm. We used Wyatt’s ASTRA 6 to align the measurements from the detectors, take band-broadening into account and calculate masses of the components of the sample. The mass calculation requires the knowledge of dn/dc of each component. For the protein part, this was set to 0.185 ml/g and for the DDM/CHS part, we measured the value of 0.1618 ml/g. As we could not find a reference value of dn/dc for DDM/CHS mixture in the literature, we measured dn/dc of DDM and DDM/CHS. 1 g of saltless samples of DDM and CHS were dried overnight and weighted to measure the amount of water in the samples. Taking this correction into account, 1% stock solutions of DDM and DDM/CHS were prepared in the 50 mM, HEPES pH 7.4; 200 mM NaCl buffer. From this, we prepared the series dilutions to 1%, 0.8%, 0.6%, 0.4% and 0.2% concentrations. 4 ml of each concentration, followed by 1.5 ml of pure buffer was injected directly to refractometer, and the measurements were fit using ASTRA 6. We obtained dn/dc of DDM = 0.1378 ± 0.75% ml/g, which is well within range of values typically cited in the literature, and dn/dc of DDM/CHS = 0.1618 ± 0.19% ml/g. R2 of both fits was above 0.99.
cAMP assay
Measurements of cAMP production were performed by Homogeneous Time-Resolved FRET (HTRF)-based assay using the commercially available cAMP-femto-Tb kit (Cisbio, Codolet, France), according to the manufacturer’s instructions. HEK293 cells stably expressing A2A were distributed to 384-well plate and treated with the indicated compounds for 30 minutes at room temperature (test of agonistic effect). Alternatively, cells were pre-incubated with the compounds (15 minutes) followed by addition of the agonists CGS-21680 (1 μM, 30 minutes; test of antagonistic effect). After the incubation time, cells were lysed, incubated with the kit reagents (1 h, RT) and the measurements were done in the plate reader Tecan Infinite F500 (Tecan, Switzerland).
Electronic supplementary material
Acknowledgements
We thank Emmanuel DEJEAN for continuous support, Kelly GARNIER for help with thermal-shift assay and the CALIXAR team for helpful discussions. We would like to thank Patrick SCHULTZ for initial electron microscopy work and discussions and Alice ROTHNIE for critical reading of the manuscript. We thank Dr. Francisco CIRUELA ALFEREZ (University of Barcelona) for kindly providing HEK-293 cells stably expressing the A2AR-Nanoluc fusion protein and Ivan GUDELJ for help with mass spectrometry measurements. M.B. was supported by the Horizon 2020 Program of the European Union (iNEXT grant, project 653706).
Author Contributions
S.I. performed the expression, solubilization and purification and radiobinding of A2AR. S.I. and A.J. designed the experiments and analyzed the results. M.P.B. and G.L. designed and performed the glycans determination study. M.B. and J.P. performed the SEC-MALS analysis. C.R., O.C. performed the N.M.R. study designed by I.K. E.C. and R.J. designed and performed the cAMP assays. A.J. wrote the manuscript. All authors helped to improve the text.
Competing Interests
The authors S.I. and A.J. are employees of CALIXAR that have patents applications that covers CALX-R10 described in this manuscript. Apart from that, all authors have no competing interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Footnotes
Sébastien Igonet, Claire Raingeval and Erika Cecon contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-26113-0.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
6/14/2018
In the original version of this Article, there was an erroneous Supplementary Material file entitled ‘Supplementary Figures legend with changes marked’. This file has now been removed from the HTML of this Article; the PDF version was correct at time of publication.
References
- 1.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–30. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 2.Sriram K, Insel PA. G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol Pharmacol. 2018;93:251–258. doi: 10.1124/mol.117.111062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guerram M, Zhang LY, Jiang ZZ. G-protein coupled receptors as therapeutic targets for neurodegenerative and cerebrovascular diseases. Neurochem Int. 2016;101:1–14. doi: 10.1016/j.neuint.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Hauser AS, et al. Pharmacogenomics of GPCR Drug Targets. Cell. 2018;172:41–54 e19. doi: 10.1016/j.cell.2017.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutchings CJ, Koglin M, Olson WC, Marshall FH. Opportunities for therapeutic antibodies directed at G-protein-coupled receptors. Nat Rev Drug Discov. 2017;16:787–810. doi: 10.1038/nrd.2017.91. [DOI] [PubMed] [Google Scholar]
- 6.Christopoulos A, Kenakin T. G protein-coupled receptor allosterism and complexing. Pharmacol Rev. 2002;54:323–74. doi: 10.1124/pr.54.2.323. [DOI] [PubMed] [Google Scholar]
- 7.Kobilka BK. G protein coupled receptor structure and activation. Biochim Biophys Acta. 2007;1768:794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magnani F, Shibata Y, Serrano-Vega MJ, Tate CG. Co-evolving stability and conformational homogeneity of the human adenosine A2a receptor. Proc Natl Acad Sci USA. 2008;105:10744–9. doi: 10.1073/pnas.0804396105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Conformational thermostabilization of the beta1-adrenergic receptor in a detergent-resistant form. Proc Natl Acad Sci USA. 2008;105:877–82. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heydenreich FM, Vuckovic Z, Matkovic M, Veprintsev DB. Stabilization of G protein-coupled receptors by point mutations. Front Pharmacol. 2015;6:82. doi: 10.3389/fphar.2015.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strege A, Carpenter B, Edwards PC, Tate CG. Strategy for the Thermostabilization of an Agonist-Bound GPCR Coupled to a G Protein. Methods Enzymol. 2017;594:243–264. doi: 10.1016/bs.mie.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Allison TM, et al. Quantifying the stabilizing effects of protein-ligand interactions in the gas phase. Nat Commun. 2015;6:8551. doi: 10.1038/ncomms9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta K, et al. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature. 2017;541:421–424. doi: 10.1038/nature20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng H, et al. Palmitoylation and membrane cholesterol stabilize mu-opioid receptor homodimerization and G protein coupling. BMC Cell Biol. 2012;13:6. doi: 10.1186/1471-2121-13-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hussain H, et al. Accessible Mannitol-Based Amphiphiles (MNAs) for Membrane Protein Solubilisation and Stabilisation. Chemistry. 2016;22:7068–73. doi: 10.1002/chem.201600533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kean J, Bortolato A, Hollenstein K, Marshall FH, Jazayeri A. Conformational thermostabilisation of corticotropin releasing factor receptor 1. Sci Rep. 2015;5:11954. doi: 10.1038/srep11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye L, Van Eps N, Zimmer M, Ernst OP, Prosser RS. Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nature. 2016;533:265–8. doi: 10.1038/nature17668. [DOI] [PubMed] [Google Scholar]
- 18.Glukhova A, et al. Structure of the Adenosine A1 Receptor Reveals the Basis for Subtype Selectivity. Cell. 2017;168:867–877 e13. doi: 10.1016/j.cell.2017.01.042. [DOI] [PubMed] [Google Scholar]
- 19.Cromie KD, Van Heeke G, Boutton C. Nanobodies and their Use in GPCR Drug Discovery. Curr Top Med Chem. 2015;15:2543–57. doi: 10.2174/1568026615666150701113549. [DOI] [PubMed] [Google Scholar]
- 20.Hino T, et al. G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature. 2012;482:237–40. doi: 10.1038/nature10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manglik, A., Kobilka, B. K. & Steyaert, J. Nanobodies to Study G Protein-Coupled Receptor Structure and Function. Annu Rev Pharmacol Toxicol (2016). [DOI] [PMC free article] [PubMed]
- 22.Pardon E, et al. A general protocol for the generation of Nanobodies for structural biology. Nat Protoc. 2014;9:674–93. doi: 10.1038/nprot.2014.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–80. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ring AM, et al. Adrenaline-activated structure of beta2-adrenoceptor stabilized by an engineered nanobody. Nature. 2013;502:575–9. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staus DP, et al. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature. 2016;535:448–52. doi: 10.1038/nature18636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steyaert J, Kobilka BK. Nanobody stabilization of G protein-coupled receptor conformational states. Curr Opin Struct Biol. 2011;21:567–72. doi: 10.1016/j.sbi.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brea RJ, et al. In Situ Reconstitution of the Adenosine A2A Receptor in Spontaneously Formed Synthetic Liposomes. J Am Chem Soc. 2017;139:3607–3610. doi: 10.1021/jacs.6b12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frauenfeld J, et al. A saposin-lipoprotein nanoparticle system for membrane proteins. Nat Methods. 2016;13:345–51. doi: 10.1038/nmeth.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison KA, et al. Membrane protein extraction and purification using styrene-maleic acid (SMA) copolymer: effect of variations in polymer structure. Biochem J. 2016;473:4349–4360. doi: 10.1042/BCJ20160723. [DOI] [PubMed] [Google Scholar]
- 30.Dore AS, et al. Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure. 2011;19:1283–93. doi: 10.1016/j.str.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–7. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaakola VP, Ijzerman AP. The crystallographic structure of the human adenosine A2A receptor in a high-affinity antagonist-bound state: implications for GPCR drug screening and design. Curr Opin Struct Biol. 2010;20:401–14. doi: 10.1016/j.sbi.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Lebon G, et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474:521–5. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasmussen SG, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–7. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 35.Zheng Y, et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature. 2016;540:458–461. doi: 10.1038/nature20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erlanson DA, Fesik SW, Hubbard RE, Jahnke W, Jhoti H. Twenty years on: the impact of fragments on drug discovery. Nat Rev Drug Discov. 2016;15:605–19. doi: 10.1038/nrd.2016.109. [DOI] [PubMed] [Google Scholar]
- 37.Congreve M, Oswald C, Marshall FH. Applying Structure-Based Drug Design Approaches to Allosteric Modulators of GPCRs. Trends Pharmacol Sci. 2017;38:837–847. doi: 10.1016/j.tips.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 38.Aristotelous T, et al. Discovery of beta2 Adrenergic Receptor Ligands Using Biosensor Fragment Screening of Tagged Wild-Type Receptor. ACS Med Chem Lett. 2013;4:1005–1010. doi: 10.1021/ml400312j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christopher JA, et al. Biophysical fragment screening of the beta1-adrenergic receptor: identification of high affinity arylpiperazine leads using structure-based drug design. J Med Chem. 2013;56:3446–55. doi: 10.1021/jm400140q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navratilova I, Hopkins AL. Emerging role of surface plasmon resonance in fragment-based drug discovery. Future Med Chem. 2011;3:1809–20. doi: 10.4155/fmc.11.128. [DOI] [PubMed] [Google Scholar]
- 41.Congreve M, et al. Fragment screening of stabilized G-protein-coupled receptors using biophysical methods. Methods Enzymol. 2011;493:115–36. doi: 10.1016/B978-0-12-381274-2.00005-4. [DOI] [PubMed] [Google Scholar]
- 42.Chen D, et al. Fragment screening of GPCRs using biophysical methods: identification of ligands of the adenosine A(2A) receptor with novel biological activity. ACS Chem Biol. 2012;7:2064–73. doi: 10.1021/cb300436c. [DOI] [PubMed] [Google Scholar]
- 43.Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 2001;123:6108–17. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 44.Cala O, Krimm I. Ligand-Orientation Based Fragment Selection in STD NMR Screening. J Med Chem. 2015;58:8739–42. doi: 10.1021/acs.jmedchem.5b01114. [DOI] [PubMed] [Google Scholar]
- 45.Hardy, D., Desuzinges Mandon, E., Rothnie, A. & Jawhari, A. The yin and yang of solubilization and stabilization for wild-type and full-length membrane protein. Methods (2018). [DOI] [PubMed]
- 46.Desuzinges Mandon E, Agez M, Pellegrin R, Igonet S, Jawhari A. Novel systematic detergent screening method for membrane proteins solubilization. Anal Biochem. 2017;517:40–49. doi: 10.1016/j.ab.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 47.Hardy D, Bill RM, Jawhari A, Rothnie AJ. Overcoming bottlenecks in the membrane protein structural biology pipeline. Biochem Soc Trans. 2016;44:838–44. doi: 10.1042/BST20160049. [DOI] [PubMed] [Google Scholar]
- 48.Desuzinges Mandon E, et al. Expression and purification of native and functional influenza A virus matrix 2 proton selective ion channel. Protein Expr Purif. 2017;131:42–50. doi: 10.1016/j.pep.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 49.Rosati A, et al. BAG3 promotes pancreatic ductal adenocarcinoma growth by activating stromal macrophages. Nat Commun. 2015;6:8695. doi: 10.1038/ncomms9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agez M, et al. Molecular architecture of potassium chloride co-transporter KCC2. Sci Rep. 2017;7:16452. doi: 10.1038/s41598-017-15739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fredholm BB, AP IJ, Jacobson KA, Linden J, Muller CE. International Union of Basic and Clinical Pharmacology. LXXXI. Nomenclature and classification of adenosine receptors–an update. Pharmacol Rev. 2011;63:1–34. doi: 10.1124/pr.110.003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salamone JD. Preladenant, a novel adenosine A(2A) receptor antagonist for the potential treatment of parkinsonism and other disorders. IDrugs. 2010;13:723–31. [PubMed] [Google Scholar]
- 53.Allard D, Turcotte M, Stagg J. Targeting A2 adenosine receptors in cancer. Immunol Cell Biol. 2017;95:333–339. doi: 10.1038/icb.2017.8. [DOI] [PubMed] [Google Scholar]
- 54.Popoli P, et al. Modulation of glutamate release and excitotoxicity by adenosine A2A receptors. Neurology. 2003;61:S69–71. doi: 10.1212/01.WNL.0000095216.89483.A2. [DOI] [PubMed] [Google Scholar]
- 55.Ferre S, et al. An update on adenosine A2A-dopamine D2 receptor interactions: implications for the function of G protein-coupled receptors. Curr Pharm Des. 2008;14:1468–74. doi: 10.2174/138161208784480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weiss HM, Grisshammer R. Purification and characterization of the human adenosine A(2a) receptor functionally expressed in Escherichia coli. Eur J Biochem. 2002;269:82–92. doi: 10.1046/j.0014-2956.2002.02618.x. [DOI] [PubMed] [Google Scholar]
- 57.Desuzinges Mandon E, et al. Expression and purification of native and functional influenza A virus matrix 2 proton selective ion channel. Protein Expr Purif. 2016;131:42–50. doi: 10.1016/j.pep.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 58.Ashok Y, Nanekar R, Jaakola VP. Defining thermostability of membrane proteins by western blotting. Protein Eng Des Sel. 2015;28:539–42. doi: 10.1093/protein/gzv049. [DOI] [PubMed] [Google Scholar]
- 59.Robertson N, et al. The properties of thermostabilised G protein-coupled receptors (StaRs) and their use in drug discovery. Neuropharmacology. 2011;60:36–44. doi: 10.1016/j.neuropharm.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 60.Liu W, et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–6. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Atwood BK, Lopez J, Wager-Miller J, Mackie K, Straiker A. Expression of G protein-coupled receptors and related proteins in HEK293, AtT20, BV2, and N18 cell lines as revealed by microarray analysis. BMC Genomics. 2011;12:14. doi: 10.1186/1471-2164-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thal DM, et al. Recent advances in the determination of G protein-coupled receptor structures. Curr Opin Struct Biol. 2018;51:28–34. doi: 10.1016/j.sbi.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 63.Serrano-Vega MJ, Tate CG. Transferability of thermostabilizing mutations between beta-adrenergic receptors. Mol Membr Biol. 2009;26:385–96. doi: 10.3109/09687680903208239. [DOI] [PubMed] [Google Scholar]
- 64.Eddy MT, Didenko T, Stevens RC, Wuthrich K. beta2-Adrenergic Receptor Conformational Response to Fusion Protein in the Third Intracellular Loop. Structure. 2016;24:2190–2197. doi: 10.1016/j.str.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Magalhaes AC, Dunn H, Ferguson SS. Regulation of GPCR activity, trafficking and localization by GPCR-interacting proteins. Br J Pharmacol. 2012;165:1717–36. doi: 10.1111/j.1476-5381.2011.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fredriksson K, et al. Nanodiscs for INPHARMA NMR Characterization of GPCRs: Ligand Binding to the Human A2A Adenosine Receptor. Angew Chem Int Ed Engl. 2017;56:5750–5754. doi: 10.1002/anie.201612547. [DOI] [PubMed] [Google Scholar]
- 67.Cox BD, et al. Structural analysis of CXCR4 - Antagonist interactions using saturation-transfer double-difference NMR. Biochem Biophys Res Commun. 2015;466:28–32. doi: 10.1016/j.bbrc.2015.08.084. [DOI] [PubMed] [Google Scholar]
- 68.Gater DL, et al. Two classes of cholesterol binding sites for the beta2AR revealed by thermostability and NMR. Biophys J. 2014;107:2305–12. doi: 10.1016/j.bpj.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pereira A, Pfeifer TA, Grigliatti TA, Andersen RJ. Functional cell-based screening and saturation transfer double-difference NMR have identified haplosamate A as a cannabinoid receptor agonist. ACS Chem Biol. 2009;4:139–44. doi: 10.1021/cb800264k. [DOI] [PubMed] [Google Scholar]
- 70.Assadi-Porter FM, et al. Direct NMR detection of the binding of functional ligands to the human sweet receptor, a heterodimeric family 3 GPCR. J Am Chem Soc. 2008;130:7212–3. doi: 10.1021/ja8016939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bartoschek S, et al. Drug design for G-protein-coupled receptors by a ligand-based NMR method. Angew Chem Int Ed Engl. 2010;49:1426–9. doi: 10.1002/anie.200905102. [DOI] [PubMed] [Google Scholar]
- 72.Fronik P, Gaiser BI, Sejer Pedersen D. Bitopic Ligands and Metastable Binding Sites: Opportunities for G Protein-Coupled Receptor (GPCR) Medicinal Chemistry. J Med Chem. 2017;60:4126–4134. doi: 10.1021/acs.jmedchem.6b01601. [DOI] [PubMed] [Google Scholar]
- 73.Sabbadin D, Ciancetta A, Moro S. Bridging molecular docking to membrane molecular dynamics to investigate GPCR-ligand recognition: the human A(2)A adenosine receptor as a key study. J Chem Inf Model. 2014;54:169–83. doi: 10.1021/ci400532b. [DOI] [PubMed] [Google Scholar]
- 74.Mashalidis EH, Sledz P, Lang S, Abell C. A three-stage biophysical screening cascade for fragment-based drug discovery. Nat Protoc. 2013;8:2309–24. doi: 10.1038/nprot.2013.130. [DOI] [PubMed] [Google Scholar]
- 75.Azzi M, et al. Beta-arrestin-mediated activation of MAPK by inverse agonists reveals distinct active conformations for G protein-coupled receptors. Proc Natl Acad Sci USA. 2003;100:11406–11. doi: 10.1073/pnas.1936664100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kenakin T. Functional selectivity through protean and biased agonism: who steers the ship? Mol Pharmacol. 2007;72:1393–401. doi: 10.1124/mol.107.040352. [DOI] [PubMed] [Google Scholar]
- 77.Akmacic IT, et al. High-throughput glycomics: optimization of sample preparation. Biochemistry (Mosc) 2015;80:934–42. doi: 10.1134/S0006297915070123. [DOI] [PubMed] [Google Scholar]
- 78.Lebon G, Bennett K, Jazayeri A, Tate CG. Thermostabilisation of an agonist-bound conformation of the human adenosine A(2A) receptor. J Mol Biol. 2011;409:298–310. doi: 10.1016/j.jmb.2011.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.