Abstract
As one of the most fatal malignancies, pancreatic ductal adenocarcinoma (PDAC) has significant resistance to the currently available treatment approaches. Gemcitabine, the standard chemotherapeutic agent for locally advanced and metastatic PDAC, has limited efficacy, which is attributed to innate/acquired resistance and the activation of prosurvival pathways. Here, we investigated the in vitro efficacy of I-BET762, an inhibitor of the bromodomain and extraterminal (BET) family of proteins, in treating PDAC cell lines alone and in combination with gemcitabine (GEM). The effect of these two agents was also examined in xenograft PDAC tumors in mice. We found that I-BET762 induced cell cycle arrest in the G0/G1 phase and cell death and suppressed cell proliferation and metastatic stem cell factors in PDAC cells. In addition, the BH3-only protein Bim, which is related to chemotherapy resistance, was upregulated by I-BET762, which increased the cell death triggered by GEM in PDAC cells. Moreover, GEM and I-BET762 exerted a synergistic effect on cytotoxicity both in vitro and in vivo. Furthermore, Bim is necessary for I-BET762 activity and modulates the synergistic effect of GEM and I-BET762 in PDAC. In conclusion, we investigated the effect of I-BET762 on PDAC and suggest an innovative strategy for PDAC treatment.
Subject terms: Cancer, Drug discovery
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the 12th most prevalent malignancy worldwide1. The prevalence of PDAC varies from six to eight people in every 100,000 men in developed countries1,2. Despite its comparatively low incidence, PDAC is the 4th most fatal malignancy1,3. The five-year PDAC-specific mortality is over ninety-five percent, resulting from its asymptomatic features in the early stage1,4. Diagnosis is often made at the terminal stage of PDAC4,5.
Surgical excision is the desired strategy for treating PDAC, but the commonly late diagnosis renders the majority of PDAC cases inoperable5,6. Only twenty percent of patients receiving a diagnosis of PDAC qualify for surgery6. Metastasis is common even when an operation has been performed7. Contemporary radiotherapies and chemotherapies are usually ineffective in PDAC. Consequently, numerous studies have explored innovative therapeutic strategies targeting PDAC8–10. Fluorouracil (5-FU) has served as a conventional first-line drug for chemotherapy8. However, as another kind of nucleoside analog, gemcitabine (GEM) displays better efficacy and has replaced its predecessor as the standard drug for chemotherapy11,12. However, the five-year survival rates for patients with PDAC after surgical resection are only approximately twenty percent, even with GEM supplementation13.
The bromodomain and extraterminal (BET) family participates in recognizing ε-N-acetylated lysine residues in histone tails14. As an essential member of the BET family that serves as a transcriptional coactivator, BET domain containing protein (BRD)-4 (BRD4) draws P-TEFb to chromatin, which undergoes acetylation14,15. Other mediators besides BRD4 co-occupy the promoters and enhancers of stimulated genes16. These mediators are enriched at numerous enhancer sequences, commonly named superenhancers17. Notably, the above mentioned enhancers modulate essential oncogene expression in various human malignancies, indicating the therapeutic application of BET bromodomain inhibitors14. In particular, the benzodiazepine JQ-1 was revealed to be effective against lymphoma, myeloma, and ALL, both in vivo and in vitro18,19. I-BET762 is a novel benzodiazepine compound that selectively binds the acetyl-recognizing BET pocket with nanomolar affinity20. I-BET762 has good pharmacological properties as an oral agent and inhibits the proliferation of myeloma cells, resulting in survival advantages in a systemic myeloma xenograft model21. I-BET762 is currently being used in phase I/II clinical trials for nuclear protein in testis (NUT) midline carcinoma and other cancers22. The effect of I-BET762 against ALL such as AML associated with mixed lineage leukemia was also previously reported in preclinical settings22. Previously study has shown that I-BET762 downregulates c-Myc, and dephosphorylation of ERK1/2 leading to proliferation inhibition in pancreatic cancer cells23. However, the influence of I-BET762 on PDAC is not well understood. In the present study, the influence of BET inhibitors together with GEM on PDAC was explored both in vitro and in vivo.
Results
The effect of I-BET762 on the cell death, survival, and cell cycle of PDAC cells
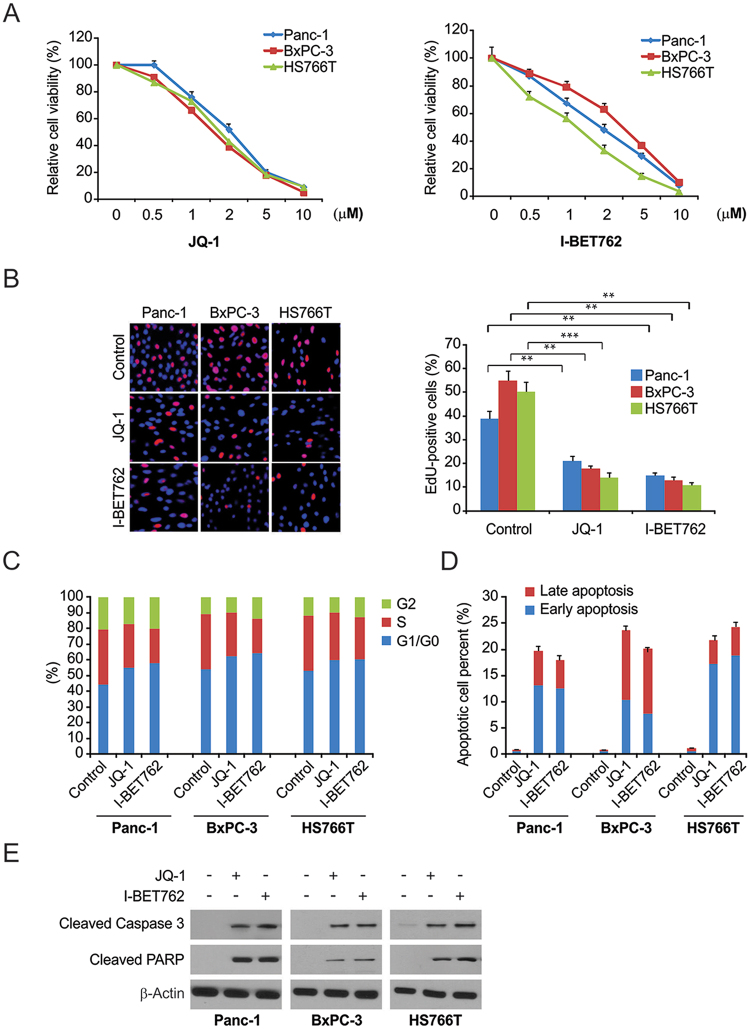
To determine the influence of BET inhibitors, PDAC cells were treated with JQ-1 and I-BET762. Both JQ-1 and I-BET762 remarkably decreased cell survival compared with that in the control group at 72 h (Fig. 1A). Both JQ-1 and I-BET762 noticeably suppressed DNA synthesis, as observed by EdU incorporation (Fig. 1B). Flow cytometry showed that both JQ-1 and I-BET762 triggered cell cycle arrest in HS766T, Panc-1, and BxPC-3 cells (Fig. 1C). Stimulation of cell death was analyzed by annexin V/PI staining to detect apoptosis in both the early and late stages. The results revealed that I-BET762 and JQ-1 noticeably triggered apoptosis in PDAC cells (Fig. 1D). Analyses of essential modulators of cell death, including PARP cleavage and caspase 3, were performed to verify the cell death induction observed in Fig. 1E. The BET inhibitors significantly promoted caspase 3 activation and PARP cleavage in PDAC cells. These findings indicate that I-BET762 suppressed proliferation and induced cell cycle arrest and death in PDAC cells.
Figure 1.
The effects of BET inhibitor on PDAC cells. (A) Panc-1, BxPC-3, and HS766T cells were treated with increasing does of JQ-1 or I-BET762 for 72 h. Cell viability was determined by the CCK-8 assay. (B) Indicated cell lines were treated with 1 μM JQ-1 or 1 μM I-BET762. DNA synthesis was analyzed by the Cell-light EdU Apollo 488 in vitro assay. (C) Panc-1, BxPC-3, and HS766T cells were treated with 1 μM JQ-1 or 1 μM I-BET762 for 24 h. The percentage of cells in different phases of cell cycle was analyzed by flow cytometry. (D) PDAC cells were treated with 1 μM JQ-1 or 1 μM I-BET762 for 24 h. Early and late apoptotic cells were analyzed by flow cytometry. (E) PDAC cells were treated with the 1 μM JQ-1 or 1 μM I-BET762for 24 h. The indicated protein levels were analyzed by western blotting. The results of (B) are expressed as the means ± SD of 3 independent experiments. **P < 0.01; ***P < 0.001.
I-BET762 suppressed migration, invasion, and colony formation in PDAC cells
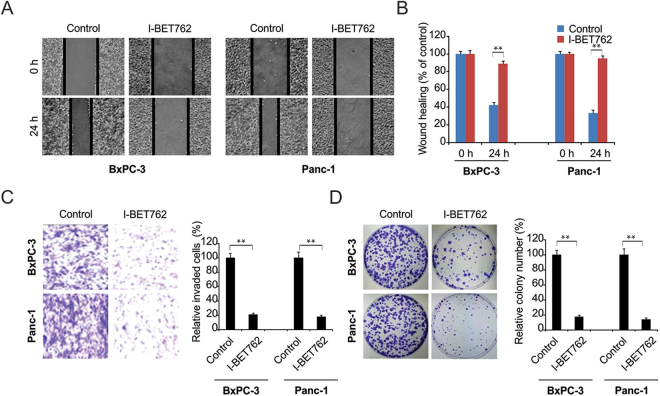
Subsequently, we examined the effect of I-BET762 in counteracting the in vitro migration and invasion of PDAC cells through functional evaluation. I-BET762 remarkably suppressed migration in BxPC-3 and Panc-1 PDAC cells compared to that in the control group (Fig. 2A and B). I-BET762 also significantly suppressed invasion in BxPC-3 and Panc-1 PDAC cells compared with that in the control group (Fig. 2C and D). Colony formation was evaluated in terms of one thousand cells seeded in 6-well plates. After cell attachment, the cells were treated with I-BET762. Colony formation was significantly suppressed in Panc-1 and BxPC-3 cells at 14 days (Fig. 2E and F), indicating that I-BET762 suppresses invasion, colony formation, and migration in PDAC cells.
Figure 2.
I-BET762 possesses anti-migratory and anti-invasive properties. (A) Scratch wound healing assays showed that 1 μM I-BET762 inhibits migration of BxPC-3 and Panc-1 cells. (B) The distance migrated by BxPC-3 and Panc-1 cells after treatment was quantified. The migrated distance was quantified by measuring the difference at time 0 and 24 h and was normalized to control. (C) I-BET762 at 1 μM inhibits the invasion of BxPC-3 and Panc-1 cells. The invaded PDAC cells were quantified by counting the cells at the bottom of the inserts. (D) I-BET762 at 1 μM significantly inhibits colony formation in BxPC-3 and Panc-1 cells. Colony formation assays were repeated at least three times and were normalized to control. The results of (B,C and D) are expressed as the means ± SD of 3 independent experiments. **P < 0.01.
I-BET762 downregulated stem cell factors and decreased sphere generation in PDAC cells
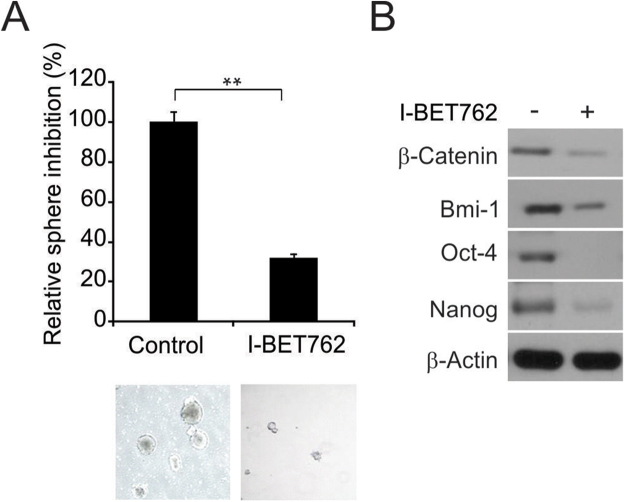
The spheroid generation experiment was modified from previous studies. Two hundred cells in sphere-generating medium (1:1 DMEM/F12 medium containing B-27 and N-2; Invitrogen) were seeded in 24-well plates with ultralow adherent conditions. The cells were treated with I-BET762 for 14 days. The compounds and medium were renewed once. The generated spheres were then counted. As shown in Fig. 3A, I-BET762 noticeably reduced spheroid generation in Panc-1 cells. Analysis of the protein expression revealed remarkable downregulation of stem cell factors (Nanog, BMI-1, β-catenin, and Oct-4) in Panc-1 cells treated with I-BET762 (Fig. 3B), which supports the proliferation-counteracting effect of I-BET762.
Figure 3.
I-BET762 downregulates stem cell factors and inhibits sphere formation in PDAC cells. (A) A single cell suspension of 200 cells in media to was seeded in ultralow adherent 24-well plates. After sphere formation, cells were treated with 1 μM I-BET762 for 14 days. The spheroids were analyzed by light microscopy and imaged. (B) I-BET762 depletes the protein expression of putative stem cell factors in PDAC cells. β-Catenin, Oct-4, Nanog, and BMI-1 were downregulated after 24 h of treatment. The results of (A) are expressed as the means ± SD of 3 independent experiments. **P < 0.01.
The effect of GEM and I-BET762 on PDAC cells
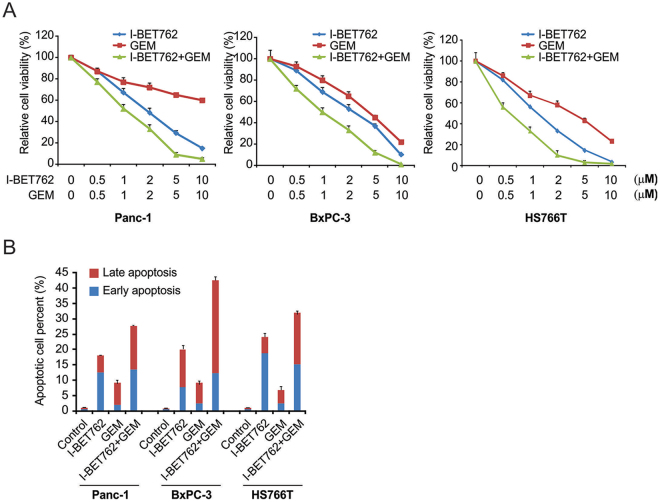
Next, we investigated the combined effect of I-BET762 and GEM on PDAC cells. A CCK-8 assay demonstrated that the combination of GEM and I-BET762 displayed stronger cytotoxicity in 3 cell lines than did either compound alone due to a synergistic effect (Fig. 4A). Evaluation of apoptosis showed that I-BET762 enhanced the apoptotic effect induced by GEM (Fig. 4B). These findings indicated that combining I-BET762 with GEM might be a promising candidate for enhancing treatment efficacy compared with that of GEM treatment alone.
Figure 4.
Effects of I-BET762 and gemcitabine (GEM) on PDAC cell lines. (A) Indicated cells were treated with I-BET762 and GEM. CCK-8 analysis showing that the combination of I-BET762 and GEM was more cytotoxic than either compound alone. (B) Indicated cells were treated with 1 μM I-BET762, 5 μM GEM or their combination. Flow cytometry analysis showing that I-BET762 enhances GEM-induced apoptosis.
Bim is required for I-BET762 function in PDAC
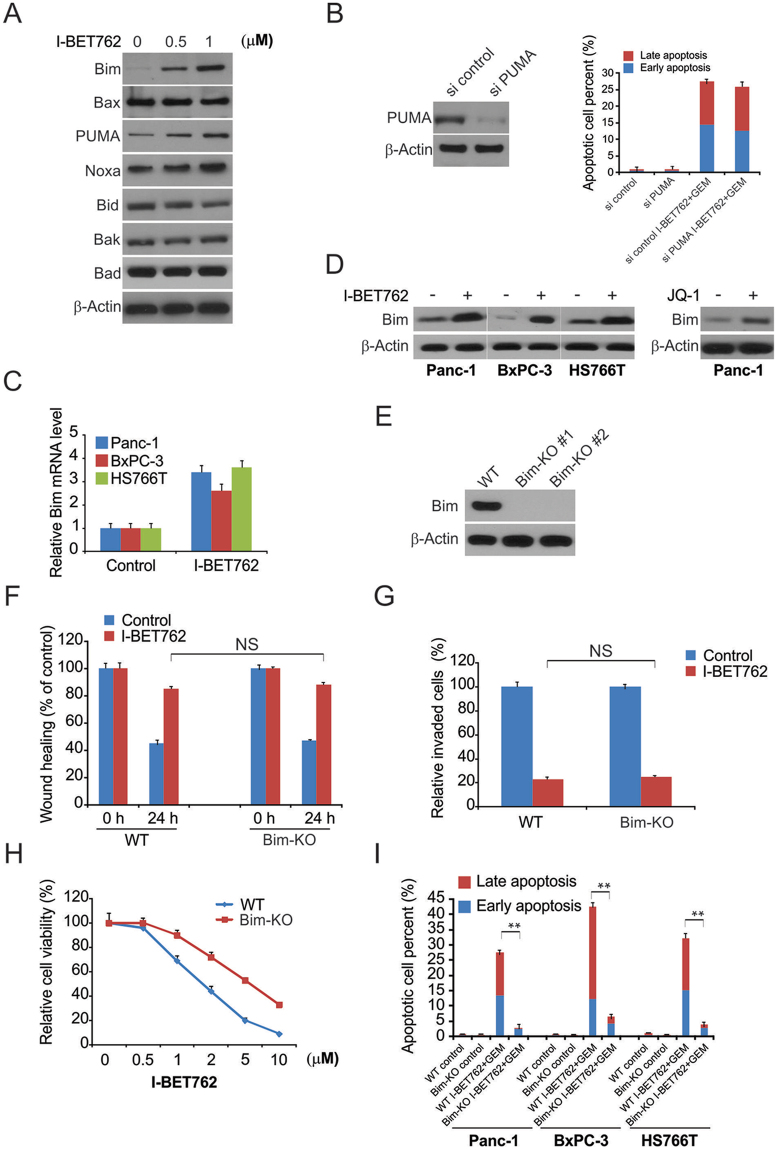
Next, we examined the mechanisms underlying the I-BET762-mediated apoptosis in PDAC. As shown in Fig. 5A, I-BET762 induced Bim and PUMA expression in PDAC. In contrast, I-BET762 treatment did not alter the expression of other Bcl-2 family members. Moreover, knockdown of PUMA did not abrogate the effect of the I-BET762 and GEM combination treatment. Therefore, next, we examined the role of Bim in I-BET762- and GEM-treated PDAC (Fig. 5B). The real-time PCR and western blotting results demonstrated remarkable upregulation of Bim mRNA and protein levels after I-BET762 treatment (Fig. 5C and D).
Figure 5.
Bim mediated the efficacy of I-BET762 and gemcitabine (GEM) action in PDAC cells. (A) Panc-1 cells were treated with I-BET762 at indicated concentration, indicated protein level were analyzed by western blotting. (B) Panc-1 cells transfected with si control or si PUMA were treated with the combination of 1 μM I-BET762 and 5 μM GEM. Apoptosis was detected by flow cytometry. (C) Indicated cell lines were treated with 1 μM I-BET762 for 24 h. Bim mRNA level was analyzed by real-time PCR. (D) Indicated cell lines were treated with 1 μM I-BET762 or 5 μM JQ1 for 24 h, and Bim protein level was analyzed by western blotting. (E) Bim protein level in Bim-KO cells was analyzed by western blotting. (F) The distance migrated by WT and Bim-KO Panc-1 cells after treatment was quantified. The migrated distance was quantified by measuring the difference at time 0 and 24 h and was normalized to control. (G) I-BET762 at 1 μM inhibits the invasion of WT and Bim-KO Panc-1 cells. The invaded PDAC cells were quantified by counting the cells at the bottom of the inserts. (H) WT and Bim-KO Panc-1 cells were treated with 1 μM I-BET762, 5 μM GEM, and their combination for 72 h. Cell viability was determined by the CCK-8 assay. (I) WT and Bim-KO Panc-1 cells were treated with 1 μM I-BET762, 5 μM GEM, and their combination for 24 h. Apoptosis was detected by flow cytometry. The results of (B,C) and (G) are expressed as the means ± SD of 3 independent experiments. **P < 0.01.
Next, we generated Bim knockout Panc-1 cells using the CRISPR-Cas9 system (Fig. 5E). Bim knockout did not affect the I-BET762-induced suppression of migration and invasion (Fig. 5F and G). Furthermore, the synergistic effect of I-BET762 and GEM in PDAC cells was suppressed by Bim knockout (Fig. 5H and I). Our findings thus indicate that Bim is required for the I-BET762-induced apoptosis in PDAC and the effects of I-BET762 on cell migration and invasion are independent of its effects on cell viability.
The effect of GEM and I-BET762 treatment on PDAC xenografts in mice
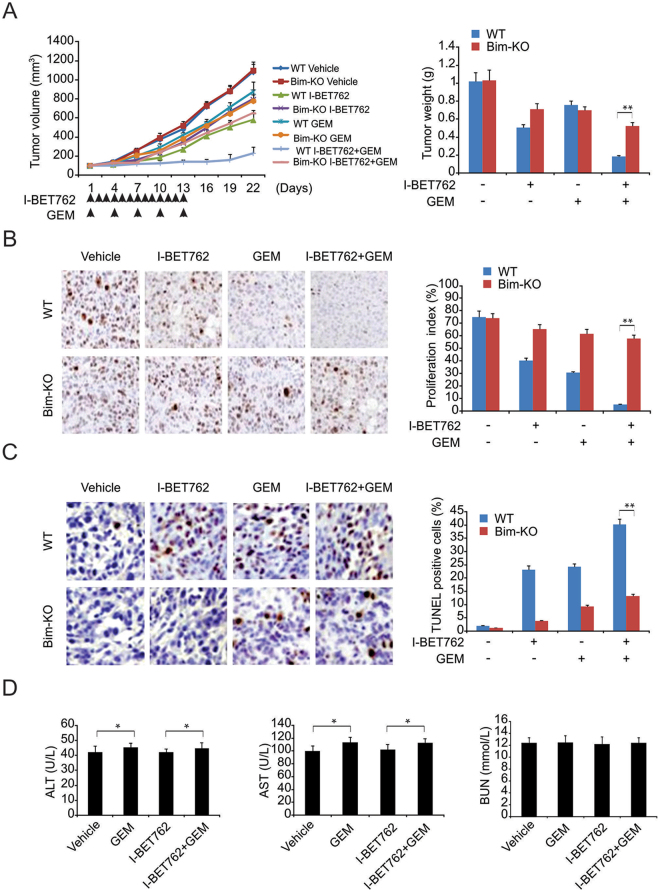
We then examined the necessity of the cell death modulated by Bim for the anticancer function of GEM and I-BET762 in xenograft mice. In Panc-1 tumor-bearing mice, GEM and I-BET762 decreased the tumor weight and volume. The combination of GEM and I-BET762 triggered a remarkable decline in tumor weight and volume compared with that of either agent alone (Fig. 6A). TUNEL and Ki67 assays indicated that I-BET762 and GEM induced less apoptosis when used alone than did the combination treatment (Fig. 6B and C). In contrast, compared with the parental tumors, Bim-KD tumors showed noticeably weaker growth suppression in response to the combination therapy (Fig. 6A–C). Furthermore, to evaluate the toxicity effects of I-BET762 and the combination of I-BET762 and GEM on mice, we measured ALT, AST and BUN levels after treatment. We found that I-BET762 did not influence the ALT or AST in serum samples or their GEM-induced elevation. BUN was not affected by any therapy mentioned above (Fig. 6D).
Figure 6.
Antitumor effects of gemcitabine (GEM), I-BET762, and their combination in a pancreatic xenograft mouse model. (A) Tumor volume over time indicating that combination therapy was more effective than either treatment alone. (B) Ki67 staining revealing that both I-BET762 and GEM inhibited the proliferation of xenograft tumor cells and that combined treatment with I-BET762 and GEM showed a greater capacity to inhibit proliferation. (C) TUNEL assay showing that I-BET762 enhanced the apoptosis induced by GEM, which was blocked in Bim-KO cells. (D) I-BET762 had no effect on the AST or ALT levels and did not enhance the AST and ALT increase induced by GEM. Both I-BET762 and GEM had no effects on BUN. The results of (A,B,C and D) are expressed as the means ± SD of 3 independent experiments. *P < 0.05.
Discussion
PDAC is a fatal malignancy without promising therapeutic alternatives24. Furthermore, only twenty percent of PDAC patients qualify for surgery, which serves as a comparatively optimal approach for treating the disease25. Despite advances in innovative therapeutic strategies including GEM, more approaches are required urgently. Previously, studies demonstrated that I-BET762 disrupts the function of BRD422,26. In addition, it has been reported that BET family proteins (BRD2, BRD3, and BRD4) in PDAC are increased in preneoplastic lesions and frank tumors of the Ptf1a+/Cre; Kras+/LSL-G12D (Kras) mutant mice compared with wild-type pancreas. BET protein inhibition suppresses PDAC growth and improves survival in a PDAC mouse model27. Furthermore, I-BET762 is considered a potential candidate for treating cancers such as breast and bladder cancer28,29. In this study, we investigated the in vitro and in vivo effects of I-BET762 in pancreatic cancer cells and a PDAC xenograft mouse model.
GEM, GEM/erlotinib, and FOLFIRINOX are chemotherapeutic candidates for PDAC30,31. However, these agents only display weak promotion of survival and enhanced toxicity, indicating the necessity of exploring innovative drugs with less toxicity that provide a better effect of counteracting oncogenes that trigger resistance in PDAC32. Previous studies showed that BET bromodomain inhibitors noticeably suppress MYC expression in lymphoma, leukemia, glioblastoma, and neuroblastoma cells15,33,34. However, excessive c-MYC expression in leukemia and glioblastoma cells could not counteract the influence of JQ-1 treatment, indicating that inhibitors of the BET bromodomain act with or without c-MYC involvement27. In the present study, we demonstrated the PDAC-counteracting effects of I-BET762. Previous studies revealed that c-Myc malfunction is prevalent during the development and initial stages of pancreatic cancer35. Excessive c-Myc expression triggered by gli2 is also reported to participate in I-BET151 and JQ-1 resistance in pancreatic cancer36. One study showed that BET bromodomain inhibition sensitizes intestinal crypts to gemcitabine-induced apoptosis37. In addition, combination therapy with gemcitabine plus JQ1 showed greater efficacy than did gemcitabine monotherapy in a mouse model38. Our results proved that I-BET762 suppresses proliferation in 3 PDAC cell lines. The effect of I-BET762 combined with GEM on PDAC treatment was explored and was found to be synergistic both in vitro and in vivo. I-BET762 treatment resulted in upregulation of Bim in vitro and subsequently enhanced apoptosis. Apart from promoting the efficiency of GEM cytotoxicity, I-BET762 also shows promise in postponing the development of drug resistance. However, further experiments are necessary to verify this hypothesis.
The most important finding of our research is the effect of I-BET762 in vivo. In Panc-1 xenograft mice, the combination of I-BET762 and GEM remarkably decreased the tumor volume and weight compared to those of mice treated with either agent alone. Although the characterized side effects of GEM manifested as trouble with eating and loss of weight, no extra loss of weight was observed with the combination of GEM and I-BET762.
Our study examines the essential influence of BET inhibitor on PDAC proliferation and demonstrates the efficacy of the innovative agent I-BET762 in PDAC. Finally, the combination of I-BET762 and GEM displays stronger cytotoxicity and is a promising approach for PDAC treatment, which requires further studies.
Materials and Methods
Cell culture
Human PDAC cells (HS766T, BxPC-3, and Panc-1) were obtained from ATCC and were cultured according to standard procedures in DMEM (Gibco, Carlsbad, CA, USA) with a high concentration of glucose, 10% FBS (Gibco, Carlsbad, CA, USA), and 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO, USA). Cells were grown in an atmosphere of 5% CO2 at 37 °C. Cells were observed every week using phase contrast microscopy to ensure the logarithmic growth phase. GEM, JQ-1, and I-BET762 were obtained from Sigma-Aldrich (St. Louis, MO, USA).
Cell proliferation assay
Cells were seeded in 96-well plates at a density of 1 × 104 cells/well. After 24 h, the medium was replaced with serum-free medium supplemented with JQ-1 or I-BET762. Cytotoxicity and IC50 were evaluated using the CCK-8 cell proliferation assay. To measure the cytotoxicity of JQ-1 and I-BET762 at various time points, media without serum were renewed via complete media supplemented with JQ-1 or I-BET762.
Cell cycle
The influence of JQ-1 and I-BET762 on the cell cycle was determined using flow cytometry. PDAC cells were treated with JQ-1 and I-BET762 for 24 h. The cells were then washed in cold PBS, stained with propidium iodide (PI), and analyzed by flow cytometry. Quantitative evaluation of the cell cycle was conducted using ModFit software (Verity Software House, Topsham, ME, USA).
Western blotting
Western blotting was carried out as described previously39,40 using antibodies against BMI-1 (Santa Cruz Biotechnology, TX, USA), Bim, Nanog, Oct-4, cleaved PARP (Cell Signaling), cleaved caspase 3, β-catenin, and β-actin (Sigma-Aldrich).
Cell motility
PDAC cells were cultured in 24-well plates to form a single layer. The cells were then incubated in medium without serum for 12 h. A scratch was made on the cell layer using a 200-µl pipette tip. The cells were then treated with specific agents and incubated for 12 h at 37 °C in common medium. The cells were observed at time zero after treatment and at 12 h with a microscope to determine the migration distance.
Invasion assay
Transwell inserts precoated with BME were incubated for 2 h at 37 °C. Cells (1 × 104) in serum-free medium were seeded in the uppermost chamber along with or without specific concentrations of the treatment compounds. The bottom chamber contained 1 ml of culture medium with 10% FBS as a chemoattractant. The inserts were incubated for 24 h at 37 °C. The cells in the uppermost chamber were then removed with cotton swabs. The cells that migrated to the bottom were fixed in cold methanol and stained with 0.05% crystal violet.
Colony formation assay
Cells were seeded in 6-well plates and allowed to adhere overnight. The cells were then treated with the test compounds at specific concentrations. The medium with the compounds was renewed every three days for fourteen days. The resulting colonies were then washed with PBS, followed by 30 min of staining with 0.05% crystal violet. Quantification was carried out with the help of ImageJ Colony Counter. Every procedure was conducted no less than three times. The results are expressed as the means ± SD.
Sphere formation assay
To assess the capability of BET inhibitors to suppress pancreatic cancer (PC) stem cells, protocols from previous studies were used. In short, 24-well plates were used to seed the cell suspension in ultralow adherent conditions. Cells at a density of 200 cells/well were seeded in serum-free DMEM/F12 (1:1) containing N-2 and B27 (Life Technologies, Gaithersburg, MD). The cells were incubated for 10 days at 37 °C before pancreatospheres were generated. The spheres were subsequently incubated for 14 days with or without I-BET762 in fresh medium. The generated pancreatospheres were quantified using light microscopy.
CRISPR-cas9-mediated Bim knockout cells
To generate Bim knockout cells, two gRNA sequences targeting Bim were selected. Single-stranded complementary oligos with BsmBI overhangs were generated. The LentiCRISPR v2 (Addgene) lentiviral vector was digested using FastDigest BsmBI obtained from Fermentas. The digested product was purified using a QIAquick Gel Extraction Kit, followed by elution in EB buffer. Phosphorylation and annealing of the oligos were carried out using T4 polynucleotide kinase in T4 ligation buffer (NEB). The reaction system was incubated at 37 °C for 30 min, followed by 90 °C for 5 min, and then cooled to 25 °C at a rate of 5 °C/min. The ligation reaction was carried out by mixing the oligos to be annealed, the digested LentiCRISPR v2 vector, and the Quick Ligase enzyme included in the Quick Ligase Buffer before transformation into Stbl3 bacteria. 293 T cells (2 × 106) were seeded on tissue culture plates (60 mm) at 24 h prior to transfection. Subsequently, 1 µg of lentiviral products was mixed with pMD2G and psPAX plasmids and the PolyJet reagent in serum-free media. After 15 min of incubation at room temperature, the mixture was slowly added to the cells. Medium containing lentiviral particles was obtained after 2 days of transfection. For lentivirus infection, 6-well plates were seeded with Panc-1 cells (4–5 × 104 cells/well). The infected cells were selected with puromycin at a concentration of 2 µg/ml after 1 day of infection and then incubated at 37 °C in 5% CO2. For selecting a single clone, the surviving cells were seeded on a 96-well plate. Western blotting was used to confirm the knockout.
Apoptosis assay
Flow cytometry analysis was used to evaluate cell death. PDAC cells were treated with I-BET762, GEM or both for 24 h. The treated cells were washed with PBS and stained using the Annexin V/PI Apoptosis Kit (BD Biosciences; Franklin Lakes, USA). The stained cells were analyzed on a BD FACS Calibur flow cytometer with BD Cell Quest software.
qRT-PCR
Total RNA was extracted and reverse transcribed using TRIzol Reagent (Invitrogen; Shanghai, China) and a Prime Script RT Kit (Dalian, People’s Republic of China; Takara Biotechnology), respectively. qRT-PCR was carried out on an ABI Prism 7900HT Real-Time System (Applied Biosystems Inc; Shanghai, China). The result is presented as Ct. Relative quantification of the target transcripts was performed using the ΔΔCt method to evaluate the associated alterations in expression. A control group without reverse transcription was included to exclude genomic DNA contamination. β-Actin served as the internal reference gene.
Animal models
All procedures and experiments involving animals in this study were approved by the Committee on the Ethics of Animal Experiments of Department of General Surgery, all methods were performed in accordance with the relevant guidelines and regulations, and a statement to this effect is included in the methods section. BALB/c nude mice (SLAC Laboratory Animal Co., Ltd., Shanghai, China) were subcutaneously injected with pancreatic cancer cells in their right flanks. When the tumor volume reached 150–200 mm3, 24 tumor-bearing mice were randomly divided into 4 groups (I-BET762, GEM, both, and control). The mice in the GEM group were injected with GEM (25 mg/kg/day) through the caudal vein every 3 days for 13 days, and those in the I-BET762 group received an intraperitoneal injection of I-BET762 (30 mg/kg/day) daily for 13 days. The mice in the combination group were treated with both I-BET762 (30 mg/kg/day) and GEM (25 mg/kg/day). In the control group, mice were treated with an equivalent amount of vehicle. Changes in body weight were monitored throughout the experiment. Tumor growth was measured every other day according to the following formula: tumor volume = length × width2/2. Mice were sacrificed on day 22 of the treatment. The tumors were excised and weighed, and the tumor volume was measured. Finally, 0.5 ml of blood was drawn from every mouse by cardiac puncture and was sent to clinical laboratories to evaluate the hepatic and renal activities.
TUNEL assay and immunohistochemical (IHC) examination
Tumor samples were fixed in 10% formalin prior to paraffin embedding, and sections of 4 µm thickness were cut. Cell death in the tumors was evaluated using the In Situ Cell Death Detection Kit, POD (Roche Molecular Biochemicals; Indianapolis, USA) and was characterized by brown staining. For IHC examination, the sections were incubated with rabbit anti-human Ki67 (Sigma Aldrich, USA) (1:400) antibodies followed by incubation with HRP-conjugated anti-rabbit IgG antibodies, and the detection was performed using the Histostain-Plus Kit (Haoran-Bio; Shanghai, China). Finally, the sections were counterstained with hematoxylin. The negative control was incubated with PBS instead of a specific primary antibody. The assessment was conducted for 5 slices per tumor.
Statistical analysis
Each experiment was performed no less than 3 times. The results are displayed as the means ± SD. Statistical analyses were conducted using Prism 5 (GraphPad, San Diego, CA, USA). The results were considered significant at P < 0.05.
Author Contributions
In this work, Fang X. and Huang Q. conceived the study and designed the experiments. Huang M., Lin X.S., Liu C.H., Liu Z., Meng F.T. and Wang C. contributed to the data collection, performed the data analysis and interpreted the results. Fang X. wrote the manuscript; Huang Q. contributed to the critical revision of the article. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41598-023-48298-9
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
11/28/2023
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41598-023-48298-9
References
- 1.Makohon-Moore A, Iacobuzio-Donahue CA. Pancreatic cancer biology and genetics from an evolutionary perspective. Nature reviews. Cancer. 2016;16:553–565. doi: 10.1038/nrc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teague A, Lim KH, Wang-Gillam A. Advanced pancreatic adenocarcinoma: a review of current treatment strategies and developing therapies. Therapeutic advances in medical oncology. 2015;7:68–84. doi: 10.1177/1758834014564775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muniraj T, Jamidar PA, Aslanian HR. Pancreatic cancer: a comprehensive review and update. Disease-a-month: DM. 2013;59:368–402. doi: 10.1016/j.disamonth.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Ansari D, et al. Pancreatic cancer: yesterday, today and tomorrow. Future oncology. 2016;12:1929–1946. doi: 10.2217/fon-2016-0010. [DOI] [PubMed] [Google Scholar]
- 5.Spadi R, et al. Current therapeutic strategies for advanced pancreatic cancer: A review for clinicians. World journal of clinical oncology. 2016;7:27–43. doi: 10.5306/wjco.v7.i1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong J, Tuli R, Shinde A, Hendifar AE. Meta-analyses of treatment standards for pancreatic cancer. Molecular and clinical oncology. 2016;4:315–325. doi: 10.3892/mco.2015.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thota R, Pauff JM, Berlin JD. Treatment of metastatic pancreatic adenocarcinoma: a review. Oncology. 2014;28:70–74. [PubMed] [Google Scholar]
- 8.Cid-Arregui A, Juarez V. Perspectives in the treatment of pancreatic adenocarcinoma. World journal of gastroenterology. 2015;21:9297–9316. doi: 10.3748/wjg.v21.i31.9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellenrieder V, Konig A, Seufferlein T. Current Standard and Future Perspectives in First- and Second-Line Treatment of Metastatic Pancreatic Adenocarcinoma. Digestion. 2016;94:44–49. doi: 10.1159/000447739. [DOI] [PubMed] [Google Scholar]
- 10.Sulkowski U. Standards and perspectives in the diagnosis and treatment of pancreatic adenocarcinoma. Digestion. 1996;57(Suppl 1):34–35. doi: 10.1159/000201390. [DOI] [PubMed] [Google Scholar]
- 11.Hong SP, Wen J, Bang S, Park S, Song SY. CD44-positive cells are responsible for gemcitabine resistance in pancreatic cancer cells. International journal of cancer. 2009;125:2323–2331. doi: 10.1002/ijc.24573. [DOI] [PubMed] [Google Scholar]
- 12.Park JK, et al. The anti-fibrotic effect of GV1001 combined with gemcitabine on treatment of pancreatic ductal adenocarcinoma. Oncotarget. 2016;7:75081–75093. doi: 10.18632/oncotarget.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koay EJ, et al. Transport properties of pancreatic cancer describe gemcitabine delivery and response. The Journal of clinical investigation. 2014;124:1525–1536. doi: 10.1172/JCI73455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padmanabhan B, Mathur S, Manjula R, Tripathi S. Bromodomain and extra-terminal (BET) family proteins: New therapeutic targets in major diseases. Journal of biosciences. 2016;41:295–311. doi: 10.1007/s12038-016-9600-6. [DOI] [PubMed] [Google Scholar]
- 15.Taniguchi, Y. The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins. International journal of molecular sciences17, 10.3390/ijms17111849 (2016). [DOI] [PMC free article] [PubMed]
- 16.Chaidos A, Caputo V, Karadimitris A. Inhibition of bromodomain and extra-terminal proteins (BET) as a potential therapeutic approach in haematological malignancies: emerging preclinical and clinical evidence. Therapeutic advances in hematology. 2015;6:128–141. doi: 10.1177/2040620715576662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raux B, et al. Exploring Selective Inhibition of the First Bromodomain of the Human Bromodomain and Extra-terminal Domain (BET) Proteins. Journal of medicinal chemistry. 2016;59:1634–1641. doi: 10.1021/acs.jmedchem.5b01708. [DOI] [PubMed] [Google Scholar]
- 18.Fu LL, et al. Inhibition of BET bromodomains as a therapeutic strategy for cancer drug discovery. Oncotarget. 2015;6:5501–5516. doi: 10.18632/oncotarget.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mertz JA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, Yang CY, Wang S. The making of I-BET762, a BET bromodomain inhibitor now in clinical development. Journal of medicinal chemistry. 2013;56:7498–7500. doi: 10.1021/jm4014407. [DOI] [PubMed] [Google Scholar]
- 21.Chaidos A, et al. Potent antimyeloma activity of the novel bromodomain inhibitors I-BET151 and I-BET762. Blood. 2014;123:697–705. doi: 10.1182/blood-2013-01-478420. [DOI] [PubMed] [Google Scholar]
- 22.Mirguet O, et al. Discovery of epigenetic regulator I-BET762: lead optimization to afford a clinical candidate inhibitor of the BET bromodomains. Journal of medicinal chemistry. 2013;56:7501–7515. doi: 10.1021/jm401088k. [DOI] [PubMed] [Google Scholar]
- 23.Leal AS, et al. Bromodomain inhibitors, JQ1 and I-BET 762, as potential therapies for pancreatic cancer. Cancer letters. 2017;394:76–87. doi: 10.1016/j.canlet.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 24.Seton-Rogers S. Pancreatic cancer: Dodging immunosuppression. Nature reviews. Cancer. 2016;16:480–481. doi: 10.1038/nrc.2016.80. [DOI] [PubMed] [Google Scholar]
- 25.Kim SK, Wu CC, Horowitz DP. Stereotactic body radiotherapy for the pancreas: a critical review for the medical oncologist. Journal of gastrointestinal oncology. 2016;7:479–486. doi: 10.21037/jgo.2015.10.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai X, et al. Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nature medicine. 2017;23:1063–1071. doi: 10.1038/nm.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazur PK, et al. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nature medicine. 2015;21:1163–1171. doi: 10.1038/nm.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alluri PG, Asangani IA, Chinnaiyan AM. BETs abet Tam-R in ER-positive breast cancer. Cell research. 2014;24:899–900. doi: 10.1038/cr.2014.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rickman DS, Beltran H, Demichelis F, Rubin MA. Biology and evolution of poorly differentiated neuroendocrine tumors. Nature medicine. 2017;23:1–10. doi: 10.1038/nm.4341. [DOI] [PubMed] [Google Scholar]
- 30.Klapdor R, Klapdor S, Bahlo M. Combination therapy with gemcitabine (GEM) and erlotinib (E) in exocrine pancreatic cancer under special reference to RASH and the tumour marker CA19-9. Anticancer research. 2012;32:2191–2197. [PubMed] [Google Scholar]
- 31.Shin S, Park CM, Kwon H, Lee KH. Erlotinib plus gemcitabine versus gemcitabine for pancreatic cancer: real-world analysis of Korean national database. BMC cancer. 2016;16:443. doi: 10.1186/s12885-016-2482-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ardavanis A, et al. Biweekly gemcitabine (GEM) in combination with erlotinib (ERL): an active and convenient regimen for advanced pancreatic cancer. Anticancer research. 2009;29:5211–5217. [PubMed] [Google Scholar]
- 33.Jung M, Gelato KA, Fernandez-Montalvan A, Siegel S, Haendler B. Targeting BET bromodomains for cancer treatment. Epigenomics. 2015;7:487–501. doi: 10.2217/epi.14.91. [DOI] [PubMed] [Google Scholar]
- 34.Tong J, Tan S, Zou F, Yu J, Zhang L. FBW7 mutations mediate resistance of colorectal cancer to targeted therapies by blocking Mcl-1 degradation. Oncogene. 2017;36:787–796. doi: 10.1038/onc.2016.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hessmann E, Schneider G, Ellenrieder V, Siveke JT. MYC in pancreatic cancer: novel mechanistic insights and their translation into therapeutic strategies. Oncogene. 2016;35:1609–1618. doi: 10.1038/onc.2015.216. [DOI] [PubMed] [Google Scholar]
- 36.Kumar K, et al. GLI2-dependent c-MYC upregulation mediates resistance of pancreatic cancer cells to the BET bromodomain inhibitor JQ1. Scientific reports. 2015;5:9489. doi: 10.1038/srep09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakagawa A, et al. Selective and reversible suppression of intestinal stem cell differentiation by pharmacological inhibition of BET bromodomains. Scientific reports. 2016;6:20390. doi: 10.1038/srep20390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto K, et al. Stromal remodeling by the BET bromodomain inhibitor JQ1 suppresses the progression of human pancreatic cancer. Oncotarget. 2016;7:61469–61484. doi: 10.18632/oncotarget.11129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong J, et al. FBW7-Dependent Mcl-1 Degradation Mediates the Anticancer Effect of Hsp90 Inhibitors. Molecular cancer therapeutics. 2017;16:1979–1988. doi: 10.1158/1535-7163.MCT-17-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong J, et al. Mcl-1 Degradation Is Required for Targeted Therapeutics to Eradicate Colon Cancer Cells. Cancer research. 2017;77:2512–2521. doi: 10.1158/0008-5472.CAN-16-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]